Abstract
We examined the molecular determinants for sustained high-level expression of “privileged” genes, defined as the 0.03% most highly expressed genes within any specific cell. We identified histone modifications by chromatin immunoprecipitation analyses on Keratin 8, the most highly expressed gene in the human breast cancer cell line, MCF-7, based on serial analysis of gene expression. Quantitative comparisons to the “normal” counterpart cell line, MCF-10A, expressing 350-fold lower levels of Keratin 8 and other breast cancer cell lines expressing higher levels were performed using real-time PCR. Extraordinarily high levels of trimethyl histone H3 lysine 4 (H3K4) were found primarily in the first intron of the Keratin 8 gene stretching from 400 to 2000 bp downstream from the promoter in all breast cancer cells lines but not in MCF-10A cells. The highest levels of histone H3K4 trimethylation in MCF-7 cells ranged from 70% to 80% over input within 1200 bp of this region. Knockdown of mixed-lineage leukemia (MLL), the specific methyltransferase for histone H3K4, with MLL-specific siRNA decreased histone H3K4 trimethylation on the Keratin 8 gene and decreased Keratin 8 mRNA levels. Histone H3K4 trimethylation mediates approximately 86% of the elevated, sustained expression of the Keratin 8 gene in MCF-7 cells.
Key words: Histone modifications, H3K4 trimethylation, Gene expression, SAGE, Mixed-lineage leukemia (MLL)
INTRODUCTION
We are interested in determining the mechanism(s) for expression of the most highly expressed genes that are apparently not shut down in specific cells. Each of the 0.03% most highly expressed genes produces approximately 2% of the total 300,000–400,000 mRNA transcripts present in a human cell (26). We call this extraordinarily high level of sustained gene expression “privileged.” Surprisingly, the privileged genes together produce about 20% of the total mRNA transcripts on average, setting these genes apart as a distinct class of genes. In contrast, the vast majority of genes produce fewer than 10 mRNA transcripts per cell. Generally, cell nuclear components serve to keep gene expression “in check” and do not allow continued high expression of any particular gene. Therefore, the word privileged seems appropriate to describe this phenomenon. We wish to investigate the mechanism(s) by which privileged genes escape the tight surveillance. To date, gene expression has simply been defined as active or inactive when correlated to the identification of histone modifications. However, an enormous dynamic range exists for gene expression ranging from 1× to 10,000×, and therefore we investigated the mechanism for gene expression at the highest dynamic range.
A majority of genes are kept repressed, and several mechanisms for gene silencing have been reported. The heterochromatin protein 1 (HP1) plays a major role in gene repression and is broadly conserved in eukaryotes with the exception of yeast (7). HP1 primarily associates with pericentric heterochromatin and is essential for repression of heterochromatic genes. However, it can also be found at euchromatin sites where it promotes gene repression often in association with histone H3K9 methylation (7,17,24). However, HP1 can also repress genes in the absence of histone H3K9 methylation (7) in a context-dependent manner. In other highly coordinated gene regulatory systems (e.g., nuclear receptors) gene expression is shut down by specific corepressors (13).
Serial Analyses of Gene Expression (SAGE) has provided a method to precisely quantify the number of mRNA transcripts produced by genes in specific cells (26). Furthermore, the National Cancer Institute (NCI) has generated SAGE databases for several cancer cell lines and tissues and their normal counterparts. We have interests in human MCF-7 breast cancer cells and found that the Keratin 8 gene is the most highly expressed gene in MCF-7 cells with 12,622 mRNA transcripts per million transcripts reported in the NCI SAGE database (12). Furthermore, the Keratin 8 gene was expressed at far lower levels in normal human mammary epithelium and normal human luminal epithelial cells. Therefore, the privileged gene pool may also differ in diseased versus normal cells, and the Keratin 8 gene appears to be aberrantly expressed in several breast cancer cells. Keratin 8 plays an important role in several invasive breast cancers. Keratin 8 is a member of the intermediate filament gene family and is found on the surface of breast cancer cells, including MCF-7 cells (4,10). Keratin 8 is the major plasminogen receptor that is required for accelerated activation of cell-associated plasminogen by tissue-type plasminogen activator (5,10,11), which is important for cellular migration, including tumor invasion and metastasis (8,9). Keratin 8 has also been used as a diagnostic marker for invasive breast carcinoma (25) and node-positive metastases (2,3). Recently, Keratin 8 has also been found in the nucleus and is colocalized with nuclear proteins containing O-linked N-acetylglucosamine residues including Epitope H, hnRNPs, G and A1, c-myc, RNA pol II, and its transcription factors in breast cancer cells and biopsy material from infiltrating ductal breast carcinomas and fibroadenomas (6). However, the role of nuclear Keratin 8 has not been identified. Interestingly, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene is also a privileged gene in MCF-7 cells and is the seventh most highly expressed gene (12).
To understand the contribution of chromatin and its components to privileged gene expression, we examined histone modifications on the Keratin 8 gene including acetylations and methylations. We chose a highly quantitative approach using real-time PCR to evaluate the results of our chromatin immunoprecipitation (ChIP) assays. Several laboratories have demonstrated histone acetylation and trimethylation of histone H3K4 associated with active gene expression (20,21). The results of our work reported here complement that of prior publications regarding trimethylation of histone H3K4 associated with active gene expression (1,19,21); however, our work provides the first detailed quantitative assessment of the specific contribution of histone modifications to privileged gene expression.
MATERIALS AND METHODS
Cell Culture
The cell lines were purchased from the American Type Culture Collection (Manassas, VA) and cultured at 37°C with 5% CO2. MCF-7 cells were cultured in DMEM/F12 with 2 mM l-glutamine and 10% fetal bovine serum (FBS). MCF-10A cells were cultured in DMEM/F12 with 0.1 μg/ml cholera toxin, 0.5 μg/ml hydrocortisone, 10 μg/ml bovine insulin, 20 ng/ml EGF, and 5% horse serum. SK-BR-3 cells were cultured in McCoy’s 5a medium (modified) with 1.5 mM l-glutamine, 2.2 g/L sodium bicarbonate, and 10% FBS. T-47D cells were cultured in RPMI-1640 medium with 2 mM l-glutamine, 1.5 g/ L sodium bicarbonate, 4.5 g/L glucose, 10 mM HEPES, 1 mM sodium pyruvate, 10 μg/ml bovine insulin, and 10% FBS.
Antibodies
Anti-acetyl histone H4, anti-acetyl histone H2B, anti-dimethyl histone H3 K9, anti-acetyl histone H3 K9, normal rabbit IgG, and anti-MLLc were purchased from Upstate Ltd. (Lake Placid, NY). Anti-trimethyl K4 histone H3 was purchased from Abcam (Cambridge, MA). Anti-β-actin was purchased from Sigma (St. Louis, MO).
Mixed-Lineage Leukemia (MLL) Knockdown in Human Cells
Three human MLL siRNA duplexes (#107890, #107891, and #107892) were purchased from Ambion (Austin, TX). Cells were transfected with an siRNA pool consisting of three siRNA duplexes by using siPORT Amine Transfection Agent (Ambion) according to the manufacturer’s instructions. Cells were harvested 72 h after transfection and used for experiments. Silencer Negative Control siRNA (Ambion) was used for control transfections.
Real-Time Quantitative RT-PCR
Primers were designed using the Primer Express 2.0 software. The primers for the human keratin 8 mRNA are: sense, 5′-ATC AGC TCC TCG AGC TTC TC-3′; antisense, 5′-GGC TCT GGT TGA CCG TAA CT-3′. The primers for the human mixed lineage leukemia (MLL) mRNA are: sense, 5′-GGA AGT CAA GCA AGC AGG TC-3′; antisense, 5′-TGC TTT TTC TTG GGC TCA CT-3′. The primers for housekeeping gene human β-actin are: sense, 5′-CTG GAA CGG TGA AGG TGA CA-3′; antisense, 5′-AAG GGA CTT CCT GTA ACA ATG CA-3′. Total RNA was extracted from the cells with Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer’ s protocol and treated with DNase I (Invitrogen). One microgram of total RNA was used for reverse transcription. The mRNAs were reverse-transcribed into cDNAs with an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) containing mixture of oligo(dT) and random primers. Real-time PCR was performed using an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA) with a DyNAmo HS SYBR Green qPCR kit (Finnzymes, Espoo, Finland, supplied by New England BioLabs) with the program running for 40 cycles at 95°C for 15 s and 60°C for 1 min. The PCR efficiency for the primers was examined by serially diluting the cDNA templates. PCR-specific products were determined as a clear single peak, at the melting curves, of >80°C. The specificity of primers was also confirmed as a single band with the correctly amplified fragment size through agarose. Each gene mRNA level was acquired from the value of the threshold cycle (CT) of the real-time PCR as related to that for β-actin using the formula 2ΔCT (ΔCT = β-actin CT − gene of interest CT).
Chromatin Immunoprecipitation (ChIP) Assay
In brief, ∼2 × 108 cells were used per ChIP assay. Cells were cross-linked with 1% formaldehyde for 10 min at room temperature and the cross-linking reaction was stopped by 0.1 M glycine. Cells were harvested by scraping and a brief centrifugation. Cell pellets were washed twice with ice-cold phosphate-buffered saline (PBS), once with solution 1 (10 mM HEPES, pH 7.5, 10 mM EDTA, 0.5 mM EGTA, 0.75% Triton X-100), and once with solution 2 (10 mM HEPES, pH 7.5, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA). Cell pellets were resuspended in lysis buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholate and protease inhibitors). Sonication was performed on ice using a Fisher model 60 Sonic Dismembrator (Fisher, Pittsburgh, PA) 10 times for 15 s each at 40% output (output power in the range of about 7–11 W) with microtip to achieve chromatin fragments enriched for 500 bp in size followed by centrifugation at 15,000 × g for 20 min at 4°C. Supernatants were collected and divided into three aliquots with equal volume: one aliquot served as an input sample, the second aliquot used the experimental antibody, and the third aliquot used the negative control antibody. After preimmunoprecipitation clearing with the control antibody and protein A at 4°C, immunoprecipitation was carried out with 4 μg of antibody at 4°C overnight with rotation. After immunoprecipitation, 35 μl 50% slurry of protein A beads was added and incubated at 4°C with rotation for 1 h and followed by brief centrifugation. The precipitates were washed once with RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% NP40, 0.1% SDS, 0.5% deoxycholate), once with high salt buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 1 mM EDTA, 1% NP40, 0.1% SDS, 0.5% deoxycholate), and once with LiCl buffer (50 mM Tris-HCl, pH 8.0, 250 mM LiCl, 1 mM EDTA, 1% NP40, 0.5% deoxycholate). Then the precipitates were washed again with TE buffer twice for 10 min each. The immune complexes and the input were treated with 50 μg/ml RNase A at 37°C for 30 min and then with 0.25 mg/ml Proteinase K for 4 h at 37°C. After the samples were heated at 65°C for at least 6 h to reverse the cross-link, DNA was extracted by phenol/ chloroform solution and precipitated with ethanol. The recovered DNA was resuspended in 50 μl of H2O, and 0.5 μl was used for real-time PCR. We designed a series of primers to amplify the regions within the 5′ Keratin 8 gene locus (Table 1) from 1098 bp upstream of TATA box to 2103 bp downstream of TATA box. Primers were designed with Primer Express 2.0 software (Applied Biosystems). The amount of genomic DNA coprecipitated with antibody was calculated as a percent input the following way: % input = 2ΔCT × 100 [ΔCT = CT(input) − CT (ChIP)]. Normal Rabbit IgG served as the negative control in the assay.
TABLE 1.
5′ KERATIN 8 GENOMIC REGION
| Genomic Site (#) | Region of 5′ Keratin 8 Gene |
|---|---|
| +1 | TATA box start site |
| +12 | 5′ end of Keratin 8 mRNA |
| +107 | First codon start for the Keratin 8 protein |
| +431 | Intron 1—5′ end |
| +3015 | Intron 1—3′ end |
Western Blot
Cells were lysed in protein lysis buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% NP40, 0.1% SDS, 0.5% deoxycholate, 1 mM NaF, 1 mM Na3VO4, protease inhibitors) for 30 min on ice. Forty micrograms of protein samples was subjected to 4–12% SDS-PAGE and electrophoretically transferred to nitrocellulose membrane. The membrane was blocked in 3% nonfat milk in TBS-T (50 mM Tris-HCI, pH 7.5, 150 mM NaCl, 0.05% Tween 20) for 1 h at room temperature. After incubation with primary antibody in TBS-T containing 3% milk overnight at 4°C, the membrane was washed extensively with TBS-T and then incubated with secondary anti-mouse horseradish peroxidase-conjugated antibody (Sigma) for 1 h at room temperature. After extensive washes with TBS-T, the membrane was visualized with ECL plus reagents (Amersham Biosciences, Piscataway, NJ) according to the manufacturer’s instructions.
RESULTS
Comparisons of Keratin 8 mRNA Levels in Different Cell Lines
Real-time reverse transcriptase (RT) PCR was performed to quantitatively compare levels of Keratin 8 mRNAs isolated from human breast cancer cell lines including MCF-7, SK-BR-3, and T-47D and the “normal” counterpart breast epithelial cell line, MCF-10A. MCF-10A cells are often used as the near normal control cell line because they are human mammary gland cells that have a normal or near-normal karyotype (23). MCF-7 cells are human breast adenocarcinoma cells that make tumors in immune-deficient mice in response to estrogen. Our results showed that MCF-7 cells had about 350-fold higher levels of Keratin 8 mRNA compared to MCF-10A cells (Fig. 1). T-47D and SK-BR-3 breast cancer cells had approximately 325-fold and 260-fold greater levels of Keratin 8 mRNA, respectively, compared to MCF-10A cells. Based on data presented in Figure 1, we predicted that these cell lines would be useful to study differences in histone modifications on the Keratin 8 gene.
Figure 1.
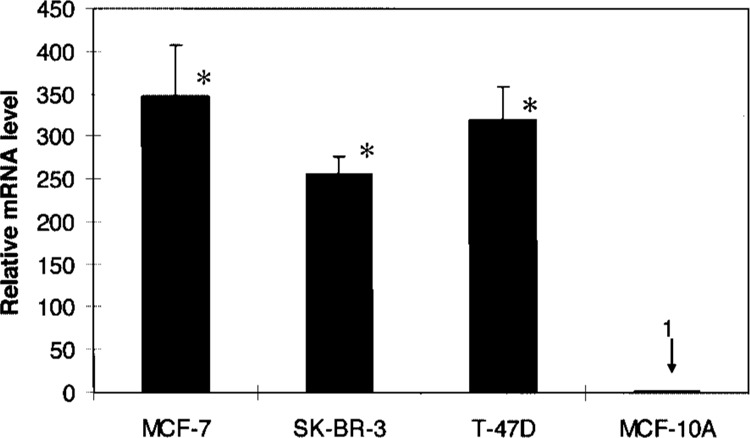
Keratin 8 mRNA levels in MCF-7, SK-BR-3, T-47D, and MCF-10A cells. The expression levels of Keratin 8 mRNA were quantified by real-time RT-PCR and normalized to the housekeeping gene β-actin. Results are expressed as the ratio relative to Keratin 8 mRNA level in MCF-10A cells. All data are presented as the means ± SD of three independent experiments. *p < 0.01 versus MCF-10A by Student’s t-test.
Trimethylation of Histone H3K4 on the Keratin 8 Gene in MCF-7 Versus MCF-10A Cells
Results generated from our ChIP assays using the anti-trimethyl histone H3K4 antibody followed by real-time PCR with primers spanning from 1.1 kb upstream and 2.1 kb downstream of the Keratin 8 start site of transcription are presented in Figure 2. Numbers along the x-axis indicate the location along the 5′ end of the Keratin 8 gene with “+12” designating the start site of transcription (Table 1). Trimethylation of histone H3K4 on the Keratin 8 gene was compared in MCF-7 versus MCF-10A cells. Histone H3K4 trimethylation was higher in MCF-7 cells and was exceptionally high in regions spanning +406 to +2013, downstream of the Keratin 8 promoter. The highest levels reached 80% over input. Prior to this study, the highest level of histone H3K4 trimethylation reported on an active gene was approximately 20% over input (21). Furthermore, elevated histone H3K4 trimethylation was found only in short, scattered regions at the start site and sites downstream of the start site of transcription of the active genes examined (21). No widespread, high levels of histone H3K4 trimethylation were reported. Interestingly, for the Keratin 8 gene in MCF-7 cells, the highest levels of histone H3K4 trimethylation were far downstream of the Keratin 8 gene start site of transcription compared to the start site that showed histone H3K4 trimethylation levels at 10% over input (Fig. 2, from −94/+25).
Figure 2.
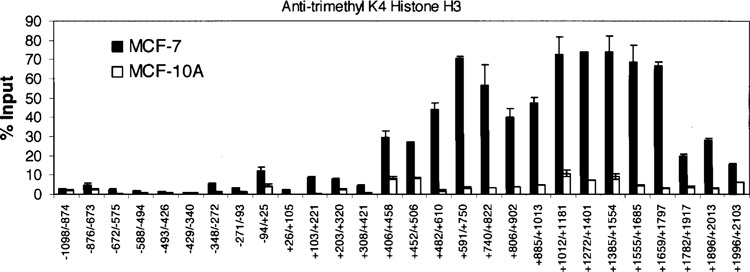
Histone H3 K4 trimethylation within the Keratin 8 gene in MCF-7 and MCF-10A cells. ChIP assays were performed with anti-trimethyl K4 Histone H3. The amount of precipitated DNA was quantified relative to input as described in Materials and Methods. The data are presented as the means ± SD of two independent experiments. The initiation site of TATA box is designated at +1.
Comparisons of Other Histone Modifications on the Keratin 8 Gene in MCF-7 Versus MCF-10A Cells
We examined histone acetylations (Fig. 3A–C) that are associated with active gene expression and histone H3K9 dimethylation that is associated with gene repression (Fig. 3D). No obvious pattern difference between MCF-7 and MCF-10A Keratin 8 genes was observed for histone H4 acetylation (Fig. 3A). Surprisingly, the MCF-10A Keratin 8 gene showed slightly elevated levels of acetylation at 13 regions upstream and downstream of the promoter, whereas the MCF-7 Keratin 8 gene showed slight elevations in histone H4 acetylation in only four regions. In addition, the MCF-10A Keratin 8 gene showed more regions with minor elevations in histone H2B acetylation compared to that in MCF-7 cells (Fig. 3B). Levels of histone H3K9 acetylation were the highest, reaching 25% over input, for the Keratin 8 gene in MCF-7 cells (Fig. 3C). However, histone H3K9 acetylation in MCF-10A Keratin 8 was only diminished about twofold in eight regions and fourfold in one short region of 138 bp (+1659 to +1797), whereas acetylation levels were significantly higher in MCF-10A Keratin 8 in four regions (Fig. 3C). Both MCF-7 and MCF-10A Keratin 8 genes showed high levels of histone H3K9 acetylation mainly in the region downstream of the transcription start site, mostly in the first intron, +406 to +2103, the same region that showed even greater levels of trimethylated histone H3K4 in the MCF-7 Keratin 8 gene. Although no obvious pattern difference was detected between the Keratin 8 gene in MCF-7 versus MCF-10A cells with respect to histone H3K9 acetylation, these data suggest that perhaps this acetylation may be necessary but not sufficient to mediate privileged gene expression.
Figure 3.
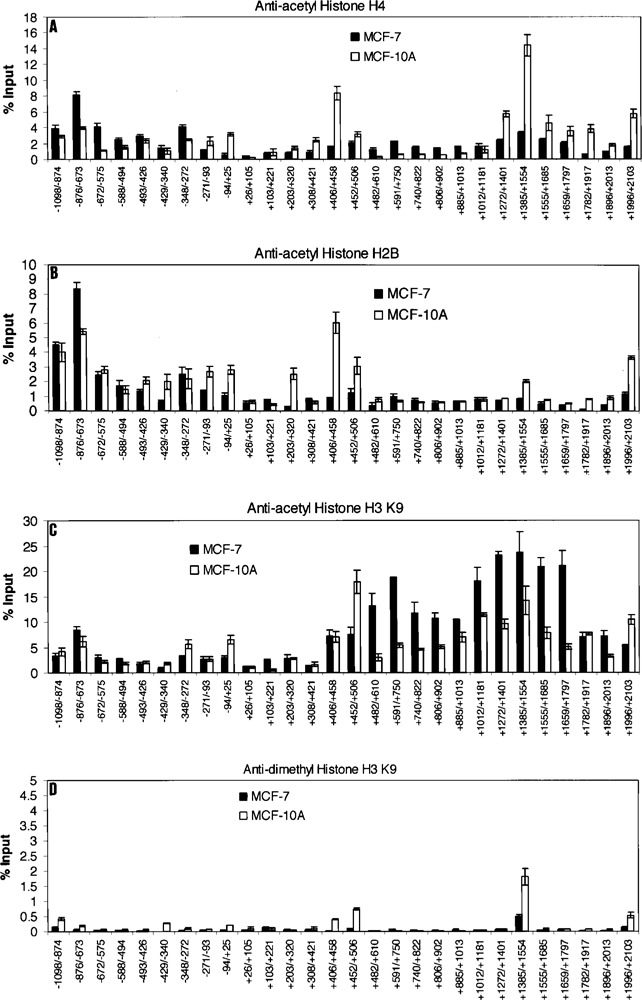
Histone modifications within the Keratin 8 gene locus in MCF-7 and MCF-10A cells. ChIP assays were performed with anti-acetyl histone H4 (A), anti-acetyl histone H2B (B), anti-acetyl histone H3 K9 (C), or anti-dimethyl histone H3 K9 (D). Normal Rabbit IgG served as the negative control (E). The amount of precipitated DNA was quantified relative to input as described in Materials and Methods. All data are presented as the means ± SD of two independent experiments. The initiation site of TATA box is designated at +1. (F) Summary and comparison of the histone modifications examined on the Keratin 8 gene in MCF-7 cells.
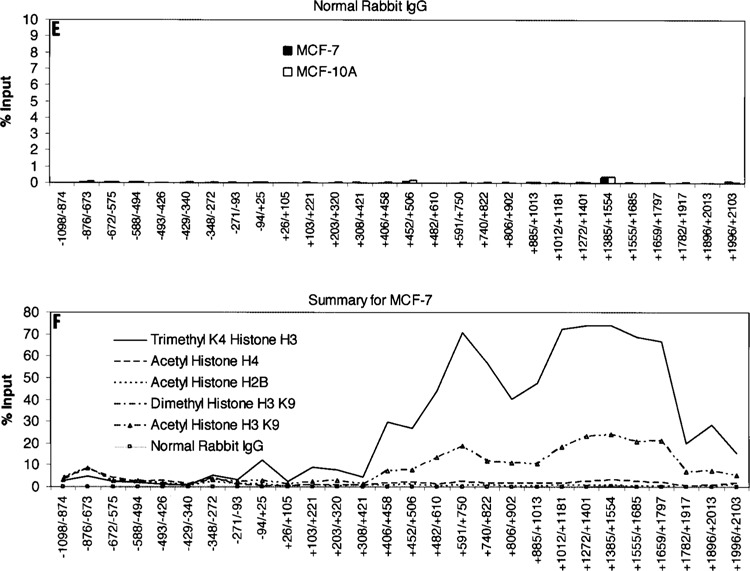
Data generated from ChIP assays using the anti-dimethyl histone H3K9 antibody showed no obvious pattern difference for this histone modification on the Keratin 8 genes in either MCF-7 or MCF-10A cells. These data suggest that histone H3K9 dimethylation is not associated with the low levels of Keratin 8 expression in MCF-10A cells. Our negative control data (Fig. 3E) using normal rabbit IgG show no background in the detection of histone modifications on the Keratin 8 genes in MCF-7 and MCF-10A cells.
In summary (Fig. 3F), in the 5′ end of the first intron of Keratin 8 gene in MCF-7 cells, the trimethylated histone H3K4 modification was approximately 3–5-fold, 15–40-fold, and 30–80-fold higher than acetylated histone H3K9, acetylated histone H4, and acetylated histone H2B, respectively. The highest levels of all histone modifications, those above 12% over input, were found only in this intron region and not in the region approximately 1100 bp upstream to 420 bp downstream of the Keratin 8 start site of transcription. At the Keratin 8 promoter, the trimethylated histone H3K4 level was fourfold higher in MCF-7 versus MCF-10A cells, about 12% versus 3% over input. Surprisingly, all histone acetylations were higher at the Keratin 8 gene promoter in MCF-10A versus MCF-7 cells; 3% versus 1% over input for acetylated histone H4 and acetylated histone H2B, and 7% versus 3% over input for acetylated histone H3K9.
Trimethylation of Histone H3K4 in Other Breast Cancer Cell Lines
We investigated other breast cancer cell lines, SK-BR-3 and T-47D, which showed high levels of Keratin 8 mRNA (Fig. 1). The Keratin 8 genes in both cell lines showed highly elevated, widespread histone H3K4 trimethylation in the same first intron region as that in MCF-7, +406 to +2013 (Fig. 4A, B). The highest levels of this modification were about 85% and 60% over input for SK-BR-3 and T-47D Keratin 8 genes, respectively. These data support those in Figure 2 showing that highly elevated levels of Keratin 8 mRNA are associated with widespread, exceptionally high levels of histone H3K4 trimethylation far downstream of the promoter.
Figure 4.
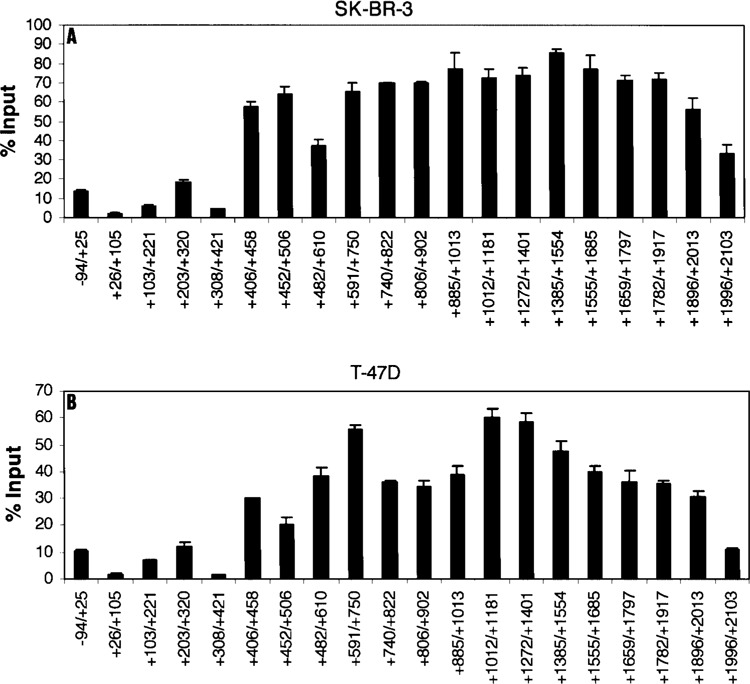
Histone H3 K4 trimethylation within the Keratin 8 gene in SK-BR-3 and T-47D cells. ChIP assays were performed with anti-trimethyl K4 histone H3 in SK-BR-3 cells (A) or in T-47D cells (B). The amount of precipitated DNA was quantified relative to input as described in Materials and Methods. The data are presented as the means ± SD of two independent experiments. The initiation site of TATA box is designated at +1.
Knockdown of MLL Substantially Decreases Keratin 8 mRNA Levels in the Breast Cancer Cell Lines
The C-terminal SET domain of the MLL protein family is a specific histone methyltransferase that methylates only histone H3K4 (14,16). Furthermore, MLL associates with some transcriptionally active genes (15) and regulates gene expression at the stage of elongation (15,22). Most likely, MLL travels with RNA pol II during transcription elongation (22). To quantify the contribution of the histone H3K4 tri-methylation to the expression of the Keratin 8 gene in MCF-7 cells, we performed knockdown experiments of MLL using siRNAs. All cell lines studied had similar levels of MLL mRNA and protein (Fig. 5). In all cell lines, MLL mRNA levels were decreased between 87% and 83% (Fig. 5A) in response to transfection with MLL siRNA. MLL was knocked down by 86% in MCF-7 cells. These knockdown results were further supported by Western blot analyses to detect MLL protein in all cell lines transfected with MLL siRNA (+) or with unrelated control siRNA (−) (Fig. 5B).
Figure 5.
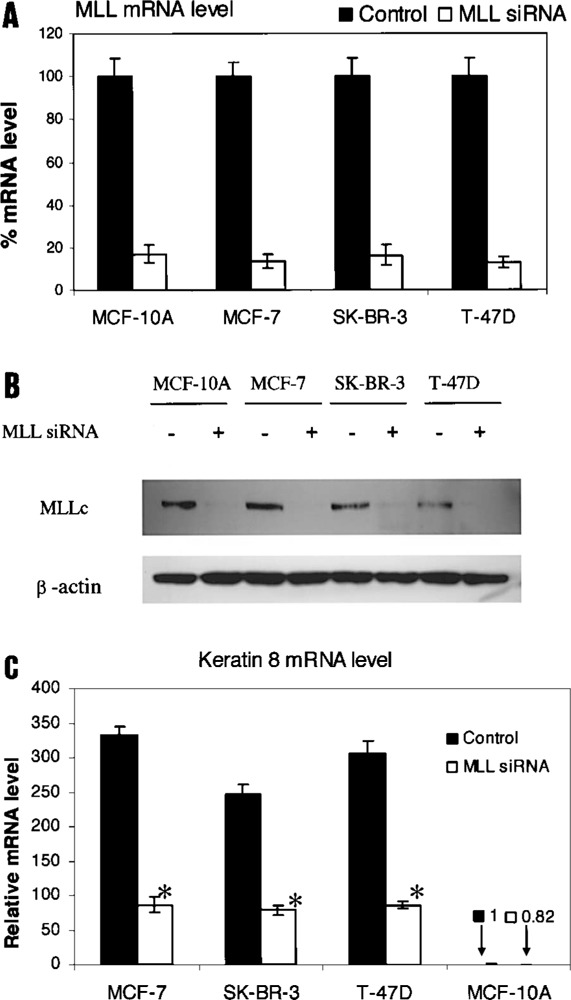
Effect of MLL knockdown on Keratin 8 mRNA levels in MCF-7, SK-BR-3, T-47D, and MCF-10A cells. Cells were transfected with MLL siRNA (+) or with control siRNA (−) and harvested 72 h after transfection. (A) Treatment with MLL siRNA results in a decrease of MLL mRNA levels compared to controls. Results are expressed as the percent ratio relative to Keratin 8 mRNA level in control cells. All data are presented as the means ± SD of three independent experiments. (B) Treatment with MLL siRNA results in a decrease of MLLc protein levels compared to controls. Whole cell lysate was subjected to Western blotting using anti-MLLc antibody. β-Actin served as the loading control. (C) Analyses of the effect of MLL knockdown on Keratin 8 mRNA levels. Results are expressed as the ratio relative to the Keratin 8 mRNA level in control MCF-10A cells. All data are presented as the means ± SD of three independent experiments. *p < 0.0001 versus control by Student’s t-test.
Our results also showed large decreases in Keratin 8 mRNA levels in MCF-7, SK-BR-3, and T-47D cells associated with MLL knockdown (Fig. 5C). These decreases were detected and quantified by realtime RT-PCR on mRNA from cell lines transfected with MLL siRNA or control siRNA. Keratin 8 mRNA levels were decreased by 74%, 68%, and 72% in MCF-7, SK-BR-3, and T-47D cells, respectively. Typically, siRNA does not knockdown gene expression by 100%. Therefore, if MLL siRNA knockdown in MCF-7 cells was 100% effective, we predict that the Keratin 8 mRNA levels would be reduced by 86%.
Knockdown of MLL Decreases Histone H3K4 Trimethylation of the Keratin 8 Gene in all Cell Lines
To support data presented in Figure 5, we confirmed that knockdown of MLL did significantly reduce histone H3K4 trimethylation levels in all Keratin 8 regions in all cell lines (Fig. 6A–D). This histone modification was obviously most greatly reduced in the region from +406 to +2103 where the histone H3K4 trimethylation levels are extraordinarily high in cell lines treated with control siRNA.
Figure 6.
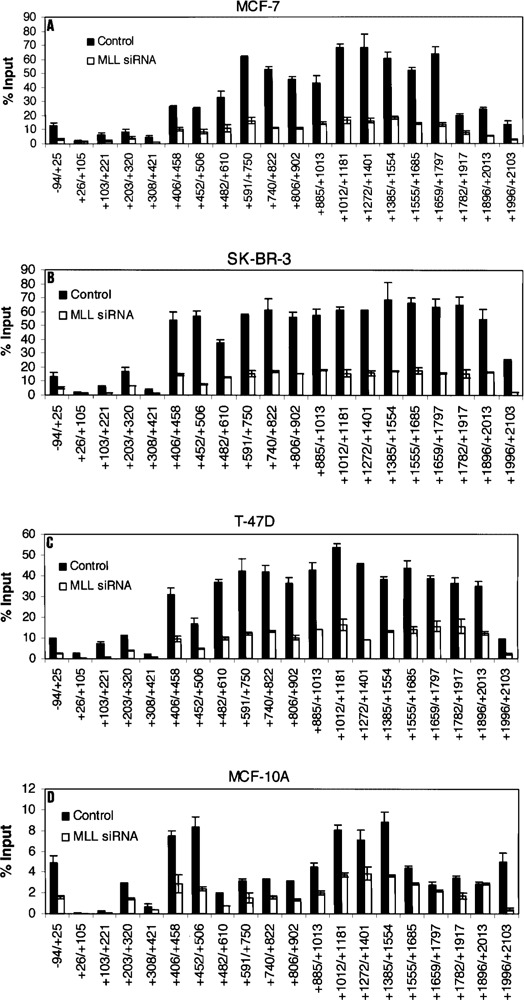
Effect of MLL knockdown on Histone H3 K4 trimethylation within the Keratin 8 gene. MCF-7 (A), SK-BR-3 (B), T-47D (C), and MCF-10A (D) cells were transfected with MLL siRNA or unrelated control siRNA and used for ChIP assays 72 h after transfection. The amount of precipitated DNA was quantified relative to input as described in Materials and Methods. All data are presented as the means ± SD of two independent experiments. The initiation site of TATA box is designated at +1.
DISCUSSION
We investigated and extensively quantified the role of histone modifications in privileged gene expression. We used the Keratin 8 gene as a model system, which is the gene that has the highest sustained expression in MCF-7 cells. We found extremely high levels of histone H3K4 trimethylation throughout 1.6 kb of the 5′ end of the first intron located about 400 bp downstream of the Keratin 8 promoter. Histone acetylations, commonly associated with active gene expression, and histone H3K9 dimethylation associated with gene repression, showed no outstanding differences between the Keratin 8 genes in MCF-7 cells versus the control MCF-10A cells that have about 350-fold reduced levels of Keratin 8 mRNA. Through quantitative MLL knockdown experiments, we estimated that approximately 86% of the Keratin 8 privileged gene expression observed in MCF-7 cells is contributed by histone H3K4 trimethylation. MLL regulates gene expression at the elongation stage by an unknown mechanism, and experiments performed in null MLL knockout cells showed that RNA pol II seemed to be stalled on MLL responsive genes (15, 22). Although these published studies suggest that sequence-specific transcription factors may exist that recruit MLL to MLL target genes, our present work comparing MCF-10A to human breast cancer cells demonstrates that sequence specificity is not sufficient. Instead, recruitment of MLL to genes is also cell type specific, and, therefore, is not dependent on gene-specific sequences alone. Further work on the MLL complex found at the Keratin 8 gene in MCF-7 cells could substantially contribute to understanding how histone H3K4 trimethylation contributes to the most highly efficient transcription elongation.
MLL has been shown to specifically associate with only a few transcriptionally active promoters (15). Furthermore, somewhat elevated levels of histone H3K4 trimethylation have been found associated with active gene expression, up to approximately 20% over input, in short regions around and downstream of promoters for genes that are turned on and off during development (21). We have found further elevated levels of histone H3K4 trimethylation, up to 80% over input, in a long stretch downstream of the promoter for the Keratin 8 gene in MCF-7 cells. Work from other groups involving tightly focused ChIP assays have demonstrated loci of histone H3K4 methylation associated with active gene expression and separated by boundaries to prevent gene inactivation associated with histone H3K9 methylation (18, 27). In the chicken β-globin gene, insulator elements are the barriers that separate this gene locus from neighboring genes (27), and thus is not affected by surrounding chromosomal silencing. The insulator elements perform this function by constitutively recruiting histone H3K4 methylation and histone acetylation in order to prevent propagation of condensed chromatin. In fission yeast, the mating-type locus consists of heterochromatic and euchromatic regions that are separated by boundary elements consisting of two inverted repeats (18). Histone H3K9 methylation and the Swi6 protein are localized to repressed gene expression in heterochromatin (18). Conversely, his-tone H3K4 methylation is present on active genes in the euchromatic region.
Our identification of highly elevated levels of his-tone H3K4 trimethylation downstream of the Keratin 8 promoter supports transcription elongation as the key contributor to the ability of privileged genes to produce the highest levels of gene expression. Further understanding of highly efficient transcription elongation would be useful. Perhaps the Keratin 8 gene in MCF-7 cells produces a far greater number of full-length transcripts than all other genes in those cells. Production of far more full-length transcripts could be mediated by factors that bind to or associate with trimethylated histone H3K4 downstream of the promoter. These factors could assist in RNA pol II transcription elongation and ensure completion of mRNA transcription. Higher levels of transcription reinitiation at the Keratin 8 promoter in MCF-7 cells may also be enhanced by histone H3K4 trimethylation and could contribute to privileged gene expression. No outstanding pattern differences in histone modifications were noted upstream of the Keratin 8 promoters in MCF-7 versus MCF-10A. Apparently, a higher order of histone modifications exists within this model system in which histone H3K4 trimethylation far downstream of the promoter dictates the vast majority of privileged gene expression.
ACKNOWLEDGMENT
We thank David D. Roberts, National Cancer Institute, NIH, Bethesda, MD for review of the manuscript and for helpful advice.
REFERENCES
- 1. Bernstein B. E.; Kamal M.; Lindblad-Toh K.; Bekiranov S.; Bailey D. K.; Huebert D. J.; McMahon S.; Karlsson E. K.; Kulbokas E. J. III; Gingeras T. R.; Schreiber S. L.; Lander E. S. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120:169–181; 2005. [DOI] [PubMed] [Google Scholar]
- 2. Brotherick I.; Robson C. N.; Browell D. A.; Shenfine J.; White M. D.; Cunliffe W. J.; Shenton B. K.; Egan M.; Webb L. A.; Lunt L. G.; Young J. R.; Higgs M. J. Cytokeratin expression in breast cancer: Phenotypic changes associated with disease progression. Cytometry 32:301–308; 1998. [PubMed] [Google Scholar]
- 3. Cianga C.; Cianga P.; Cozma L.; Diaconu C.; Carasevici E. Detection of lymph nodes micrometastases in breast carcinoma using immunohistochemistry for cytokeratin 8. Rev. Med. Chir. Soc. Med. Nat. Iasi 106:720–724; 2002. [PubMed] [Google Scholar]
- 4. Godfroid E.; Geuskens M.; Dupressoir T.; Parent I.; Szpirer C. Cytokeratins are exposed on the outer surface of established human mammary carcinoma cells. J. Cell Sci. 99:595–607; 1991. [DOI] [PubMed] [Google Scholar]
- 5. Gonias S. L.; Hembrough T. A.; Sankovic M. Cytokeratin 8 functions as a major plasminogen receptor in select epithelial and carcinoma cells. Front. Biosci. 6:D1403–D1411; 2001. [DOI] [PubMed] [Google Scholar]
- 6. Havaki S.; Voloudakis-Baltatzis I.; Goutas N.; Arvanitis L. D.; Vassilaros S. D.; Arvanitis D. L.; Kittas C.; Marinos E. Nuclear localization of cytokeratin 8 and the O-linked N-acetylglucosamine-containing epitope H in epithelial cells of infiltrating ductal breast carcinomas: a combination of immunogold and EDTA regressive staining methods. Ultrastructur. Pathol. 30:177–186; 2006. [DOI] [PubMed] [Google Scholar]
- 7. Hediger F.; Gasser S. M. Heterochromatin protein 1: Don’t judge the book by its cover! Curr. Opin. Genet. Dev. 16:143–150; 2006. [DOI] [PubMed] [Google Scholar]
- 8. Hembrough T. A.; Vasudevan J.; Allietta M. M.; Glass W. F.; Gonias S. L. A cytokeratin 8-like protein with plasminogen-binding activity is present on the external surfaces of hepatocytes, HepG2 cells and breast carcinoma cell lines. J. Cell Sci. 108:1071–1082; 1995. [DOI] [PubMed] [Google Scholar]
- 9. Hembrough T. A.; Kralovich K. R.; Li L.; Gonias S. L. Cytokeratin 8 released by breast carcinoma cells in vitro binds plasminogen and tissue-type plasminogen activator and promotes plasminogen activation. Biochem. J. 317:763–769; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hembrough T. A.; Li L.; Gonias S. L. Cell-surface cytokeratin 8 is the major plasminogen receptor on breast cancer cells and is required for the accelerated activation of cell-associated plaminogen by tissue-type plasminogen activator. J. Biol. Chem. 271:25684–25691; 1996. [DOI] [PubMed] [Google Scholar]
- 11. Kralovich K. R.; Li L.; Hembrough T. A.; Webb D. J.; Karns L. R.; Gonias S. L. Characterization of the binding sites for plasminogen activator in cytokeratin 8 and cytokeratin 18. J. Protein Chem. 17:845–854; 1998. [DOI] [PubMed] [Google Scholar]
- 12. Lu H.; Zhang Y.; Roberts D. D.; Osborne C. K.; Templeton N. S. Enhanced gene expression in breast cancer cells in vitro and tumors in vivo. Mol. Ther. 6:783–792; 2002. [DOI] [PubMed] [Google Scholar]
- 13. McKenna N. J.; O’Malley B. W. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465–474; 2002. [DOI] [PubMed] [Google Scholar]
- 14. Milne T. A.; Briggs S. D.; Brock H. W.; Martin M. E.; Gibbs D.; Allis C. D.; Hess J. L. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10:1107–1117; 2002. [DOI] [PubMed] [Google Scholar]
- 15. Milne T. A.; Dou Y.; Martin M. E.; Brock H. W.; Roeder R. G.; Hess J. L. MLL associates specifically with a subset of transcriptionally active target genes. Proc. Natl. Acad. Sci. USA 102:14765–14770; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakamura T.; Mori T.; Tada S.; Krajewski W.; Rozovskaia T.; Wassell R.; Dubois G.; Mazo A.; Croce C. M.; Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcription regulation. Mol. Cell 10:1119–1128; 2002. [DOI] [PubMed] [Google Scholar]
- 17. Nguyen T. T.; Cho K.; Stratton S. A.; Barton M. C. Transcription factor interactions and chromatin modifications associated with p53-mediated, developmental repression of the alpha-fetoprotein gene. Mol. Cell. Biol. 25:2147–2157; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Noma K.-I.; Allis C. D.; Grewal S. I. S. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150–1155; 2001. [DOI] [PubMed] [Google Scholar]
- 19. Pokholok D. K.; Harbison C. T.; Levine S.; Cole M.; Hannett N. M.; Lee T. I.; Bell G. W.; Walker K.; Rolfe P. A.; Herbolsheimer E.; Zeitlinger J.; Letwitter F.; Gifford D. K.; Young R. A. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122:517–527; 2005. [DOI] [PubMed] [Google Scholar]
- 20. Santos-Rosa H.; Schneider R.; Bannister A. J.; Sherriff J.; Bernstein B. E.; Emre N. C. T.; Schreiber S. L.; Mellor J.; Kouzarides T. Active genes are trimethylated at K4 of histone H3. Nature 419:407–411; 2002. [DOI] [PubMed] [Google Scholar]
- 21. Schneider R.; Bannister A. J.; Myers F. A.; Thorne A. W.; Crane-Robinson C.; Kouzarides T. Histone H3 lysine 4 methylation patterns in eukaryotic genes. Nat. Cell Biol. 6:465–469; 2004. [DOI] [PubMed] [Google Scholar]
- 22. Slany R. K. Chromatin control of gene expression: Mixed-lineage leukemia methyltransferase SETs the stage for transcription. Proc. Natl. Acad. Sci. USA 102:14481–14482; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soule H. D.; Maloney T. M.; Wolman S. R.; Peterson W. D. J.; Brenz R.; McGrath C. M.; Russo J.; Pauley R. J.; Jones R. F.; Brooks S. C. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50:6075–6086; 1990. [PubMed] [Google Scholar]
- 24. Stewart M. D.; Li J.; Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin 1 recruitment. Mol. Cell. Biol. 25:2525–2538; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takei H.; Iino Y.; Horiguchi J.; Kanoh T.; Takao Y.; Ovama T.; Morishita Y. Immunohistochemical analysis of cytokerastin #8 as a prognostic factor in invasive breast carcinoma. Anticancer Res. 15:1101–1105; 1995. [PubMed] [Google Scholar]
- 26. Velculescu V. E.; Zhang L.; Vogelstein B.; Kinzler K. W. Serial analysis of gene expression. Science 270:484–487; 1995. [DOI] [PubMed] [Google Scholar]
- 27. West A. G.; Huang S.; Gaszner M.; Litt M. D.; Felsenfeld G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol. Cell 16:453–463; 2004. [DOI] [PubMed] [Google Scholar]


