Abstract
Activation of Wnt signaling induces Connexin43 (Cx43) expression via the transcriptional activity of β-catenin, and results in the enhanced accumulation of the Cx43 protein and the formation of gap junction channels. In response to Wnt signaling, β-catenin co-localizes with the Cx43 protein itself as part of a complex at the gap junction plaque. Work from several labs have also shown indirect evidence of this interaction via reciprocal co-immunoprecipitation. Our goal for the current study was to identify whether β-catenin directly interacts with Cx43, and if so, the location of that direct interaction. Identifying residues involved in direct protein–protein interaction is of importance when they are correlated to the phosphorylation of Cx43, as phosphorylation can modify the binding affinities of Cx43 regulatory protein partners. Therefore, combining the location of a protein partner interaction on Cx43 along with the phosphorylation pattern under different homeostatic and pathological conditions will be crucial information for any potential therapeutic intervention. Here, we identified that β-catenin directly interacts with the Cx43 carboxyl-terminal domain, and that this interaction would be inhibited by the Src phosphorylation of Cx43CT residues Y265 and Y313.
Keywords: Cx43, β-catenin, phosphorylation
1. Introduction
Gap junctions are intercellular channels that permit the passage of ions, small metabolites, and signaling molecules between neighboring cells [1]. They are important in a number of physiological processes, including cellular development, growth, and differentiation. In the heart, gap junctions mediate the propagation of cardiac action potentials and the maintenance of a regular beating rhythm [2]. Dysfunctional intercellular communication via gap junctions has been implicated in causing many human diseases [3]. Gap junctions are formed by the apposition of connexons from adjacent cells, where six connexin proteins form each connexon. Although the 21-connexin isoforms (e.g., 43-kDa isoform, Cx43) share significant sequence homology, differences in the amino acid sequence occur in the cytoplasmic loop and carboxyl terminal (CT) domains.
The CT domain is involved in regulating the trafficking of connexons to and from the plasma membrane, as well as the level of gap junction intercellular communication via a number of post-translational modifications and interactions with protein partners [4,5,6,7,8,9,10,11]. The CT domain is predominately unstructured (i.e., intrinsically disordered), making it an ideal substrate for the regulation of intercellular signaling by facilitating both high specificity and low affinity interaction with many different binding partners to allow the rapid feedback to cytoplasmic signals [12,13,14,15]. Over 20 protein partners have been identified to directly interact with Cx43, which can be categorized according to those that can promote or inhibit intercellular communication (for review see [16,17]). For example, connexin localization and stability at the gap junction plaques are strongly determined by interaction with cytoskeletal-associated proteins; these interactions are modulated by specific phosphorylation events [18,19]. The demonstration that Src phosphorylates Y247 and Y265 of Cx43 [20,21] enabled subsequent findings that pY247 inhibits the Cx43 interaction with β-tubulin [22] and that pY265 inhibits the Cx43 interaction with drebrin [23]. At the gap junction plaque, inhibiting the β-tubulin interaction could be a mechanism involved in the disassembly process. If this inhibition occurs after connexon formation in the trans-Golgi network, trafficking may be re-routed for degradation or to the lateral membrane [24]. The depletion of drebrin results in impaired gap junction intercellular communication, also by targeting Cx43 for degradation [25]. Further, while Cx43CT phosphorylation by Src does not inhibit Zonula occludens-1 (ZO-1) binding, active Src directly competes with Cx43 to bind ZO-1 [26]. Studies from the Gourdie and Lampe labs suggest that interaction with ZO-1 transitions Cx43 from the non-junctional membrane into the gap junction plaque, and that Src inhibits this process [27,28]. Thus, even though our knowledge is no doubt incomplete, it is clear that for Cx43, there exists a network of integrated processes involving phosphorylations and binding partners that control junctional Cx43 [19]. Another protein that has been identified to modulate both Cx43 expression and gap junction intercellular communication is β-catenin [29].
β-catenin is a critical protein in the canonical Wnt signaling transduction cascade. β-catenin (781 amino acids) consists of a well-structured central region made up of 12 armadillo repeats that are flanked by intrinsically disordered N-terminal and C-terminal domains [30,31]. β-catenin is a multi-functional protein whose activity depends on its subcellular localization. β-catenin at the plasma membrane is a component of cell adhesion junctions, while cytosolic accumulation leads to increased nuclear localization and transcriptional activity [32]. The activation of Wnt signaling induces Cx43 expression via the transcriptional activity of β-catenin, which leads to the increased formation of gap junction channels [29,33,34,35,36,37,38]. In response to Wnt signaling, β-catenin co-localizes with the Cx43 protein itself as part of a complex at the gap junction plaque [29]. Unfortunately, none of these studies was able to determine whether the β-catenin and Cx43 interaction is direct, or instead requires other protein partners.
2. Results
2.1. β-Catenin CT Domain Directly Interacts with the Cx43CT
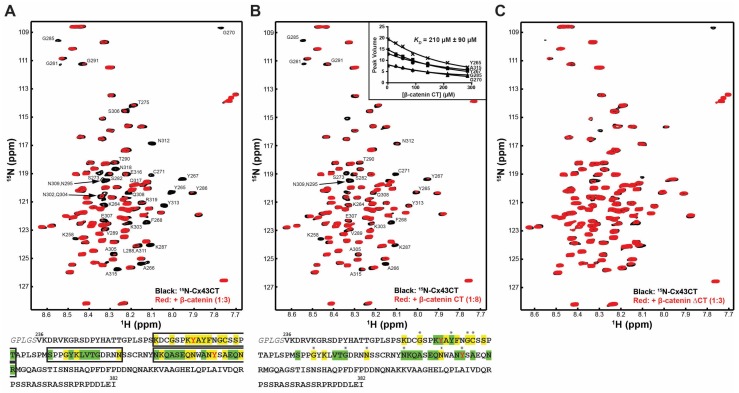
The domain architecture of β-catenin includes a disordered N-terminal domain (~150 residues; binds α-catenin, glycogen synthase kinase 3β (GSK3β), and is phosphorylated by casein kinase I), a well-structured central armadillo repeat domain (~530 residues; major protein partner binding domain; e.g., binds axin, adenomatous polyposis coli protein, 14-3-3ζ, and E-cadherin), and a disordered C-terminal domain (~100 residues; binds several transcriptional coactivators) [39,40]. To determine whether β-catenin directly interacts with the Cx43CT domain, we performed a 15N-heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR) experiment using purified 15N-lableled Cx43CT (V236–I382) and unlabeled full-length β-catenin (Figure 1A, top). Each chemical shift (or peak) in this two-dimensional experiment corresponds to one amide group; thus, the number of peaks should correspond to the number of Cx43CT residues (except proline). We have previously published the 15N-HSQC assignment for the Cx43CT domain [41]. These chemical shifts are sensitive to their environment and small changes in structure and/or dynamics, such as those that would occur from a direct protein–protein interaction, can influence the chemical shift (i.e., change the location or broaden beyond detection) of an amino acid. The advantage of using NMR to study direct protein–protein interactions over cellular assays such as immunoprecipitation is its specificity. As only two proteins are present in the solution, any detected interaction is the result of a direct interaction, as opposed to the possibility that both are part of a larger molecular complex (limits of immunoprecipitation). Moreover, because the chemical shifts of the affected amino acids drift or diminish, the specific residues involved in the interaction can be determined. The addition of β-catenin affected a subset of Cx43CT residues (Figure 1A, top). When mapped onto the Cx43CT sequence, they were located within three areas: K259–T275, S282–N295, and N302–R319 (Figure 1A, bottom). The data indicates that the Cx43CT domain and β-catenin directly interact.
Figure 1.
Nuclear magnetic resonance spectra showing the direct interaction between the Cx43CT and β-catenin. 15N- heteronuclear single quantum coherence (HSQC) spectra of Cx43CT alone (black) and in the presence of (A) full-length β-catenin (red); (B) the β-catenin carboxyl-terminal (CT) domain (red); or (C) the β-catenin ΔCT domains (red). Molar ratio for each experiment is indicated in the figure. In panel B, provided is a subset of residues used to calculate the KD of the interaction by fitting their decrease in signal intensity according to β-catenin CT concentration. Below each 15N-HSQC spectra is the Cx43CT amino acid sequence. Highlighted are the affected residues (yellow—peaks broadened beyond detection; green—peaks decreased in intensity). Black boxes delimitate the three areas of interaction with β-catenin. Asterisks denote that amino acids that were used to calculate the binding affinity for the β-catenin CT. Two of the residues phosphorylated by Src and affected by β-catenin are also highlighted (red letters).
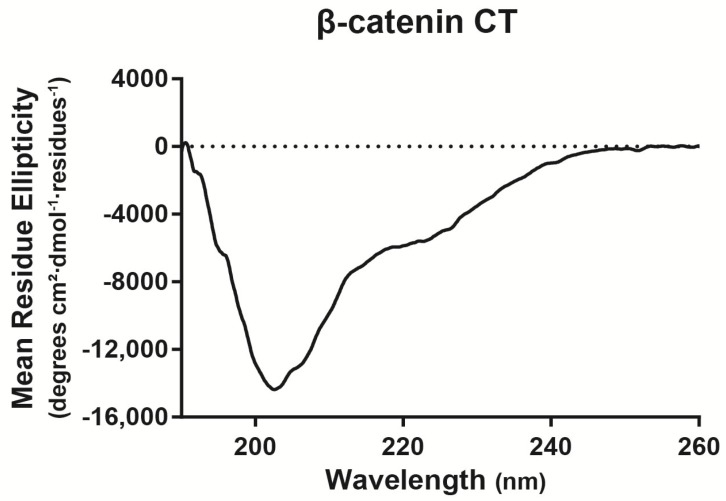
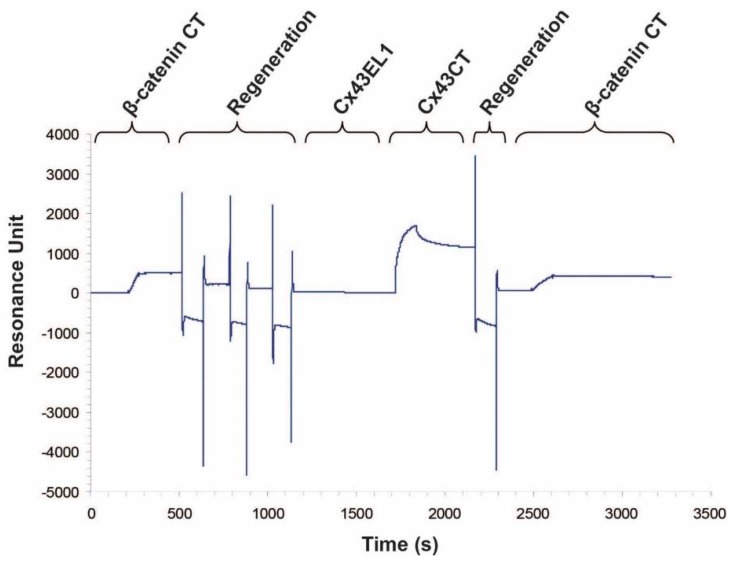
To identify the β-catenin domain mediating the direct interaction with Cx43CT, we initially focused on the β-catenin CT domain (S681–L781). The N-terminal domain is the primary locus for Wnt signaling (GSK3β/E3 ubiquitin ligase), and armadillo repeat domains have been well characterized for binding partners involved in cell adhesion. Moreover, most of the binding partners to the C-terminal domain occur in the nucleus, thus potentially leaving the CT domain free to associate with a different set of proteins in the cytoplasm [42]. Upon purification, circular dichroism was used to determine whether the β-catenin CT domain contains any secondary structure (Figure 2). Analysis of the circular dichroism spectrum by Dichroweb (London, UK) determined that the protein is predominately intrinsically disordered with between 18–21% α-helical content [43,44]. The low amount of secondary structure is consistent with the low peak dispersion (<1 1H ppm) that was previously seen in the β-catenin CT domain 15N-HSQC [32]. The addition of the unlabeled β-catenin CT domain affected the same subset of 15N-Cx43CT residues as full-length β-catenin (Figure 1B, top). These have been highlighted on the Cx43CT sequence (Figure 1B, bottom). Next, a titration of the unlabeled β-catenin CT domain was performed to determine the binding affinity (KD). The decrease in signal intensity caused by increasing the β-catenin CT domain concentration was fit according to the nonlinear least-square method (Figure 1B, inset). The KD was determined to be 210 μM (±90 μM). A surface plasmon resonance (SPR) experiment confirmed the NMR results (Figure 3). When the Cx43CT domain was immobilized onto a carboxymethyl-dextran 5 chip, the addition of the β-catenin CT domain resulted in a direct interaction. A peptide to the Cx43 first extracellular loop (EL1, residues G38–R76) served as a negative control, and the Cx43CT domain itself served as a positive control (specific areas of dimerization include M281–N295, R299–Q304, S314–I327, and Q342–A348, [45]). To ensure that the β-catenin CT is the only β-catenin domain interacting with the Cx43CT, we purified a β-catenin construct containing the N-terminal and armadillo repeat domains (β-catenin ΔCT, i.e., deleted the CT domain). The addition of the unlabeled β-catenin ΔCT had no effect on the 15N-Cx43CT residues (Figure 1C). Of note, the N-terminal domain of β-catenin has poor expression and degraded during purification, which prevented any attempt to test this domain only. Altogether, the data indicate that the β-catenin CT is the domain that directly interacts with Cx43CT.
Figure 2.
Circular dichroism spectrum showing the secondary structure of the β-catenin CT domain. The spectrum is represented as mean residue ellipticity as a function of wavelength. Data were analyzed using Dichroweb.
Figure 3.
Surface plasmon resonance spectra showing direct interaction between the Cx43CT and β-catenin CT domains. Cx43CT was immobilized onto a CM5 chip by amine coupling (Biacore; GE Healthcare, Uppsala, Sweden) and either β-catenin CT (500 response units), Cx43EL1 (negative control), or Cx43CT (residues S255–I382, positive control) were flown over the chip as indicated on the top of the graph. The chip was regenerated after an interaction was observed. Repeat of an injection of β-catenin CT was performed to confirm the interaction.
2.2. Phosphorylation of Y265 and Y313 Inhibits Cx43 Binding with β-Catenin
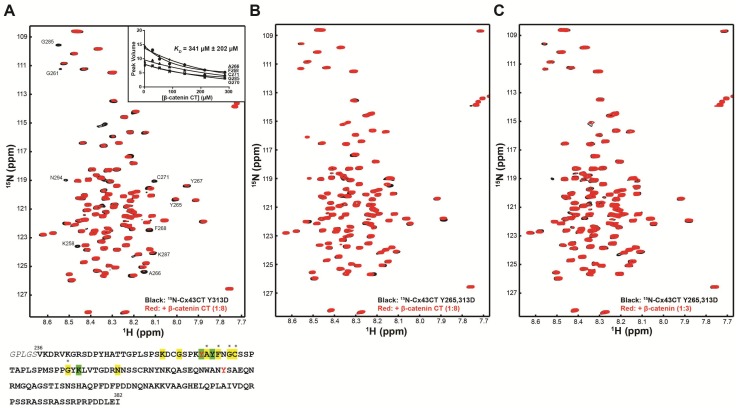
Previous studies have identified that Src phosphorylates Cx43CT residues Y247 and Y265 [21,46]. Additional studies have identified that Src also phosphorylates Cx43CT residues Y313 (Li et al., 2018, Journal of Molecular and Cellular Cardiology, publication under revision; PhosphoSitePlus, [47]). Since the β-catenin CT domain affected Cx43CT residues Y265 and Y313, we addressed whether the phosphorylation of both sites could dissociate β-catenin from Cx43. A number of gap junction studies have determined that an aspartic acid can mimic a phosphate for Cx43 [41,48,49] and responds to the need to purify enough protein for biophysical studies. Therefore, NMR titration experiments were performed using purified soluble 15N-labeled Cx43CT single or double phosphomimetics (Y313D or Y265,313D) and different concentrations of either an unlabeled β-catenin CT domain (Figure 4A,B) or full-length β-catenin (Figure 4C). The 15N-HSQC spectrum of each control (no β-catenin, black) has been overlaid with spectra when either the β-catenin CT domain or full-length β-catenin (red) were added at a single molar ratio. For the single phosphomimetic construct, among the three areas where the β-catenin CT domain interacted with Cx43CT wild type (WT), binding was completely inhibited for area three (N302–R319), significantly reduced in area two (S282–N295), and mostly preserved in area one (K259–T275). A titration of the unlabeled β-catenin CT domain was performed to determine the KD. The interaction with β-catenin decreased by approximately two-fold compared to WT (KD = 341 μM vs. 202 μM) (Figure 4A, inset). When both Cx43CT Y265 and Y313 sites were mutated to mimic Src phosphorylation, the Cx43CT interaction with β-catenin was completely inhibited. These results confirm the direct interaction between Cx43CT and CT portion of β-catenin, and strongly suggest that Src phosphorylation of Cx43CT regulates this interaction.
Figure 4.
Mimicking phosphorylation of Cx43CT residues Y265 and Y313 inhibits the interaction with the β-catenin CT domain. 15N-HSQC spectra of (A) Cx43CT Y313D alone (black) and in the presence of the β-catenin CT domain (red) or Y265,313D alone (black), and in the presence of the (B) β-catenin CT domain and (C) full-length β-catenin (red). The molar ratio for each experiment is indicated in the figure. In panel A, provided is a subset of residues used to calculate the KD of the interaction by fitting their decrease in signal intensity according to β-catenin CT concentration. Also represented below the 15N-HSQC spectra is the Cx43CT amino acid sequence. Highlighted are the affected residues (yellow—peaks broadened beyond detection; green—peaks decreased in intensity). Asterisks denote amino acids used to calculate the binding affinity for the β-catenin CT interaction with the Cx43CT. Tyrosine residues 265 and 313 phosphorylated by Src are indicated in red.
3. Discussion
The Cx43CT domain binds multiple proteins, many of which have been shown to regulate Cx43 function through altering trafficking to the gap junction plaque, opening/closing gap junction channels, disassembly, and degradation (for review see [16,50]). Numerous excellent reviews have summarized the functional significance of these Cx43-interacting proteins [50,51,52,53]. These proteins can be partitioned into those that directly interact with the Cx43CT, and those proteins that can affect all aspects of the Cx43 life cycle, but no current evidence exists that they directly interact with the Cx43CT. One of the proteins in the latter category is β-catenin. Work from several labs has shown indirect evidence of this interaction, including reciprocal co-immunoprecipitation as well as co-localization of Cx43 with β-catenin [29,35]. Additionally, β-catenin segregates in triton insoluble fractions with Cx43 [29]. Our goal here was to identify whether β-catenin directly interacts with Cx43, and the location of that direct interaction.
β-catenin is an intracellular signal transducer in the Wnt signaling pathway that is involved in the regulation and coordination of cell–cell adhesion and gene transcription [54]. Ai et al. first identified that in response to Wnt signaling by the addition of Li+, β-catenin interacts with the Cx43 gene GJA1 (contains three transcription factor 4 (TCF)/lymphoid enhancer binding factor binding sites; [55]) to increase transcription expression [29]. The accumulating Cx43 in the junctional membrane increased neonatal rat cardiomyocyte cell-to-cell coupling and co-localization with β-catenin [29]. Cx43 and β-catenin co-localization also occurs at the intercalated discs of adult rat cardiomyocytes [56]. A similar response to Li+ elicited Wnt/β-catenin signaling, which increased Cx43 expression and gap junction intercellular communication in skeletal myoblasts [57]. Additionally, the rapid electrical stimulation of cardiomyocytes had a similar effect as Li+, leading to the increased nuclear localization of β-catenin and subsequent Cx43 expression [58]. The suggested sequestering of β-catenin by Cx43 would serve to reduce the transactivation potential of β-catenin [29]. This observation is consistent with the increase in the active form of β-catenin with the knockdown of Cx43 seen in human neural progenitor cells [35], and the decrease in the nuclear localization of β-catenin with Cx43 overexpression in breast adenocarcinoma cell lines [36]. Conversely, Moorer et al. observed in a Cx43CT truncation (K258stop) mouse model that osteoblasts had reduced active β-catenin (along with protein kinase C δ and extracellular signal-regulated kinase 1/2), leading to altered proliferation, differentiation, collagen processing, and organization [34]. While the phenotype observed from loss of the Cx43CT matches that from the complete loss of Cx43 in bone cells, the same is not true in the cardiovascular system [34,59]. This suggests the influence of Cx43 on the activity and cellular localization of β-catenin may be tissue specific [34,59].
Shaw et al. put forth a model for connexin trafficking to the plasma membrane [56]. Cx43 oligomerizes into connexons in the trans-Golgi network [60]. Upon exiting, they use microtubules to travel to adherens junctions, which capture the microtubules allowing for connexon offloading to the plasma membrane [56,61,62,63,64]. Based upon the co-localization of Cx43 and β-catenin at the gap junction plaque, at some point in the trafficking, either to or at the adherens junctions, the interaction occurs. The β-catenin interaction occurs at Cx43 residues K259–T275, S282–N295, and N302–R319. Interestingly, similar residues directly interact with drebrin [23]. A commonality with these proteins is they would both help Cx43 indirectly interact with F-actin (β-catenin indirectly through α-catenin; drebrin directly) and stabilize gap junctions to favor intercellular communication. Conversely, they both cannot interact at the same time. Therefore, the available data would suggest that β-catenin binds first, and then at some point in the maturation of the gap junction plaque, Cx43CT switches to interact with drebrin. Since the phosphorylation of Y265 and Y313 also inhibits the Cx43 interaction with drebrin, this would not be the mechanism [23]. The possibility exists that regulation from the β-catenin perspective inhibits the interaction with Cx43. Consistent with this is that: (1) the phosphorylation of β-catenin at S552 by protein kinase B increases the association between β-catenin and 14-3-3ζ, leading to β-catenin translocation into the cytosol and nucleus [65]; (2) the phosphorylation of β-catenin by casein kinase 1 is necessary for subsequent glycogen synthase kinase-3 phosphorylation and then degradation [66]; (3) the protein kinase A phosphorylation of β-catenin leads to the nuclear localization of β-catenin [67]. Interestingly, protein kinase B, casein kinase 1, and protein kinase A also phosphorylate Cx43 to promote synthesis, trafficking to the gap junction plaque, and channel opening.
The importance of identifying if and where a direct protein interaction occurs is in relationship to the phosphorylation of Cx43, because phosphorylation modifies the binding affinities of the Cx43 protein partners that regulate assembly, disassembly, and channel function [68]. For example, we demonstrated that mitogen-activated protein kinase phosphorylation of Cx43 increases the binding affinity for the E3 ubiquitin ligase neural precursor cell expressed, developmentally down-regulated 4 [69], which leads to Cx43 degradation [70,71]. Therefore, combining the location of a protein partner interaction on Cx43 along with the phosphorylation pattern under different homeostatic and pathological conditions will be crucial information for any potential therapeutic intervention. Here, we identified that β-catenin directly interacts with the Cx43CT domain, and that this interaction would be inhibited by Src phosphorylation of Cx43CT residues Y265 and Y313.
4. Material and Methods
4.1. Expression and Purification of Recombinant Proteins
The rat Cx43CT (V236–I382) (or (S255–I382) for the SPR study) polypeptide (unlabeled or 15N-labeled) cloned into the bacterial expression vector pGEX-6P-2 (GST-tagged; Amersham Biosciences, Little Chalfont, UK) was expressed and purified in 1× phosphate-buffered saline (PBS), as previously described [72,73]. Y313D and Y265,313D mutations in the Cx43CT plasmid were incorporated using the Quick Change Lightning kit (Qiagen, Hilden, Germany). Human β-catenin in pET-28a (+) was purchased from Addgene, expressed (unlabeled), and purified by Nickel affinity column (Buffer A: 50 mM Tris, 150 mM NaCl, 10 mM Imidazole, 2 mM β-mercaptoethanol (BME), pH 8.0; Buffer B: 50 mM Tris, 150 mM NaCl, 600 mM Imidazole, 2 mM BME, pH 8.0) followed by Anion exchange (Buffer A: 50 mM Tris, 2 mM BME, pH 8; Buffer B: 50 mM Tris, 1 M NaCl, 2 mM BME, pH 8). β-catenin ΔCT (M1-S680) was obtained by introducing a stop codon after serine 680 using the Quick Change Lightning kit (Agilent, Santa Clara, CA, USA). Purification was identical to the full-length β-catenin. β-catenin CT (S681–L781) was subcloned into the pET-16b vector, expressed (unlabeled), and purified by Nickel affinity column similarly to the full-length β-catenin. Purity and analysis for degradation was assessed by SDS-PAGE, and all of the polypeptides were equilibrated by dialysis in Slide-A-Lyzer G2 Dialysis Cassettes (Thermo Scientific, Waltham, MA, USA) in 1× PBS at pH 7.8 in presence of 2 mM dithiothreitol.
4.2. Nuclear Magnetic Resonance (NMR)
NMR data were acquired at 7 °C using a 600-MHz Varian INOVA spectrometer (Agilent, Palo Alto, CA, USA) upgraded with a Bruker Avance-III HD console (Bruker, Billerica, MA, USA) and outfitted with a Bruker z-axis PFG “inverse” triple-resonance cryogenic (cold) probe (Bruker). Gradient-enhanced two-dimensional 15N-HSQC experiments were used to obtain the binding isotherms of the 15N-labeled Cx43CT WT, Y313D, and Y265,313D at a constant concentration (35 μM) in the absence or presence of increasing amounts (up to 285 μM) of β-catenin, β-catenin CT, or β-catenin ΔCT. Data acquisition, processing, and analysis, including calculation of the dissociation constants (KD), have been previously described [14,46,69].
4.3. Circular dichroism (CD)
The CD experiment was performed on a JASCO J-815 CD spectrometer (JASCO, Mary’s Court, Easton, MD, USA) at 7 °C in the far UV (260–190 nm). Spectra of the β-catenin CT were collected in 1× PBS at pH 7.4, with a 0.1-mm path length quartz cell, using a bandwidth of 1 nm, an integration time of 1 s, and a scan rate of 50 nm/min. The final spectrum was obtained from the average of five scans. All of the spectra were corrected by subtracting the solvent spectrum acquired under identical conditions. CD data were processed and converted to mean residue ellipticity using Spectra Analysis from the Jasco Spectra Manager software, Version 2.05.01 (JASCO, Mary’s Court, Easton, MD, USA).
4.4. Surface plasmon resonance (SPR)
The SPR experiments were performed on a Biacore (GE Healthcare) 1000 at 25 °C. The Cx43CT (S255–I382) was immobilized onto a CM5 sensor chip by amine coupling, and the flow cell was equilibrated with the reaction buffer at a flow rate of 5 μL/min (213 mM phosphate buffer, pH 7.1). Then, 5 μL of either the β-catenin CT (4 μM), the Cx43EL1 (residues G38–R76, 10 μM, negative control), or the Cx43CT (10 μM, positive control) were injected over the chip, and the responses were recorded as resonance units (RU).
Acknowledgments
P.L.S. was supported by National Institutes of Health Grants GM072631, GM319613, and GM103427.
Author Contributions
P.L.S. and G.S. conceived and designed the experiments; G.S., A.J.T., L.Z., M.G., I.B., C.S., G.M. and M.C. performed the experiments; G.S., A.J.T. and L.Z., analyzed the data; P.L.S., G.S. and A.J.T. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Goodenough D.A., Goliger J.A., Paul D.L. Connexins, connexons, and intercellular communication. Annu. Rev. Biochem. 1996;65:475–502. doi: 10.1146/annurev.bi.65.070196.002355. [DOI] [PubMed] [Google Scholar]
- 2.Severs N.J., Bruce A.F., Dupont E., Rothery S. Remodelling of gap junctions and connexin expression in diseased myocardium. Cardiovasc. Res. 2008;80:9–19. doi: 10.1093/cvr/cvn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans W.H., Martin P.E. Gap junctions: Structure and function. Mol. Membr. Biol. 2002;19:121–136. doi: 10.1080/09687680210139839. [DOI] [PubMed] [Google Scholar]
- 4.Laird D.W. The gap junction proteome and its relationship to disease. Trends Cell Biol. 2010;20:92–101. doi: 10.1016/j.tcb.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Lampe P.D., Lau A.F. The effects of connexin phosphorylation on gap junctional communication. Int. J. Biochem. Cell Biol. 2004;36:1171–1186. doi: 10.1016/S1357-2725(03)00264-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herve J., Bourmeyster N., Sarrouilhe D., Duffy H. Gap junctional complexes: From partners to functions. Prog. Biophys. Mol. Biol. 2007;94:29–65. doi: 10.1016/j.pbiomolbio.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Thevenin A.F., Kowal T.J., Fong J.T., Kells R.M., Fisher C.G., Falk M.M. Proteins and mechanisms regulating gap-junction assembly, internalization, and degradation. Physiology. 2013;28:93–116. doi: 10.1152/physiol.00038.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno A.P., Chanson M., Elenes S., Anumonwo J., Scerri I., Gu H., Taffet S.M., Delmar M. Role of the carboxyl terminal of connexin43 in transjunctional fast voltage gating. Circ. Res. 2002;90:450–457. doi: 10.1161/hh0402.105667. [DOI] [PubMed] [Google Scholar]
- 9.Morley G.E., Taffet S.M., Delmar M. Intramolecular interactions mediate pH regulation of connexin43 channels. Biophys. J. 1996;70:1294–1302. doi: 10.1016/S0006-3495(96)79686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anumonwo J.M., Taffet S.M., Gu H., Chanson M., Moreno A.P., Delmar M. The carboxyl terminal domain regulates the unitary conductance and voltage dependence of connexin40 gap junction channels. Circ. Res. 2001;88:666–673. doi: 10.1161/hh0701.088833. [DOI] [PubMed] [Google Scholar]
- 11.Revilla A., Castro C., Barrio L.C. Molecular dissection of transjunctional voltage dependence in the connexin-32 and connexin-43 junctions. Biophys. J. 1999;77:1374–1383. doi: 10.1016/S0006-3495(99)76986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorgen P.L., Duffy H.S., Sahoo P., Coombs W., Delmar M., Spray D.C. Structural changes in the carboxyl terminus of the gap junction protein connexin43 indicates signaling between binding domains for c-Src and zonula occludens-1. J. Biol. Chem. 2004;279:54695–54701. doi: 10.1074/jbc.M409552200. [DOI] [PubMed] [Google Scholar]
- 13.Bouvier D., Kieken F., Kellezi A., Sorgen P.L. Structural changes in the carboxyl terminus of the gap junction protein connexin 40 caused by the interaction with c-Src and zonula occludens-1. Cell Commun. Adhes. 2008;15:107–118. doi: 10.1080/15419060802014347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stauch K., Kieken F., Sorgen P. Characterization of the structure and intermolecular interactions between the connexin 32 carboxyl-terminal domain and the protein partners synapse-associated protein 97 and calmodulin. J. Biol. Chem. 2012;287:27771–27788. doi: 10.1074/jbc.M112.382572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson T.K., Sorgen P.L., Burt J.M. Carboxy terminus and pore-forming domain properties specific to Cx37 are necessary for Cx37-mediated suppression of insulinoma cell proliferation. Am. J. Physiol. Cell Physiol. 2013;305:C1246–C1256. doi: 10.1152/ajpcell.00159.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilleron J., Carette D., Chevallier D., Segretain D., Pointis G. Molecular connexin partner remodeling orchestrates connexin traffic: From physiology to pathophysiology. Crit. Rev. Biochem. Mol. Biol. 2012;47:407–423. doi: 10.3109/10409238.2012.683482. [DOI] [PubMed] [Google Scholar]
- 17.Sorgen P.L., Trease A.J., Spagnol G., Delmar M., Nielsen M.S. Protein–Protein Interactions with Connexin 43: Regulation and Function. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19051428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basheer W., Shaw R. The “tail” of Connexin43: An unexpected journey from alternative translation to trafficking. Biochim. Biophys. Acta. 2016;1863:1848–1856. doi: 10.1016/j.bbamcr.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epifantseva I., Shaw R.M. Intracellular trafficking pathways of Cx43 gap junction channels. Biochim. Biophys. Acta. 2017;1860:40–47. doi: 10.1016/j.bbamem.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau A.F., Kurata W.E., Kanemitsu M.Y., Loo L.W., Warn-Cramer B.J., Eckhart W., Lampe P.D. Regulation of connexin43 function by activated tyrosine protein kinases. J. Bioenerg. Biomembr. 1996;28:359–368. doi: 10.1007/BF02110112. [DOI] [PubMed] [Google Scholar]
- 21.Lin R., Warn-Cramer B.J., Kurata W.E., Lau A.F. v-Src phosphorylation of connexin 43 on Tyr247 and Tyr265 disrupts gap junctional communication. J. Cell Biol. 2001;154:815–827. doi: 10.1083/jcb.200102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saidi Brikci-Nigassa A., Clement M.J., Ha-Duong T., Adjadj E., Ziani L., Pastre D., Curmi P.A., Savarin P. Phosphorylation controls the interaction of the connexin43 C-terminal domain with tubulin and microtubules. Biochemistry. 2012;51:4331–4342. doi: 10.1021/bi201806j. [DOI] [PubMed] [Google Scholar]
- 23.Ambrosi C., Ren C., Spagnol G., Cavin G., Cone A., Grintsevich E.E., Sosinsky G.E., Sorgen P.L. Connexin43 Forms Supramolecular Complexes through Non-Overlapping Binding Sites for Drebrin, Tubulin, and ZO-1. PLoS ONE. 2016;11:e0157073. doi: 10.1371/journal.pone.0157073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. Molecular Biology of the Cell. Garland Science; New York, NY, USA: 2002. Transport from the Trans Golgi Network to Lysosomes. [Google Scholar]
- 25.Butkevich E., Hulsmann S., Wenzel D., Shirao T., Duden R., Majoul I. Drebrin is a novel connexin-43 binding partner that links gap junctions to the submembrane cytoskeleton. Curr. Biol. 2004;14:650–658. doi: 10.1016/j.cub.2004.03.063. [DOI] [PubMed] [Google Scholar]
- 26.Kieken F., Mutsaers N., Dolmatova E., Virgil K., Wit A.L., Kellezi A., Hirst-Jensen B.J., Duffy H.S., Sorgen P.L. Structural and molecular mechanisms of gap junction remodeling in epicardial border zone myocytes following myocardial infarction. Circ. Res. 2009;104:1103–1112. doi: 10.1161/CIRCRESAHA.108.190454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhett J.M., Jourdan J., Gourdie R.G. Connexin 43 connexon to gap junction transition is regulated by zonula occludens-1. Mol. Biol. Cell. 2011;22:1516–1528. doi: 10.1091/mbc.e10-06-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solan J.L., Lampe P.D. Connexin 43 in LA-25 cells with active v-src is phosphorylated on Y247, Y265, S262, S279/282, and S368 via multiple signaling pathways. Cell Commun. Adhes. 2008;15:75–84. doi: 10.1080/15419060802014016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ai Z., Fischer A., Spray D.C., Brown A.M., Fishman G.I. Wnt-1 regulation of connexin43 in cardiac myocytes. J. Clin. Investig. 2000;105:161–171. doi: 10.1172/JCI7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prakash S., Swaminathan U., Nagamalini B.R., Krishnamurthy A.B. β-catenin in disease. J. Oral Maxillofacc. Pathol. 2016;20:289–299. doi: 10.4103/0973-029X.185938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenzon A., Calore M., Poloni G., De Windt L.J., Braghetta P., Rampazzo A. Wnt/β-catenin pathway in arrhythmogenic cardiomyopathy. Oncotarget. 2017;8:60640–60655. doi: 10.18632/oncotarget.17457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xing Y., Takemaru K., Liu J., Berndt J.D., Zheng J.J., Moon R.T., Xu W. Crystal structure of a full-length β-catenin. Structure. 2008;16:478–487. doi: 10.1016/j.str.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson D.J., Christian J.L., Moon R.T. Effect of wnt-1 and related proteins on gap junctional communication in Xenopus embryos. Science. 1991;252:1173–1176. doi: 10.1126/science.252.5009.1173. [DOI] [PubMed] [Google Scholar]
- 34.Moorer M.C., Hebert C., Tomlinson R.E., Iyer S.R., Chason M., Stains J.P. Defective signaling, osteoblastogenesis and bone remodeling in a mouse model of connexin 43 C-terminal truncation. J. Cell Sci. 2017;130:531–540. doi: 10.1242/jcs.197285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rinaldi F., Hartfield E.M., Crompton L.A., Badger J.L., Glover C.P., Kelly C.M., Rosser A.E., Uney J.B., Caldwell M.A. Cross-regulation of Connexin43 and β-catenin influences differentiation of human neural progenitor cells. Cell Death Dis. 2014;5:e1017. doi: 10.1038/cddis.2013.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talhouk R.S., Fares M.B., Rahme G.J., Hariri H.H., Rayess T., Dbouk H.A., Bazzoun D., Al-Labban D., El-Sabban M.E. Context dependent reversion of tumor phenotype by connexin-43 expression in MDA-MB231 cells and MCF-7 cells: Role of β-catenin/connexin43 association. Exp. Cell Res. 2013;319:3065–3080. doi: 10.1016/j.yexcr.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Swope D., Cheng L., Gao E., Li J., Radice G.L. Loss of cadherin-binding proteins β-catenin and plakoglobin in the heart leads to gap junction remodeling and arrhythmogenesis. Mol. Cell. Biol. 2012;32:1056–1067. doi: 10.1128/MCB.06188-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du W.J., Li J.K., Wang Q.Y., Hou J.B., Yu B. Lithium chloride preconditioning optimizes skeletal myoblast functions for cellular cardiomyoplasty in vitro via glycogen synthase kinase-3β/β-catenin signaling. Cells Tissues Organs. 2009;190:11–19. doi: 10.1159/000167699. [DOI] [PubMed] [Google Scholar]
- 39.Xu W., Kimelman D. Mechanistic insights from structural studies of β-catenin and its binding partners. J. Cell Sci. 2007;120:3337–3344. doi: 10.1242/jcs.013771. [DOI] [PubMed] [Google Scholar]
- 40.Kim W., Kim M., Jho E.H. Wnt/β-catenin signalling: From plasma membrane to nucleus. Biochem. J. 2013;450:9–21. doi: 10.1042/BJ20121284. [DOI] [PubMed] [Google Scholar]
- 41.Grosely R., Kopanic J.L., Nabors S., Kieken F., Spagnol G., Al-Mugotir M., Zach S., Sorgen P.L. Effects of phosphorylation on the structure and backbone dynamics of the intrinsically disordered connexin43 C-terminal domain. J. Biol. Chem. 2013;288:24857–24870. doi: 10.1074/jbc.M113.454389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valenta T., Hausmann G., Basler K. The many faces and functions of β-catenin. EMBO J. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitmore L., Wallace B.A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitmore L., Wallace B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 45.Sorgen P.L., Duffy H.S., Spray D.C., Delmar M. pH-dependent dimerization of the carboxyl terminal domain of Cx43. Biophys. J. 2004;87:574–581. doi: 10.1529/biophysj.103.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H., Spagnol G., Naslavsky N., Caplan S., Sorgen P.L. TC-PTP directly interacts with connexin43 to regulate gap junction intercellular communication. J. Cell Sci. 2014;127:3269–3279. doi: 10.1242/jcs.145193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hornbeck P.V., Kornhauser J.M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solan J.L., Marquez-Rosado L., Sorgen P.L., Thornton P.J., Gafken P.R., Lampe P.D. Phosphorylation at S365 is a gatekeeper event that changes the structure of Cx43 and prevents down-regulation by PKC. J. Cell Biol. 2007;179:1301–1309. doi: 10.1083/jcb.200707060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Remo B.F., Qu J., Volpicelli F.M., Giovannone S., Shin D., Lader J., Liu F.Y., Zhang J., Lent D.S., Morley G.E., et al. Phosphatase-resistant gap junctions inhibit pathological remodeling and prevent arrhythmias. Circ. Res. 2011;108:1459–1466. doi: 10.1161/CIRCRESAHA.111.244046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trease A.J., Capuccino J.M.V., Contreras J., Harris A.L., Sorgen P.L. Intramolecular signaling in a cardiac connexin: Role of cytoplasmic domain dimerization. J. Mol. Cell. Cardiol. 2017;111:69–80. doi: 10.1016/j.yjmcc.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leithe E., Mesnil M., Aasen T. The connexin 43 C-terminus: A tail of many tales. Biochim. Biophys. Acta. 2018;1860:48–64. doi: 10.1016/j.bbamem.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 52.Solan J.L., Lampe P.D. Spatio-temporal regulation of connexin43 phosphorylation and gap junction dynamics. Biochim. Biophys. Acta. 2018;1860:83–90. doi: 10.1016/j.bbamem.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falk M.M., Bell C.L., Kells Andrews R.M., Murray S.A. Molecular mechanisms regulating formation, trafficking and processing of annular gap junctions. BMC Cell Biol. 2016;17:S22. doi: 10.1186/s12860-016-0087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deb A. Cell-cell interaction in the heart via Wnt/β-catenin pathway after cardiac injury. Cardiovasc. Res. 2014;102:214–223. doi: 10.1093/cvr/cvu054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van der Heyden M.A., Rook M.B., Hermans M.M., Rijksen G., Boonstra J., Defize L.H., Destree O.H. Identification of connexin43 as a functional target for Wnt signalling. Pt 12J. Cell Sci. 1998;111:1741–1749. doi: 10.1242/jcs.111.12.1741. [DOI] [PubMed] [Google Scholar]
- 56.Shaw R.M., Fay A.J., Puthenveedu M.A., von Zastrow M., Jan Y.N., Jan L.Y. Microtubule plus-end-tracking proteins target gap junctions directly from the cell interior to adherens junctions. Cell. 2007;128:547–560. doi: 10.1016/j.cell.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du W.J., Li J.K., Wang Q.Y., Hou J.B., Yu B. Lithium chloride regulates connexin43 in skeletal myoblasts in vitro: Possible involvement in Wnt/β-catenin signaling. Cell Commun. Adhes. 2008;15:261–271. doi: 10.1080/15419060802198587. [DOI] [PubMed] [Google Scholar]
- 58.Nakashima T., Ohkusa T., Okamoto Y., Yoshida M., Lee J.K., Mizukami Y., Yano M. Rapid electrical stimulation causes alterations in cardiac intercellular junction proteins of cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2014;306:H1324–H1333. doi: 10.1152/ajpheart.00653.2013. [DOI] [PubMed] [Google Scholar]
- 59.Reaume A.G., de Sousa P.A., Kulkarni S., Langille B.L., Zhu D., Davies T.C., Juneja S.C., Kidder G.M., Rossant J. Cardiac malformation in neonatal mice lacking connexin43. Science. 1995;267:1831–1834. doi: 10.1126/science.7892609. [DOI] [PubMed] [Google Scholar]
- 60.Musil L.S., Goodenough D.A. Multisubunit assembly of an integral plasma membrane channel protein, gap junction connexin43, occurs after exit from the ER. Cell. 1993;74:1065–1077. doi: 10.1016/0092-8674(93)90728-9. [DOI] [PubMed] [Google Scholar]
- 61.Lauf U., Giepmans B.N., Lopez P., Braconnot S., Chen S.C., Falk M.M. Dynamic trafficking and delivery of connexons to the plasma membrane and accretion to gap junctions in living cells. Proc. Natl. Acad. Sci. USA. 2002;99:10446–10451. doi: 10.1073/pnas.162055899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giepmans B.N., Verlaan I., Hengeveld T., Janssen H., Calafat J., Falk M.M., Moolenaar W.H. Gap junction protein connexin-43 interacts directly with microtubules. Curr. Biol. 2001;11:1364–1368. doi: 10.1016/S0960-9822(01)00424-9. [DOI] [PubMed] [Google Scholar]
- 63.Fort A.G., Murray J.W., Dandachi N., Davidson M.W., Dermietzel R., Wolkoff A.W., Spray D.C. In vitro motility of liver connexin vesicles along microtubules utilizes kinesin motors. J. Biol. Chem. 2011;286:22875–22885. doi: 10.1074/jbc.M111.219709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smyth J.W., Vogan J.M., Buch P.J., Zhang S.S., Fong T.S., Hong T.T., Shaw R.M. Actin cytoskeleton rest stops regulate anterograde traffic of connexin 43 vesicles to the plasma membrane. Circ. Res. 2012;110:978–989. doi: 10.1161/CIRCRESAHA.111.257964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fang D., Hawke D., Zheng Y., Xia Y., Meisenhelder J., Nika H., Mills G.B., Kobayashi R., Hunter T., Lu Z. Phosphorylation of β-catenin by AKT promotes β-catenin transcriptional activity. J. Biol. Chem. 2007;282:11221–11229. doi: 10.1074/jbc.M611871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu C., Li Y., Semenov M., Han C., Baeg G.H., Tan Y., Zhang Z., Lin X., He X. Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/S0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 67.Zhang M., Mahoney E., Zuo T., Manchanda P.K., Davuluri R.V., Kirschner L.S. Protein kinase A activation enhances β-catenin transcriptional activity through nuclear localization to PML bodies. PLoS ONE. 2014;9:e109523. doi: 10.1371/journal.pone.0109523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Solan J.L., Lampe P.D. Specific Cx43 phosphorylation events regulate gap junction turnover in vivo. FEBS Lett. 2014;588:1423–1429. doi: 10.1016/j.febslet.2014.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spagnol G., Kieken F., Kopanic J.L., Li H., Zach S., Stauch K.L., Grosely R., Sorgen P.L. Structural Studies of the Nedd4 WW Domains and Their Selectivity for the Connexin43 (Cx43) Carboxyl Terminus. J. Biol. Chem. 2016;291:7637–7650. doi: 10.1074/jbc.M115.701417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Girao H., Catarino S., Pereira P. Eps15 interacts with ubiquitinated Cx43 and mediates its internalization. Exp. Cell Res. 2009;315:3587–3597. doi: 10.1016/j.yexcr.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 71.Leykauf K., Salek M., Bomke J., Frech M., Lehmann W.D., Durst M., Alonso A. Ubiquitin protein ligase Nedd4 binds to connexin43 by a phosphorylation-modulated process. J. Cell Sci. 2006;119:3634–3642. doi: 10.1242/jcs.03149. [DOI] [PubMed] [Google Scholar]
- 72.Duffy H.S., Sorgen P.L., Girvin M.E., O’Donnell P., Coombs W., Taffet S.M., Delmar M., Spray D.C. pH-dependent intramolecular binding and structure involving Cx43 cytoplasmic domains. J. Biol. Chem. 2002;277:36706–36714. doi: 10.1074/jbc.M207016200. [DOI] [PubMed] [Google Scholar]
- 73.Hirst-Jensen B.J., Sahoo P., Kieken F., Delmar M., Sorgen P.L. Characterization of the pH-dependent interaction between the gap junction protein connexin43 carboxyl terminus and cytoplasmic loop domains. J. Biol. Chem. 2007;282:5801–5813. doi: 10.1074/jbc.M605233200. [DOI] [PubMed] [Google Scholar]