Abstract
Calcium plays an important role in plant growth, development, and response to environmental stimuli. Copines are conserved plasma membrane-localized calcium-binding proteins which regulate plant immune responses and development. In this study, we found that copine proteins BON2 and BON3, the paralogs of BON1, physically interact with calcium pumps ACA8 and ACA10 in Arabidopsis. Notably, ACA9, the closest homologue of ACA8 and ACA10 functioning in pollen tube growth, interacts with all three copines. This is consistent with the protein–protein interactions between the two protein families, the aca8, aca10, aca8/aca10, bon1/2/3 mutants as well as aca9 mutant exhibited defects on pollen germination and seed production. Taken together, plasma membrane-localized interacting calcium pumps and copines coordinately control pollen tube growth, likely through manipulating calcium efflux.
Keywords: autoinhibited Ca2+ ATPase (ACA), calcium, calcium pump, copine, fertility, pollen germination
1. Introduction
Ca2+ is essential for eukaryotic cells as an important secondary messenger or a structural component of enzymes and macromolecular complexes [1,2,3]. Calcium carries specific information response to environmental and endogenous cues through the amplitude, frequency, and duration of calcium spikes [2,4]. This Ca2+ signature is shaped by the coordinated actions of membrane transport proteins in Ca2+ influx and efflux systems including channels, pumps, and exchangers [3,5]. Several ion channels, such as Ca2+ permeable cyclic nucleotide-gated channels and voltage-gated channels, locate at the plasma membrane (PM), endoplasmic reticulum (ER), vacuole, or mitochondria and control calcium influx [6,7,8]. Calcium efflux can be mediated by Ca2+ ATPase pumps and Ca2+/H+ exchangers [5]. In Arabidopsis thaliana, there are 14 putative Ca2+ ATPase pumps that can be divided into two distinct subfamilies, the autoinhibited calcium ATPases (ACAs) and the endoplasmic reticulum-type Ca2+ ATPases (ECAs) [9,10]. The autoinhibitory domain at the N-terminal ACA represses its ion pump activity. When calmodulin binding to the autoinhibitory domain, the calcium pump activity was released [11,12,13]. Among the 10 ACAs in Arabidopsis, ACA8, ACA9, ACA10 belong to one clade and localize on the plasma membrane [14,15,16].
ACA genes are involved in multiple biological processes, including plant growth, development, and immunity. ACA7, ACA9, and ACA13, which are located at the plasma membrane, have been shown to be important for pollen development [15,17,18]. ACA9 specifically expresses in pollen and plays a key role in the regulation of pollen tube growth and fertilization [15]. The loss of function mutant of ACA7 produces significant levels of dead pollen grains in mature flowers, suggesting its important role in pollen development [17]. The mutation in ACA13 causes defects both in pollination and seed production [18]. ACA10 and ACA8 play major roles in vegetative growth and immunity [9,19]. In addition, ACA8 interacts with FLS2 and functions in disease resistance response [10]. ACA13 and ACA12 are also involved in disease resistance and growth, especially when the function of ACA10 and/or ACA8 is compromised [9]. In addition, the mutation of ACA13 and ACA10, together with a reduced function of ACA8, result in death at bolting time, revealing the essential roles of their collective function in plant growth [9]. ACA4 and ACA11 reside at vacuole, the double mutant of aca4aca11 displays a high frequency of hypersensitive response-like lesions associated with the activation of the salicylic acid pathway [6,20].
Copines are evolutionarily conserved proteins in protozoa, plants, nematodes, and mammals [21]. The BON1/CPN1 in Arabidopsis was the first reported copine gene which acts as a repressor of plant autoimmunity [22,23]. There are two additional copine genes in the Arabidopsis genome, designated as BON2 and BON3 [22]. The three genes have overlapped functions which are essential for the viability of plants [22]. The loss of function of BON1, combined with BON2 or BON3, leads to extensive cell death, resembling the hypersensitive response seen in defense responses; however, no abnormal growth or development phenotypes are found in single bon2 or bon3 mutants under normal conditions [22].
Recently, we found that the von Willebrand factor type A (VWA)domain of BON1 physically interacts with the autoinhibited domain (ID) of ACA10 and ACA8, suggesting another regulatory mechanism of calcium pump activity except calmodulin binding [19]. Consistent with physical interaction between BON1 and ACA8/10, the mutation in BON1, ACA8, and ACA10 compromised the production of calcium signature in guard cell upon external calcium treatment [19]. Consequently, the mutants exhibited defects on stomatal closure and inhibition of constitutive defense response. This uncovered a critical role for ACA8 and ACA10 in calcium signature generation as well as their regulation by an evolutionarily conserved BON1 protein in Arabidopsis [19].
In this study, we also found that BON2 and BON3 physically interact with ACA8 and ACA10. Interestingly, BON1/2/3 also physically interacts with ACA9, the closest homologue with ACA8/10. Consistent with the protein–protein interaction, the aca8, aca9, aca10, aca8/aca10, and bon1/2/3 mutants exhibited defects on pollen germination and seed production. These results demonstrate the coordination between ACA pumps and copines in pollen growth, likely via calcium signaling.
2. Results and Discussion
Given that BON1 genetically and physically interacts with ACA8/10 [19], it is possible that the other two copines interact with plasma membrane-localized ACA pumps. To test the hypothesis, we performed split-LUC assay, a widely used method to test protein–protein interaction in tobacco leaves [24]. The coding regions of BON2 and BON3 without stop codon were respectively ligated into the pCAMBIA-NLuc vector [24], and the amino-terminal part of ACA8 (1-106aa, Supplementary Materials, Figure S1A) was ligated into the pCAMBIA-CLuc vector [24]. When the fused protein of BON2 and BON3 with the amino-terminus of firefly luciferase coexpressed with the fused protein of segment I of ACA8 (1–106aa, Supplementary Materials Figure S1A) with carboxyl-terminus of luciferase, it showed strong luminescence signal, suggesting the protein–protein interaction between ACA8 and BON2 and BON3 (Supplementary Materials Figure S2A). In addition, the split-LUC assay also demonstrated the protein–protein interaction between N-terminal of ACA10 (1–72aa, Supplementary Materials Figure S1A) and BON2 and BON3 (Supplementary Materials Figure S2B). Given that the segment I of ACA8 and ACA10 contained the autoinhibited domain, the interaction between BON2 and BON3 with ACA8 and ACA10 released the pump’s activity [19].
To confirm the interaction and further locate the domain responsible for the physical interaction, we cloned segment I of ACA8 (1–106aa), ACA10 (1–72aa, Supplementary Materials Figure S1A) into the activation domain vector (AD) and the VWA domain of BON2 (328–586aa), BON3 (326–584aa, Supplementary Materials Figure S1A) into the DNA-binding domain vector (BD) to perform yeast-two-hybrid assay. As we found previously, the BON1-VWA-BD with ACA8-I-AD and BON1-VWA-BD with ACA10-I-AD (Supplementary Materials Figure S1A) yeasts grew well on synthetic defined (SD) medium which lacked four kinds of amino acids (Trp, Leu, His, and Ade) (Supplementary Materials Figure S3); this confirmed that the protein–protein interaction happens on the VWA domain of BON1 (324–578aa) and the autoinhibitory domain of ACA8 and ACA10 (Supplementary Materials Figure S1A). Yeast-two hybrid assay also demonstrated that the VWA domains of BON2-VWA-BD (328–586aa) and BON3-VWA-BD (326–584aa) interact with the autoinhibitory domains of ACA8-I-AD (1–106aa) and ACA10-I-AD (1–72aa, Supplementary Materials Figure S1A; Supplementary Materials Figure S3). The physical interactions between BON1, BON2, BON3 and ACA8, ACA10 raise the possibility that the cooperative role of five proteins in regulating calcium signature, immunity and stomatal movement.
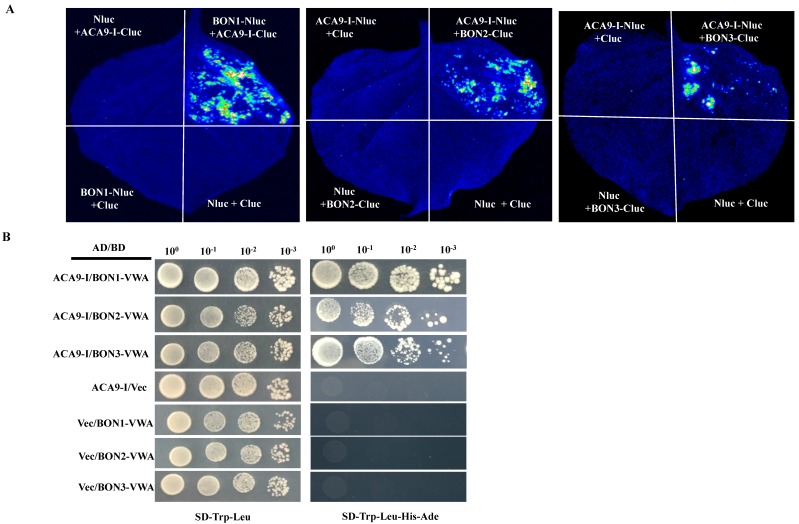
ACA9 is an essential factor for pollen development and fertilization [15]; its possible interactions with copines were also examined. As seen in ACA8 and ACA10, the fused protein of the autoinhibitory domain of ACA9 (1-101aa, Supplementary Materials Figure S1A) with C-terminus of luciferase produced a strong luminescence signal when coexpressed with fused protein of BON1 with N-terminus of luciferase in N. benthamiana (Figure S1A). When the fused protein of the autoinhibitory domain of ACA9 (Supplementary Materials Figure S1A) with N-terminus of luciferase coexpressed with the fused protein of BON2 and BON3 with C-terminus of luciferase, a similar strong luminescence signal was observed (Figure 1A). Subsequently, this interaction was further verified by the yeast two-hybrid assay; we found that the C-terminal VWA domain of BON1 (324–578aa), BON2 (328–586aa), BON3 (326–584aa, Supplementary Materials Figure S1A) interact with the segment I of ACA9 (1–101aa, Supplementary Materials Figure S1A,B).
Figure 1.
Protein-protein interaction between ACA9 and BON1,BON2,BON3. (A). Split-LUC assay of the interaction of BON1, BON2 and BON3 with the first segment of ACA9. The indicated combinations were used as controls. (B). Yeast-two-hybrid assay between the first segment of ACA9 and the VWA domain of BON1, BON2 and BON3.
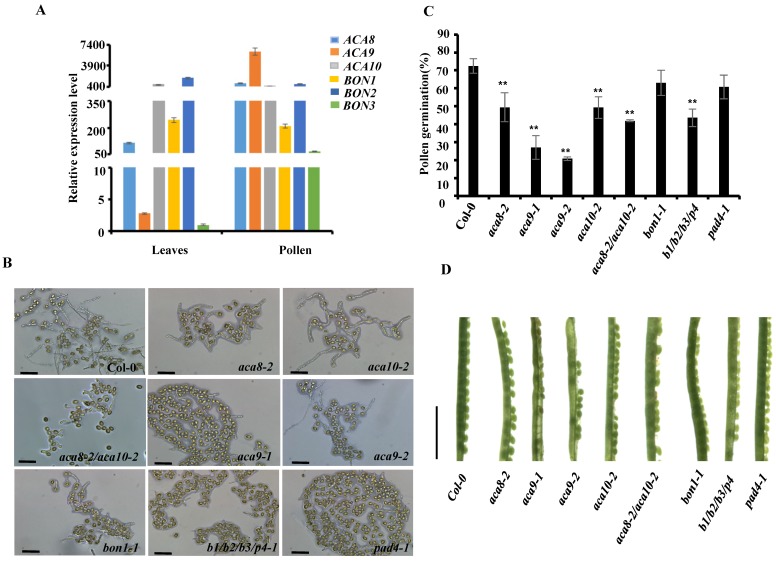
On the basis of the protein–protein interaction, it was speculated that BON1, BON2, and BON3 coordinate with ACA calcium pumps to control pollen development and fertilization. Real-time assay demonstrated that the six genes express both in pollens and leaves with ACA9 predominantly expressing in pollens (Figure 2A). To test the hypothesis, we characterized the mutants of ACA pumps and copines, focusing on pollen germination. In addition to aca9 mutant, the aca8, aca10 and aca8/aca10 mutants exhibited lower pollen germination ratio, suggesting the specific role of different calcium pumps in pollen function (Figure 2B,C). The pollen germination in bon1 mutant was comparable to wild-type control, suggesting the redundancy between copine genes (Figure 2B,C). Because the triple mutant of bon1/bon2/bon3 is lethal at seedling stage, we took advantage of bon1/bon2/bon3/pad4 mutant, in which the autoimmunity is largely rescued by mutation in PAD4 [22]. Indeed, the pollen germination ratio was largely reduced in bon1/bon2/bon3/pad4 mutant as compared to wild type (Figure 2B,C), suggesting that all three copines coordinately control pollen germination. Consistent with the defects in pollen germination, the seed production was compromised in aca8, aca10, aca8/aca10, and bon1/bon2/bon3/pad4 mutants as well as aca9 mutant (Figure 2C). In summary, two physically interacting family proteins, calcium pumps, and copines coordinately regulated pollen germination and fertility.
Figure 2.
The mutation in ACAs and BONs affect pollen germination. (A). Expression levels of ACA8/9/10 and BON1/2/3 in leaves and pollens. (B). The phenotype of pollen germination in wild type Col-0 and indicated genotypes (scale bars: 50 μm); (C). The pollen germination ratio in various genotypes. The average germination rate was generated from three biological replicates, each replicate with more than 300 pollens. Genotypes: b1/b2/b3/p4=bon1-1/bon2-2/bon3-3/pad4-1. ** represents significant differences compared to WT (p < 0.01). (D). Seed setting was affected in aca and bon mutants (scale bar 0.5 cm).
3. Materials and Methods
3.1. Plant Materials
The seeds of bon1-1/bon2-2/bon3-3/pad4-1 were kindly provided by Jian Hua [22]. The aca8-2 (GK_688H09), aca9-1 (SALK_045408), aca9-2 (SALK_108766), and aca10-2 (GK_044H01) mutants were obtained from the Arabidopsis Stock Centre (http://arabidopsis.org).
3.2. Plant Growth Conditions
Arabidopsis seedlings were first grown on half Murashige and Skoog (1/2MS) plates at 21 °C with 1% sucrose for 14 days under 16 h/8 h day/night cycle. The plants were then transferred to soil. The Arabidopsis plants were grown at 21 °C under conditions of 16 h/8 h day/night cycle and relative humidity at 50% to 70%.
3.3. Split-Luc Assay
The opening reading frames of BON1, BON2, and BON3 were amplified using the oligos listed in Table S1. The PCR fragments of BON1, BON2, and BON3 were ligated into the pCAMBIA-NLuc vector digested by BamHI and SalI using the ClonExpress MultiS One Step Cloning Kit (Vazyme, Nanjing, China) to generate BON1-NLuc, BON2-Nluc, and BON3-Nluc. The PCR fragment of BON2 and BON3 was ligated into the pCAMBIA-CLuc vector, digested by KpnI and SalI, using the ClonExpress MultiS One Step Cloning Kit (Vazyme, Nanjing, China) to generate BON2-CLuc and BON3-CLuc. The N-terminal parts of ACA8, ACA9 and ACA10 were amplified using the oligos listed in Table S1. The PCR fragments corresponding to the N-terminals of ACA8, ACA9, and ACA10 were inserted into the CLuc vector cleaved by KpnI and SalI to produce recombinant plasmids of ACA8-I-CLuc, ACA9-I-Cluc, and ACA10-I-CLuc. The PCR fragments corresponding to the N-terminal of ACA9 were inserted into the NLuc vector [24] cleaved by BamHI and SalI to produce recombinant plasmid of ACA9-I-NLuc. The above recombinant plasmids were transformed into Agrobacterium GV3101.
The agrobacterium with recombinant plasmid was cultured in liquid Luria-Bertani medium with antibiotics (50 mg/L rifampicin and 50 mg/L kanamycin) for about two days. The bacterium was then pelleted by centrifuge and then diluted with Murashige and Skoog medium (10 mM MES and 200 mM acetosyringone) into the concentration of OD600 = 0.6–0.8. After 2 h of induction, the agrobacterium with NLuc and CLuc recombinant constructs were mixed in equal volumes and co-transformed into Nicotiana benthamiana young leaves. After two days of infiltration under darkness, the leaves were transferred to light for 16 h. The leaves were then sprayed with 1 mM luciferin in 0.01% Triton X100 and kept in dark for 5 min. The luciferase activity was measured using a luminescence imaging system (Sony ICX694, Tanon, Shanghai, China) with a 3 min exposure time.
3.4. Pollen Germination Assay
The medium for in vitro pollen germination was modified from a previous report [25] and contained 18% (w/v) sucrose, 1.6 mM boric acid, 10 mM CaCl2, 1 mM Ca(NO3)2, 1 mM MgSO4, and 0.5% (w/v) agarose, pH adjusted to 6.4 with 0.5 M KOH. In vitro pollen germination experiments were conducted at 22 °C under a 100% relative humidity for 6 h [26] and then photographed using a fluorescence microscope (Olypums BX53, Tokyo, Japan). In each replicate, about 300 pollen grains were counted.
3.5. Yeast-Two-Hybrid Assay
The DNA fragments corresponding to the N-terminal segment I of ACA8, ACA9, ACA10 were respectively cloned into the pGADT7 vector (Clontech, Palo Alto, CA, USA). The DNA fragments corresponding to the VWA domain of BON1, BON2, BON3 were respectively cloned into the pGBKT7 vector (Clontech, Palo Alto, CA, USA). To examine the protein–protein interactions, the pGADT7 and pGBKT7 recombinant constructs were co-transformed into the yeast strain AH109. The yeast strains were selected on the SD-Trp-Leu medium and then transferred to the SD-Ade-Trp-Leu-His medium. The colony was photographed after three days growth.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/6/1774/s1.
Author Contributions
D.-L.Y. conceived the project and designed the experiment. Y.L. conducted most of the experiments. J.G. and Z.Y. contributed the materials and agents. D.-L.Y. and Y.L. wrote the manuscript.
Funding
This work was supported by the Natural Science Foundation of Jiangsu (BK20170027, BK20150659), the National Key Research and Development Program of China (2016YFD0100600) the Fundamental Research Funds for the Central Universities (KYZ201503, KJJQ201801), National Natural Science Foundation of China (31671340), National Key Transformation Program (2016ZX08001002), the Innovative Project of State Key Laboratory of Crop Genetics and Germplasm Enhancement (ZZ2017009) and Jiangsu Collaborative Innovation Center for Modern Crop Production to D.-L.Y.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.White P.J., Broadley M.R. Calcium in plants. Ann. Bot. 2003;92:487–511. doi: 10.1093/aob/mcg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dodd A.N., Kudla J., Sanders D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010;61:593–620. doi: 10.1146/annurev-arplant-070109-104628. [DOI] [PubMed] [Google Scholar]
- 3.Kudla J., Batistic O., Hashimoto K. Calcium signals: the lead currency of plant information processing. Plant Cell. 2010;22:541–563. doi: 10.1105/tpc.109.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonza M.C., De Michelis M.I. The plant Ca2+—ATPase repertoire: biochemical features and physiological functions. Plant Biol. (Stuttg. Ger.) 2011;13:421–430. doi: 10.1111/j.1438-8677.2010.00405.x. [DOI] [PubMed] [Google Scholar]
- 5.Steinhorst L., Kudla J. Calcium—A central regulator of pollen germination and tube growth. Biochim. Biophys. Acta. 2013;1833:1573–1581. doi: 10.1016/j.bbamcr.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Geisler M., Axelsen K.B., Harper J.F., Palmgren M.G. Molecular aspects of higher plant P-type Ca(2+)-ATPases. Biochim. Biophys. Acta. 2000;1465:52–78. doi: 10.1016/S0005-2736(00)00131-0. [DOI] [PubMed] [Google Scholar]
- 7.Sze H., Liang F., Hwang I., Curran A.C., Harper J.F. Diversity and regulation of plant Ca2+ pumps: Insights from expression in yeast. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:433–462. doi: 10.1146/annurev.arplant.51.1.433. [DOI] [PubMed] [Google Scholar]
- 8.Boursiac Y., Harper J.F. The origin and function of calmodulin regulated Ca2+ pumps in plants. J. Bioenerg. Biomembr. 2007;39:409–414. doi: 10.1007/s10863-007-9104-z. [DOI] [PubMed] [Google Scholar]
- 9.Yu H., Yan J., Du X., Hua J. Overlapping and differential roles of plasma membrane calcium ATPase ACAs in Arabidopsis growth and environmental responses. J. Exp. Bot. 2018;69:2693–2703. doi: 10.1093/jxb/ery073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frei dit Frey N., Mbengue M., Kwaaitaal M., Nitsch L., Altenbach D., Haweker H., Lozano-Duran R., Njo M.F., Beeckman T., Huettel B., et al. Plasma membrane calcium ATPases are important components of receptor-mediated signaling in plant immune responses and development. Plant Physiol. 2012;159:798–809. doi: 10.1104/pp.111.192575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa A., Luoni L., Marrano C.A., Hashimoto K., Koster P., Giacometti S., De Michelis M.I., Kudla J., Bonza M.C. Ca2+-dependent phosphoregulation of the plasma membrane Ca2+-ATPase ACA8 modulates stimulus-induced calcium signatures. J. Exp. Bot. 2017;68:3215–3230. doi: 10.1093/jxb/erx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giacometti S., Marrano C.A., Bonza M.C., Luoni L., Limonta M., De Michelis M.I. Phosphorylation of serine residues in the N-terminus modulates the activity of ACA8, a plasma membrane Ca2+-ATPase of Arabidopsis thaliana. J. Exp. Bot. 2012;63:1215–1224. doi: 10.1093/jxb/err346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tidow H., Poulsen L.R., Andreeva A., Knudsen M., Hein K.L., Wiuf C., Palmgren M.G., Nissen P. A bimodular mechanism of calcium control in eukaryotes. Nature. 2012;491:468–472. doi: 10.1038/nature11539. [DOI] [PubMed] [Google Scholar]
- 14.Bonza M.C., Morandini P., Luoni L., Geisler M., Palmgren M.G., De Michelis M.I. At-ACA8 encodes a plasma membrane-localized calcium-ATPase of Arabidopsis with a calmodulin-binding domain at the N terminus. Plant Physiol. 2000;123:1495–1506. doi: 10.1104/pp.123.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schiott M., Romanowsky S.M., Baekgaard L., Jakobsen M.K., Palmgren M.G., Harper J.F. A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc. Natl. Acad. Sci. USA. 2004;101:9502–9507. doi: 10.1073/pnas.0401542101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George L., Romanowsky S.M., Harper J.F., Sharrock R.A. The ACA10 Ca2+-ATPase regulates adult vegetative development and inflorescence architecture in Arabidopsis. Plant Physiol. 2008;146:716–728. doi: 10.1104/pp.107.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucca N., Leon G. Arabidopsis ACA7, encoding a putative auto-regulated Ca(2+)-ATPase, is required for normal pollen development. Plant Cell Rep. 2012;31:651–659. doi: 10.1007/s00299-011-1182-z. [DOI] [PubMed] [Google Scholar]
- 18.Iwano M., Igarashi M., Tarutani Y., Kaothien-Nakayama P., Nakayama H., Moriyama H., Yakabe R., Entani T., Shimosato-Asano H., Ueki M., et al. A pollen coat-inducible autoinhibited Ca2+-ATPase expressed in stigmatic papilla cells is required for compatible pollination in the Brassicaceae. Plant Cell. 2014;26:636–649. doi: 10.1105/tpc.113.121350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang D.L., Shi Z., Bao Y., Yan J., Yang Z., Yu H., Li Y., Gou M., Wang S., Zou B., et al. Calcium Pumps and Interacting BON1 Protein Modulate Calcium Signature, Stomatal Closure, and Plant Immunity. Plant Physiol. 2017;175:424–437. doi: 10.1104/pp.17.00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boursiac Y., Lee S.M., Romanowsky S., Blank R., Sladek C., Chung W.S., Harper J.F. Disruption of the vacuolar calcium-ATPases in Arabidopsis results in the activation of a salicylic acid-dependent programmed cell death pathway. Plant Physiol. 2010;154:1158–1171. doi: 10.1104/pp.110.159038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Creutz C.E., Tomsig J.L., Snyder S.L., Gautier M.C., Skouri F., Beisson J., Cohen J. The copines, a novel class of C2 domain-containing, calcium-dependent, phospholipid-binding proteins conserved from Paramecium to humans. J. Biol. Chem. 1998;273:1393–1402. doi: 10.1074/jbc.273.3.1393. [DOI] [PubMed] [Google Scholar]
- 22.Yang S., Yang H., Grisafi P., Sanchatjate S., Fink G.R., Sun Q., Hua J. The BON/CPN gene family represses cell death and promotes cell growth in Arabidopsis. Plant J. 2006;45:166–179. doi: 10.1111/j.1365-313X.2005.02585.x. [DOI] [PubMed] [Google Scholar]
- 23.Yang S., Hua J. A Haplotype-Specific Resistance Gene Regulated by BONZAI1 Mediates Temperature-Dependent Growth Control in Arabidopsis. Plant Cell. 2004;16:1060–1071. doi: 10.1105/tpc.020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.-M. Firefly Luciferase Complementation Imaging Assay for Pro tein-Protein Interactions in Plants. Plant Physiol. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H., Lin Y., Heath R.M., Zhu M.X., Yang Z. Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell. 1999;11:1731–1742. doi: 10.1105/tpc.11.9.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boavida L.C., McCormick S. Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 2007;52:570–582. doi: 10.1111/j.1365-313X.2007.03248.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.