Abstract
The evolutionarily conserved Aurora family kinases, a family of mitotic serine/threonine kinases, has three members in humans (Aurora-A, -B and -C). Overexpression of Aurora family members, particularly Aurora-A, has been reported in many human cancers and cell lines. In this study, we present evidence based on comparative gene expression analysis via quantitative RT-PCR to delineate the relative contributions of these kinases in 60 cell lines and statistical analysis in five different human cancer microarray datasets. The analysis demonstrated the selective upregulation of these Aurora members in various cancers. In general, Aurora-A exhibited the highest expression levels, with substantially decreased quantities of the Aurora-C transcript detected relative to Aurora-A and -B. Moreover, to characterize the roles of each Aurora member, which share many similarities, we investigated the expression profiles of the family in normal tissues and a panel of different phases of the HeLa cell cycle. Finally, both Aurora-A and -B were overexpressed in a majority of esophageal tumor tissues in comparison to the normal variants. Taken together, the results show that each Aurora member exhibits distinct expression patterns, implying that they are engaged in different biological processes to accomplish more elaborate cell physiological functions in higher organisms.
Key words: Aurora family, Aurora-A, Aurora-B, Aurora-C, Esophageal carcinoma, Quantitative RT-PCR
AURORA, an evolutionarily conserved family of serine/threonine kinases, has drawn intense attention recently for its association with human cancer development and its role in mitotic progression, including control of chromosome condensation, centrosome separation and maturation, bipolar-spindle assembly, mitotic checkpoint, chromosome alignment, and cytokinesis (7,20). Phylogenetic classification reveals that there are three family members in humans, namely Aurora-A, -B, and -C. A conserved kinase domain at the C-terminus characterizes these kinases, while the N-terminal domains are of variable lengths and share low sequence identity. All three Aurora family kinases are highly expressed in the G2/M phase at the transcription and translation levels, where they exhibit subtle differences in subcellular localization (7,20).
Aurora-A was mapped to chromosome region 20q13.2, in which DNA amplification has frequently been observed in various human cancers (24,33,34). Ectopic expression of Aurora-A transforms Rat-1 and NIH3T3 cells (5,41), suggesting that the kinase possesses oncogenic potential. In human tumors, gene amplification and/or overexpression of Aurora-A have been detected in a significant fraction of pancreatic (16), breast (18,32), esophageal (31), prostate (3), gastric (23), bladder (26), ovarian (11), colorectal (30), and hepatocellular carcinomas (HCC) (28,37). Aurora-B was mapped to 17p13.1, in which a DNA deletion is associated with various human cancers (9). Moreover, overexpression of Aurora-B causes multinuclei, thereby leading to genomic instability (35), a major factor in the predisposition for tumor development. Upregulation of Aurora-B was observed in colorectal tumors (1), astrocytomas (2), and seminomas (8). Aurora-C was mapped to chromosome 19q13.43, with several kinds of DNA alterations documented in this chromosome region in human cancers (4,15). The expression level of Aurora-C is increased in several cancer cell lines, including NB1RGB, HeLa, MDZ-MB-453, HepG2, and HuH7 (15) and in primary colorectal cancer (30). Taken together, expanding the observation of gene expression profiles for the Aurora family, particularly Aurora-B or -C, to additional human cancer samples would certainly be worthwhile. Furthermore, systematic comparisons of gene expressions profiling of each family member are essential for addressing whether the Aurora family members are selectively overexpressed in different human cancers.
There is only one Aurora family member in yeast. By contrast, there are two in Drosophila, Caenorhabditis elegans, and Xenopus, and three in mice and humans, implying that the functions of Aurora family should have been fine-tuned to meet complicated cellular activities in higher organisms. A systematic comparison of Aurora members in humans and various model organisms has revealed the conservation of their biological networks (36). However, common characters shared by the family, such as a highly conserved kinase domain and high expression in mitosis, raise a question concerning whether Aurora-A, -B, or -C might possess distinct functions for carrying out elaborate cell physiological functions in mammals. Recently, the functions of the Aurora family has begun to be unraveled, and differences in how Aurora-A or -B work in mitosis have been reported (17). Nevertheless, a systematic comparison among the Aurora family members to depict their expression levels and to distinguish the roles they play would be helpful in understanding why the Aurora family evolved from one member in yeast to three members in mammals.
Genome-wide microarray analysis has provided insights into the mechanisms of transcriptional regulation of these Aurora kinases. Examples from several recent microarray studies have greatly expanded our knowledge of their expression patterns in different tissues (http://expression.gnf.org/cgi-bin/index.cgi/) (29), cell lines (examples at http://genome-www.stanford.edu/nci60) (22), and cell cycle progression (examples at http://genome-www.stanford.edu/Human-CellCycle/Hela/) (39). In addition to the detailed but dispersed reports regarding the expression patterns for these three Aurora genes, there is a lack of statistical analysis of the patterns of these Aurora family members. In addition, simultaneous and quantitative investigation of three Aurora kinases is either inadequate or completely lacking with respect to the following issues: (i) transcripts of all three Aurora genes show periodic variations during the cell cycle; however, the exact temporal expressions of these Aurora genes through the cell cycle have not been quantitatively compared; and (ii) all three Aurora genes are mapped to loci with DNA abnormalities that have been associated with many cancers (6,10), yet, except for Aurora-A, the gene expression levels of Aurora-B and -C in different cancers are inadequate.
To elucidate how the three Aurora family members are transcriptionally regulated and to gain comparative insights into the gene expression signatures of the Aurora family members, it is imperative to obtain their quantitative gene expression patterns. In this study, therefore, RNAs from 23 human tissues, 60 cell lines, and 10 pairs of esophageal carcinoma tissues were collected and analyzed using quantitative RT-PCR to delineate the gene expression patterns of each Aurora member. Furthermore, the addition of comprehensive statistical analysis of the gene expression patterns for each Aurora member from five cancer-related microarray datasets further expands our knowledge of the physiological and pathological scope of the Aurora kinases.
MATERIALS AND METHODS
Cell Cultures
Most of the cell lines used in this study were obtained from American Type Culture Collection except those primary culture normal nasal mucosal cells, primary culture normal cervical epithelial cells, nasopharyngeal carcinoma (NPC) cell lines, bladder cancer cell lines (E6, UB09, UB37, UB40, and UB47), esophageal cancer cell lines, and hepatoma cell lines (Tong, T59, HCC36, HA22T/VGH, PLL5, and Malaru). In this study, we collected three skin fibroblast cell lines (HS-68, CCD-966SK, and Detroit551), mammary epithelial cell line (M10), primary culture normal cervical epithelial cells, three primary culture normal nasal mucosal cells (NNM9, NNM11, and NNM13), six nasopharyngeal carcinoma cell lines (NPC094, NPC112, NPC113, NPC119, NPC121, and NPC-CNE1), six cervical cancer cell lines (172, SiHa, z183A, Cx, CaSki and HeLa), 10 hepatoma cell lines (Huh-7, HepG2, Hep3B, SK-Hep1, Tong, T59, HCC36, HA22T/VGH, PLC5, and Malaru), four oral cancer cell lines (OECM1, CAL27, SAS, and Ca9-22), four lung cancer cell lines (H1155, H522, P13, and CL1-0), two neuroblastoma cell lines (IMR32 and Gbm), two colon cancer cell lines (SW620 and HCT-116), two esophageal cancer cell lines (CE81T/VGH and CE146T/VGH), two breast cancer cell lines (Bcm1 and MCF-7), kidney transformed cell line (293), leukemia cancer cell line (Jurkat), stomach cancer cell line (Scm1), uroepithelial cell line (E7), and 10 bladder cancer cell lines (E6, RT4, TSGH8301, UB09, UB37, UB40, UB47, TCC-sup, J82, and T24). Among these bladder cancer cell lines, three of them [UB37: stage B2 (Duke classification), infiltrative; UB40: stage A, papillary; UB47: stage B1, papillary] were from the TCC patients of “black foot disease” high incident area (unpublished data). These cells were maintained at 37°C in a 5% CO2 incubator and grown in DMEM or RPMI medium supplemented with 10% heat-inactivated fetal calf serum and 100 μg/ml penicillin/streptomycin (BRL). CE81T/VGH and CE146T/VGH were derived from human squamous cell carcinoma of the esophagus (12).
Cell Cycle Synchronization
HeLa cells were synchronized at the G1/S boundary by double-thymidine. Cells were cultured for 14 h in the presence of 2 mM thymidine (Sigma), released for 12 h in fresh medium, and arrested for 14 h in the presence of 2 mM thymidine. Cells were released from double-thymidine block by changing to fresh culture medium. Cells were harvested by centrifugation at 1000 rpm for 5 min at 4°C, followed by washing twice with cold PBS, and slowly fixed by adding 500 μl 70% ice-cold ethanol gently. Fixed cells were centrifuged at 1000 rpm for 5 min at room temperature, and then stained with 1 ml Triton X-100 (0.1%)/propidium iodide (50 μg/ml)/RNase (100 μg/ ml) solution in the dark for 15 min at room temperature. The percentage of cells at different phases of the cell cycle was determined by flow cytometry (Becton Dickinson FACStar Plus flow cytometer). Each measurement was based on analysis of 30,000 cells.
Quantitative Reverse-Transcription Polymerase Chain Reaction (RT-PCR)
The mRNA expression levels of Aurora-A, -B, and -C in various tissues and cell lines were examined using RT-PCR analysis according to the manufacturer’s instruction (Applied Biosystems). The specificity of the amplification reaction was demonstrated by using Aurora-A, -B, and -C plasmids, which were constructed on pCMV2 vector as described previously (40). The primer sequences, designed by Primer Express software (Applied Biosystems), were Aurora-A (forward and reverse primers, 5′-TTCCAGGAGGACCACTCTCTGT-3′ and 5′-TGCATCCGACCTTCAATCATT-3′), Aurora-B (forward and reverse primers, 5′-GGCGCATGCACAATGAGA-3′ and 5′-TCCCCACCAGCAGCTCATAG-3′), and Aurora-C (forward and reverse primers, 5′-GGAAATTGAGATCCAGGCTCAT-3′ and 5′-GGCGTGCATCATGGAAATAGT-3′). Twenty-three commercially available normal tissues were obtained from Clontech. Total RNA was isolated from frozen tissues or cultured cells using the RNeasy RNA extraction kit (Qiagen) plus DNase I treatment (Promega). Two micrograms of total RNA from each sample was subjected to reverse transcription using the ThermoScript RT-PCR system (Invitrogene). The cDNA products from each sample (200-fold dilution) were used to perform quantitative PCR. Each 10 μl of quantitative PCR reaction mixture contained 5 μl of twofold SYBR Green Master Mixture (Applied Biosystems), 4 μl of cDNA product mixture (200-fold dilution), 0.5 μl each of 6-μM PCR forward and reverse primers (final concentration: 300 nM). The Rn value obtained from the PCR amplification curve was used to calculate relative mRNA copy number. GAPDH mRNA was used as an internal comparison control to normalize the data. A melting curve was used to identify the temperature where only the amplicon, but not the primer dimers, accounted for the SYBR Green-bound fluorescence. Assays were performed in triplicate using Applied Biosystems Model 7700 instruments.
RESULTS AND DISCUSSION
Specificity of the Amplification Reaction
To delineate the expression profiles of the Aurora family, we employed quantitative RT-PCR to analyze a variety of biological samples, including various tissues, cell lines with different derivatives, a HeLa cell cycle panel released from the G1/S boundary, and esophageal cancer tissues and their adjacent nontumor pairs. In this study, we employed two templates for PCR amplification, namely three Aurora plasmids and cDNA generated from the bladder cancer cell line, UB37, to illustrate the specificity of each primer. Figure 1 shows that each specific primer for Aurora-A, -B, or -C could only amplify its own target regardless of which template we used. Moreover, the PCR products were subjected to DNA sequencing. The results showed that each PCR reaction only generated its own corresponding product (data not shown), indicating the PCR reaction could specifically reflect the expression pattern among Aurora members. For quantitative RT-PCR measurements, it is generally believed that there is no perfect internal control expressing equal quantities in different tissues. Therefore, in this study, the relative expression percentage of each Aurora member was reported using GAPDH as the internal control, as its expression levels remain relatively constant in different tissues (19).
Figure 1.
Primer specificity for Aurora family members. To detect the gene expression pattern of each Aurora family member, specific primers for Aurora-A, -B, and -C were designed for quantitative RT-PCR using Aurora family members that were constructed on the pCMV2 vector (A) and mRNA templates from the UB37 bladder cancer cell line as an example (B). The amplified products were then subjected to agarose gel followed by ethidium bromide staining.
Aurora Family Members Exhibited Different Expression Profiles Among Various Human Tissues
The gene expression patterns for the Aurora members in various tissues have separately been reported (13–15,21); however, these detailed but dispersed reports failed to quantitatively and simultaneously provide comparative views of the relative expression patterns. Analysis of the expression levels for the three human Aurora family members using quantitative RT-PCR across a wide spectrum of human tissues revealed that the general expression profiles for the Aurora family were quite distinct (Fig. 2). Of the 23 normal tissues examined, Aurora-A was expressed at higher levels than Aurora-B and -C in 16 samples (the pancreas, liver, salivary gland, small intestine, lung, adrenal gland, thymus, trachea, cerebellum, colon, kidney, uterus, bone marrow, prostate, placenta, and mammary gland). Abundant expression of the Aurora-B transcript was detected at levels higher than those of Aurora-A and -C in seven tissues (testis, thyroid, heart, fetal brain, brain, spleen, and skeletal muscle). Gene expression levels of Aurora-C in the tissues examined were lower than those of Aurora-A and -B. In addition, Aurora-C was predominantly expressed in testes followed by the salivary gland, cerebellum, and placenta. Such abundant testicular expression of Aurora-C transcripts suggests that Aurora-C may play a unique role in this reproductive organ. Although it has been demonstrated that the Aurora kinases act on basic cellular physiology related to mitotic progression, the simultaneous quantitative measurement of three Aurora transcripts in normal human tissues implies that the Aurora kinases may have roles in different tissues, meriting further investigation.
Figure 2.
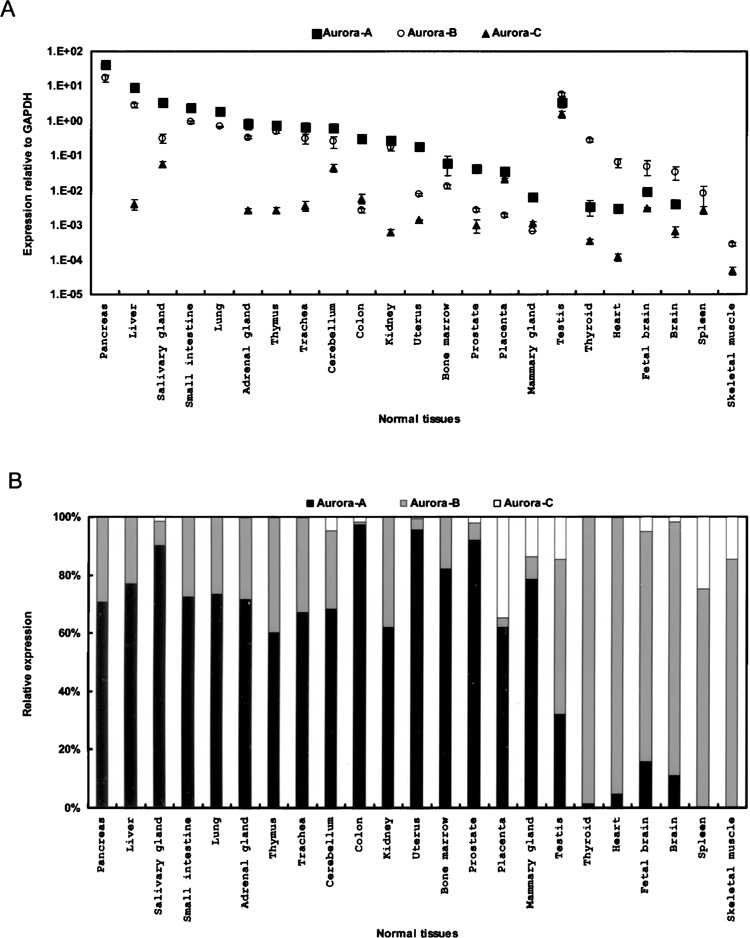
Tissue-specific distribution of Aurora family members. (A) The gene expression profiles for Aurora-A (squares), -B (circles), and -C (triangles) were examined in 23 human tissues using quantitative RT-PCR, with data normalized to the GAPDH expression level. Values are presented as the mean ± SD. (B) To display the relative expression ratio, the expression of each Aurora member was normalized to that of the entire family.
Expression Profiles in 60 Human Cell Lines, Distinctive From NCI60 Collections
The cellular and molecular mechanisms that elicit the role of three Aurora members rely on distinct cell model systems. Analysis of the gene expression patterns for the three human Aurora members was carried out in the 60 cell lines. Our cell line collections had less than 10% overlap with those reported in the NCI60 microarray dataset (22). By quantitative RT-PCR analysis, the expression levels could be roughly classified into three subgroups, with Aurora-A having the highest, followed by Aurora-B and -C in descending order (Fig. 3A, B). There were exceptions such as in primary nasal mucosal, NPC, and oral cancer cell lines where Aurora-B gene expression was higher than that of Aurora-A (Fig. 3B). The expression levels of Aurora-B in cancer cell lines of those tissues were much higher than those tissues themselves tested, implying that Aurora-B is selectively upregulated in nasal/nasopharyngeal tissues. Interestingly, of the ten bladder cancer cell lines collected, extraordinarily higher expression patterns were observed for all three Aurora members in arsenic-treated variants (UB37, UB40, and UB47) compared to those in other bladder cancer cell lines tested, implying the possible involvement of arsenic in inducing Aurora family expression. Taken together, the analytical results promise an extension of the gene expression profiles of each Aurora member in the NCI60 dataset and further refinement of the roles of each family member across a diversity of cell lines.
Figure 3.
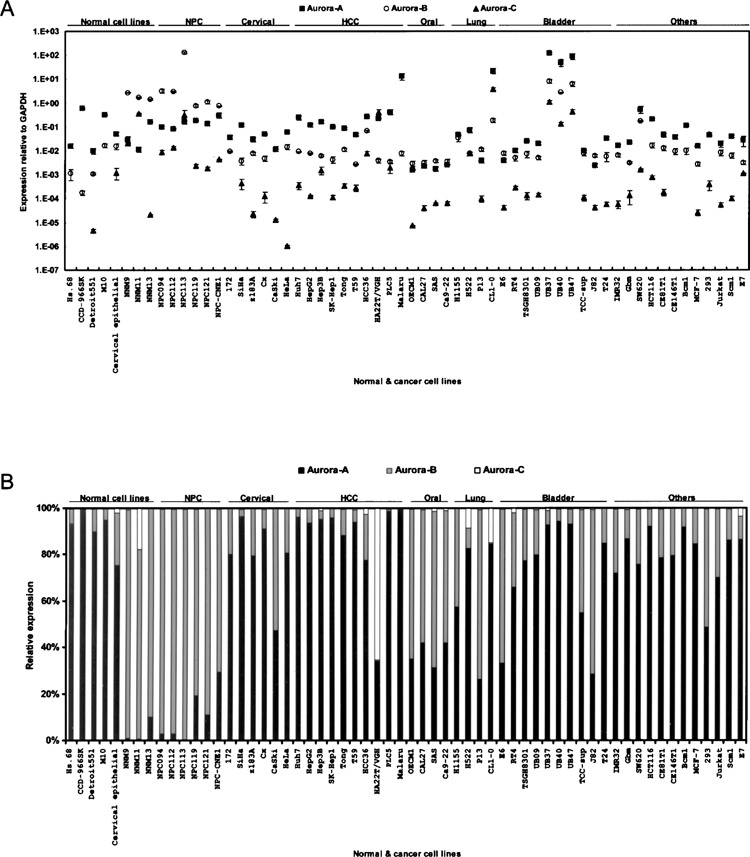
Gene expression profiles of Aurora family members in various human cell lines. (A) Gene expression profiles for Aurora-A (squares), -B (circles), and -C (triangles) were examined in 60 human cell lines using quantitative RT-PCR analysis. Data were normalized to the GAPDH expression level. Values are presented as the mean ± SD. (B) To present the relative expression ratio, the expression of each Aurora member was normalized to that of the entire family.
Aurora Family Members Were Periodically Expressed in the M Phase of the HeLa Cell Cycle
Both HeLa cell cycle microarray analysis (http://genome-www.stanford.edu/Human-CellCycle/Hela/) and Northern blotting indicated that Aurora members are predominately expressed in mitosis (15,21,27,32). However, the exact temporal expression patterns of the Aurora family members have not been clearly defined and compared. Thus, the gene expression patterns of all three Aurora members in synchronized HeLa cells were examined. HeLa cells were synchronized at the G1/S boundary by a double thymidine block, with total RNA extracted from cells harvested at various times after release from the G1/S transition for quantitative RT-PCR analysis. FACS analysis indicated that cells entered the S phase at 3 h and reached the G2/M phase at 6–10 h (Fig. 4A). In addition, cyclin B1 expression was elevated at 6–11 h with the peak at 9 h (Fig. 4A), indicating that mitosis had occurred within that period. In general, the expression levels of Aurora-C were substantially decreased, by two to seven orders of magnitude, compared to those of Aurora-A and -B (Fig. 4C). The expression of Aurora-A suddenly rose at 7 h, and was maintained for another 3 h, but declined at 11 h. There was no evident peak within the expression window ranging from 7 to 11 h for Aurora-A (Fig. 4B). On the contrary, expression of Aurora-B steadily increased to 7 h, reached a peak at 9 h, and then declined at 10 h (Fig. 4B). Expression of Aurora-C was first elevated at 8 h, peaked at 9 h, and had abruptly decreased at 12 h (Fig. 4C). Interestingly, the dramatic increase in Aurora-C transcripts with a 100,000-fold difference between the interphase and mitosis indicates tight but efficient regulation with cell cycle progression. The expression plateau of Aurora-A (at 7–8 h) was slightly ahead of the peaks of Aurora-B or -C (both at 9 h), and the expression duration for Aurora-C lasted longer (until 11 h) than that of Aurora-B (until 9 h), suggesting possible differential roles for Aurora family members in regard to the timescale along mitosis. Collectively, although previous findings pointed out that Aurora family members are highly expressed in mitosis, our analysis further revealed subtle but distinct differences in temporal transcriptional regulation for the Aurora family.
Figure 4.
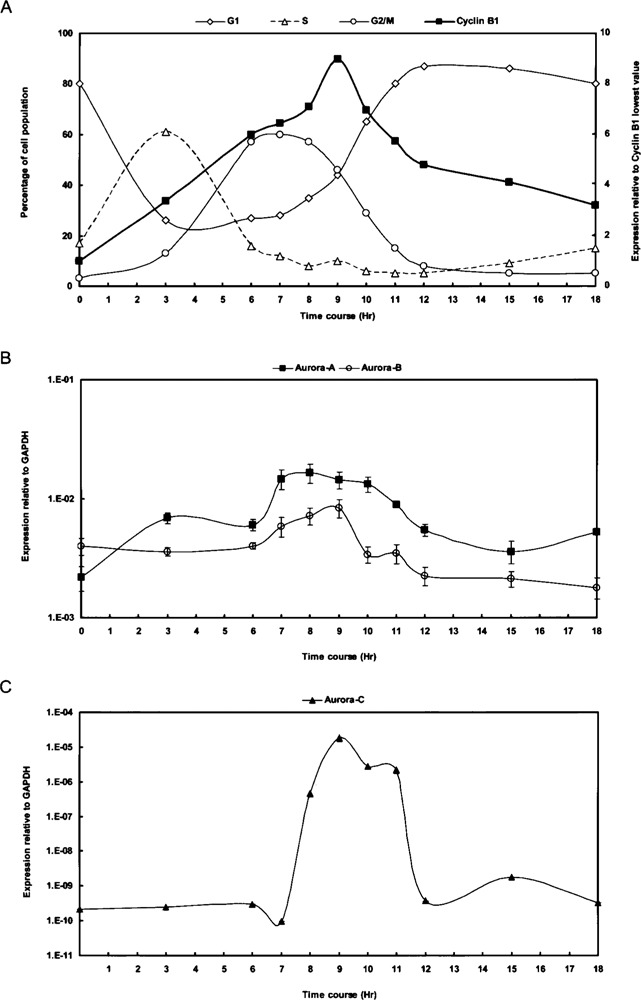
Aurora family members exhibit periodical expression patterns in the M phase of the HeLa cell cycle. (A) A HeLa cell cycle panel was prepared by synchronizing cells at the G1/S border using double thymidine treatment. Cells were then released from the G1/S block by removing the chemicals. RNA samples were collected at various time points and used as templates for quantitative RT-PCR analysis. The percentages of cells at different phases of the cell cycle are plotted on the left (Y-axis; G1 phase, diamonds; S phase, triangles; G2/M phase, circles). The relative expression of cyclin B1 (squares), indicative of the G2/M phase, is plotted on the right (Y-axis). FACS analysis, in accordance with the expression of the cell cycle marker cyclin B1, indicates that the cells reached the G2/M phase at 6–10 h, with the peak of mitosis occurring at 9 h. (B, C) Gene expression patterns of Aurora-A (squares) and Aurora-B (circles) are shown in (B), whereas that of Aurora-C (triangles) is shown in (C) due to its low expression level. Data were normalized to the GAPDH expression level. Values are presented as the mean ± SD.
Overexpression of Aurora-A and -B in Esophageal Carcinoma
Overexpression of Aurora-A is associated with many human tumors, while there are fewer reports concerning upregulation of Aurora-B or -C in human cancers as described in the introduction section. To investigate the prevalence of human cancers with elevated expression of Aurora family members, and scrutinize the possible selective overexpression of Aurora family members in malignancies, we extended our study to examine the expressions of Aurora family members in esophageal carcinoma and five additional human cancers (see below). Primary esophageal tumor tissues (T) and the corresponding adjacent noncancerous esophageal tissues (N) were obtained from 10 patients. Quantitative RT-PCR analysis revealed significantly elevated expression of both Aurora-A and -B in all 10 pairs of esophageal carcinoma tissues relative to their adjacent noncancerous tissues (p < 0.0001 for Aurora-A and -B) (Fig. 5). By contrast, no such association was demonstrated for Aurora-C. In fact, an inverse association between Aurora-C expression and esophageal cancer was demonstrated even though statistical significance was not achieved (p = 0.42).
Figure 5.
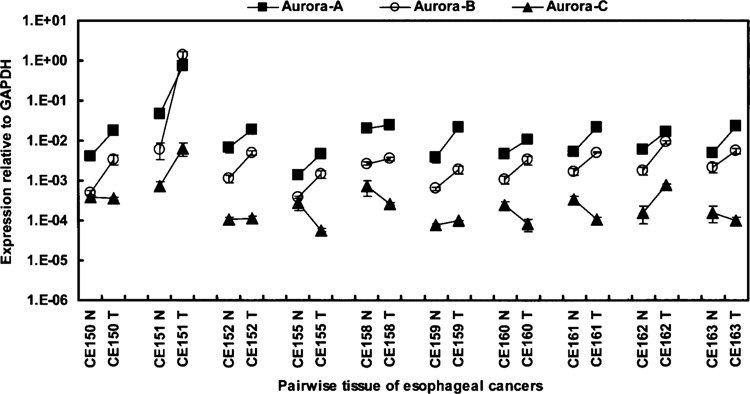
Overexpression of Aurora-A and -B in esophageal carcinoma. The gene expression profiles of Aurora-A (squares), -B (circles), and -C (triangles) in 10 pairs of tumor (T) and adjacent normal (N) human esophageal tissues were examined using quantitative RT-PCR. Data were normalized to the GAPDH expression level. Values are presented as the mean ± SD.
Differential Upregulation by Statistical Analysis of the Gene Expression Profiles for the Aurora Members in Five Cancer Microarray Datasets
Numerous cell cycle-regulated genes are also highly expressed in more proliferative tumors, as exemplified in the case of Aurora-A in HCC (28). To provide a comprehensive understanding of the association between these cell cycle-regulated Aurora genes and human tumors, the microarray datasets for HCC, and lung, breast, kidney, and gastric cancers were downloaded from the Stanford Microarray Database (http://genome-www5.stanford.edu/), followed by statistical analysis using SAS software (SAS Institute; version 8e). One-way ANOVA was used to analyze the gene expression profiles (38) of the Aurora genes, comparing normal and tumor groups for different cancers (Table 1). Results showed that Aurora-A is significantly overexpressed in HCC, and lung and gastric cancers (p < 0.05), whereas no significant difference was demonstrated for breast (p = 0.12) or kidney cancers (p = 0.63), although overexpression of Aurora-A has been reported to be associated with breast cancers (18,32) and related cell lines (25). In addition, overexpression of Aurora-B and -C was respectively observed in gastric cancer and HCC (Table 1), implying roles for Aurora-B or -C in the corresponding cancer development.
TABLE 1.
STATISTICAL ANALYSIS OF THE DIFFERENTIAL UPREGULATION OF AURORA FAMILY MEMBERS IN VARIOUS CANCERS
| Disease | Variable | Gene | Sample Number | p Value |
|---|---|---|---|---|
| Hepatoma | disease | Aurora A | 180 | 0.0001 |
| Aurora C | 0.05 | |||
| Lung cancer | disease | Aurora A | 72 | 0.032 |
| Aurora B | 0.56 | |||
| Aurora C | 0.076 | |||
| Kidney cancer | disease | Aurora A | 42 | 0.63 |
| Aurora B | 0.09 | |||
| Aurora C | 0.98 | |||
| Breast cancer | disease | Aurora A | 74 | 0.12 |
| Aurora B | 0.98 | |||
| Gastric cancer | disease | Aurora A | 124 | 0.0001 |
| Aurora B | 0.0001 | |||
| Aurora C | 0.56 |
The gene expression data of Aurora members were downloaded from five publicly accessible microarray databases (HCC, and lung, kidney, breast, and gastric cancers; http://genome-www5.stanford.edu/). The raw data were further analyzed using one-way ANOVA. Statistical significance (p < 0.05) is indicated in bold.
In summary, we have demonstrated the following. (i) The surprising differences in gene expression levels of Aurora members suggest that each member possesses unique tissue-specific functions. The relative expression levels of Aurora-A, -B, and -C in descending order, in various cell lines, is probably a reflection of specific physiological needs, indicating basic mechanisms operating to modulate the transcription of each kinase. (ii) The extraordinary overexpression of the Aurora family in arsenic-treated cell lines may reveal a possible activity of arsenic in interfering with transcription of the Aurora family, which may represent a common feature of transcriptional regulation for the Aurora family. (iii) The cell cycle-dependent oscillation in Aurora-C transcription has been demonstrated for the first time, implying tight regulation of transcriptional control. The expression window for Aurora-B was the narrowest, followed by Aurora-C, and Aurora-A was expressed throughout mitosis. Furthermore, the sequential timing of the initial rise in transcription during mitosis is in the order of Aurora-A, -B, and then -C, implying that these kinases may act within different but precise periods to guarantee mitotic progression. (iv) Aurora members are associated with different cancers. Some of the novel observations include overexpression of Aurora-A or -B in esophageal carcinoma and elevated expressions of Aurora-B in gastric cancer and Aurora-C in HCC. These analyses suggest a general role for the Aurora family in the development of certain cancers. In addition, each Aurora member exhibits a distinct expression profile, suggesting their functions may have expanded from Ipl1, the only Aurora family member in yeast, to achieve more complicated cellular functions in mammals. However, the M phase-dependent expressions of Aurora-A, -B, and -C are similar to that of Ipl1. These observations highlight the possibility that the roles of Aurora family members in mitosis progression are selectively conserved, and this certainly becomes the expression signature of the family. Accordingly, this comprehensive analysis of gene expression profiles not only provides mechanistic insights into the transcriptional regulation of these Aurora kinases, but also serves as a heuristic guide for further biochemical and molecular studies.
ACKNOWLEDGMENTS
We thank Drs. Shiou-Hwei Yeh, Chin-Tarng Lin, Cheng-Yang Chou, Nan-Haw Chow, Jenn-Han Chen, Li-Wha Wu, A-Min Huang, and King-Song Jeng for providing cell lines. This work was supported in part by grants from the National Health Research Institutes, National Science Council (DOH94-TD-G-111-007, DOH94-TD-G-111-009, DOH94-TD-G-111-023, and NSC93-3112-B-010-026), and the Taichung Veteran General Hospital (TCVGH-937317D).
REFERENCES
- 1. Adams R. R.; Eckley D. M.; Vagnarelli P.; Wheatley S. P.; Gerloff D. L.; Mackay A. M.; Svingen P. A.; Kaufmann S. H.; Earnshaw W. C. Human INCENP colocalizes with the Aurora-B/AIRK2 kinase on chromosomes and is overexpressed in tumour cells. Chromosoma 110:65–74; 2001. [DOI] [PubMed] [Google Scholar]
- 2. Araki K.; Nozaki K.; Ueba T.; Tatsuka M.; Hashimoto N. High expression of Aurora-B/Aurora and Ipll-like midbody-associated protein (AIM-1) in astrocytomas. J. Neurooncol. 67:53–64; 2004. [DOI] [PubMed] [Google Scholar]
- 3. Bar-Shira A.; Pinthus J. H.; Rozovsky U.; Goldstein M.; Sellers W. R.; Yaron Y.; Eshhar Z.; Orr-Urtreger A. Multiple genes in human 20q13 chromosomal region are involved in an advanced prostate cancer xenograft. Cancer Res. 62:6803–6807; 2002. [PubMed] [Google Scholar]
- 4. Bicher A.; Ault K.; Kimmelman A.; Gershenson D.; Reed E.; Liang B. Loss of heterozygosity in human ovarian cancer on chromosome 19q. Gynecol. Oncol. 66:36–40; 1997. [DOI] [PubMed] [Google Scholar]
- 5. Bischoff J. R.; Anderson L.; Zhu Y.; Mossie K.; Ng L.; Souza B.; Schryver B.; Flanagan P.; Clairvoyant F.; Ginther C.; Chan C. S.; Novotny M.; Slamon D. J.; Plowman G. D. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 17:3052–3065; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bischoff J. R.; Plowman G. D. The Aurora/Ipl1p kinase family: Regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 9:454–459; 1999. [DOI] [PubMed] [Google Scholar]
- 7. Carmena M.; Earnshaw W. C. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 4:842–854; 2003. [DOI] [PubMed] [Google Scholar]
- 8. Chieffi P.; Troncone G.; Caleo A.; Libertini S.; Linardopoulos S.; Tramontano D.; Portella G. Aurora B expression in normal testis and seminomas. J. Endocrinol. 181:263–270; 2004. [DOI] [PubMed] [Google Scholar]
- 9. Eiriksdottir G.; Barkardottir R. B.; Agnarsson B. A.; Johannesdottir G.; Olafsdottir K.; Egilsson V.; Ingvarsson S. High incidence of loss of heterozygosity at chromosome 17p13 in breast tumours from BRCA2 mutation carriers. Oncogene 16:21–26; 1998. [DOI] [PubMed] [Google Scholar]
- 10. Giet R.; Prigent C. Aurora/Ipl1p-related kinases, a new oncogenic family of mitotic serine-threonine kinases. J. Cell Sci. 112(Pt. 21):3591–3601; 1999. [DOI] [PubMed] [Google Scholar]
- 11. Gritsko T. M.; Coppola D.; Paciga J. E.; Yang L.; Sun M.; Shelley S. A.; Fiorica J. V.; Nicosia S. V.; Cheng J. Q. Activation and overexpression of centrosome kinase BTAK/Aurora-A in human ovarian cancer. Clin. Cancer Res. 9:1420–1426; 2003. [PubMed] [Google Scholar]
- 12. Hu C. P.; Hsieh H. G.; Chien K. Y.; Wang P. Y.; Wang C. I.; Chen C. Y.; Lo S. J.; Wuu K. D.; Chang C. M. Biologic properties of three newly established human esophageal carcinoma cell lines. J. Natl. Cancer Inst. 72:577–583; 1984. [PubMed] [Google Scholar]
- 13. Kimura M.; Kotani S.; Hattori T.; Sumi N.; Yoshioka T.; Todokoro K.; Okano Y. Cell cycle-dependent expression and spindle pole localization of a novel human protein kinase, Aik, related to Aurora of Drosophila and yeast Ipl1. J. Biol. Chem. 272:13766–13771; 1997. [DOI] [PubMed] [Google Scholar]
- 14. Kimura M.; Matsuda Y.; Yoshioka T.; Sumi N.; Okano Y. Identification and characterization of STK12/Aik2: A human gene related to aurora of Drosophila and yeast IPL1. Cytogenet. Cell Genet. 82:147–152; 1998. [DOI] [PubMed] [Google Scholar]
- 15. Kimura M.; Matsuda Y.; Yoshioka T.; Okano Y. Cell cycle-dependent expression and centrosome localization of a third human aurora/Ipl1-related protein kinase, AIK3. J. Biol. Chem. 274:7334–7340; 1999. [DOI] [PubMed] [Google Scholar]
- 16. Li D.; Zhu J.; Firozi P. F.; Abbruzzese J. L.; Evans D. B.; Cleary K.; Friess H.; Sen S. Overexpression of oncogenic STK15/BTAK/Aurora A kinase in human pancreatic cancer. Clin. Cancer Res. 9:991–997; 2003. [PubMed] [Google Scholar]
- 17. Meraldi P.; Honda R.; Nigg E. A. Aurora kinases link chromosome segregation and cell division to cancer susceptibility. Curr. Opin. Genet. Dev. 14:29–36; 2004. [DOI] [PubMed] [Google Scholar]
- 18. Miyoshi Y.; Iwao K.; Egawa C.; Noguchi S. Association of centrosomal kinase STK15/BTAK mRNA expression with chromosomal instability in human breast cancers. Int. J. Cancer 92:370–373; 2001. [DOI] [PubMed] [Google Scholar]
- 19. Moore D. J.; Chambers J. K.; Wahlin J. P.; Tan K. B.; Moore G. B.; Jenkins O.; Emson P. C.; Murdock P. R. Expression pattern of human P2Y receptor subtypes: A quantitative reverse transcription-polymerase chain reaction study. Biochim. Biophys. Acta 1521:107–119; 2001. [DOI] [PubMed] [Google Scholar]
- 20. Nigg E. A. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer 2:815–825; 2002. [DOI] [PubMed] [Google Scholar]
- 21. Niwa H.; Abe K.; Kunisada T.; Yamamura K. Cell-cycle-dependent expression of the STK-1 gene encoding a novel murine putative protein kinase. Gene 169:197–201; 1996. [DOI] [PubMed] [Google Scholar]
- 22. Ross D. T.; Scherf U.; Eisen M. B.; Perou C. M.; Rees C.; Spellman P.; Iyer V.; Jeffrey S. S.; Van de Rijn M.; Waltham M.; Pergamenschikov A.; Lee J. C.; Lashkari D.; Shalon D.; Myers T. G.; Weinstein J. N.; Botstein D.; Brown P. O. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 24:227–235; 2000. [DOI] [PubMed] [Google Scholar]
- 23. Sakakura C.; Hagiwara A.; Yasuoka R.; Fujita Y.; Nakanishi M.; Masuda K.; Shimomura K.; Nakamura Y.; Inazawa J.; Abe T.; Yamagishi H. Tumour-amplified kinase BTAK is amplified and overexpressed in gastric cancers with possible involvement in aneuploid formation. Br. J. Cancer 84:824–831; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Savelieva E.; Belair C. D.; Newton M. A.; DeVries S.; Gray J. W.; Waldman F.; Reznikoff C. A. 20q gain associates with immortalization: 20q13.2 amplification correlates with genome instability in human papillomavirus 16 E7 transformed human uroepithelial cells. Oncogene 14:551–560; 1997. [DOI] [PubMed] [Google Scholar]
- 25. Sen S.; Zhou H.; White R. A. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene 14:2195–2200; 1997. [DOI] [PubMed] [Google Scholar]
- 26. Sen S.; Zhou H.; Zhang R. D.; Yoon D. S.; Vakar-Lopez F.; Ito S.; Jiang F.; Johnston D.; Grossman H. B.; Ruifrok A. C.; Katz R. L.; Brinkley W.; Czerniak B. Amplification/overexpression of a mitotic kinase gene in human bladder cancer. J. Natl. Cancer Inst. 94:1320–1329; 2002. [DOI] [PubMed] [Google Scholar]
- 27. Shindo M.; Nakano H.; Kuroyanagi H.; Shirasawa T.; Mihara M.; Gilbert D. J.; Jenkins N. A.; Copeland N. G.; Yagita H.; Okumura K. cDNA cloning, expression, subcellular localization, and chromosomal assignment of mammalian aurora homologues, aurora-related kinase (ARK) 1 and 2. Biochem. Biophys. Res. Commun. 244:285–292; 1998. [DOI] [PubMed] [Google Scholar]
- 28. Smith M. W.; Yue Z. N.; Geiss G. K.; Sadovnikova N. Y.; Carter V. S.; Boix L.; Lazaro C. A.; Rosenberg G. B.; Bumgarner R. E.; Fausto N.; Bruix J.; Katze M. G. Identification of novel tumor markers in hepatitis C virus-associated hepatocellular carcinoma. Cancer Res. 63:859–864; 2003. [PubMed] [Google Scholar]
- 29. Su A. I.; Cooke M. P.; Ching K. A.; Hakak Y.; Walker J. R.; Wiltshire T.; Orth A. P.; Vega R. G.; Sapinoso L. M.; Moqrich A.; Patapoutian A.; Hampton G. M.; Schultz P. G.; Hogenesch J. B. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. USA 99:4465–4470; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takahashi T.; Futamura M.; Yoshimi N.; Sano J.; Katada M.; Takagi Y.; Kimura M.; Yoshioka T.; Okano Y.; Saji S. Centrosomal kinases, HsAIRK1 and HsAIRK3, are overexpressed in primary colorectal cancers. Jpn. J. Cancer Res. 91:1007–1014; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tanaka E.; Hashimoto Y.; Ito T.; Okumura T.; Kan T.; Watanabe G.; Imamura M.; Inazawa J.; Shimada Y. The clinical significance of Aurora-A/STK15/BTAK expression in human esophageal squamous cell carcinoma. Clin. Cancer Res. 11:1827–1834; 2005. [DOI] [PubMed] [Google Scholar]
- 32. Tanaka T.; Kimura M.; Matsunaga K.; Fukada D.; Mori H.; Okano Y. Centrosomal kinase AIK1 is overexpressed in invasive ductal carcinoma of the breast. Cancer Res. 59:2041–2044; 1999. [PubMed] [Google Scholar]
- 33. Tanner M. M.; Tirkkonen M.; Kallioniemi A.; Collins C.; Stokke T.; Karhu R.; Kowbel D.; Shadravan F.; Hintz M.; Kuo W. L.; Waldman F. M.; Isola J. J.; Gray J. W.; Kallioniemi O. P. Increased copy number at 20q13 in breast cancer: Defining the critical region and exclusion of candidate genes. Cancer Res. 54:4257–4260; 1994. [PubMed] [Google Scholar]
- 34. Tanner M. M.; Tirkkonen M.; Kallioniemi A.; Holli K.; Collins C.; Kowbel D.; Gray J. W.; Kallioniemi O. P.; Isola J. Amplification of chromosomal region 20q13 in invasive breast cancer: Prognostic implications. Clin. Cancer Res. 1:1455–1461; 1995. [PubMed] [Google Scholar]
- 35. Tatsuka M.; Katayama H.; Ota T.; Tanaka T.; Odashima S.; Suzuki F.; Terada Y. Multinuclearity and increased ploidy caused by overexpression of the aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 58:4811–4816; 1998. [PubMed] [Google Scholar]
- 36. Tien A. C.; Lin M. H.; Su L. J.; Hong Y. R.; Cheng T. S.; Lee Y. C.; Lin W. J.; Still I. H.; Huang C. Y. Identification of the substrates and interaction proteins of aurora kinases from a protein–protein interaction model. Mol. Cell Proteomics 3:93–104; 2004. [DOI] [PubMed] [Google Scholar]
- 37. Yu C. T.; Hsu J. M.; Lee Y. C.; Tsou A. P.; Chou C. K.; Huang C. Y. Phosphorylation and stabilization of HURP by Aurora-A: Implication of HURP as a transforming target of Aurora-A. Mol. Cell Biol. 25:5789–5800; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yue H.; Eastman P. S.; Wang B. B.; Minor J.; Doctolero M. H.; Nuttall R. L.; Stack R.; Becker J. W.; Montgomery J. R.; Vainer M.; Johnston R. An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res. 29:E41; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Whitfield M. L.; Sherlock G.; Saldanha A. J.; Murray J. I.; Ball C. A.; Alexander K. E.; Matese J. C.; Perou C. M.; Hurt M. M.; Brown P. O.; Botstein D. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13:1977–2000; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu J. C.; Chen T. Y.; Yu C. T.; Tsai S. J.; Hsu J. M.; Tang M. J.; Chou C. K.; Lin W. J.; Yuan C. J.; Huang C. Y. Identification of V23RalA-Ser194 as a critical mediator for Aurora-A-induced cellular motility and transformation by small pool expression screening. J. Biol. Chem. 280:9013–9022; 2005. [DOI] [PubMed] [Google Scholar]
- 41. Zhou H.; Kuang J.; Zhong L.; Kuo W. L.; Gray J. W.; Sahin A.; Brinkley B. R.; Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 20:189–193; 1998. [DOI] [PubMed] [Google Scholar]


