Abstract
Physic nut (Jatropha curcas L.) is a species of flowering plant with great potential for biofuel production and as an emerging model organism for functional genomic analysis, particularly in the Euphorbiaceae family. DNA binding with one finger (Dof) transcription factors play critical roles in numerous biological processes in plants. Nevertheless, the knowledge about members, and the evolutionary and functional characteristics of the Dof gene family in physic nut is insufficient. Therefore, we performed a genome-wide screening and characterization of the Dof gene family within the physic nut draft genome. In total, 24 JcDof genes (encoding 33 JcDof proteins) were identified. All the JcDof genes were divided into three major groups based on phylogenetic inference, which was further validated by the subsequent gene structure and motif analysis. Genome comparison revealed that segmental duplication may have played crucial roles in the expansion of the JcDof gene family, and gene expansion was mainly subjected to positive selection. The expression profile demonstrated the broad involvement of JcDof genes in response to various abiotic stresses, hormonal treatments and functional divergence. This study provides valuable information for better understanding the evolution of JcDof genes, and lays a foundation for future functional exploration of JcDof genes.
Keywords: Jatropha curcas, Dof gene family, transcription factor, phylogenetic analysis, gene expression analysis
1. Introduction
Physic nut (Jatropha curcas L.) is a perennial small tree in the spurge family, Euphorbiaceae, with a high seed-oil content (40–50%). It can grow easily in barren soil and endure drought and saline environments, thus, having a broad adaptability in various agro-climatic conditions. Given its great potential for biofuel production, nowadays, Physic nut is attracting much attention due to the gradual depletion and a cost increase of fossil energy resources [1,2]. However, there are still a number of challenges in physic nut industries. For example, most physic nut germplasms are monoecious with very low ratios of female to male flowers (approximately female:male = 1:13–29), which considerably reduces the seed yield in physic nut [3,4]. Another serious drawback to the use of physic nut is the presence of toxic components, such as lectin, trypsin inhibitor, and phorbol esters, in all parts of the plant [5]. Therefore, in-depth understanding of the structure and function of key gene families and metabolic pathways of physic nut is essential for improving its crop productivity and commercialization.
Additionally, physic nut is a potential model organism for functional genomic analysis, particularly in Euphorbiaceae. Physic nut is a diploid species (2n = 22) [6], with a relatively small genome size (approximately 416Mbp) compared to other members of the Euphorbiaceae [7,8]. According to the most recently updated version, the assembled genome has a total length of 320.5 Mbp consisting of 27,127 putative protein-coding genes, and most of the scaffolds have been anchored on the genetic linkage map [9]. In addition, high-throughput sequencing has mushroomed over the past decade, stimulating the transcriptome profiling analyses. Gene expression profiles of physic nut from different tissues, developmental stages, and biotic/abiotic stresses were constructed [10,11]. Additionally, more than 170 biosamples of physic nut are publicly available from NCBI (National Coalition Building Institute) (as of 5 February 2018). All these genomic and transcriptional data provided a valuable resource for basic and applied studies in physic nut.
Transcription factors (TFs), also known as trans-acting elements, are DNA-binding proteins that specifically bind cis-acting elements in the eukaryotic promoters to activate or inhibit transcriptional regulation [12,13]. As its important roles in the regulation of plant gene expression [14], the structure and function of TFs are now becoming research hotspots in plant molecular biology. The gene expression involves different classes of TFs, which have evolved to regulate a variety of plant-specific genes or signals [15,16]. The Dof gene family is a typical example of such TFs. The Dof proteins typically have 200–400 amino acids and contain two main functional domains. One is the Dof domains in the N-terminal domain includes a highly-conserved single zinc-finger structure formed by a CX2CX21CX2C motif, where one Zn2+ can covalently combine with four Cys residues. The other one is a regulatory C-terminal domain [17,18,19]. For instance, the transcription activation domain of the maize ZmDof1 gene is formed by 44 amino acid residues located at the C-terminal domain [20]. In spite of high level homology in the Dof domain, the rest of the amino acid sequences in the proteins are divergent, coinciding with their expected diverse functions [19].
Previous studies disclosed a variety of roles of Dof proteins in gene expression regulation when associated with plant-specific phenomena, including metabolism, photoperiodic regulation, phytohormone responses, defense responses, and other aspects of plant development [21,22,23]. Maize Dof1 (ZmDof1) is the first member of the Dof gene family identified in plants which may be involved in novel molecular mechanisms underlying tissue-specific and light-regulated gene expression in plants [24]. In addition, recent studies revealed that two closely-related Arabidopsis thaliana L. Dof genes, AtDof3.7 (DAG1) and AtDof2.5 (DAG2), are maternal genes involved in the control of seed germination, although their actions are opposite [25]. A rice Dof protein, OsDof3, affected the DNA binding of GAMYB to GARE, which is important for combinational regulation of the transcriptional response to Gibberellic acid (GA), and might be a mediator for GA signaling during germination [19,26]. Although none of roles has been confirmed conclusively, Dof proteins apparently participate in the regulation of various processes.
Until now, the Dof genes have been identified and characterized in different plant species, such as A. thaliana, Oryza sativa L. [27], Glycine max L. [28], Triticuma estivum L. [29], and Ricinus communis L. [30]. However, understanding of the Dof gene family members and their evolutionary and functional characteristics in physic nut is limited. Based on the genomic and transcriptional data, we focused on the identification, characterization and functional exploration of the Dof genes in physic nut. In total, 33 JcDof proteins (encoded by 24 JcDof genes) were identified and characterized. These JcDof genes were further divided into three major groups by comparison to their orthologs/paralogs in castor bean (Ricinus communis L.) and A. thaliana. The gene structure, motif, and phylogenetic analysis revealed that genes within each group exhibited similar gene structure and protein motif arrangements. Segmental duplication probably played crucial roles in the expansion of the JcDof gene family, and the gene expansion was mainly subjected to positive selection. The expression profile demonstrated the broad involvement of JcDof genes in response to various abiotic stresses and hormonal treatments and their possible functional divergence. Taken together, our results provide valuable information for understanding the JcDof genes’ evolution, and lay a foundation for future functional analysis of the JcDof genes.
2. Results
2.1. Identification and Characterization of Dof Genes in Physic Nut
To extensively identify all the Dof candidate members in the physic nut genome, we used a whole-genome scanning to identify genes that encode proteins containing the Dof DNA-binding domain by both BLASTP and HMM profile search. Initially, the Dof protein sequences from Arabidopsis thaliana and their HMM profiles of the Dof domain were used as the BLASTP and HMMER query sequences to screen the physic nut genome. Subsequently, it was examined for the presence of the Dof domain using the SMART software and NCBI Conserved Domain database for all the Dof candidate sequences. Eventually, we identified 24 candidates of Dof genes in total, represented by 33 transcripts in physic nut (Table S1). Based on their gene loci, we designated each Dof protein uniquely as JcDof-1, and JcDof-2 to JcDof-24.
In addition, we systematically evaluated the basic properties of JcDof protein, including domain position, protein length, molecular weight (Mw), isoelectric point (pI), instability coefficient, and orthologous genes (Table 1). The average length of these Dof protein sequences was 339 amino acid residues and the length mainly centered on the range of 160–518 amino acid residues. Correspondingly, the molecular weights were mainly distributed from 18.2 kDa (JcDof-1) to 55.7 kDa (JcDof-6). The predicted isoelectric point of Dof proteins varied from 4.65 (JcDof-21) to 9.42 (JcDof-3). The instability coefficient of JcDof protein showed a variation from 39.4 (JcDof-17) to 61.74 (JcDof-7.3-5). The location of JcDof protein conserved domain was analyzed by SMART. It was found that the domain positions of JcDof proteins encoded by the same gene (i.e., JcDof proteins that are generated by alternative splicing of the same gene model) were similar, but quite different for those encoded by different genes.
Table 1.
The information of the JcDof gene family.
| Gene ID | Protein Name | Protein Model | Position of Dof Domain | Protein Length | Mw | pI | Instability Index | Orthologous |
|---|---|---|---|---|---|---|---|---|
| 105649666 | JcDof-1 | XP_012091770.1 | 40–98 | 160 | 18,217 | 9.4 | 49.94 | AT1G29160.1 |
| 105649561 * | JcDof-2.1 | XP_012091631.1 | 67–125 | 331 | 36,503 | 8.74 | 58.25 | AT1G28310.2 |
| JcDof-2.2 | XP_012091630.1 | 67–125 | 365 | 40,036 | 8.72 | 60.68 | AT1G28310.2 | |
| 105649560 * | JcDof-3.1 | XP_012091629.1 | 39–97 | 321 | 34,862 | 9.42 | 61.1 | AT5G60850.1 |
| JcDof-3.2 | XP_012091628.1 | 39–97 | 321 | 34,862 | 9.42 | 61.1 | AT5G60850.1 | |
| 105649506 | JcDof-4 | XP_012091561.1 | 48–106 | 283 | 31,234 | 8.87 | 58.45 | AT1G28310.2 |
| 105645621 | JcDof-5 | XP_012086658.1 | 39–97 | 302 | 32,787 | 8.69 | 56.53 | AT4G24060.1 |
| 105644078 | JcDof-6 | XP_012084716.1 | 148–206 | 518 | 55,717 | 5.88 | 43.82 | AT5G62430.1 |
| 105642820 * | JcDof-7.1 | XP_012083166.1 | 38–96 | 256 | 27,889 | 9.04 | 58.51 | AT3G61850.4 |
| JcDof-7.2 | XP_012083165.1 | 41–99 | 259 | 28,222 | 9.04 | 57.94 | AT3G61850.4 | |
| JcDof-7.3 | XP_012083164.1 | 23–81 | 272 | 29,648 | 8.89 | 61.74 | AT3G61850.4 | |
| JcDof-7.4 | XP_012083162.1 | 23–81 | 272 | 29,648 | 8.89 | 61.74 | AT3G61850.4 | |
| JcDof-7.5 | XP_012083161.1 | 23–81 | 272 | 29,648 | 8.89 | 61.74 | AT3G61850.4 | |
| JcDof-7.6 | XP_012083160.1 | 38–96 | 287 | 31,275 | 8.87 | 59.86 | AT3G61850.4 | |
| JcDof-7.7 | XP_012083159.1 | 41–99 | 290 | 31,607 | 8.87 | 59.35 | AT3G61850.4 | |
| 105641716 | JcDof-8 | XP_012081705.1 | 32–90 | 344 | 36,852 | 8.97 | 52.37 | AT5G65590.1 |
| 105640671 | JcDof-9 | XP_012080436.1 | 31–89 | 334 | 36,528 | 6.86 | 57.05 | AT5G60850.1 |
| 105640546 | JcDof-10 | XP_012080278.1 | 121–179 | 471 | 51,491 | 6.61 | 54.99 | AT5G39660.1 |
| 105640379 | JcDof-11 | XP_012080063.1 | 52–110 | 312 | 34,986 | 6.75 | 39.5 | AT5G62940.1 |
| 105639962 | JcDof-12 | XP_012079559.1 | 26–84 | 236 | 24,419 | 9.17 | 48.6 | AT3G50410.1 |
| 105639655 | JcDof-13 | XP_012079164.1 | 63–121 | 326 | 34,900 | 9.08 | 48.8 | AT1G07640.3 |
| 105639642 | JcDof-14 | XP_012079148.1 | 26–84 | 246 | 25,139 | 8.72 | 41.9 | AT3G50410.1 |
| 105636282 * | JcDof-15.1 | XP_012074917.1 | 75–133 | 353 | 36,578 | 9.14 | 50.3 | AT2G37590.1 |
| JcDof-15.2 | XP_012074916.1 | 84–142 | 362 | 37,605 | 9.14 | 50.73 | AT2G37590.1 | |
| 105635894 | JcDof-16 | XP_012074418.1 | 10–68 | 282 | 30,892 | 5.14 | 48.76 | AT3G52440.1 |
| 105633699 | JcDof-17 | XP_012071724.1 | 21–79 | 245 | 25,883 | 8.52 | 39.4 | AT1G47655.1 |
| 105632564 | JcDof-18 | XP_012070363.1 | 101–159 | 497 | 53,629 | 7.78 | 43.27 | AT3G47500.1 |
| 105631489 | JcDof-19 | XP_012069011.1 | 18–76 | 249 | 26,497 | 8.26 | 47.16 | AT3G21270.1 |
| 105630455 | JcDof-20 | XP_012067660.1 | 129–187 | 465 | 51,091 | 6.8 | 47.74 | AT5G39660.1 |
| 105629142 | JcDof-21 | XP_012066060.1 | 28–86 | 287 | 32,762 | 4.65 | 51.26 | AT1G21340.1 |
| 105628246 | JcDof-22 | XP_012065018.1 | 71–129 | 315 | 33,921 | 9.23 | 51.88 | AT2G28810.1 |
| 105628152 | JcDof-23 | XP_012064896.1 | 36–94 | 290 | 32,430 | 6.65 | 41.41 | AT2G28510.1 |
| 105647749 | JcDof-24 | XP_012089351.1 | 70–128 | 338 | 35,691 | 9.19 | 50.23 | AT3G55370.3 |
* These genes are regulated by alternative splicing mechanisms. Mw: Molecular weight; pI: Isoelectric point.
2.2. DNA-Binding Domain Conservation Analysis of JcDof Protein
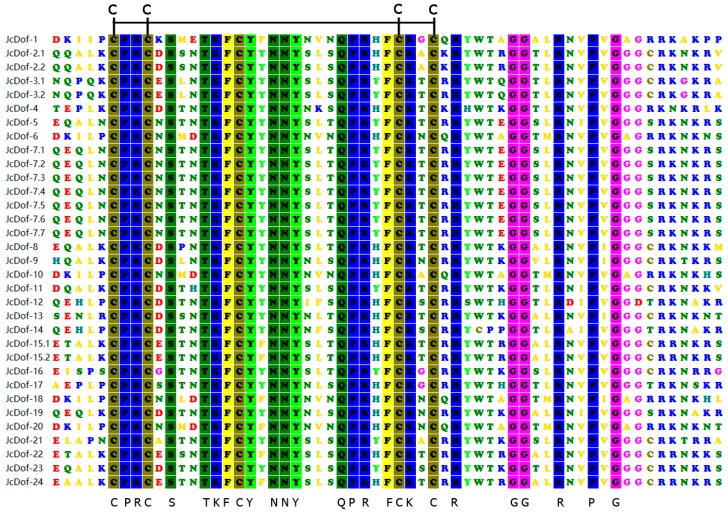
Dof protein usually has a DNA-binding domain of approximate 40–60 amino acid residues in the N-terminus. This domain contains a highly-conserved CX2CX21CX2C single zinc-finger structure, which is essential for the zinc finger configuration and loop stability. In this study, the conservation of DNA-binding domain of JcDof proteins was analyzed. Multiple protein sequence alignments against Dof DNA-binding domain of JcDof proteins revealed that all of them were highly conserved. Especially, we found 20 highly-conserved (100% identical in all 33 JcDof proteins) amino acids CPRC-S–TKFCY-NNY—QPR-FCK-C in the 29 amino acid-long region which corresponded to the CX2CX21CX2C single zinc-finger structure (Figure 1).
Figure 1.
Multiple protein sequences alignments against Dof DNA-binding domain in JcDof genes. The identical amino acids are shown in bottom and the four cysteine residues are indicated on top.
2.3. Phylogenetic Analysis and Classification of JcDof Proteins
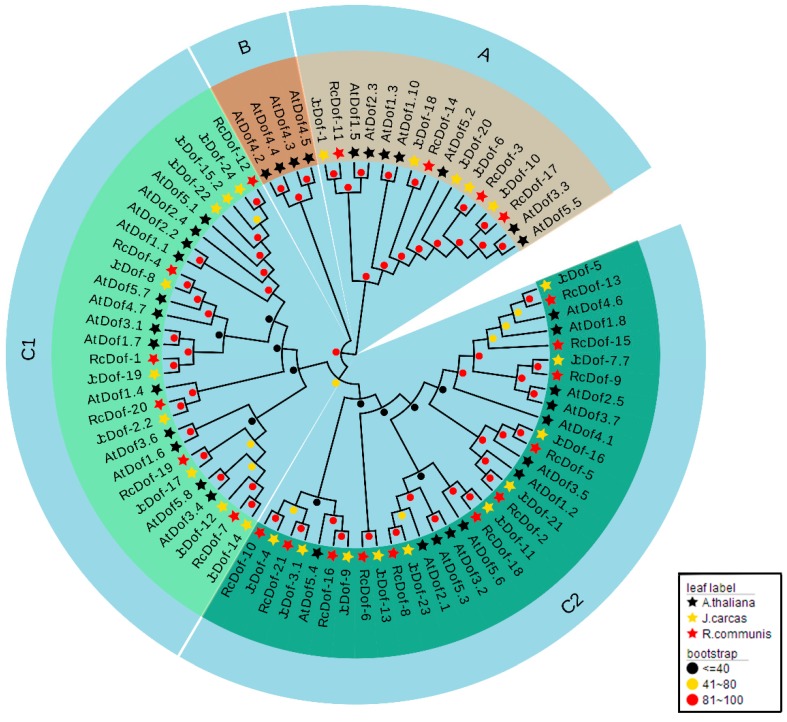
To explore the phylogenetic relationships of JcDof proteins, we carried out phylogenetic analysis on Dof proteins from physic nut and other two plant species, including Ricinus communis, also from the Euphorbiaceae family, and A. thaliana, as an outgroup (detailed information on all of the Dof proteins is listed in Supplementary Table S2). A phylogenetic tree was reconstructed including 24 physic nut, 21 R. communis and 36 A. thaliana Dof proteins (Figure 2). For each gene, we chose the longest protein formed by alternative splicing. The resulting phylogenetic tree was clustered into three major groups (A, B, and C), and they were considered to be evidentfor distinct phylogenetic lineages, which were supported by a bootstrap value over 80%. The two external nodes at the end of the same clades of phylogenetic tree were likely to represent the closest homologous gene pairs.
Figure 2.
Phylogenetic relationships among J. curcas, A. thaliana, and R. communis Dof proteins. The neighbor-joining tree was created using the MEGA6.0 program (bootstrap value set at 1000). Thirty-six (36) AtDof proteins marked with black pentacle, 24 JcDof proteins marked with yellow pentacle, and 21RcDof proteins marked with red pentacle. The resulting phylogenetic tree was clustered into three major groups (A, B, and C), which were supported by a bootstrap value over 80%. The Dof proteins in Group C were further divided into two sub-groups, C1 and C2, supported by a bootstrap value over 40%. The detailed information of all the Dof proteins is listed in Supplementary Table S2.
Of the three major groups, Group C was the first main clade, containing 19 physic nut Dof proteins, 17 R. communis Dof proteins, and 25 A. thaliana Dof proteins, which were further divided into two sub-groups, C1 and C2, supported by a bootstrap value over 40%. Group A was the second major clade with five physic nut Dof proteins, four R. Communis Dof proteins, and seven A. Thaliana Dof proteins. Group B was the minimal clade, with only four proteins. Distinguishingly, the Group B Dof proteins were only found in Arabidopsis, which could be explained by species/lineage-specific gene gain or loss events. We further checked the GO (Gene Ontology) annotations of these four Arabidopsis Dof genes, and found that comparing with the Arabidopsis Dof genes in other groups, two of these four genes (At4g21030, At4g21050) have some specific annotations, such as “cotyledon development”, “mucilage metabolic process involved in seed coat development”, “regulation of secondary shoot formation”, and “fruit development”, which implied the possible function divergence of Dof genes in group B (Supplementary Table S3 for detailed information). The phylogenetic tree showed that Dofs in the Group A and C were duplicated several times before the divergence of these three species, and were highly conserved among J. curcas, R. communis, and A. thaliana. In addition, the physic nut Dof proteins were more closely related, evolutionarily, to R. communis than to the Arabidopsis Dof proteins.
2.4. JcDof Gene Structures and Conserved Motifs in JcDof Proteins
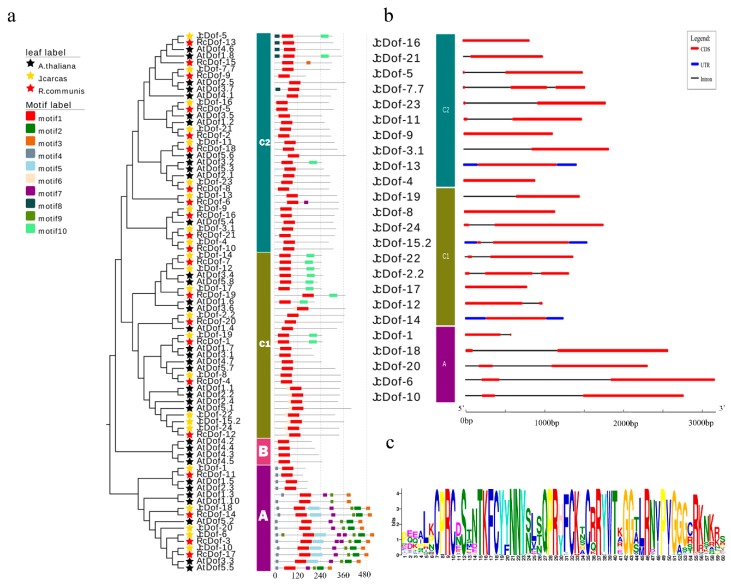
Introns and exons are the backbones of genes. Their numbers and distribution patterns are an evolutionary mark for a gene family. We, therefore, compared the intron-exon structure of each JcDof gene. The results revealed that the gene structure pattern was consistent with the phylogenetic analysis. Based on the exon-intron structures, the number of introns varied from one to three in J. curcas (Figure 3b). There are ten JcDof genes with one intron (41.7%), 12 JcDof genes with two introns (50%), and two JcDof genes with three introns (8.3%). All of the JcDof genes in subfamily A possessed two introns, while the number of introns of the JcDof gene in subfamily C varied from one to three.
Figure 3.
JcDof gene structures and conserved motifs in JcDof proteins. (a) The distribution of 10 conserved motifs in Dof proteins; (b) Gene structures of JcDof genes. CDS, UTR and introns were depicted by filled red boxes, blue boxes, and single black lines; and (c) Motif-1, corresponding to theCX2CX21CX2C single zinc-finger structure. The detailed motif’s sequences are shown in Figure S1 and Table S4.
Our classification of Dof genes was also verified by the conserved motif analysis. All of the Dof protein sequences were loaded into the MEME analysis tool to identify the conserved motifs. As a result, a total of ten conserved motifs were observed, which were statistically-significant with E-values less than 1× 10−40 (Figure 3a, described in detail in Supplementary Figure S1 and Table S4). The motifs of Dof proteins identified by MEME were between 13–43 amino acids in length. Among them, Motif-1 is a common motif in all Dof proteins, corresponding to the CX2CX21CX2C single zinc-finger structure in the Dof domain, which was the highly-homologous core region of Dof family (Figure 3c). While all of the Group B proteins and many of the Group C1 and C2 proteins only contain Motif-1, some Dof proteins have extra specific motifs, which may be relevant to different functions. The Dof proteins from Group A had the most complicated motif patterns, and Motif-2, Motif-4, Motif-5, and Motif-9 were specific for them. While Group C members have relatively simple motif patterns compared with Group A, they also had group-specific motifs, such as Motif-6, Motif-8, and Motif-10, but not all the group members have these specific motifs. For further elucidation of the potential roles of the Group A specific motifs, we checked the GO annotations of the Group A genes in Arabidopsis. Interestingly, we found that comparing with the Arabidopsis Dof genes in other groups, most of the genes in Group A (5 out of 7) have some flower-development-related annotations, such as “flower development”, “negative regulation of long-day photo periodism”, “flowering”, “negative regulation of short-day photo periodism”, “regulation of timing of transition from vegetative to reproductive phase”, and “vegetative to reproductive phase transition of meristem”, which implied the possible function divergence of the Dof genes in group A (see Supplementary Table S3 for detailed information).
2.5. Chromosomal Locations and Gene Duplication Events of JcDof Genes
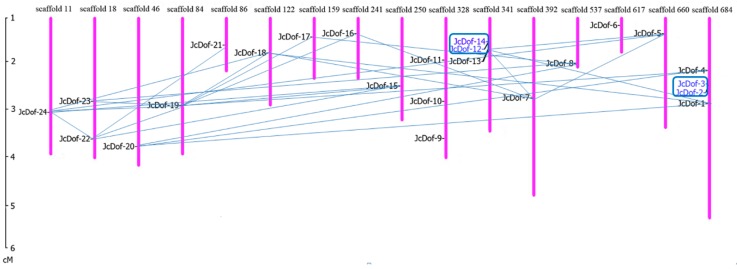
In order to explore the mechanism of evolution and amplification of JcDof gene, the chromosomal locations and gene duplication events of JcDof genes were further analyzed. The chromosomal distribution of JcDof genes was plotted using Map Inspect software (Figure 4). The duplication events of JcDof genes were also examined, and Dof gene-pairs arising from segmental and tandem duplication were marked with light blue line and dark blue rectangles, respectively. From Figure 4 we can find that some Dof genes, such as JcDof-19, have been duplicated several times to form more than one duplicated gene-pair with other genes; and some JcDof genes, such as JcDof-15, JcDof-22, and JcDof-24, are evolutionarily too close to resolve their gene duplication order (the duplication pairs are described in detail in Supplementary Table S5). The gene expansion of the Dof family in physic nut mainly resulted from segmental duplication, and tandem duplication also played a minor role. In total, 26 pairs of segmental duplicated JcDof genes (93% of all duplicated genes) and two pairs of tandem duplicated JcDof genes (7% of all duplicated genes) were found. For most of the duplicated gene pairs (22 out of 28), the pairwise JcDof genes often came from the same phylogenetic group, with very high sequence similarities. Specifically, tandem duplicated genes have higher sequence similarity than segmental duplicated genes (Table S5).
Figure 4.
Chromosomal locations and gene duplication events of JcDof. Respective scaffold numbers are indicated at the top of each bar. The scale on the left is in centimorgan (cM). The JcDof gene pairs of segmental and tandem duplication are linked by pale blue lines and marked in dark blue rectangles, respectively. The detailed information of duplication pairs are described in Supplementary Table S5.
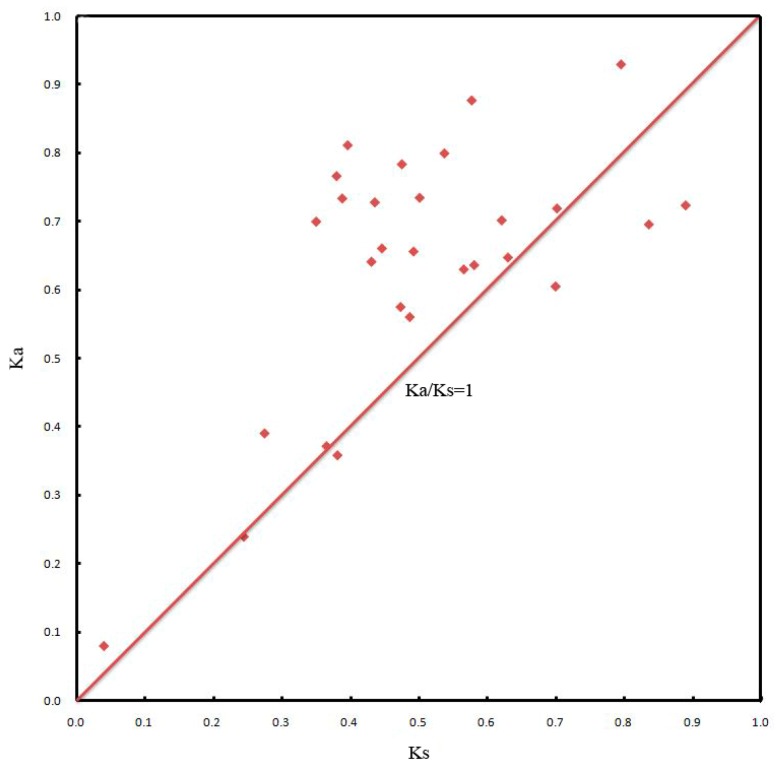
To further understand the evolutionary constraints acting on all of the duplicated JcDof genes, we calculated the non-synonymous substitution rate (Ka), synonymous substitution rate (Ks) and Ka/Ks for all of the 28 pairs duplicated genes (Figure 5 and Table S5). We found 23 pairs duplicated genes whose Ka/Ks were more than one (accounting for 82% of all the duplicated genes) and five pairs duplicated genes whose Ka/Ks ratio were less than one (accounting for 18% of all the duplicated gene pairs) (Table S5). This implied that most of the Dof duplicated gene pairs tended to be subjected to positive selection, which may play important roles in the origin of adaptive phenotypes and the possible function divergence in JcDof genes.
Figure 5.
The Ka/Ks value of duplicated JcDof gene pairs. 28 pairs of duplicated JcDof genes and 23 pairs duplicated genes with Ka/Ks more than one. The detailed Ka/Ks information of duplication pairs are described in Supplementary Table S5.
2.6. Expression Patterns of JcDof Genesunder Different Abiotic Stress and Hormone Treatments
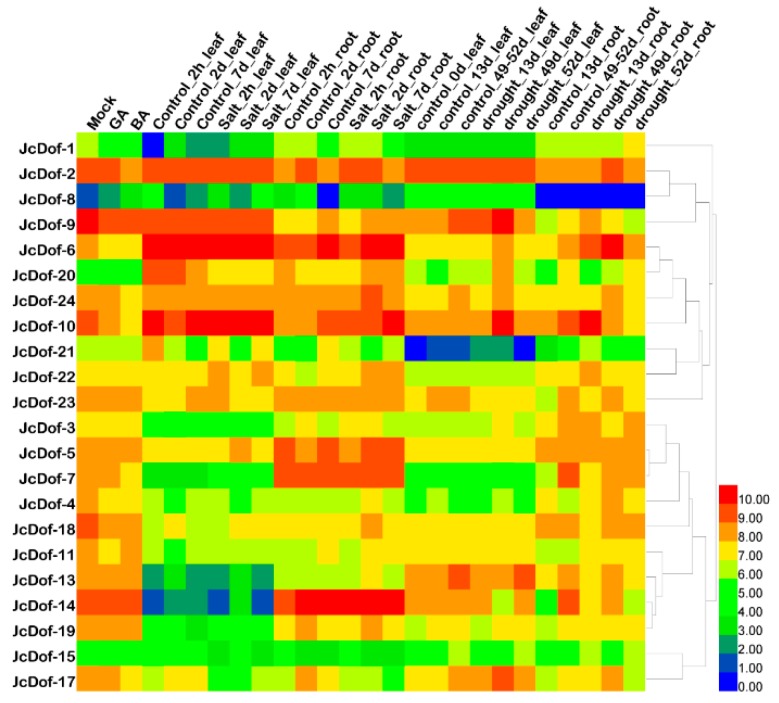
In order to further study the possible function divergence of JcDof genes, we investigated the expression level of JcDof genes under various abiotic stresses and hormonal treatments by using the public transcriptome data from NCBI SRA database (Supplementary Tables S6 and S7 for detailed information). We employed a heatmap to visualize a global transcription profile of the JcDof genes. As shown in Figure 6, JcDof genes showed diverse responses to various treatments, and significant differences were found in response to 6-Benzylaminopurine (BA), salt, and drought treatments (two-fold increases or decreases compared to controls).
Figure 6.
Expression patterns of JcDof genes under different treatments. The heatmap was generated by HemI software using the expression data of the JcDof genes, and normalized log2 transformed values were used with hierarchical clustering represented by the color scale (0–10). Blue indicates low expression, and red indicates high expression. The samples were: roots and leaves (salt- and drought-stressed at different times), and roots (BA treatment). The detailed information of expression data are described in Supplementary Tables S6 and S7.
In the BA treatment experiments (gene expression data collected from roots), compared with the negative control (mock), three genes (JcDof-1, JcDof-8, and JcDof-10) exhibited significant responses. Among them, JcDof-1 and JcDof-10 showed reduced expression when responding to BA treatment, with more than two-fold (JcDof-1) and nearly four-fold (JcDof-10) decreasing, respectively. Meanwhile, JcDof-8 showed a significantly up-regulated expression with more than four-fold increase. We further checked the GO annotations of their Arabidopsis orthologs, and found they were annotated as “seed coat development” (JcDof-1, AT1G29160.1), “guard cell differentiation, positive regulation of transcription, regulation of cell wall pectin metabolic process, stomatal movement” (JcDof-8, AT5G65590.1), and “flower development” (JcDof-10, AT5G39660.1) respectively, which may imply the possible roles of these three genes (Supplementary Table S3 for detailed information).
We further analyzed the expression patterns of the JcDof genes in salt- and drought-stressed roots and leaves at different times: 2 h, 2 days, and 7 days (salt-stressed); 13 days, 49 days, and 52 days (drought-stressed). The fold changes of gene expression were calculated between abiotic stress treatments and controls. Many JcDof genes exhibited significant responses, and some of them showed significant up- or down-regulation in both roots and leaves, such as JcDof-8, JcDof-17, and JcDof-20 in salt-stressed treatments, and JcDof-6, JcDof-8, JcDof-10, JcDof-14, JcDof-17, and JcDof-21 in drought-stressed treatments. Most of these significantly up- or down-regulated genes (seven out of nine) tended to show similar expression changes (up- or down-regulation) in both roots and leaves. The only two exceptions were JcDof-20 and JcDof-14. JcDof-20 showed significantly reduced expression in leaves (from 2 h to 7 days) when responding to salt treatment, while JcDof-20 expression in salt-treated roots first decreased (at 2 h), and then increased significantly (two days and seven days). Another gene, JcDof-14, showed significantly reduced expression in leaves (in 49 days) when responding to drought treatment, while JcDof-14 expression in drought-treated roots first increased (in 13 days), and then decreased significantly (49 days and 52 days).
We have also checked the differential expression patterns of the duplicated JcDof gene pairs, and found that if JcDof genes differentially expressed in some stress treatments, and their duplicated counterparts were more likely not to show differential expression (27 pairs vs. 20 pairs, Supplementary Table S8 for detailed information). We think these results are consistent with our Ka/Ks results, that most of the duplicated JcDof genes tended to be subjected to positive selection, and implied the possible function divergence in JcDof genes.
3. Discussion
The Euphorbiaceae family includes some of the most efficient biomass accumulators, such as physic nut, castor bean, cassava, and rubber tree [9,31]. Crop improvement in Euphorbiaceae for sustainable industrial raw materials and food production requires more extensive genome-wide studies on these species. Notably, physic nut has become an ideal model organism in Euphorbiaceae for further functional genomics analysis due to its sequenced genome, genetic linkage map, and abundance of high-throughput transcriptome data. Studies on physic nut will provide insights into the investigation of other Euphorbiaceae organisms.
Genome-wide gene family analysis is abasic and a key step to understanding the gene structure, function, and evolution [32]. The Dof gene family has been shown to play crucial roles in the regulatory network of plant defense, including responses to diverse biotic and abiotic stresses [22,23,33,34]. Until now, the Dof genes have been identified and characterized in different plant species, but not in the promising energy plant physic nut yet. Therefore, we conducted a comprehensive analysis of the JcDof family in physic nut, along with their homologs in R. communis and A. thaliana, to study their phylogenetic relationships and potential functions.
In total, we identified 24 JcDof genes in the physic nut genome. Compared with the number of Dof genes in A. thaliana (36 genes from TAIR), the size of physic nut Dof gene family is much smaller [35], although the assembled genome size of physic nut is approximately three times larger than the A. thaliana genome (320.5 Mbp vs. 125 Mbp) [9,36]. Correspondingly, we had discovered that the members from Group B, one of the major groups in the phylogenetic tree, all pertained to AtDof genes. In addition, Subgroup C1 contained 13 AtDof genes; while only nine JcDof genes were noted. Subgroup C2 had 12 AtDof genes and 10 JcDof genes. These results suggested that JcDof and AtDof genes should arise through different duplication events, and might have undergone species/lineage-specific gene gain or loss.
Both tandem duplication and segmental duplication contributed to the variation in gene family number and distribution [37,38]. In total, 26 gene-pairs from segmental duplication and two from tandem duplication were found in physic nut. We calculated the Ka/Ks ratios for these duplicated JcDof paralog genes, and found most of the duplicated genes pairs had Ka/Ks ratios over 1, implying that positive selection played an important role in the evolution of JcDof genes, and high-throughput expression data analysis further confirmed the functional diversity of JcDof genes. JcDof genes showed diverse responses to various treatments, and might participate in different stress/hormone-responding regulatory processes. This work provides valuable information for understanding the evolution of JcDof genes and lays a foundation for future functional analysis of Dof genes in the process of growth, development, and Dof-mediated regulation in physic nut.
4. Materials and Methods
4.1. Data Sources
The physic nut genomic and proteomic sequences were downloaded from the NCBI database (Available online: https://www.ncbi.nlm.nih.gov/, Assembly JatCur_1.0). The Dof protein sequences of A. thaliana were obtained from the Arabidopsis genome database (TAIR 9.0 release, Available online: http://www.arabidopsis.org/) [35]. The Dof protein sequences of castor bean were obtained from the PlantTFDB database (Available online: http://planttfdb.cbi.pku.edu.cn/) [16]. The physic nut gene expression data were collected from the SRA database (Available online: https://www.ncbi.nlm.nih.gov/) [39].
4.2. Dof Gene Identification and Characterization
To identify all the possible Dof genes in physic nut, both local BLASTP [40] and Hidden Markov model (HMM) searches were performed [41]. For BLASTP, the known Dof proteins from Arabidposis were taken as queries and the E-value was set to 1 × 10−10. For the HMM search, the HMM profile of the Dof domain was used as query and the E-value was set to 1 [24]. All the retrieved sequences were further scanned and tested using SMART (Available online: http://smart.embl-heidelberg.de/) [42] and NCBI Conserved Domains database (Available online: http://www.ncbi.nih.gov/Structure/cdd/cdd.shtml) for authentication of the presence of Dof domain [43]. We manually removed redundant sequences that do not have Dof domain or have incomplete encoding frame. Parameters, such as protein length, molecular weight, isoelectric point, and instability coefficient of all the Dof proteins in physic nut were predicted using ExPASy Proteomics Server (Available online: http://prosite.expasy.org/) [44]. The orthologous genes of JcDof proteins in A. thaliana were predicted by BLASTP.
4.3. DNA-Binding Domain Conservation Analysis of JcDof Protein
The conserved regions of JcDof proteins were extracted by DNAMAN tool (version 2.6 Lynnon Biosoft, Quebec City, QC, Canada) [45]. We then identified highly-conserved Dof domain for all Dof proteins by multiple sequence alignment analysis using ClustalW MEGA integration software [46].
4.4. Phylogenetic Analysis
Physic nut, A. thaliana, and R. communis Dof protein sequences were pretreated by GUIDANCE2 online tool to remove unreliable columns [47]. The phylogenetic relationship among the Dof proteins was analyzed using ClustalW and the dendrogram was constructed using MEGA (v6.0, Tokyo Metropolitan University, Tokyo, Japan) by neighbor-joining method, with the following parameters: Poisson correction, pairwise deletion, and 1000-bootstrap replicates [48].
4.5. Gene Structure of Dof Proteins
Positional information for both the gene sequences and the corresponding coding sequences was loaded into the Gene Structure Display Server (GSDS v2.0, Available online: http://gsds.cbi.pku.edu.cn/) to obtain information on intron/CDS structure [49]. The coordinates of the Dof domain in each protein were recalculated into the coordinates in the corresponding gene sequence and featured in the gene structure.
4.6. Detection of Additional Conserved Motifs
To identify additional conserved motifs outside the Dof domain of physic nut Dof proteins, we used Multipel Expectation Maximization for Motif Elucidation (MEME v4.11.2, Available online: http://meme.nbcr.net/meme/) [50]. The limits on maximum width, minimum width, and maximum number of motifs were specified as 5, 150, and 10, respectively. The motifs were numbered serially according to their order in MEME. Those motifs common to genes in one of the three similarity groups were designated as the group-specific signatures.
4.7. Chromosomal Localization
According to the chromosomal positions of genes, we drew a map of the distribution of Dof genes throughout the physic nut genome using MapInspect software (Available online: http://mapinspect.software.informer.com/) [51]. The Dof gene pairs resulting from segmental or tandem duplication were linked by lines and marked in blue rectangle, respectively.
4.8. Detection of Gene Duplication Events and Estimation of Synonymous (Ks) and Nonsynonymous (Ka) Substitutions per Site and Their Ratio
Duplicated gene pairs derived from segmental or tandem duplication were identified in physic nut genome based on the method described in the Plant Genome Duplication Database [52,53]. An all-against-all BLASTP comparison (E-value ≤ 1 × 10−20) provided the gene pairs for syntenic clustering determined by MCScanX (E-value ≤ 1 × 10−20) [54]. Tandem duplication arrays were identified using BLASTP with a threshold of E-value < 1× 10−20, and one unrelated gene among cluster members was tolerated, as described for A. thaliana. Pairs from segmental and tandem duplications were used to estimate Ka, Ks, and their ratio. Coding sequences from segmentally and tandemly duplicated Dof gene pairs were aligned by PRANK [55] and trimmed by Gblocks. The software DnaSP (Available online: http://www.softpedia.com/get/Science-CAD/DnaSP.shtml) [56] was then used to compute Ka and Ks values for each pair following the YN model (a simple model of voting) [57]. If Ka/Ks > 1, there is positive selection pressure; if Ka/Ks = 1, there is neutral selection or natural selection pressure; if Ka/Ks < 1, there is a purification selection effect [58,59].
4.9. Expression Analysis of Physic Nut Dof Genes
The original expression data for JcDof genes under different treatments (including gibberellins [GA], 6-Benzylaminopurine[BA], high salt concentration and drought) were retrieved from NCBI SRA database (Available online: https://www.ncbi.nlm.nih.gov/). All the data were analyzed using Tuxedo suite (TopHat and Cufflinks, http://post.queensu.ca/~rc91/NGS/TuxedoTutorial.html) and then upper-quartile normalized and log transformed. Heat maps were generated by means of the HemI toolkit (Available online: http://hemi.biocuckoo.org/) with average linkage hierarchical clustering [60,61].
5. Conclusions
In conclusion, a total of 24 Dof genes were identified from physic nut, and these Dof genes were further divided into three major groups based on the phylogenetic inference. The gene structures, conserved motifs, gene duplicated events, selection pressures, and expression profiling of these JcDof genes were analyzed. A genome comparison discovered that the expansion of the Dof gene family in physic nut mainly resulted from segmental duplication, and this expansion was mainly subjected to positive selection. The expression profile demonstrated the broad involvement of JcDof genes in different hormonalor abiotic stressed treatments. Among them, three genes (JcDof-1, JcDof-8, and JcDof-10) exhibited significant responses to the BA treatment. Furthermore, many JcDof genes were significantly responsive to the salt and drought treatments. On the whole, this study provides an extensive resource for understanding the Dof genes in physic nut.
Acknowledgments
This work was supported by the following grants: National Natural Science Foundation of China (grant nos. 31471220, 91440113), Start-up Fund from Xishuangbanna Tropical Botanical Garden, and ‘Top Talents Program in Science and Technology’ from Yunnan Province.
Abbreviations
| TF | Transcription factor |
| HMM | Hidden Markov model |
| Mw | Molecular weight |
| pI | Isoelectric point |
| GA | Gibberellic acid |
| BA | 6-Benzylaminopurine |
Supplementary Materials
The following are available online at http://www.mdpi.com/1422-0067/19/6/1598/s1.
Author Contributions
C.L. conceived and supervised this study. C.L. and P.W. designed the experiments. P.W. carried out the experiments, and analyzed and interpreted the data. J.L., X.G., D.Z. and A.L. participated in the discussion and provided valuable advice and practical contributions. C.L., P.W. and J.L. wrote the manuscript. All authors reviewed, edited, and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bhasanutra R., Sutiponpeibun S. Jatropha curcas oil as a substitute for diesel engine oil. Int. Energy J. 2017;4:56–70. [Google Scholar]
- 2.Openshaw K. A review of Jatropha curcas: An oil plant of unfulfilled promise. Biomass Bioenergy. 2000;19:1–15. doi: 10.1016/S0961-9534(00)00019-2. [DOI] [Google Scholar]
- 3.Chen M.-S., Pan B.-Z., Fu Q., Tao Y.-B., Martínez-Herrera J., Niu L., Ni J., Dong Y., Zhao M.-L., Xu Z.-F. Comparative transcriptome analysis between gynoecious and monoecious plants identifies regulatory networks controlling sex determination in Jatropha curcas. Front. Plant Sci. 2017;7:1953. doi: 10.3389/fpls.2016.01953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan B.-Z., Xu Z.-F. Benzyladenine treatment significantly increases the seed yield of the biofuel plant Jatropha curcas. J. Plant Growth Regul. 2011;30:166–174. doi: 10.1007/s00344-010-9179-3. [DOI] [Google Scholar]
- 5.Makkar H., Becker K., Sporer F., Wink M. Studies on nutritive potential and toxic constituents of different provenances of Jatropha curcas. J. Agric. Food Chem. 1997;45:3152–3157. doi: 10.1021/jf970036j. [DOI] [Google Scholar]
- 6.Dehgan B. Phylogenetic significance of interspecific hybridization in Jatropha (Euphorbiaceae) Syst. Bot. 1984;9:467–478. doi: 10.2307/2418796. [DOI] [Google Scholar]
- 7.Carvalho C.R., Clarindo W.R., Praça M.M., Araújo F.S., Carels N. Genome size, base composition and karyotype of Jatropha curcas L., an important biofuel plant. Plant Sci. 2008;174:613–617. doi: 10.1016/j.plantsci.2008.03.010. [DOI] [Google Scholar]
- 8.Rahman A.Y.A., Usharraj A.O., Misra B.B., Thottathil G.P., Jayasekaran K., Feng Y., Hou S., Ong S.Y., Ng F.L., Lee L.S. Draft genome sequence of the rubber tree Hevea brasiliensis. BMC Genom. 2013;14:75. doi: 10.1186/1471-2164-14-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu P., Zhou C., Cheng S., Wu Z., Lu W., Han J., Chen Y., Chen Y., Ni P., Wang Y. Integrated genome sequence and linkage map of physic nut (Jatropha curcas L.), a biodiesel plant. Plant J. 2015;81:810–821. doi: 10.1111/tpj.12761. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L., Zhang C., Wu P., Chen Y., Li M., Jiang H., Wu G. Global analysis of gene expression profiles in physic nut (Jatropha curcas L.) seedlings exposed to salt stress. PLoS ONE. 2014;9:e97878. doi: 10.1371/journal.pone.0097878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C., Zhang L., Zhang S., Zhu S., Wu P., Chen Y., Li M., Jiang H., Wu G. Global analysis of gene expression profiles in physic nut (Jatropha curcas L.) seedlings exposed to drought stress. BMC Plant Biol. 2015;15:17. doi: 10.1186/s12870-014-0397-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi M.U., Nakagawa S. Evolution of the Human Genome I. Springer; Berlin, Germany: 2017. Transcription Factor Genes; pp. 241–263. [Google Scholar]
- 13.Riaño-Pachón D.M., Ruzicic S., Dreyer I., Mueller-Roeber B. PlnTFDB: An integrative plant transcription factor database. BMC Bioinform. 2007;8:42. doi: 10.1186/1471-2105-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noguero M., Atif R.M., Ochatt S., Thompson R.D. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 2013;209:32–45. doi: 10.1016/j.plantsci.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 15.Franco-Zorrilla J.M., López-Vidriero I., Carrasco J.L., Godoy M., Vera P., Solano R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA. 2014;111:2367–2372. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin J., Tian F., Yang D.-C., Meng Y.-Q., Kong L., Luo J., Gao G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016;45:D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yanagisawa S., Schmidt R.J. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999;17:209–214. doi: 10.1046/j.1365-313X.1999.00363.x. [DOI] [PubMed] [Google Scholar]
- 18.Yanagisawa S. Dof domain proteins: Plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 2004;45:386–391. doi: 10.1093/pcp/pch055. [DOI] [PubMed] [Google Scholar]
- 19.Yanagisawa S. The Dof family of plant transcription factors. Trends Plant Sci. 2002;7:555–560. doi: 10.1016/S1360-1385(02)02362-2. [DOI] [PubMed] [Google Scholar]
- 20.Chen X., Wang D., Liu C., Wang M., Wang T., Zhao Q., Yu J. Maize transcription factor Zmdof1 involves in the regulation of Zm401 gene. Plant Growth Regul. 2012;66:271–284. doi: 10.1007/s10725-011-9651-5. [DOI] [Google Scholar]
- 21.Gupta S., Malviya N., Kushwaha H., Nasim J., Bisht N.C., Singh V., Yadav D. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta. 2015;241:549–562. doi: 10.1007/s00425-014-2239-3. [DOI] [PubMed] [Google Scholar]
- 22.Venkatesh J., Park S.W. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol. Biochem. 2015;94:73–85. doi: 10.1016/j.plaphy.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Yanagisawa S. Plant Transcription Factors. Elsevier; New York, NY, USA: 2015. Structure, Function, and Evolution of the Dof Transcription Factor Family; pp. 183–197. [Google Scholar]
- 24.Shu Y., Song L., Zhang J., Liu Y., Guo C. Genome-wide identification and characterization of the Dof gene family in Medicago truncatula. Genet. Mol. Res. 2015;14:10645–10657. doi: 10.4238/2015.September.9.5. [DOI] [PubMed] [Google Scholar]
- 25.Wang T., Yue J.-J., Wang X.-J., Xu L., Li L.-B., Gu X.-P. Genome-wide identification and characterization of the Dof gene family in moso bamboo (Phyllostachys heterocycla var. pubescens) Genes Genom. 2016;38:733–745. doi: 10.1007/s13258-016-0418-2. [DOI] [Google Scholar]
- 26.Wu Q., Li D., Li D., Liu X., Zhao X., Li X., Li S., Zhu L. Overexpression of OsDof12 affects plant architecture in rice (Oryza sativa L.) Front. Plant Sci. 2015;6:833. doi: 10.3389/fpls.2015.00833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lijavetzky D., Carbonero P., Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 2003;3:17. doi: 10.1186/1471-2148-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Y., Qiu L.-J. Retraction: Genome-Wide Analysis of the Dof Transcription Factor Gene Family Reveals Soybean-Specific Duplicable and Functional Characteristics. PLoS ONE. 2016;11:e0167019. doi: 10.1371/journal.pone.0167019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw L.M., McIntyre C.L., Gresshoff P.M., Xue G.-P. Members of the Dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation. Funct. Integr. Genom. 2009;9:485. doi: 10.1007/s10142-009-0130-2. [DOI] [PubMed] [Google Scholar]
- 30.Jin Z., Chandrasekaran U., Liu A. Genome-wide analysis of the Dof transcription factors in castor bean (Ricinus communis L.) Genes Genom. 2014;36:527–537. doi: 10.1007/s13258-014-0189-6. [DOI] [Google Scholar]
- 31.Chan A.P., Crabtree J., Zhao Q., Lorenzi H., Orvis J., Puiu D., Melake-Berhan A., Jones K.M., Redman J., Chen G. Draft genome sequence of the oilseed species Ricinus communis. Nat. Biotechnol. 2010;28:951. doi: 10.1038/nbt.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marquez Y., Höpfler M., Ayatollahi Z., Barta A., Kalyna M. Unmasking alternative splicing inside protein-coding exons defines exitrons and their role in proteome plasticity. Genome Res. 2015;25:995–1007. doi: 10.1101/gr.186585.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh K.B., Foley R.C., Oñate-Sánchez L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002;5:430–436. doi: 10.1016/S1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 34.Ma J., Li M.-Y., Wang F., Tang J., Xiong A.-S. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genom. 2015;16:33. doi: 10.1186/s12864-015-1242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2011;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Initiative A.G. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 37.Cannon S.B., Mitra A., Baumgarten A., Young N.D., May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leister D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 2004;20:116–122. doi: 10.1016/j.tig.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 39.Leinonen R., Sugawara H., Shumway M., Collaboration I.N.S.D. The sequence read archive. Nucleic Acids Res. 2010;39(Suppl. 1):D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mount D.W. Using the basic local alignment search tool (BLAST) Cold Spring Harb. Protoc. 2007;2007 doi: 10.1101/pdb.top17. [DOI] [PubMed] [Google Scholar]
- 41.Schuster-Böckler B., Bateman A. An introduction to hidden Markov models. Curr. Protoc. Bioinform. 2007;18:A.3A.1–A.3A.9. doi: 10.1002/0471250953.bia03as18. [DOI] [PubMed] [Google Scholar]
- 42.Letunic I., Doerks T., Bork P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2011;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2014;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woffelman C. DNAMAN for Windows, Version 2.6. Lynon Biosoft, Institute of Molecular Plant Sciences, Leiden University; Leiden, The Netherlands: 1994. [Google Scholar]
- 46.Thompson J.D., Gibson T., Higgins D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002 doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 47.Sela I., Ashkenazy H., Katoh K., Pupko T. GUIDANCE2: Accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 2015;43:W7–W14. doi: 10.1093/nar/gkv318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu B., Jin J., Guo A.-Y., Zhang H., Luo J., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2014;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Suppl. 2):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He H., Dong Q., Shao Y., Jiang H., Zhu S., Cheng B., Xiang Y. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012;31:1199–1217. doi: 10.1007/s00299-012-1241-0. [DOI] [PubMed] [Google Scholar]
- 52.Lee T.-H., Tang H., Wang X., Paterson A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2012;41:D1152–D1158. doi: 10.1093/nar/gks1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohler A., Rinaldi C., Duplessis S., Baucher M., Geelen D., Duchaussoy F., Meyers B.C., Boerjan W., Martin F. Genome-wide identification of NBS resistance genes in Populus trichocarpa. Plant Mol. Biol. 2008;66:619–636. doi: 10.1007/s11103-008-9293-9. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T.-H., Jin H., Marler B., Guo H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Löytynoja A., Goldman N. webPRANK: A phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinform. 2010;11:579. doi: 10.1186/1471-2105-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Librado P., Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 57.Yang Z., Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 58.Hurst L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002;18:486–487. doi: 10.1016/S0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- 59.Ito T.M., Trevizan C.B., dos Santos T.B., de Souza S.G.H. Genome-Wide Identification and Characterization of the Dof Transcription Factor Gene Family in Phaseolus vulgaris L. Am. J. Plant Sci. 2017;8:3233. doi: 10.4236/ajps.2017.812218. [DOI] [Google Scholar]
- 60.Deng W., Wang Y., Liu Z., Cheng H., Xue Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE. 2014;9:e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song S., Zhou H., Sheng S., Cao M., Li Y., Pang X. Genome-Wide Organization and Expression Profiling of the SBP-Box Gene Family in Chinese Jujube (Ziziphus jujuba Mill.) Int. J. Mol. Sci. 2017;18:1734. doi: 10.3390/ijms18081734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.