Abstract
Intracellular deposits of α‐synuclein in the form of Lewy bodies are major hallmarks of Parkinson's disease (PD) and a range of related neurodegenerative disorders. Post‐translational modifications (PTMs) of α‐synuclein are increasingly thought to be major modulators of its structure, function, degradation and toxicity. Among these PTMs, phosphorylation near the C‐terminus at S129 has emerged as a dominant pathogenic modification as it is consistently observed to occur within the brain and cerebrospinal fluid (CSF) of post‐mortem PD patients, and its level appears to correlate with disease progression. Phosphorylation at the neighboring tyrosine residue Y125 has also been shown to protect against α‐synuclein toxicity in a Drosophila model of PD. In the present study we address the potential roles of C‐terminal phosphorylation in modulating the interaction of α‐synuclein with other protein partners, using a single domain antibody fragment (NbSyn87) that binds to the C‐terminal region of α‐synuclein with nanomolar affinity. The results reveal that phosphorylation at S129 has negligible effect on the binding affinity of NbSyn87 to α‐synuclein while phosphorylation at Y125, only four residues away, decreases the binding affinity by a factor of 400. These findings show that, despite the fact that α‐synuclein is intrinsically disordered in solution, selective phosphorylation can modulate significantly its interactions with other molecules and suggest how this particular form of modification could play a key role in regulating the normal and aberrant function of α‐synuclein.
Keywords: Parkinson's disease, α‐synuclein, protein misfolding, phosphorylation, single‐domain antibody (sdAb, nanobody), nuclear magnetic resonance, isothermal titration calorimetry, surface plasmon resonance
Abbreviations
- ITC
isothermal titration calorimetry
- NMR
nuclear magnetic resonance
- sdAb, nanobody
single‐domain antibody
- SPR
surface plasmon resonance
Introduction
The presence of intracellular aggregated forms of α‐synuclein in dopaminergic neurons and other brain regions are hallmarks of a series of neurodegenerative disorders, collectively known as synucleinopathies, of which the most common is Parkinson's disease (PD).1, 2, 3, 4, 5 In PD these intraneuronal proteinaceous inclusion bodies, referred to as Lewy bodies, are composed primarily of fibrillar and aggregated forms of wild‐type and post‐translationally modified α‐synuclein.1, 2, 6 Various mutations in the gene coding for α‐synuclein (including A30P,7 H50Q,8, 9 G51D,10 A53T,11 and E46K12) are associated with familial forms of the disorder, although they account for only 1–5% of PD cases.13 Understanding the role of the molecular and structural determinants of the functional and aggregation properties of α‐synuclein is therefore pivotal in elucidating the role of this protein in the pathogenesis of PD, as well as identifying novel diagnostic and therapeutic strategies for the treatment of the wider family of synucleinopathies.
It is generally believed that α‐synuclein is an intrinsically disordered protein in solution that adopts predominantly a β‐sheet conformation when it self‐associates into higher order aggregates and amyloid structures.14, 15, 16 In addition, binding of the protein to lipid bilayers results in its conversion to structures with high α‐helical content.17, 18, 19 Although recent studies have suggested that native α‐synuclein could exist as a tetramer, subsequent studies have supported previous findings that α‐synuclein exists predominantly as a disordered monomer.15, 20
The sequence of α‐synuclein can be divided into three distinct regions or domains based on the distribution of charged residues and their functional roles (Fig. 1). First, a positively charged N‐terminal domain (residues 1–60), which contains the sites of many PD associated mutations, comprises imperfect repeats of a conserved sequence motif (KTKEGV). This N‐terminal region has a high propensity to form amphipathic α‐helical structures upon interaction with lipid membranes and is thought to play a role in regulating the secretion and internalization of α‐synuclein.19, 22, 24 Second, a central amyloidogenic region, termed the nonamyloid β component (NAC), which includes residues 61–95, is highly hydrophobic and is thought to play a key role in aggregation and amyloid formation of α‐synuclein.19, 25 Third, a negatively charged and highly disordered C‐terminal region (residues 96–140), has been implicated in regulating α‐synuclein nuclear transport26 and is also thought to play a role in the assembly of SNARE‐complexes.27 Interestingly, most of the binding partners identified for α‐synuclein, including proteins,28, 29, 30 metal ions,31, 32 and other ligands33 (such as polyamines), interact with its C‐terminal domain. In addition, most known PTMs, including disease‐associated modifications, are located in the C‐terminal domain, suggesting that this region plays a key role in modulating the functions of α‐synuclein in vivo.34
Figure 1.

Schematic illustration of the sequence of α‐synuclein and its significance for the binding of NbSyn87. (A) Sequence of human α‐synuclein showing the seven imperfect repeats in the N‐terminal region, and the binding epitope of NbSyn87.21 The solid red line indicates the binding region of NbSyn87, while the dotted line marks the region experiencing resonance broadening in HSQC spectra that is indicative of a conformational change upon binding of the nanobody [Fig. S1(A), Supporting Information]. (B) The three distinct domains within the primary sequence of α‐synuclein are the N‐terminal domain, the NAC region, and the C‐terminal region. The arrows show the phosphorylation sites located within the C‐terminal region.
PTMs are increasingly recognized as important modulators of the structural, aggregational, and functional properties of α‐synuclein, both in health and disease.35, 36, 37 Although most of the known α‐synuclein PTMs have been identified in Lewy bodies, biophysical studies of site‐specifically modified proteins have shown that these modifications do not universally enhance α‐synuclein aggregation, suggesting that they may be involved in the regulation of normal physiological functions of α‐synuclein rather than its aggregation.38
The majority of published studies have focused primarily on elucidating the effects of phosphorylation on the structure, aggregation, and toxicity of α‐synuclein.35, 39, 40, 41, 42 In this study, we have used strategies that combine recombinant expression, chemical synthesis and native chemical ligation to produce α‐synuclein that is site specifically phosphorylated at Y125 and S129, in order to investigate how these commonly detected and well‐studied C‐terminal modifications influence the interaction of α‐synuclein with a specific protein binding partner.
As an example of an α‐synuclein protein–protein interaction, we used the recently generated camelid antibody fragment (nanobody), NbSyn87.21 NbSyn87 binds with nanomolar affinity to the C‐terminal domain of α‐synuclein, near the phosphorylation sites Y125 and S129,21 enabling us to investigate the impact of these two modifications on a specific protein–protein interaction. We have studied the effects of α‐synuclein phosphorylation on binding to NbSyn87 by using isothermal calorimetry (ITC), surface plasmon resonance (SPR), and nuclear magnetic resonance (NMR) spectroscopy. Furthermore, we have sought to determine whether or not the phosphorylation‐mimicking mutation, S129E, and the phosphorylation‐blocking mutation, S129A, commonly used to probe the effects of phosphorylation of α‐synuclein in vivo, reproduce the effects of α‐synuclein phosphorylation on its interactions with NbSyn87. Our results demonstrate, that despite the proximity of these two C‐terminal modification sites, the binding of NbSyn87 is affected very differently by the addition of phosphate groups to Y125 or S129. As well as demonstrating the highly selective effects on binding of the position of phosphorylation, our findings suggest that NbSyn87 could be a useful probe to evaluate the effects of α‐synuclein PTMs on its structure and toxicity.
Results
We have shown previously that polo‐like kinases 2 and 3 (PLK2 and PLK3) can phosphorylate α‐synuclein selectively at S129 with great efficiency (>95%).43 In contrast, little is known about kinases that can potently phosphorylate α‐synuclein at Y125 in a selective manner. Therefore, α‐synuclein phosphorylated at Y125 was prepared using semi‐synthetic strategies, as described previously.36 15N labeled pY125 α‐synuclein for NMR experiments was prepared in vitro by site‐specifically phosphorylating the Y133F/Y136F α‐synuclein double mutant, using the Syk kinase, which is known to phosphorylate at three C‐terminal tyrosine residues.36
Binding of NbSyn87 to α‐synuclein by ITC
We used ITC to measure the thermodynamic parameters of the binding of NbSyn87 nanobody to the wild type, pY125, S129A, S129E, and pS129 forms of α‐synuclein. The data shown in Figure 2 demonstrate that the binding of NbSyn87 to each α‐synuclein variant is exothermic, and that the isotherm generated from the integrated heats of the injections are consistent with a 1:1 bimolecular association reaction. For pS129, S129E, S129A, and wild type α‐synuclein, the measured values of K d are respectively 9.2 (±3.1), 41.1 (±10.4), 16.8 (±5.8), and 14.2(±8.4) nM, corresponding to ΔΔG values of −0.22, +0.67, and +0.15 kcal/mol, respectively, compared to the wild type protein, with concomitantly small differences in both the binding enthalpy and entropy (Table 1). In contrast, the interaction between pY125 α‐synuclein and NbSyn87 yielded a K d of 6 µM, i.e., about 400‐fold decrease in affinity compared to the interaction between the wild type α‐synuclein and NbSyn87. This corresponds to a ΔΔG value of 3.6 kcal/mol, originating from a large decrease in the binding enthalpy (ΔΔH = 2.5 kcal/mol) with an additional decrease in the binding entropy (TΔΔS = 1.1 kcal/mol) of the interaction. To confirm that the effect seen on the phosphorylated protein is due to the phosphorylation of Y125, we also measured the ITC of the double mutant Y133F/Y136F α‐synuclein (Fig. S2, Supporting Information). The measured value of K d, 11.6(±2.2) nM (Table 1), is very similar to that of the wild type protein, and only small differences in binding enthalpy and entropy were observed.
Figure 2.
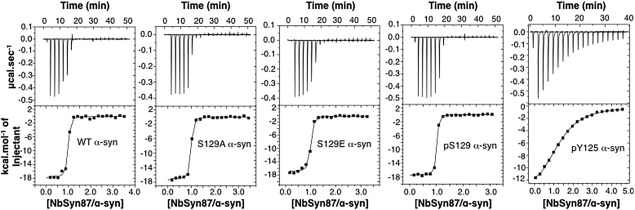
Interaction of NbSyn87 with α‐synuclein variants monitored by ITC. The top panels illustrate the ITC data for NbSyn87 binding to wild type and the S129A, S129E, pS129, and pY125 α‐synuclein variants at 25 °C in PBS buffer. The lower panels illustrate the integrated heat release at each titration point for each protein. The resulting binding isotherms were fitted to a 1:1 bimolecular binding model (see Materials and Methods section), and the values of the binding parameters are given in Table I.
Table 1.
Equilibrium Association (Ka) and Dissociation Constants (Kd) for the Binding of NbSyn87 to Different Variants of α‐Synuclein Along with Values of the Binding Enthalpy, Entropy, and Gibbs Free Energy, Determined by ITC, at 298 K in PBS Buffer
| N a | ΔH b b (kcal/mol) | –TΔS b c (kcal/mol) | ΔG b d (kcal/mol) | K a (107 M−1)e | K d (10−9 M)e | |
|---|---|---|---|---|---|---|
| Wild type | 0.94 ± 0.01 | −17.6 ± 0.4 | 6.9 ± 0.5 | −10.8 ± 0.4 | 8.5 ± 4.9 | 14.2 ± 8.4 |
| pY125 | 1.23 ± 0.01 | −15.2 ± 0.2 | 8.0 ± 0.3 | −7.2 ± 0.4 | 0.017 ± 0.001 | 6580 ± 253 |
| S129A | 0.90 ± 0.01 | −17.2 ± 0.2 | 6.6 ± 0.3 | −10.6 ± 0.2 | 6.0 ± 2.1 | 16.8 ± 5.8 |
| S129E | 0.92 ± 0.01 | −17.5 ± 0.3 | 7.4 ± 0.3 | −10.1 ± 0.2 | 2.6 ± 0.7 | 41.1 ± 10.4 |
| pS129 | 0.95 ± 0.01 | −17.1 ± 0.2 | 6.2 ± 0.3 | −11.0 ± 0.2 | 11.2 ± 3.5 | 9.2 ± 3.1 |
| Y133F/Y136F | 0.90 ± 0.01 | −19.1 ± 0.2 | 8.3 ± 0.3 | −10.8 ± 0.2 | 8.63 ± 1.6 | 11.6 ± 2.2 |
Stoichiometry of the interaction.
Enthalpy of binding.
Entropy of binding.
Gibbs free energy of binding.
Association constant.
e Dissociation constant.
Binding of NbSyn87 to α‐synuclein by SPR
In order to extend the conclusions from the ITC data and to explore the kinetics of the interaction between NbSyn87 and the α‐synuclein variants, we carried out SPR experiments (Fig. 3, Table 2). These experiments confirm that phosphorylation or mutations at S129 have little or no effect on interactions with NbSyn87 while phosphorylation of Y125 results in a dramatic reduction in affinity. The SPR traces obtained for the wild type protein and the S129A, S129E, and pS129 variants (Fig. 3) are quite similar and yield comparable values of k on, k off, and K d (Table 2). The k on values are typical of protein‐protein association reactions, which are commonly in the range of 105–106 M−1 s−1, and the k off values are found to be in the 10−2 s−1 range, which are similar to those of other nanobody‐antigen interactions.44 Only small differences were observed between the binding of the S129 variants and wild type α‐synuclein to NbSyn87, supporting the conclusion that S129 is not an important determinant of the interaction. Interestingly, all modifications at this position led to a reduction in both k on and k off, to a maximum of about 2.5‐fold for pS129. In contrast, for pY125, very weak binding to NbSyn87 precluded the accurate measurement of rate constants, although, a thermodynamic analysis of the equilibrium resonance unit values yielded a K d of greater than 1 µM, that is at least a 66‐fold reduction in affinity (Fig. 3, Table 2), confirming that phosphorylation at this site significantly weakens interactions with the nanobody.
Figure 3.

Interaction of NbSyn87 with α‐synuclein variants monitored by SPR. (A) Triplicate kinetic traces for the binding of NbSyn87 (concentrations ranging from 227 to 0 nM) to immobilized wild type (WT), S129A, S129E, and pS129 α‐synuclein. The data are corrected for the signals originating from the buffer and for nonspecific binding to the dextran matrix surface and are fitted to a 1:1 binding isotherm (solid lines), enabling the k on, k off, and K d values to be extracted for the interaction of NbSyn87 with each α‐synuclein variant. (B) A separate chip was prepared with immobilized wild type, S129A and pY125 α‐synuclein, in three different flow‐cells. Right panel: triplicate binding traces of NbSyn87 with immobilized pY125 α‐synuclein; Left panel: triplicate binding traces of NbSyn87 with S129A, immobilized in the flow‐cell adjacent to immobilized pY125 α‐synuclein. (C) Left panel: the equilibrium values of the binding traces shown in A. for the binding of NbSyn87 to wild type α‐synuclein and to the variants S129A, S129E, and pS129, as a function of the concentration of NbSyn87. Right panel: the equilibrium values of the binding traces shown in B. for the binding of NbSyn87 to pY125 α‐synuclein for concentrations ranging from 2500 to 0 nM (♦), for the binding of NbSyn87 to S129A α‐synuclein (□) for concentrations ranging from 625 to 0 nM, and for the binding of NbSyn87 to wild type α‐synuclein (▲), for concentrations ranging from 2500 to 0 nM.
Table 2.
Kinetic Rate Constants for Association (kon) and Dissociation (koff) and the Equilibrium Association Constant (Ka) for the Binding of NbSyn87 to Different Variants of α‐Synuclein, Determined by SPR, at 298 K in PBS Buffer
| k on a (106 M−1 s−1) | k off a (10−3 s−1) | K a (107 M−1)b | K d (10−9 M)b | K a (107 M−1)c | K d (10−9 M)c | |
|---|---|---|---|---|---|---|
| Wild type | 4.09 ± 0.04 | 32.0 ± 0.1 | 12.8 ± 0.1 | 7.82 ± 0.08 | 6.18 ± 0.30 | 16.2 ± 0.8 |
| pY125 | n.d.d | n.d.d | n.d.d | n.d.d | 0.093 ± 0.024 | 1070 ± 267 |
| S129A | 2.64 ± 0.02 | 21.0 ± 0.1 | 12.6 ± 0.1 | 7.95 ± 0.06 | 12.5 ± 0.5 | 8.0 ± 0.3 |
| S129E | 3.47 ± 0.03 | 24.0 ± 0.1 | 14.5 ± 0.1 | 6.92 ± 0.07 | 9.81 ± 0.38 | 10.2 ± 0.4 |
| pS129a | 1.54 ± 0.02 | 13.0 ± 0.1 | 11.8 ± 0.2 | 8.44 ± 0.12 | 8.26 ± 0.90 | 12.1 ± 1.3 |
The kinetic and affinity constants and fitting errors were estimated from global fitting of a concentration series measured in triplicate.
Affinity constant estimated from the ratio: k on /k off; dissociation constant estimated from the ratio: k off /k on.
The affinity constant (K a) was estimated using triplicate equilibrium values of binding and then fitted using a 1:1 Langmuir binding model.
Not determined.
NMR studies of the binding interaction
We have previously mapped the epitope for NbSyn87 on wild type α‐synuclein by considering the effects of the backbone amide resonances as an indication of binding.21 We observed in this study both broadening and chemical shift perturbations of the resonances of residues located at the C‐terminal region of α‐synuclein, including residues 118–140.21 To determine the effects of S129 and Y125 phosphorylation on the mode of interaction of NbSyn87 and α‐synuclein, 1H15N HSQC spectra of the wild type, the Y133F/Y136F variant and the phosphorylated Y125 and S129 forms of α‐synuclein were measured in the presence and absence of saturating concentrations of NbSyn87. Figure S1 (Supporting Information) shows that the 1H15N HSQC spectra of the Y133F/Y136F, pY125, and pS129 variants are virtually identical to that of wild type α‐synuclein, with all the resonances of the amide protons values falling between 7.8 and 8.8 ppm, a typical feature of unstructured proteins. As reported previously, α‐synuclein in aqueous solution behaves as an intrinsically disordered protein with and without phosphorylation at Y125 and S129 (Fig. S1, Supporting Information). Nevertheless, slight perturbations in chemical shifts are observed for residues in the vicinity of the phosphorylation site, which are likely to result from the change in the covalent structure of the protein upon the introduction of the phosphate group (Fig. 4, S1, Supporting Information). It is noteworthy, however, that the resonances affected, in both phosphorylated α‐synuclein variants, are located within the epitope region of α‐synuclein for NbSyn87.
Figure 4.
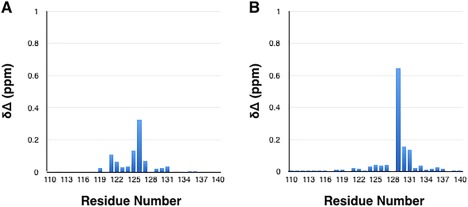
NMR chemical shift changes observed for phosphorylated α‐synuclein variants. Chemical shift changes of phosphorylated (A) pY125 and (B) pS129 variants of α‐synuclein, calculated relative to the wild type protein. The chemical shift changes are defined as [0.04 × (δ15Nwild type – δ15Nvariant)2 + (δ1Hwild type – δ1Hvariant)2]1/2.
It has been reported previously that saturation of the binding of α‐synuclein is achieved with a molar equivalent of NbSyn87.21 Therefore, to validate our ITC and SPR results and to confirm the effects of Y125 and S129 phosphorylation on the binding of NbSyn87 to α‐synuclein, we recorded 1H15N HSQC spectra of the different 15N‐labeled α‐synuclein variants containing equimolar concentrations of NbSyn87. As with the wild type protein, both pS129 and Y133F/Y136F forms of α‐synuclein were saturated with an equimolar concentration of NbSyn87, and displayed fundamentally identical spectra to that of the wild type protein bound to NbSyn87 (Fig. 5). In particular, the residues within the epitope (residues 118–140) displayed similar broadening even at equimolar concentration of NbSyn87. Interestingly, to achieve effective saturation of the pY125 α‐synuclein variant with NbSyn87, ∼1.4M equivalents of NbSyn87 were required, suggesting that pY125 α‐synuclein and NbSyn87 bind with lower affinity, consistent with the data obtained by ITC and SPR.
Figure 5.

1H‐15N HSQC NMR spectra of phosphorylated α‐synuclein variants bound to NbSyn87. 1H‐15N HSQC correlation spectra of uniformly 15N labeled (A) wild type, (B) pS129, and (C) pY125 variants of α‐synuclein upon binding to NbSyn87, shown in blue, in comparison to those of the unbound form of each α‐synuclein variant, represented in red. The spectra of the bound proteins were measured in the presence of saturating concentrations of NbSyn87.
The spectrum of the pY125 α‐synuclein variant when saturated with NbSyn87 showed identical broadening of residues 118–140 to that of the wild type protein saturated with the nanobody (Fig. 5). The fraction of protein bound to NbSyn87 was calculated using the saturation molar ratio obtained by NMR (1.4) and the value of the dissociation constant (K d) obtained by ITC (6 µM) and SPR (1 µM); the values obtained were 0.90 and 0.99, respectively. Thus, the higher concentration of the nanobody required for saturation is also consistent with the lower binding affinity observed in both the ITC and SPR experiments.
Discussion
The combination of advances in the fields of peptide synthesis and chemical ligation has made it possible to produce proteins with site‐specific PTMs, enabling the study of the effects of such modifications on the structure and binding properties of proteins.35, 36 In the present study, these recent developments have enabled site‐specific phosphorylation of α‐synuclein and hence have allowed the investigation of the effects of these modifications on its interactions with other proteins. Several kinases that phosphorylate the C‐terminal serine and residues Y125, Y133, and Y136, in vitro, have been identified, although only phosphorylation events at Y125 and pS129 have been detected in vivo.40 It has also been shown that phosphorylation at Y125 in transgenic Drosophila expression of wild type human α‐synuclein protects against neurotoxicity.40 Additionally, age‐related decreases in the levels of the pY125 variant have been identified in both human subjects and transgenic fly models of PD, and indeed this phosphorylated state has not been found in the brains of patients with Lewy body dementia.40 In contrast, phosphorylation at S129 has been consistently reported as the most commonly detected and abundant modification of α‐synuclein in Lewy bodies. In addition, the level of S129 phosphorylation correlates with pathology in animal models of PD and disease progression in human studies.10, 39, 40, 45
Phosphorylation of Y125 or S129 results in different effects on the modulation of α‐synuclein interactions with NbSyn87. Previous studies of the binding interactions of α‐synuclein have mostly relied on coimmunoprecipitation and yeast two‐hybrid screening techniques.46, 47, 48 These methodologies may, however, be insufficient to identify the physiological and pathological interactions of α‐synuclein or to probe the structural state of the protein that is involved in binding.46, 47, 48 The use of a nanobody raised to bind to the C‐terminal domain of α‐synuclein21 has enabled us to quantify the effects of phosphorylation at S129 and Y125 on a model α‐synuclein interaction in detail. The results obtained using ITC, SPR, and NMR spectroscopy have revealed that phosphorylation of Y125 affects very significantly the affinity of NbSyn87 for α‐synuclein. In contrast, phosphorylation of S129, located just four residues away in the α‐synuclein sequence and very close to the epitope for NbSyn87, has no significant effects on the binding affinity. The ITC results obtained for the interaction of pY125 compared to the wild type protein show a dramatic change in the binding enthalpy (Table 1). In contrast, no detectable changes to the binding enthalpy or entropy were observed for the S129 α‐synuclein variants (Table 1). These findings demonstrate that the impact of phosphorylation on the interaction of α‐synuclein with a given protein partner can be extremely site‐specific, despite its largely unstructured nature. This suggests that C‐terminal modifications of α‐synuclein could be a mechanism for fine‐tuning its interactions with binding partners and regulating its biological activity.
Because the binding of α‐synuclein to NbSyn87 leads to extensive broadening of the NMR cross peaks of the residues between V118 and P140 in the 1H‐15N HSQC spectrum of the protein, it has not been possible to elucidate in detail the structural effects of NbSyn87 on the phosphorylated variants within the binding region (Fig. S1, Supporting Information). Nonetheless, the NMR experiments provide further confirmation of phosphorylation‐dependent changes in nanobody affinity, via an examination of the concentration of NbSyn87 needed to saturate the panel of α‐synuclein variants. Our findings reveal no significant differences in the molar ratios needed to reach effective saturation of NbSyn87 binding to the wild type protein and the S129 α‐synuclein variants (S129A and S129E, and the phosphorylated species pS129), confirming the conclusion that phosphorylation or mutation of S129 does not detectably influence the interactions of α‐synuclein with the nanobody. Titration of the pY125 variant of α‐synuclein with NbSyn87, reached saturation only at a 2:3 α‐synuclein:nanobody ratio consistent with a significant drop in affinity.
Although no changes in the binding enthalpy or in the binding entropy are observed for the pS129 variant, the loss of the binding affinity of NbSyn87 for pY125 α‐synuclein can be attributed to the fact that addition of the phosphate group to Y125 increases the negative charge on the protein as well as the steric bulk of the tyrosine side chain. The Y133F/Y136F mutant exhibited similar binding affinity as the wild type protein, as confirmed by the ITC data (Fig. S2, Supporting Information), confirming that the effect seen by NMR spectroscopy on the pY125 α‐synuclein variant is a specific consequence of the phosphorylation of this residue.
The C‐terminal region of α‐synuclein is a hub for many binding proteins, small molecules, and metal ions32, 34, 49, 50, 51 and governs intra‐molecular long‐range contacts within α‐synuclein52, 53, 54 itself. It has furthermore been found to regulate interactions with other proteins involved in SNARE complex assembly and neurotransmitter release.27, 34, 55 Thus, by harboring two phosphorylation sites that can be discriminated by a protein partner, our findings demonstrate the potential role of PTMs in the C‐terminal domain as molecular switches for regulating α‐synuclein interactions and functions.
Implications for PD diagnostics and therapeutic strategies
Due to its ability to differentiate between the pS129 and pY125 forms of α‐synuclein, NbSyn87 has the potential to be developed as a method for discriminating pathological from physiologically‐benign states of α‐synuclein. This finding lays the groundwork for potentially important immunological and vaccine development strategies, approaches that have recently received considerable attention because of their reported potential to ameliorate neurodegenerative diseases.56, 57, 58 Recently, antibodies targeting α‐synuclein have been shown to be sensitive indicators of neurodegeneration, suggesting that they may have a role in protecting against disease and hence have the potential for therapeutic applications.59 In addition, discrimination between two sites of phosphorylation on nearby residues by antibodies has been shown to be invaluable in diagnostic and therapeutic research, for example the cardiac muscle protein phospholamban60 and the tau protein in Alzheimer's disease.61
The single domain antibody used in this study represents a novel and promising alternative to conventional antibodies for PD diagnostic and therapeutic applications. Its small molecular size (ca. 14 kDa), reflects the fact that it is composed of a single domain antigen recognition motif. Nevertheless, NbSyn87 has a binding affinity that is comparable to that of conventional antibodies.62, 63 Moreover, nanobodies are generally extremely stable and have low immunogenicity because of their high sequence similarity with the human VH family III domains.64 A number of earlier studies have reported the ability of nanobodies to modulate the conformation of proteins both in vitro and in living cells, and have established their potential as therapeutic agents, for example in rheumatoid arthritis, chronic colitis, and Alzheimer's disease.56, 65, 66, 67, 68, 69 Our findings provide new insights into the potential applications of NbSyn87 in diagnostic and therapeutic strategies based on two variants of α‐synuclein that are associated with opposite effects on the pathogenicity of α‐synuclein.
In conclusion, studies using ITC, SPR, and NMR spectroscopy have provided direct evidence that phosphorylation can significantly influence the interaction of α‐synuclein with its binding partners. Moreover, the results suggest that the impact of phosphorylation on the interaction of α‐synuclein with a specific protein partner can be very significant and highly site specific and underscore the critical importance of conducting systematic studies to investigate the role of PTMs in modulating the α‐synuclein proteome in health and disease.
Materials and Methods
Materials
All reagents were purchased from Sigma‐Aldrich (Dorset, England), unless otherwise stated. Protein concentrations were measured by UV absorbance spectroscopy using molecular extinction coefficients calculated using the ExPASy‐ProtParam method (http://web.expasy.org/protparam/). The extinction coefficients at 280 nm of the wild type, pS129, S129E, and S129A α‐synuclein variants were all taken to be 5960 M−1 cm−1. The extinction coefficients of the pY125 variant and Y133F/Y136F double‐mutant α‐synuclein were taken to be 2077 M−1 cm−1 and 2517 M−1 cm−1 at 267 nm, respectively, and that of NbSyn87 as 26025 M−1 cm−1 at 280 nm.
Generation of wild type α‐synuclein
α‐Synuclein in its unlabeled and 15N labeled forms was prepared according to previously published procedures.21, 70
Generation of semisynthetic pY125 α‐synuclein
Semisynthetic pY125 α‐synuclein was generated according to a previously published procedure.36
Generation of 15N labeled wild type and Y133F/Y136F α‐synuclein
Y133F/Y136F α‐synuclein was generated using two single‐point mutagenesis with the following primers forward: 5′CTTCTGAGGAAGGGTTTCAAGACTACGAAC‐3′ and reverse 5′‐GTTCGTAGTCTTGAAACCCTTCCTCAGAAG‐3′ and subsequent primers forward 5′‐GGAAGGGTATCAAGACTTTGAACCTGAAGCCTAAG‐3′ and reverse 5′‐CTTAGGCTTCAGGTTCAAAGTCTTGATACCCTTCC‐3′. The proteins were purified according to previously published procedure.36 Briefly, BL21 (DE3) cells were transformed with the plasmid and grown in M9 minimal media with 15N labeled NH4Cl. Protein expression was induced by the addition of 1 mM IPTG for 4 h at 37°C. After harvesting the cells by centrifugation at 6000g for 15 min, the cell pellet was resuspended in 40 mM Tris‐acetate and 5 mM EDTA (pH 8.3) in the presence of 0.3 mM PMSF and lysed by ultra‐sonication. The insoluble material was removed by centrifugation at 23,000g for 40 min at 4°C. The supernatant was boiled for 15 min in a water bath at 100°C followed by centrifugation at 23,000g for 40 min. Anion‐exchange chromatography was performed on a Pharmacia AKTA FPLC system using a 20 mL HiPrep 16/10 Q FF column (Amersham, UK). The protein was eluted with a linear gradient of 0–1M NaCl and subsequently purified on a Superdex 200 26/60 size‐exclusion chromatography column (Amersham, UK) in 50 mM Tris, 150 mM NaCl pH 7.5. The purified protein was dialyzed extensively against deionized water and lyophilized and stored at −20°C until used.
Generation of 15N labeled pS129 α‐synuclein and pY125 Y133F/Y136F α‐synuclein
Phosphorylation of α‐synuclein at S129 was carried out by incubation of 15N wild type α‐synuclein with 0.42 μg of PLK3 kinase (Invitrogen, PV3812) in 50 mM Hepes pH 7.4 in presence of 1 mM MgCl2, 1 mM EGTA, 2 mM Mg‐ATP, and 1 mM DTT for 12 h at 30°C. To insure complete conversion to phosphorylated state, 500 μg of 15N α‐synuclein were incubated in 200 μL of reaction volume. Phosphorylation of α‐synuclein Y133F/Y136F at Y125 was carried out by incubation with 0.23 μg of Syk kinase (Invitrogen, PV3857) in 50 mM Hepes pH 7.4 in presence of 1 mM MgCl2, 1 mM EGTA, 2 mM Mg‐ATP (stock solution of 0.39M ATP and 0.87M MgCl2 at pH 7.0) and 1 mM Na3VO4 for 12 h at 30 °C. To ensure complete conversion to the phosphorylated state, 500 μg of 15N α‐synuclein were incubated in 200 μL of reaction volume. The proteins were subsequently purified by reverse‐phased HPLC on a C8 semi‐preparative column (Inertsile WP300) using a linear gradient of 20–70% B over 30 min (where A is water/0.1% TFA and B is acetonitrile/0.1% TFA).
Generation of 1H15N labeled S129E and S129A α‐synuclein
15N labeled S129E and S129A α‐synuclein variants were purified according to previously published procedures.36
Isolation, expression, and purification of the antibody fragment NbSyn87
The expression and purification of NbSyn87 was performed as described previously.21 Briefly, NbSyn87, obtained from dromedary immunization and isolated through phage display selection,71, 72 was expressed in the periplasm of E. coli and purified using immobilized metal affinity and size‐exclusion chromatography according to published protocols.21, 72
ITC measurements of binding affinity
Calorimetric data were recorded using an iTC200 calorimeter (MicroCal, LLC, Northampton, MA). A solution of 40 μL of NbSyn87 at a concentration of 150 μM was titrated in 2 μL aliquots at 150 s intervals into a calorimetric cell containing a standard volume (203 μL) of 10 μM α‐synuclein in solution. Experiments were performed at 25°C in 10 mM phosphate and 150 mM sodium chloride at a pH of 7.4. The thermodynamic analysis was performed using the MicroCal analysis software (Origin 7.0) using a simple 1:1 bimolecular binding model.
Kinetic and affinity measurements using SPR
The kinetic constants of the interaction of NbSyn87 with wild type, pY125, S129A, S129E, and pS129 α‐synuclein variants were determined by SPR (Biacore 3,000, GE Healthcare, Sweden). On one CM5 chip, wild type α‐synuclein, and the S129A and S129E variants, were immobilized in flow‐cells 2, 3, and 4, respectively, at ∼50 resonance units (50 pg/mm2) via EDC/NHS chemistry and according to the manufacturer's recommendations. Flow‐cell 1 of the chip contained no protein and served as a blank reference cell. Similarly, a second CM5 chip was prepared with wild type α‐synuclein, pS129 and S129E immobilized in respectively flow‐cell 2, 3, and 4. For both chips, triplicate association traces of NbSyn87 for concentrations ranging from 0 to 227 µM were recorded for 2 min in a buffer containing 10 mM HEPES pH 7.4, 150 mM NaCl, 0.005% Tween 20 and 3 mM EDTA; triplicate dissociation traces of the complexes in the buffer were recorded for 5 min. The curves obtained after subtraction of the signals of the reference cell (the flow cell that contained no immobilized protein) and injection of buffer, were then fitted to a 1:1 Langmuir binding model with the program BIAeval (Biacore). In addition, the equilibrium values of the association binding traces were plotted versus the concentration of NbSyn87 for each variant, and fitted to a 1:1 bimolecular binding isotherm, to obtain the affinity constant for the NbSyn87:α‐synuclein interactions, independently of the binding kinetics. For the measurement of the pY125 α‐synuclein:NbSyn87 interaction, a separate chip was prepared with ∼100 RU of immobilized wild type α‐synuclein (flow‐cell 2) and S129A (flow‐cell 3), and ∼50 RU of pY125 α‐synuclein (flow‐cell 4). Binding traces of NbSyn87 concentrations ranging from 2500–0 nM were subsequently recorded and evaluated as described above.
NMR spectroscopy
1H‐15N HSQC spectra of 15N‐labeled α‐synuclein variants were acquired at 283 K on Varian INOVA 800 MHz spectrometer equipped with a cold‐probe (Varian, USA). Samples of unbound α‐synuclein (the wild type protein, the Y133F/Y136F mutant, and the phosphorylated variants pS129 and pY125) were prepared at concentrations of 70 µM in NMR buffer (25 mM Tris, 70 mM NaCl, pH 7.4, in 90% H2O, and 10% D2O). DSS (4,4‐dimethyl‐4‐silapentane‐1‐sulfonic acid) was used to calibrate the 1H chemical shifts directly; calibration of 15N chemical shifts were calculated according to their gyromagnetic ratios.73 α‐Synuclein samples in the presence of NbSyn87 were prepared by adding 1M equivalent of the unlabeled nanobody to a fresh sample of protein (final concentration 70 µM). Previous experiments demonstrated that saturation of wild type α‐synuclein is effectively achieved with an equimolar equivalent of NbSyn87.21 Therefore, spectra of the different 15N‐labeled α‐synuclein variants were first recorded in the presence or absence of 1M equivalent of unlabeled NbSyn87. Separate samples with higher than 1M equivalent of unlabeled nanobody (1.1, 1.2, 1.3, and 1.4 molar equivalents) were prepared for the pY125 α‐synuclein variant. The NMR data were subsequently processed with NMRpipe74 and the program SPARKY (http://www.cgl.ucsf.edu/home/sparky/) was used to analyze the data. Assignment of the spectra of the phosphorylated (pS129 and pY125) forms of α‐synuclein, as well as the control mutant form Y133F/Y136F of α‐synuclein, was achieved by means of three‐dimensional (3D) CBCA(CO)NH and HNCACB experiments. The 3D experiments were acquired with 72 complex data points in the 15N dimension. Chemical shift changes in the α‐synuclein variants, both unbound and bound to NbSyn87, defined relative to the wild type protein chemical shifts as [0.04 × (δ15Nwild type – δ15Nvariant)2 + (δ1Hwild type – δ1Hvariant)2]1/2.51
Conflict of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Author Contributions
HL, CMD, MV, FT, and EDG conceived the idea of the study and designed the experiments. FT performed and interpreted the NMR experiments at McGill University (AM's lab); EDG, TG, FT, and JDT the ITC experiments; and EDG, the SPR experiments. MH, BF, and AC expressed, purified, and prepared the α‐synuclein proteins used in this study. FT wrote the manuscript; EDG, CMD, HL, MV, and AM contributed to the writing and all authors approved the final version of the manuscript.
Supporting information
Supporting Information
Contributor Information
Michele Vendruscolo, Email: mv245@cam.ac.uk.
Hilal A. Lashuel, Email: hilal.lashuel@epfl.ch
Christopher M. Dobson, Email: cmd44@cam.ac.uk
References
- 1. Baba M, Nakajo S, Tu PH, Tomita T, Nakaya K, Lee VM, Trojanowski JQ, Iwatsubo T (1998) Aggregation of alpha‐synuclein in Lewy bodies of sporadic Parkinson's disease and dementia with Lewy bodies. Am J Pathol 152:879–884. [PMC free article] [PubMed] [Google Scholar]
- 2. Kahle PJ, Neumann M, Ozmen L, Muller V, Jacobsen H, Schindzielorz A, Okochi M, Leimer U, van Der Putten H, Probst A, Kremmer E, Kretzschmar HA, Haass C (2000) Subcellular localization of wild‐type and Parkinson's disease‐associated mutant alpha‐synuclein in human and transgenic mouse brain. J Neurosci 20:6365–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Galvin JE, Lee VM, Trojanowski JQ (2001) Synucleinopathies: clinical and pathological implications. Arch Neurol 58:186–190. [DOI] [PubMed] [Google Scholar]
- 4. Martí MJ, Tolosa E, Campdelacreu J (2003) Clinical overview of the synucleinopathies. Mov Disord 18:21–27. [DOI] [PubMed] [Google Scholar]
- 5. Wakabayashi K, Tanji K, Mori F, Takahashi H (2007) The Lewy body in Parkinson's disease: molecules implicated in the formation and degradation of alpha‐synuclein aggregates. Neuropathology 27:494–506. [DOI] [PubMed] [Google Scholar]
- 6. Fujiwara H, Hasegawa M, Dohmae N, Kawashima A, Masliah E, Goldberg MS, Shen J, Takio K, Iwatsubo T (2002) Alpha‐synuclein is phosphorylated in synucleinopathy lesions. Nat Cell Biol 4:160–164. [DOI] [PubMed] [Google Scholar]
- 7. Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen JT, Schols L, Riess O (1998) Ala30Pro mutation in the gene encoding alpha‐synuclein in Parkinson's disease. Nat Genet 18:106–108. [DOI] [PubMed] [Google Scholar]
- 8. Chi YC, Armstrong GS, Jones DN, Eisenmesser EZ, Liu CW (2014) Residue histidine 50 plays a key role in protecting alpha‐synuclein from aggregation at physiological pH. J Biol Chem 289:15474–15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rutherford NJ, Moore BD, Golde TE, Giasson BI (2014) Divergent effects of the H50Q and G51D SNCA mutations on the aggregation of alpha‐synuclein. J Neurochem 131:859–867. [DOI] [PubMed] [Google Scholar]
- 10. Fares MB, Ait‐Bouziad N, Dikiy I, Mbefo MK, Jovicic A, Kiely A, Holton JL, Lee SJ, Gitler AD, Eliezer D, Lashuel HA (2014) The novel Parkinson's disease linked mutation G51D attenuates in vitro aggregation and membrane binding of alpha‐synuclein, and enhances its secretion and nuclear localization in cells. Human Mol Genet 23:4491–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL (1997) Mutation in the alpha‐synuclein gene identified in families with Parkinson's disease. Science 276:2045–2047. [DOI] [PubMed] [Google Scholar]
- 12. Zarranz JJ, Alegre J, Gómez‐Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atarés B, Llorens V, Gomez Tortosa E, del Ser T, Muñoz DG, de Yebenes JG (2004) The new mutation, E46K, of alpha‐synuclein causes Parkinson and Lewy body dementia. Ann Neurol 55:164–173. [DOI] [PubMed] [Google Scholar]
- 13. Maraganore DM, de Andrade M, Elbaz A, Farrer MJ, Ioannidis JP, Kruger R, Rocca WA, Schneider NK, Lesnick TG, Lincoln SJ, Hulihan MM, Aasly JO, Ashizawa T, Chartier‐Harlin MC, Checkoway H, Ferrarese C, Hadjigeorgiou G, Hattori N, Kawakami H, Lambert JC, Lynch T, Mellick GD, Papapetropoulos S, Parsian A, Quattrone A, Riess O, Tan EK, Van Broeckhoven C (2006) Collaborative analysis of alpha‐synuclein gene promoter variability and Parkinson disease. J Am Med Assoc 296:661–670. [DOI] [PubMed] [Google Scholar]
- 14. Maries E, Dass B, Collier TJ, Kordower JH, Steece‐Collier K (2003) The role of alpha‐synuclein in Parkinson's disease: insights from animal models. Nat Rev 4:727–738. [DOI] [PubMed] [Google Scholar]
- 15. Fauvet B, Mbefo MK, Fares MB, Desobry C, Michael S, Ardah MT, Tsika E, Coune P, Prudent M, Lion N, Eliezer D, Moore DJ, Schneider B, Aebischer P, El‐Agnaf OM, Masliah E, Lashuel HA (2012) Alpha‐synuclein in central nervous system and from erythrocytes, mammalian cells, and Escherichia coli exists predominantly as disordered monomer. J Biol Chem 287:15345–15364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lashuel HA, Overk CR, Oueslati A, Masliah E (2013) The many faces of alpha‐synuclein: from structure and toxicity to therapeutic target. Nat Rev 14:38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fusco G, De Simone A, Arosio P, Vendruscolo M, Veglia G, Dobson CM (2016) Structural ensembles of membrane‐bound alpha‐synuclein reveal the molecular determinants of synaptic vesicle affinity. Sci Rep 6:27125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bodner CR, Maltsev AS, Dobson CM, Bax A (2010) Differential phospholipid binding of alpha‐synuclein variants implicated in Parkinson's disease revealed by solution NMR spectroscopy. Biochemistry 49:862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fusco G, De Simone A, Gopinath T, Vostrikov V, Vendruscolo M, Dobson CM, Veglia G (2014) Direct observation of the three regions in alpha‐synuclein that determine its membrane‐bound behaviour. Nat Commun 5:3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burre J, Sharma M, Sudhof TC (2014) Alpha‐synuclein assembles into higher‐order multimers upon membrane binding to promote SNARE complex formation. Proc Natl Acad Sci U S A 111:E4274–E4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guilliams T, El‐Turk F, Buell AK, O'Day EM, Aprile FA, Esbjorner EK, Vendruscolo M, Cremades N, Pardon E, Wyns L, Welland ME, Steyaert J, Christodoulou J, Dobson CM, De Genst E (2013) Nanobodies raised against monomeric alpha‐synuclein distinguish between fibrils at different maturation stages. J Mol Biol 425:2397–2411. [DOI] [PubMed] [Google Scholar]
- 22. Desplats P, Lee HJ, Bae EJ, Patrick C, Rockenstein E, Crews L, Spencer B, Masliah E, Lee SJ (2009) Inclusion formation and neuronal cell death through neuron‐to‐neuron transmission of alpha‐synuclein. Proc Natl Acad Sci U S A 106:13010–13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH, Stefanis L, Vekrellis K (2010) Cell‐produced alpha‐synuclein is secreted in a calcium‐dependent manner by exosomes and impacts neuronal survival. J Neurosci 30:6838–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jang A, Lee HJ, Suk JE, Jung JW, Kim KP, Lee SJ (2010) Non‐classical exocytosis of alpha‐synuclein is sensitive to folding states and promoted under stress conditions. J Neurochem 113:1263–1274. [DOI] [PubMed] [Google Scholar]
- 25. Del Mar C, Greenbaum EA, Mayne L, Englander SW, Woods VL Jr (2005) Structure and properties of alpha‐synuclein and other amyloids determined at the amino acid level. Proc Natl Acad Sci U S A 102:15477–15482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou M, Xu S, Mi J, Ueda K, Chan P (2013) Nuclear translocation of alpha‐synuclein increases susceptibility of MES23.5 cells to oxidative stress. Brain Res 1500:19–27. [DOI] [PubMed] [Google Scholar]
- 27. Burre J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, Sudhof TC (2010) Alpha‐synuclein promotes SNARE‐complex assembly in vivo and in vitro. Science 329:1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jensen PH, Hager H, Nielsen MS, Hojrup P, Gliemann J, Jakes R (1999) Alpha‐synuclein binds to Tau and stimulates the protein kinase A‐catalyzed tau phosphorylation of serine residues 262 and 356. J Biol Chem 274:25481–25489. [DOI] [PubMed] [Google Scholar]
- 29. Giasson BI, Forman MS, Higuchi M, Golbe LI, Graves CL, Kotzbauer PT, Trojanowski JQ, Lee VM (2003) Initiation and synergistic fibrillization of tau and alpha‐synuclein. Science 300:636–640. [DOI] [PubMed] [Google Scholar]
- 30. Goers J, Manning‐Bog AB, McCormack AL, Millett IS, Doniach S, Di Monte DA, Uversky VN, Fink AL (2003) Nuclear localization of alpha‐synuclein and its interaction with histones. Biochemistry 42:8465–8471. [DOI] [PubMed] [Google Scholar]
- 31. Paik SR, Shin HJ, Lee JH, Chang CS, Kim J (1999) Copper(II)‐induced self‐oligomerization of alpha‐synuclein. Biochem J 340:821–828. [PMC free article] [PubMed] [Google Scholar]
- 32. Brown DR (2007) Interactions between metals and alpha‐synuclein–function or artefact? FEBS J 274:3766–3774. [DOI] [PubMed] [Google Scholar]
- 33. Goers J, Uversky VN, Fink AL (2003) Polycation‐induced oligomerization and accelerated fibrillation of human alpha‐synuclein in vitro. Protein Sci 12:702–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eliezer D (2013) The mysterious C‐terminal tail of alpha‐synuclein: nanobody's guess. J Mol Biol 425:2393–2396. [DOI] [PubMed] [Google Scholar]
- 35. Oueslati A, Fournier M, Lashuel HA (2010) Role of post‐translational modifications in modulating the structure, function and toxicity of alpha‐synuclein: implications for Parkinson's disease pathogenesis and therapies. Prog Brain Res 183:115–145. [DOI] [PubMed] [Google Scholar]
- 36. Hejjaoui M, Butterfield S, Fauvet B, Vercruysse F, Cui J, Dikiy I, Prudent M, Olschewski D, Zhang Y, Eliezer D, Lashuel HA (2012) Elucidating the role of C‐terminal post‐translational modifications using protein semisynthesis strategies: alpha‐synuclein phosphorylation at tyrosine 125. J Am Chem Soc 134:5196–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmid AW, Fauvet B, Moniatte M, Lashuel HA (2013) Alpha‐synuclein post‐translational modifications as potential biomarkers for Parkinson disease and other synucleinopathies. Mol Cell Proteom 12:3543–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oueslati A (2016) Implication of alpha‐synuclein phosphorylation at S129 in synucleinopathies: what have we learned in the last decade? J Parkinsons Dis 6:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cavallarin N, Vicario M, Negro A (2010) The role of phosphorylation in synucleinopathies: focus on Parkinson's disease. CNS Neurol Disord Drug Targets 9:471–481. [DOI] [PubMed] [Google Scholar]
- 40. Chen L, Periquet M, Wang X, Negro A, McLean PJ, Hyman BT, Feany MB (2009) Tyrosine and serine phosphorylation of alpha‐synuclein have opposing effects on neurotoxicity and soluble oligomer formation. J Clin Invest 119:3257–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ahmad B, Chen Y, Lapidus LJ (2012) Aggregation of alpha‐synuclein is kinetically controlled by intramolecular diffusion. Proc Natl Acad Sci U S A 109:2336–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paleologou KE, Schmid AW, Rospigliosi CC, Kim HY, Lamberto GR, Fredenburg RA, Lansbury PT, Jr , Fernandez CO, Eliezer D, Zweckstetter M, Lashuel HA (2008) Phosphorylation at Ser‐129 but not the phosphomimics S129E/D inhibits the fibrillation of alpha‐synuclein. J Biol Chem 283:16895–16905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mbefo MK, Paleologou KE, Boucharaba A, Oueslati A, Schell H, Fournier M, Olschewski D, Yin G, Zweckstetter M, Masliah E, Kahle PJ, Hirling H, Lashuel HA (2010) Phosphorylation of synucleins by members of the Polo‐like kinase family. J Biol Chem 285:2807–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schlosshauer M, Baker D (2004) Realistic protein‐protein association rates from a simple diffusional model neglecting long‐range interactions, free energy barriers, and landscape ruggedness. Protein Sci 13:1660–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Saito Y, Kawashima A, Ruberu NN, Fujiwara H, Koyama S, Sawabe M, Arai T, Nagura H, Yamanouchi H, Hasegawa M, Iwatsubo T, Murayama S (2003) Accumulation of phosphorylated alpha‐synuclein in aging human brain. J Neuropathol Exp Neurol 62:644–654. [DOI] [PubMed] [Google Scholar]
- 46. Payton JE, Perrin RJ, Clayton DF, George JM (2001) Protein‐protein interactions of alpha‐synuclein in brain homogenates and transfected cells. Brain Res 95:138–145. [DOI] [PubMed] [Google Scholar]
- 47. Ahn BH, Rhim H, Kim SY, Sung YM, Lee MY, Choi JY, Wolozin B, Chang JS, Lee YH, Kwon TK, Chung KC, Yoon SH, Hahn SJ, Kim MS, Jo YH, Min DS (2002) Alpha‐synuclein interacts with phospholipase D isozymes and inhibits pervanadate‐induced phospholipase D activation in human embryonic kidney‐293 cells. J Biol Chem 277:12334–12342. [DOI] [PubMed] [Google Scholar]
- 48. Breydo L, Wu JW, Uversky VN (2012) Alpha‐synuclein misfolding and Parkinson's disease. Biochim Biophys Acta 1822:261–285. [DOI] [PubMed] [Google Scholar]
- 49. Binolfi A, Rasia RM, Bertoncini CW, Ceolin M, Zweckstetter M, Griesinger C, Jovin TM, Fernandez CO (2006) Interaction of alpha‐synuclein with divalent metal ions reveals key differences: a link between structure, binding specificity and fibrillation enhancement. J Am Chem Soc 128:9893–9901. [DOI] [PubMed] [Google Scholar]
- 50. Uversky VN, Li J, Fink AL (2001) Metal‐triggered structural transformations, aggregation, and fibrillation of human alpha‐synuclein. A possible molecular NK between Parkinson's disease and heavy metal exposure. J Biol Chem 276:44284–44296. [DOI] [PubMed] [Google Scholar]
- 51. Fernandez CO, Hoyer W, Zweckstetter M, Jares‐Erijman EA, Subramaniam V, Griesinger C, Jovin TM (2004) NMR of alpha‐synuclein‐polyamine complexes elucidates the mechanism and kinetics of induced aggregation. EMBO J 23:2039–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bernado P, Bertoncini CW, Griesinger C, Zweckstetter M, Blackledge M (2005) Defining long‐range order and local disorder in native alpha‐synuclein using residual dipolar couplings. J Am Chem Soc 127:17968–17969. [DOI] [PubMed] [Google Scholar]
- 53. Bertoncini CW, Jung YS, Fernandez CO, Hoyer W, Griesinger C, Jovin TM, Zweckstetter M (2005) Release of long‐range tertiary interactions potentiates aggregation of natively unstructured alpha‐synuclein. Proc Natl Acad Sci U S A 102:1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dedmon MM, Lindorff‐Larsen K, Christodoulou J, Vendruscolo M, Dobson CM (2005) Mapping long‐range interactions in alpha‐synuclein using spin‐label NMR and ensemble molecular dynamics simulations. J Am Chem Soc 127:476–477. [DOI] [PubMed] [Google Scholar]
- 55. Alderson TR, Markley JL (2013) Biophysical characterization of alpha‐synuclein and its controversial structure. Intrinsic Disord Prot 1:e26255–e26239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schenk D, Barbour R, Dunn W, Gordon G, Grajeda H, Guido T, Hu K, Huang J, Johnson‐Wood K, Khan K, Kholodenko D, Lee M, Liao Z, Lieberburg I, Motter R, Mutter L, Soriano F, Shopp G, Vasquez N, Vandevert C, Walker S, Wogulis M, Yednock T, Games D, Seubert P (1999) Immunization with amyloid‐beta attenuates Alzheimer‐disease‐like pathology in the PDAPP mouse. Nature 400:173–177. [DOI] [PubMed] [Google Scholar]
- 57. Bard F, Cannon C, Barbour R, Burke RL, Games D, Grajeda H, Guido T, Hu K, Huang J, Johnson‐Wood K, Khan K, Kholodenko D, Lee M, Lieberburg I, Motter R, Nguyen M, Soriano F, Vasquez N, Weiss K, Welch B, Seubert P, Schenk D, Yednock T (2000) Peripherally administered antibodies against amyloid beta‐peptide enter the central nervous system and reduce pathology in a mouse model of Alzheimer disease. Nat Med 6:916–919. [DOI] [PubMed] [Google Scholar]
- 58. DeMattos RB, Bales KR, Cummins DJ, Dodart JC, Paul SM, Holtzman DM (2001) Peripheral anti‐A beta antibody alters CNS and plasma A beta clearance and decreases brain A beta burden in a mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A 98:8850–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yanamandra K, Gruden MA, Casaite V, Meskys R, Forsgren L, Morozova‐Roche LA (2011) Alpha‐synuclein reactive antibodies as diagnostic biomarkers in blood sera of Parkinson's disease patients. PloS One 6:e18513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Drago GA, Colyer J (1994) Discrimination between two sites of phosphorylation on adjacent amino acids by phosphorylation site‐specific antibodies to phospholamban. J Biol Chem 269:25073–25077. [PubMed] [Google Scholar]
- 61. Augustinack JC, Schneider A, Mandelkow EM, Hyman BT (2002) Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer's disease. Acta Neuropath 103:26–35. [DOI] [PubMed] [Google Scholar]
- 62. Muyldermans S, Baral TN, Retamozzo VC, De Baetselier P, De Genst E, Kinne J, Leonhardt H, Magez S, Nguyen VK, Revets H, Rothbauer U, Stijlemans B, Tillib S, Wernery U, Wyns L, Hassanzadeh‐Ghassabeh G, Saerens D (2009) Camelid immunoglobulins and nanobody technology. Vet Immunol Immunopath 128:178–183. [DOI] [PubMed] [Google Scholar]
- 63. Vincke C, Loris R, Saerens D, Martinez‐Rodriguez S, Muyldermans S, Conrath K (2009) General strategy to humanize a camelid single‐domain antibody and identification of a universal humanized nanobody scaffold. J Biol Chem 284:3273–3284. [DOI] [PubMed] [Google Scholar]
- 64. De Genst E, Dobson CM (2012) Nanobodies as structural probes of protein misfolding and fibril formation. Methods Mol Biol 911:533–558. [DOI] [PubMed] [Google Scholar]
- 65. Dumoulin M, Last AM, Desmyter A, Decanniere K, Canet D, Larsson G, Spencer A, Archer DB, Sasse J, Muyldermans S, Wyns L, Redfield C, Matagne A, Robinson CV, Dobson CM (2003) A camelid antibody fragment inhibits the formation of amyloid fibrils by human lysozyme. Nature 424:783–788. [DOI] [PubMed] [Google Scholar]
- 66. Chan PH, Pardon E, Menzer L, De Genst E, Kumita JR, Christodoulou J, Saerens D, Brans A, Bouillenne F, Archer DB, Robinson CV, Muyldermans S, Matagne A, Redfield C, Wyns L, Dobson CM, Dumoulin M (2008) Engineering a camelid antibody fragment that binds to the active site of human lysozyme and inhibits its conversion into amyloid fibrils. Biochemistry 47:11041–11054. [DOI] [PubMed] [Google Scholar]
- 67. Hmila I, Abdallah RB, Saerens D, Benlasfar Z, Conrath K, Ayeb ME, Muyldermans S, Bouhaouala‐Zahar B (2008) VHH, bivalent domains and chimeric heavy chain‐only antibodies with high neutralizing efficacy for scorpion toxin AahI'. Mol Immunol 45:3847–3856. [DOI] [PubMed] [Google Scholar]
- 68. Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas‐Delucchi CS, Cardoso MC, Leonhardt H, Hopfner KP, Rothbauer U (2010) Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol 17:133–138. [DOI] [PubMed] [Google Scholar]
- 69. Vandenbroucke K, de Haard H, Beirnaert E, Dreier T, Lauwereys M, Huyck L, Van Huysse J, Demetter P, Steidler L, Remaut E, Cuvelier C, Rottiers P (2010) Orally administered L. lactis secreting an anti‐TNF nanobody demonstrate efficacy in chronic colitis. Mucosal Immunol 3:49–56. [DOI] [PubMed] [Google Scholar]
- 70. De Genst EJ, Guilliams T, Wellens J, O'Day EM, Waudby CA, Meehan S, Dumoulin M, Hsu ST, Cremades N, Verschueren KH, Pardon E, Wyns L, Steyaert J, Christodoulou J, Dobson CM (2010) Structure and properties of a complex of alpha‐synuclein and a single‐domain camelid antibody. J Mol Biol 402:326–343. [DOI] [PubMed] [Google Scholar]
- 71. Decanniere K, Desmyter A, Lauwereys M, Ghahroudi MA, Muyldermans S, Wyns L (1999) A single‐domain antibody fragment in complex with RNase A: non‐canonical loop structures and nanomolar affinity using two CDR loops. Structure 7:361–370. [DOI] [PubMed] [Google Scholar]
- 72. Conrath KE, Lauwereys M, Galleni M, Matagne A, Frere JM, Kinne J, Wyns L, Muyldermans S (2001) Beta‐lactamase inhibitors derived from single‐domain antibody fragments elicited in the camelidae. Antimicrob Agents Chemo 45:2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wishart DS, Case DA (2001) Use of chemical shifts in macromolecular structure determination. Methods Enzymol 338:3–34. [DOI] [PubMed] [Google Scholar]
- 74. Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


