Abstract
Mist1 is a tissue-specific basic helix–loop–helix (bHLH) transcription factor that plays an essential role in maintaining and organizing the exocrine pancreas. Consequently, mice lacking Mist1 exhibit disrupted acinar cellular polarity and defective zymogen granule trafficking. Despite extensive studies demonstrating a requirement for Mist1 in exocrine pancreas development and function, little is known about the molecular targets for Mist1 interaction and the mechanism(s) of how Mist1 regulates gene transcription. To address these deficiencies, a series of molecular studies was performed to identify the preferred Mist1 dimer complex and to establish the preferred DNA binding site for this bHLH factor. In vivo coimmunoprecipitation assays confirmed that the functional Mist1 complex in pancreatic acinar cells was a Mist1 homodimer that bound to a unique DNA target site known as the TA-E-box. Binding of Mist1 to a TA-E-box-regulated promoter led to transcriptional activation of the target gene. Surprisingly, Mist1 truncations containing only the central bHLH domain retained approximately 80% of transcriptional activity. Coimmunoprecipitation studies demonstrated that the bHLH domain interacted with coactivators belonging to the p300/CBP family, suggesting that Mist1 activates exocrine-specific gene transcription through an acetylation mechanism.
Key words: Coactivator, Pancreas, Gene regulation, Transcription factor, CBP
INTRODUCTION
The ability of basic helix–loop–helix (bHLH) factors to generate different functional transcriptional complexes provides cells a wide capacity to regulate numerous molecular events (11). The combinatorial effect of forming different dimer complexes with members of the bHLH family is due to differences in individual protein interacting domains, each of which contains unique variations that generate preferences in binding partner choice (17). The two major classes of bHLH transcription factors include widely expressed Class A proteins and the tissue-restricted Class B transcription factors. Typically, Class A and Class B bHLH proteins form heterodimer complexes, but on rare occasions members within the same class may also form homodimer complexes.
The predominant outcome of combining different bHLH partners is that individual dimer complexes exhibit unique specificities for DNA targets, leading to activity of different transcriptional networks. For example, Class A and Class B bHLH proteins typically interact with a hexanucleotide DNA element known as an E-box (CANNTG). The CA and TG sequences are common to all E-boxes, but the central dinucleotides are variable and provide much of the binding specificity for different bHLH protein dimers (31). Each E-box is composed of two half-sites that provide specific contact points for the individual bHLH partners. For example, Tal1/E47 heterodimers specifically bind an E-box containing GA as the core dinucleotide sequence (CAGATG) (7). The 5′ half-site (18) interacts with E47 while the 3′ half-site (ATG) binds to the basic region of Tall. As predicted, E-boxes that are highly palindromic often serve as targets for homodimer complexes. Stra13, a bHLH transcription factor that plays an important role in T-cell development, forms homodimers that specifically interact with the palindromic E-box CACGTG (28).
By forming different functional complexes that interact with a wide variety of DNA targets, bHLH transcription factors modulate a large number of developmental decisions in organ systems ranging from neuronal, skeletal, and cardiac muscle to the pancreas (4,14,22). Pancreatic bHLH factors primarily function to generate and maintain the two major functional compartments: the endocrine pancreas and the exocrine pancreas (25). The Class B bHLH transcription factors known to be involved in pancreas development include Neurogenin3 (Ngn3) (4,12,26), NeuroD/BETA2 (19), pancreas transcription factor-1-p48 (PTF1-p48) (8), and Mist1 (22). Both Ngn3 and Neu-roD are necessary for endocrine cell development as they participate in initiating and promoting the endocrine developmental program (4,19). As a result, mice lacking Ngn3 or NeuroD exhibit a significant reduction in the number of endocrine cells. In contrast, PTF1-p48 and Mist1 are involved in exocrine pancreas development (8). PTF1-p48 facilitates the establishment of the exocrine lineage and consequently, mice lacking PTF1-p48 fail to develop an exocrine pancreas (10). Similarly, Mist1 null (Mist1 KO) mice show extensive defects in the exocrine pancreas (22). Although exocrine lineages are generated, acinar cell organization is highly disrupted, zymogen granule trafficking becomes altered, and Ca2+ signaling, a critical pathway in controlling regulated exocytosis events, becomes severely compromised (15). Mist1 KO mice also fail to express the gap junction protein connexin32, which leads to the breakdown of intercellular communication (24).
Despite extensive studies showing that Mist1 is essential for exocrine pancreas cell organization and zymogen secretion, the mechanism(s) by which Mist1 regulates these events is undefined because Mist1 target genes have not been identified and the consequence of Mist1 binding to specific DNA targets remains unknown. In this study, we provide evidence that Mist1 functions as a transcriptional activator by identifying the preferred Mist1 homodimer DNA binding site and showing that Mist1 activates transcription when associated with this DNA element. Surprisingly, the transcriptional activity associated with Mist1 resides within the central bHLH domain and this activity is further enhanced by the coactivator protein CBP. We conclude that Mist1 is among the first bHLH transcription factors where homodimer complexes mediate transcriptional activation in pancreatic acinar cells.
MATERIALS AND METHODS
Cell Culture and Transfections
The pancreatic cell lines AR42J and ARIP were grown in 40% F12K (Gibco), 25% high glucose DMEM (Hyclone), 25% F12 (Gibco), and 10% FBS (Gibco). ARIPMst1 cells were maintained in growth medium with the addition of G418 (260 μg/ml) (Mediatech) and blastocidin (5 μg/ml) (Invitrogen). Similarly, ARIPTA cells were grown in growth medium containing G418.
Transfections of AR42J and ARIP cells were carried out using a BioRad Gene Pulser and 10 μg of DNA. Following electroporation, cells were immediately transferred to six-well plates containing 2 ml of complete medium. When nuclear extractions were required, 8 × 106 cells and 40 μg of DNA were used. To establish the ARIPTA stable cell line, 4 μg of the TA-E-box-luc reporter gene and 40 ng of control pcDNA3 plasmid containing a neomycin-resistant gene were transfected into ARIP cells (8 × 106) and stable clones were selected in 260 μg/ml G418. ARIPMist1 cells were established by cotransfecting ARIP cells with 18 μg pcDNA6-TR (Invitrogen) and 3 μg of pcDNA4-TO-Mist-myc. Stable clones were selected in blastocidin (5 μg/ml) and zeocin (400 μg/ml).
Plasmid Constructs
The full-length (1–197) and truncated forms (1–172, 1–156, 1–130, 1–140, 36–197, 50–197, 50–140, 73–126, and 72–130) of the rat Mist1 cDNA were cloned by PCR using specific primers containing 5′ EcoRI and 3′ HindIII sites and ligated into pcDNA3.1/Myc-His (Invitrogen). Mist1MB and Mist1MH expression plasmids were generated by standard site-directed mutagenesis (Stratagene) strategies using the pcDNA3-Mist1-myc DNA construct as the template. Mist1MB contains the basic domain amino acid substitutions RER → GGG (a.a. 80–82) while Mist1MH contains a Q^P (a.a. 93) substitution within helix1 (32). All constructs were confirmed by DNA sequencing.
Luciferase reporter gene constructs used in this study, including the TA-E-boxes and their mutant forms, were cloned upstream of the pGL2-promoter luciferase plasmid (Promega) using 5′ MluI and 3′ BglI sites. The 4× TA-E-box-luc gene construct was generated by annealing two oligonucleotides containing four tandem repeats of the TA-E-box ( MluI 4× TA-E-box: CGCGTGAGGGAACATATGTTCGTC AGGGAACATATGTTCGTCAGGGAACATATGT TCGTCAGGGAACATATGTCGTCCA; BglII 4× TA-E-box: GATCTGGACGA ACATATGTTCCCT CTGGACGAACATATGTTCCCTCTGGACGAAC ATATGTTCCCTCTGGACGAACATATGTTCCC TCA). The 1x TA-E-box reporter gene was constructed using annealed oligonucleotides containing a 5′ MluI and a 3′ BglII restriction site (5′ MluI: CGC GTGAGGAGACATATGTTCGTCCA; 3′ BglII: GA TCTGGACGAACATATGTTCCCTCA) and ligated into the pGL2-promoter. Similarly, the 1X mutant TA-E-box-luc construct was made by cloning annealed oligonucleotides that contained mutations within the consensus TA-E-box (5′ MluI: CGCGT GAGGAGAGTAACCTTCGTCCA; 3′ BglII: GATC TGGACGAAGGTTACTTCCCTCA) into the pGL2-promoter plasmid.
Coimmunoprecipitations
Coimmunoprecipitation experiments were performed using lysates derived from either ARIP or AR42J cells. Briefly, cells were washed with PBS and subsequently lysed in 400 μl of cell lysis buffer containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 0.5% Triton X-100, 10% glycerol. Coimmunoprecipitation reactions were performed by incubating 350 μl of the whole-cell lysates with 5 μg of anti-myc (9E10) for 1 h at 4°C. After the incubation, 15 μl of protein A sepharose (50% slurry) (Amersham) was added and incubated at 4°C for 2 h. The samples were washed four times with lysis buffer and analyzed by SDS-PAGE, followed by immunoblotting using anti-Mist1 (5859, 1:1000), anti-Myc (9E10, 1:500), anti-Flag (M2, 1:500) (Sigma), and anti-HA (3F10, 1:1000). Detection of specific proteins was performed using an ABC kit (Pierce), followed by autoradiography.
CASTing and Electrophoretic Mobility Shift Assays
To identify the Mist1 preferred binding site(s), a cyclic amplification selection of targets (CASTing) experiment was performed using a 14-mer random oligonucleotide flanked by two known sequences containing a 5′ EcoRI site and a 3′ ClaI site (binding oligos) [GATCGGAATTCCGACCTGG(N)14GCCA CGTCCATCGATGATC] (30). Double-stranded DNA was generated using a fill-in primer (GGTAGCTAC TTAG) that recognized the 3′ end of the binding oligos.
Electrophoretic mobility shift (EMSA) assays were performed using bacterially purified GST-Mist1 and the 32P-labeled binding probes. GST-Mist1 protein (20 ng) was incubated with 5 × 104 cpm of 32P-labeled probes in 30 μl of binding buffer [20 mM HEPES, pH 7.4, 5 mM dithiothreitol, 50 mM KCl, 0.5 mM EDTA, 5 mM MgCl2, 10% glycerol, 4 μg BSA, 2 μg poly(dI-dC)] for 10 min. The protein/DNA complexes were subsequently resolved by electrophoresis on a 4% acrylamide gel and then subjected to autoradiography. Mist1/DNA complexes were gel purified and then subjected to 15 cycles of PCR using primers specific to the 5′ or 3′ ends of the binding DNA (5′ primer: GATCGGAATTTCGACCTGG; 3′ primer: GATCATCGATGGACGTGGC). PCR products were labeled with [32P-γ]ATP to prepare for the next round of EMSA. Mist1 preferred binding sites were cloned into Bluescript (Stratagene) using MluI and BglII sites and a total of 24 independent clones were sequenced on both strands.
Transient ChIP Assays
ARIPTA cells were transfected with pcDNA3-Mist1-myc or pcDNA3-Mist1MB-myc expression plasmids. Forty-eight hours after transfection, cells were washed with PBS and then subjected to cross-linking using 1% formaldehyde and incubated at room temperature for 5 min with shaking. Cross-link reactions were stopped by the addition of 0.125 M glycine. Nuclear extractions were performed by adding 1 ml of cell lysis buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.1% NP-40) on ice for 1 h. Nuclei were collected and resuspended in 750 μl of nuclear lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS). Nuclei were then sonicated three times and soluble nuclear fractions were used for chromatin immunoprecipitation.
Briefly, precleared reactions were generated by incubating 3 μl of chromatin DNA and 50 μl of protein A Sepharose (Pharmacia) in 1 ml of binding buffer (0.1% SDS, 1.1% Triton-X 100, 167 mM NaCl, 16.7 mM Tris-HCl, pH 8.1) for 1 h. After incubation, the precleared lysates were transferred to new tubes and 5 μg of the 9E10 Myc antibody was added to each reaction. The binding reactions were incubated at 4°C overnight. The following morning, 20 μl of protein A Sepharose was added and the incubation continued for another 2 h. Bound protein/DNA complexes were washed three times with binding buffer and 100 μl of TE was added to each sample; 10 μl was used for immunoblot analysis and the rest of the samples underwent cross-link reversal by incubating at 65°C for 4 h. Proteinase K (10 μl, Promega) was added and the reactions were incubated at 56°C for 2 h. The DNA was purified using the PCR Qiagen Quick kit (Qiagen) and resuspended in 30 μl of TE. PCR reactions were set up containing 1 μl of the purified ChIP product and primers specific to the flanking sequences of the TA-E-box (5′ primer: GTATCTTAT GGTACTGTAACTG; 3′ primer: CTTTATGTTTTTGGCGTCTTCC). Products were resolved on 3% agarose gels.
RESULTS
Identification of the Preferred DNA Binding Site for Mist1 Homodimer Complexes
Mist1 is among a select group of Class B bHLH transcription factors that is expressed in the exocrine pancreas and shown to be critical to normal acinar cell development and function (25). Although purified Mist1 protein forms both homodimer and heterodimer complexes in vitro, our previous studies suggested that Mist1 homodimers may be the preferred functional complex in cells (32). To expand on these earlier studies, we utilized the exocrine pancreas cell line AR42J, in which the endogenous Mist1 gene is constitutively expressed. Using these cells, a Mist1-myc tetracycline (tet)-inducible cell line (AR42JMist1) was generated where Mist1-myc expression was dependent on the presence of tet. As shown in Figure 1A, tet-induced AR42JMist1 cells expressed both the Mist1-myc transgene as well as the endogenous Mist1 protein. In contrast, uninduced cells expressed only the endogenous gene locus. Coimmunoprecipitation experiments using anti-myc, followed by immunoblots with anti-Mist1 revealed that Mist1 homodimer complexes were present within these cells (Fig. 1A). To test if Mist1 also generated heterodimer complexes, we next examined whether Mist1 interacted with the exocrine pancreas Class A bHLH protein, E47. Coimmunoprecipitation assays were again performed as described above and Mist1-myc complexes were analyzed for the presence of endogenous Mist1 or E47 protein. As expected, Mist1-myc/Mist1 homodimer complexes were identified but we failed to similarly coimmunoprecipitate E47 with the Mist1-myc protein (Fig. 1B). Additional coimmuno-precipitation assays also failed to identify Mist1 interactions with other bHLH family members including E12 (data not shown).
Figure 1.
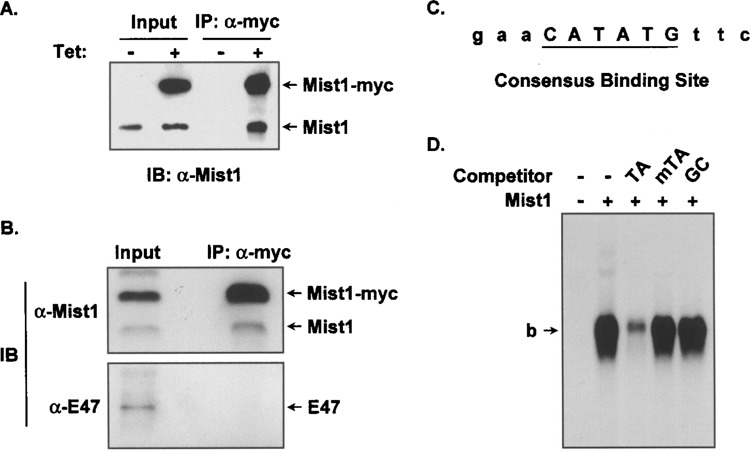
Mist1:Mist1 homodimers interact with the TA-E-box. (A) Nuclear extracts from AR42JMist1 cells grown in the presence or absence of tetracycline (tet) were subjected to coimmunoprecipitation using anti-myc, followed by immunoblots using anti-Mist1. Note that Mist1-myc was coimmunoprecipitated with the endogenous Mist1 protein when cells were induced with tetracycline. (B) AR42JMist1 cells were induced with tet and immunoprecipitation experiments were carried out using anti-myc, followed by immunoblots using anti-Mist1 or anti-E47, as in (A). Only Mist1 homodimers were detected in this assay. (C) The consensus TA-E-box sequence obtained from the Mist1 CASTing assays. (D) Competition EMSA assays using in vitro translated Mist1, 32P-labeled TA-E-box oligonucleotides, and different competitor DNAs. Mist1 specifically interacted with the TA-E-box site. TA, mTA, and GC refer to the central dinucleotides in the competitor sites. b: bound complexes. Note that the free DNA was run off the gel.
Although Mist1 homodimers are the predominant complex in acinar cells, little is known about how this complex regulates transcription since Mist1 target genes and Mist1 DNA binding elements have not been identified. To address this deficiency, we employed a series of CASTing (30) assays to identify Mist1 preferred binding sites. We focused our studies on identifying the preferred binding site for the Mist1 homodimer complex because this was the most prevalent complex in acinar cells. For these studies, a 14-mer random oligonucleotide was synthesized that also contained defined flanking nucleotides for PCR amplification. Successive cycles of Mist1-DNA incubations, separation of the DNA/protein complex by EMSA, amplification of the bound oligonucleotides, followed by cloning and sequence analysis, generated a consensus nucleotide sequence that contained a unique E-box (CANNTG) site in which the central dinucleotides were TA (Fig. 1C). We refer to this site as the TA-E-box (CATATG). As predicted, the TA-E-box site is a perfect palindrome, reflecting the symmetry of the basic domains from each single Mist1 protein interacting with the corresponding half sites of the palindrome (3).
In order to confirm the specificity of Mist1 homo-dimer interactions with the TA-E-box sequence, EMSA assays were performed using in vitro translated Mist1 protein and 32P-labeled TA-E-box oligonucleotides. As shown in Figure 1D, in the absence of competitors, a significant Mist1/TA-E-box complex was detected, confirming that Mist1 homodimers avidly bind to this identified sequence. Competition with an unlabeled TA-E-box effectively inhibited Mist1 binding but competition assays using a mutant TA-E-box site or a GC-E-box failed to influence Mist1 binding to the TA-E-box target (Fig. 1D). Thus, Mist1 homodimers specifically interact with a consensus E-box sequence that contains TA bases in the center dinucleotide positions.
To further establish the importance of individual nucleotides within the TA-E-box, a series of EMSA assays was performed using different mutant competitor oligonucleotide sequences. Competing oligonucleotides having mutations within the consensus E-box sequence, such as CTTATG, CCTATG, and CATAGG, were not able to compete for Mist1 binding (Fig. 2A). In contrast, when oligonucleotides containing 5′ or 3′ flanking alterations were used in competition assays the Mist1/TA-E-box interaction was significantly reduced. Because Mist1 prefers an E-box containing the central TA dinucleotides, EMSA assays were also performed using 32P-labeled probes containing different core dinucleotides (CATTTG, CATGTG, CAATTG). In all cases, changes to the central dinucleotides abolished Mist1 binding (Fig. 2B). These results confirm that Mist1’s DNA binding preference is a minimal consensus E-box binding site of -CATATG- and that flanking sequences are not essential to generate a specific Mist1/DNA interaction.
Figure 2.
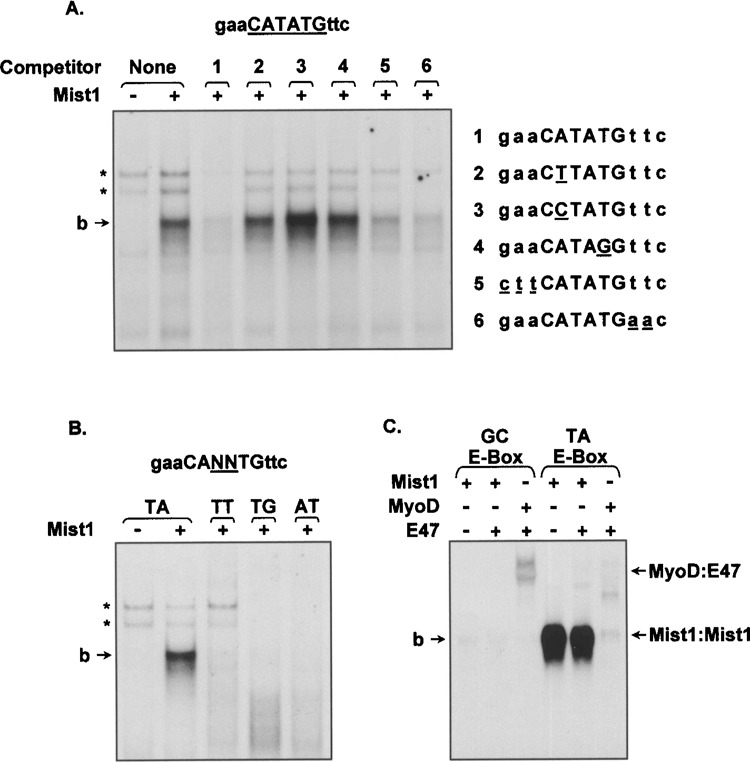
The central TA-E-box core sequence is required for Mist1 binding. (A) EMSA assays using 32P-labeled TA-E-box oligonucleotides, as well as different modifications of the flanking and core E-box nucleotides, were tested with nuclear extracts derived from Mist1-expressing cells. Competition assays were performed by adding unlabeled oligonucleotides (1–6) that contained the indicated mutations within the TA-E-box consensus sequence. Although the flanking nucleotides were dispensable, the core CATATG nucleotides were critical for Mist1 binding. (B) Similar assays as above with the exception that different modifications (TT, TG, AT) within the central dinucleotide positions of the TA-E-box were included in the 32P-labeled target oligonucleotides. The central dinucleotide TA sequence was key for Mist1 interaction. (C) EMSA analysis showing Mist1 homodimers, but not MyoD/E47 heterodimers, specifically interact with the TA-E-box site. *Nonspecific interactions.
Finally, to examine whether the TA-E-box can serve as a target for other bHLH factors, we performed EMSA assays testing the bHLH Class A protein E47 and the Class B protein MyoD. Interestingly, neither E47 homodimers (data not shown) nor a E47/MyoD heterodimer complex bound the TA-E-box sequence in these assays, whereas the MyoD/E47 complex bound its known consensus E-box site (CAGCTG) (Fig. 2C). Although rare Mist1:E47 heterodimer complexes can form with purified proteins (unpublished data), these complexes did not bind the TA-E-box DNA. Similarly, other bHLH proteins tested, including the acinarrestricted Class B factor PTF1-p48 and the Class A factor HEB, also failed to interact with the TA-E-box site (data not shown). Thus, Mist1 homodimer complexes uniquely interact with TA-E-box target sequences.
Mist1 Functions as a Transcriptional Activator of the TA-E-Box
Previous studies have suggested that Mist1 can function as a transcriptional repressor (13). However, these studies were employed using heterologous reporter genes, Gal4-Mist1 fusion proteins, and mesodermal cells, so the function of Mist1 in endodermal pancreatic acinar cells with identified target sequences remains unknown. To examine if Mist1 homodimers promote or inhibit transcription of target genes containing the TA-E-box consensus site, we generated a TA-E-box reporter gene in which four tandem repeats of the TA-E-box were cloned upstream of a luciferase reporter gene containing an SV40 minimal promoter (TA-E-box-luc). AR42J cells were transfected with the TA-E-box-luc reporter gene in the presence or absence of expression plasmids encoding wild type Mist1 or dimerization defective (Mist1MH) or DNA binding defective (Mist1MB) variants. As shown in Figure 3A, all three proteins (Mist1, Mist1MH, and Mist1MB) were produced to equivalent levels in AR42J cells. EMSA assays using AR42J nuclear extracts confirmed that Mist1 homo-dimers efficiently bound the TA-E-box oligonucleotide while the Mist1MB and Mist1MH proteins failed to interact with the TA-E-box site (Fig. 3B). Analysis of luciferase gene activity revealed that Mist1 induced high luciferase reporter gene activity when compared to the nontransfected control group (Fig. 3C). As predicted, the DNA binding defective Mist1MB protein did not activate TA-E-box-luc expression, despite its ability to dimerize and translocate to the nucleus (data not shown). Similarly, the dimerization defective Mist1MH protein also failed to activate TA-E-box-luc gene expression (Fig. 3C).
Figure 3.
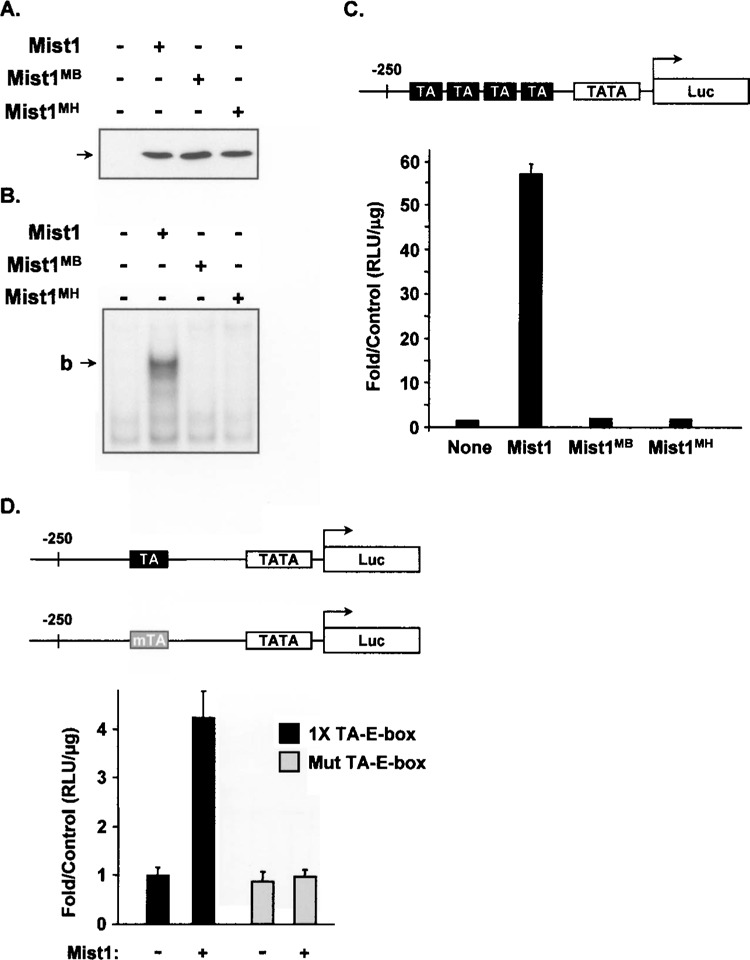
Mist1 binds and transcriptionally activates a TA-E-box reporter gene. (A) Immunoblot assay using anti-Mist1 reveals that the individual Mist1, Mist1MB, and Mist1MH proteins were expressed equivalently in transfected AR42J cells. (B) EMSA assay demonstrating that Mist1, but not Mist1MB or Mist1MH, interacted with the TA-E-box in AR42J cells. (C) AR42J cells were transfected with the 4× TA-E-box-luc gene in the presence or absence of Mist1, Mist1MB, or Mist1MH. Mist1 efficiently activated expression of this target gene. (D) Luciferase assays showing that the wild-type, but not the mut TA-E-box reporter gene, was activated by Mist1.
The above studies strongly suggested that a functional Mist1 homodimer complex interacts with the TA-E-box site to transcriptionally activate gene expression. However, to ascertain if this activation is due to Mist1 directly binding to the TA-E-box, we next performed assays in which reporter gene constructs were engineered to contain a single wild-type TA-E-box site or a single mutant TA-E-box site (Fig. 3D). As expected, cotransfection of the wild-type TA-E-box-luc gene and the Mist1 expression plasmid resulted in the predicted activation of luciferase expression (Fig. 3D). In contrast, Mist1 failed to induce luciferase gene expression from the mut TA-E-box-luc gene. We conclude that Mist1 transcriptional activity is dependent on binding to an intact TA-E-box sequence.
Mist1 Interacts With the TA-E-Box Site In Vivo
In order to investigate the ability of Mist1 to bind specific target genes in pancreatic cells, transient chromatin immunoprecipitation (ChIP) assays were carried out to monitor the in vivo interaction between Mist1 and the TA-E-box site. This assay was performed using the TA-E-box-luc gene and an exocrine pancreas cell line (ARIP) that contains a tet-inducible Mist1-myc transgene (ARIPMist1) but that does not express the endogenous Mist1 gene. ARIPMist1 cells maintained in the presence or absence of tet were transiently transfected with the TA-E-box-luc or the mut TA-E-box-luc gene and the induced and uninduced cell groups were subjected to ChIP assays as described in the Materials and Methods. As shown in Figure 4A, wild-type TA-E-box sequences were immunoprecipitated and amplified only under conditions in which Mist1 was induced. In contrast, ChIP assays failed to detect the mut TA-E-box sequence.
Figure 4.
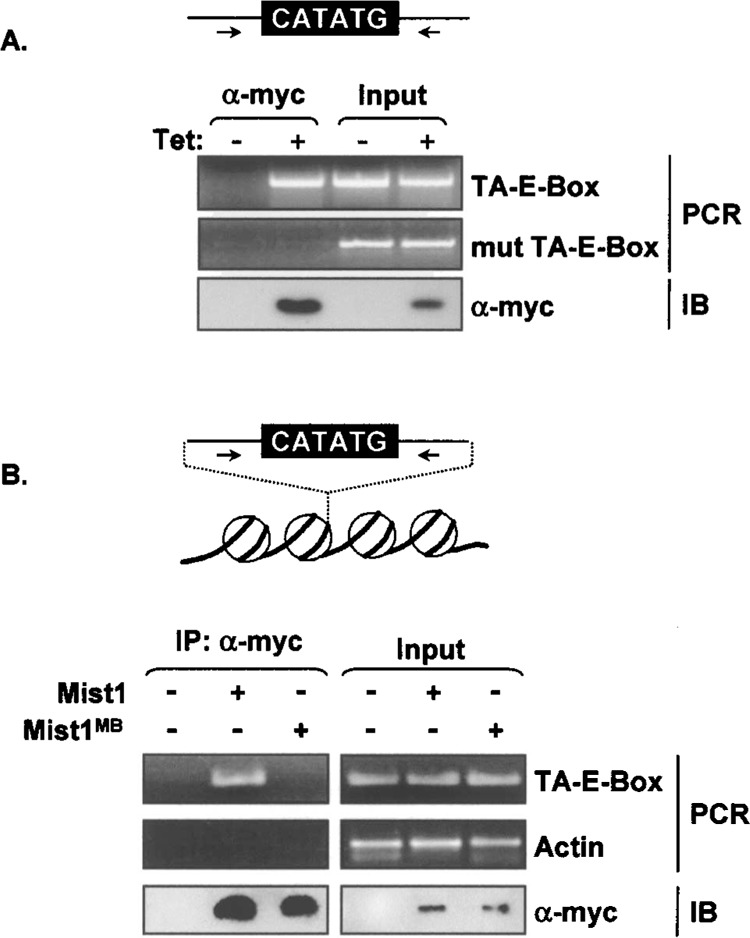
Mist1 interacts with the TA-E-box in vivo. (A) Chromatin immunoprecipitation assays using ARIPMist1 cells transfected with the TA-E-box or the mut TA-E-box gene. Chromatin immunoprecipitations were performed using anti-myc followed by PCR amplification with primers specific to the flanking sequences of the respective TA-E-box genes. Immunoblot analyses were also performed using anti-myc to detect the induced Mist1-myc protein. Mist1 was found associated only with the wild-type TA-E-box sequence in AR42J cells. (B) ChIP assays were performed using an ARIP cell line that contained an integrated copy of the TA-E-box DNA (ARIPTA). Both Mist1 and Mist1MB expression plasmids were transfected into ARIPTA cells and the nuclear lysates were used for ChIP assays. Immunoprecipitations were carried out using a Myc antibody and PCR was performed with primers specific to the flanking sequences of the TA-E-box or to the control actin gene. Mist1 associated with the TA-E-box sequence within cellular chromatin whereas Mist1MB failed to bind to this target sequence.
Although these studies established a physical interaction between Mist1 and the TA-E-box in vivo, this interaction may not represent the normal nuclear chromatin context because the TA-E-box sequence was provided through a transient DNA transfection strategy. To overcome this obstacle, a modified ChIP assay was performed. In this instance, ARIPTA cells were generated that contained an integrated copy of the TA-E-box sequence. The ARIPTA cells were then engineered to express either Mist1 or Mist1MB proteins. ChIP assays on these cells revealed the presence of the TA-E-box sequence in the ARIPTA Mist1-express-ing cells but not in the ARIPTA Mist1MB-expressing cells (Fig. 4B). These results confirm that Mist1 is capable of binding to chromatin DNA containing a TA-E-box sequence.
The N- and C-Termini of Mist1 Are Not Required for Mist1 Transcriptional Activity
Given that Mist1 activates transcription of target genes containing a TA-E-box, we next attempted to identify the region(s) of Mist1 that is essential for this activation. Ten different Mist1 truncations, all of which contained the bHLH domain but lacked various portions of the carboxyl- or amino-terminus, were generated (Fig. 5). These truncations, along with expression plasmids encoding the full-length Mist1, Mist1MB, or Mist1MH proteins, were cotransfected with the TA-E-box-luc reporter gene and tested for protein expression, subcellular location, homodimer formation, TA-E-box interaction, and for their ability to promote transcriptional activity. Immunoblot and immunohistochemistry assays confirmed that most Mist1 mutants were expressed equivalently in the AR42J pancreatic cells and that all proteins preferentially accumulated in the nuclear compartment. Coimmunoprecipitation and EMSA assays also revealed that Mist1 truncations containing an intact bHLH domain formed homodimer complexes that bound the TA-E-box sequence, whereas Mist1MH was defective in dimer formation and both Mist1MH and Mist1MB failed to bind DNA (Fig. 5).
Figure 5.
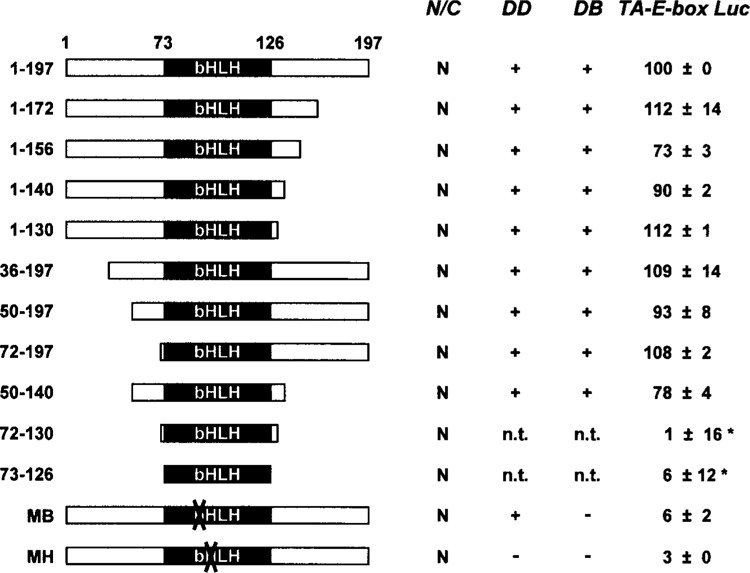
The bHLH domain of Mist1 facilitates transcriptional activity. Immunohistochemistry, protein-protein interactions, EMSA and reporter gene assays were carried out as described in Materials and Methods to establish the region(s) of the Mist1 protein that was responsible for Mist1 transcriptional activity. A series of Mist1 truncations (a.a. 1–197, 1–172, 1–156, 1–140, 1–130, 36–197, 50–197, 72–197, 50–140, 72–130, 73–126) was generated, transfected into ARIP cells, and tested for protein localization (N/C: nucleus/cytoplasm), protein–protein interactions (DD), DNA binding activity (DB), and the ability to activate the TA-E-box luciferase reporter gene (TA-E-box Luc). Additionally, Mist1MB (MB) and Mist1MH (MH) were tested as negative controls. All luciferase values were normalized to the full-length (1–197) Mist1 protein, which was set at 100 (+/− indicate standard error of the mean). Mist1 (50–140) retains 78% of normal transcriptional activity. *Mist1 truncations having very low expression levels in ARIP cells. n.t.: not tested.
When the C-terminal truncation proteins (a.a. 1– 172, 1–156, 1–140, 1–130) were tested for transcriptional activity, they all retained their ability to activate the TA-E-box reporter gene, suggesting that the region between amino acids 130 and 197 is not essential for Mist1 transcriptional activity (Fig. 5). Likewise, similar transcriptional activity was observed when N-terminal truncations (a.a. 36–197, 50–197, 72–197) were tested, demonstrating that the N-terminus amino acid residues were not critical for Mist1 transcriptional activity. Finally, we truncated both the N- and C-termini (a.a. 50–140, 72–130, 73–126) and tested the ability of these proteins to promote Mist1 transcriptional activity. Whereas Mist1 (50–140) retained 78% of wild-type Mist1 activity, the 72–130 and the 73–126 mutants were completely inactive (Fig. 5). Unfortunately, both of these truncations exhibited greatly reduced expression levels when compared to wild-type Mist1 or to any other Mist1 truncations tested (Fig. 5 and data not shown). Thus, we were not able to fully examine the true activities of these two mutants. However, because most of the Mist1 mutants, including Mist1 (50–140), promoted TA-E-box reporter gene expression, we conclude that the N- and C-termini of Mist1 are dispensable for transcriptional activity and that the key functional transcriptional domain of Mist1 lies within the bHLH region.
Mist1 Activation Is Associated With CBP
Our mutagenesis studies suggested that the Mist1 bHLH domain alone functions in promoting transcription. Because this domain is primarily responsible for protein dimerization and DNA binding, we wondered if the transcriptional activity ascribed to this region could be mediated through specific coactivators such as CBP and p/CAF (CBP associated factor) (5). For these studies, pancreatic ARIP cells were cotransfected with the TA-E-box-luc gene and various combinations of Mist1, CBP, and p/CAF expression plasmids. As previously shown, Mist1 induced high levels of TA-E-box-luc gene expression (Fig. 6A). CBP or p/CAF alone did not influence expression of the TA-E-box-luc reporter gene. Similarly, coexpression of p/CAF with Mist1 did not influence Mist1’s ability to activate the TA-E-box-luc gene. However, when Mist1 and CBP were coexpressed, Mist1 transcriptional activity was significantly increased (Fig. 6A). These results demonstrate that CBP, but not p/CAF, positively influences Mist1 transcriptional activity in this model.
Figure 6.
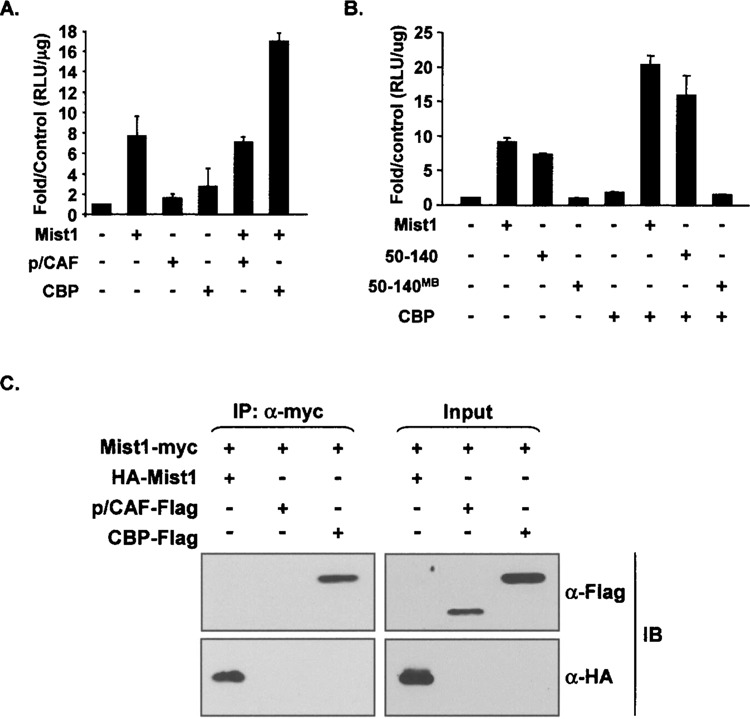
CBP enhances Mist1 transcriptional activity. (A) Luciferase reporter gene assays showing that CBP, but not p/CAF, increased Mist1 transcriptional activity. (B) Reporter gene assays testing the effect of CBP on Mist1 (50–140) transcriptional activity. The Mist1(50–140)MB protein served as a negative control. (C) Immunoprecipitation assays were carried out to establish the interaction between Mist1 and CBP. ARIP cells were transfected with Mist1-Myc in combination with either HA-Mist1, p/CAF-flag, or CBP-flag expression plasmids and nuclear extracts were subjected to coimmunoprecipitation assays using anti-myc, followed by immunoblots with anti-HA or anti-Flag. CBP was coprecipitated with Mist1, indicating a Mist1/CBP complex formed in vivo.
Because the truncated Mist1(50–140) protein retained high transcriptional activity (Fig. 5), we next tested its activity in the presence of CBP. Luciferase assays were again performed with cells transfected with the TA-E-box-luc construct and different combinations of Mist1, Mist1 (50–140), and CBP expression plasmids. In addition, we tested Mist1(50–140)MB, which contains amino acids 50–140 but with a mutated basic domain. As expected, Mist1 and Mist1(50–140), but not CBP alone or Mist1(50–140)MB, efficiently promoted transcription of the TA-E-box-luc gene (Fig. 6B). Coexpression of CBP and Mist1 or CBP and Mist1(50–140) similarly elevated expression of the TA-E-box-luc reporter gene. In contrast, cotransfection of CBP with Mist1(50–140)MB did not lead to TA-E-box-luc expression. Thus, the Mist1 bHLH domain (50–140) is sufficient for CBP enhancement.
To further examine the role of CBP in mediating Mist1 transcriptional activity, we examined if Mist1 and CBP interacted with each other in vivo. A coimmunoprecipitation (co-IP) experiment was performed where the Mist1-myc expression plasmid was cotransfected with either HA-Mist1, p/CAF-flag, or CBP-flag expression plasmids. Nuclear lysates were subjected to coimmunoprecipitations using anti-myc, followed by immunoblots with anti-HA or anti-Flag. As predicted, anti-HA detected the HA-Mist1 protein in both the input and the co-IP samples, confirming the Mist1-myc:HA-Mist1 homodimer interaction (Fig. 6C). Both p/CAF and CBP in the input groups were also detected by the Flag antibody. However, p/CAF protein was not detected in the co-IP samples whereas high levels of CBP were found to coimmunoprecipitate with Mist1 (Fig. 6C). These results demonstrate that Mist1 and CBP interact in vivo and suggest that Mist1’s transcriptional activity is mediated by CBP through a direct interaction with the Mist1 bHLH domain.
DISCUSSION
During pancreas development, specification and terminal differentiation events are controlled by members of the bHLH protein family, which have distinct roles in facilitating and ensuring proper development of the endocrine and exocrine pancreas compartments. For example, Class A bHLH factors (E12, E47, HEB) form functional heterodimer complexes with select Class B bHLH factors to regulate pancreatic exocrine cell development (23). To date, only two Class B bHLH factors have been shown to play important roles in exocrine pancreas development, PTF1-p48 and Mist1 (8,20,22). PTF1-p48 is essential to acinar cell specification because PTF1-p48 null mice fail to develop an exocrine pancreas (19). PTF1-p48 also has been shown to promote gene expression of the major exocrine pancreas enzymes, which are critical to the function of acinar cells (1,29). In contrast, Mist1 is not essential for acinar cell specification but is critical for normal acinar cell function. Mist1 KO mice develop an exocrine pancreas but the typical apical-basal organization essential to acinar cells does not develop (22). Indeed, Mist1 is the only known bHLH transcription factor that is essential for establishing and maintaining exocrine pancreas cellular polarity.
Previous studies from our laboratory have revealed several genes that are misexpressed in Mist1 KO mice (15,24). However, it is unclear if these genes are direct targets for Mist1 or whether changes in gene expression reflect secondary events that manifest from deletion of the Mist1 gene. For instance, the gap junction gene connexin32 (Cx32) is downregulated in Mist1 KO mice and Mist1 protein activates expression of the Cx32 promoter in reporter gene assays (24). The proximal Cx32 promoter (680 bp) is complex, containing five E-box sites. Surprisingly, Mist1 activates Cx32 reporter genes even when all five E-box sites are destroyed (Rukstalis and Konieczny, unpublished results), suggesting that Mist1 can function through a TA-E-box-dependent mechanism (this study) as well as through an E-box-independent mechanism (Cx32 gene), possibly by interacting with non-bHLH transcription factors. We are continuing to examine this latter activation mechanism as well as attempting to identify true Mist1 TA-E-box-dependent target genes by performing ChIP-on-Chip assays from pancreas chromatin.
In the absence of known Mist1 target genes, we undertook studies to examine the transcriptional Mist1 complex and possible DNA target sites using exocrine pancreatic cells. Our initial coimmunoprecipitation studies confirmed that Mist1 functions as a homodimer complex. These results support our earlier findings with transgenic mice in which a Mist1 MB transgene was expressed under the regulation of the exocrine-specific elastase1 promoter (32). Expression of Mist1MB inhibited endogenous Mist1 activity by forming DNA binding defective Mist1MB/Mist1 homodimer complexes but defective Mist1MB/PTF1-p48 or Mist1MB/E47 heterodimer complexes were not generated. The Mist1MB/Mist1 homodimer generated near identical cellular disorganization as observed in Mist1 KO mice. Thus, genetic and direct in vivo coimmunoprecipitation studies strongly suggest that Mist1 functions as a bHLH homodimer complex.
Although accumulating evidence indicates that Mist1 homodimers are essential to acinar cell functions, it has been difficult to study individual transcriptional components of the Mist1 protein complex because specific target genes have not been defined. We resolved this problem by first identifying a Mist1-preferred binding site (TA-E-box) and then examined the ability of Mist1 to regulate this DNA binding site. The identified TA-E-box is unique for bHLH proteins because the center two nucleotides are TA. Typically, E-box core dinucleotides contain at least one G or C residue, which determines the binding specificity of bHLH dimers to E-box targets. For example, MyoD/E47 heterodimers prefer to interact with a GC-E-box (16) while Tal1/E47 heterodimers specifically bind a GA-E-box (GA) (2,21). The fact that Mist1 homodimers selectively bind to the TA-E-box site and not to other E-box sites underscores the importance of the TA core nucleotides to Mist1:DNA interactions.
The identification of the TA-E-box has provided an important tool to further study how Mist1 regulates transcription and has demonstrated that Mist1 can function as a transcriptional activator of genes containing TA-E-box elements. As predicted, this activity depends on Mist1’s ability to form homodimers and to bind to intact TA-E-box sites. These observations identify Mist1 as one of the first examples of a Class B bHLH transcription factor that promotes transcriptional activity as a homodimer complex. To date, only a handful of bHLH proteins have been shown to form homodimers, including Stra13 (28), Hand1 (6), and E47 (27). However, with the exception of the Class A E47 homodimers found exclusively in B lymphocytes (27), most Class B homodimer complexes function as transcriptional repressors. Indeed, previous studies from our laboratory implicated Mist1 as a transcriptional repressor and this activity was shown to be driven by the N-terminal portion of the protein (13). However, these assays were performed employing a heterologous system where Gal4-Mist1 fusion proteins and Mist1 truncation proteins were used to test their ability to repress an active reporter gene construct driven by LexA-VP16 in mesodermal C3H10T1/2 cells. Based on these assays, the LexA-VP16-induced activation was reduced up to 10-fold when Mist1 was coexpressed. Thus, although Mist1 did not physically bind DNA through the bHLH domain, positioning Mist1 on the Gal4 promoter did repress LexA-VP16-induced transcription. These data are in contrast to our current studies using a true Mist1 binding site target and appropriate exocrine pancreas cell lines where Mist1 clearly functions as an activator. However, because both studies utilized specific DNA targets that require the recruitment of Mist1 to the regulatory regions of the target genes, it is possible that Mist1 functions as both an activator and repressor depending on the cellular context. In this regard, Mist1 does not activate TA-E-box genes in nonpancreatic cells such as C3H10T1/2 (unpublished data), suggesting that Mist1 transcriptional activity may be specific to pancreatic acinar cells where this transcription factor is normally found. Future studies will focus on determining if distinct Mist1 protein complexes assemble in different cell lineages.
DNA binding transcriptional regulators are typically modular in that they contain discrete functional domains. One well-known example is E47, which contains an N-terminal activation domain, a protein– protein interaction domain (HLH), and a DNA binding domain (basic) (9). Surprisingly, Mist1 is unique in that it does not follow this general rule. Mist1 transcriptional activity depends solely on the basic and helix–loop–helix domains and the interaction with the coactivator protein CBP. While this property of Mist1 is interesting, it has presented a challenge in further investigating the function of this region. The bHLH domain is essential for both homodimer formation and DNA binding, so alterations within this region to map transcriptional activity are difficult to generate because defects in dimerization or DNA binding activity inactivate the protein. Nonetheless, efforts are under way to identify individual residues within the bHLH domain that may be important for CBP interaction and for Mist1 transcriptional activity without affecting the ability of Mist1 to dimerize and bind DNA. Understanding how Mist1/CBP complexes regulate genes that control acinar cell organization should facilitate the identification of these gene targets.
ACKNOWLEDGMENTS
We thank Ryan Bettencourt, Michael Collingwood, and Peichuan Sun for providing excellent technical assistance in generating the Mist1 truncation expression plasmids. We also thank Dr. Stephen Brandt for generously providing the p/CAF and CBP plasmids used in this study. This work was supported by a grant to S.F.K. from the National Institutes of Health (DK55489). T.T. and D.J. were supported by Purdue Research Foundation Graduate Fellowships.
REFERENCES
- 1. Beres T. M.; Masui T.; Swift G. H.; Shi L.; Henke R. M.; MacDonald J. R. PTF1 is an organ-specific and Notch-independent basic helix–loop–helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol. Cell. Biol. 26:117–130; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berkes C. A.; Tapscott S. J. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 16:585–595; 2005. [DOI] [PubMed] [Google Scholar]
- 3. Blackwell T. K.; Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science 250:1104–1110; 1990. [DOI] [PubMed] [Google Scholar]
- 4. Gradwohl G.; Dierich A.; LeMeur M.; Guillemot F. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. USA 97:1607–1611; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gregory P. D.; Wagner K.; Horz W. Histone acetylation and chromatin remodeling. Exp. Cell Res. 265:195–202; 2001. [DOI] [PubMed] [Google Scholar]
- 6. Hill A. A.; Riley P. R. Differential regulation of Hand1 homodimer and Hand1-E12 heterodimer activity by the cofactor FHL2. Mol. Cell. Biol. 24:9835–9847; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hsu H. L.; Huang L.; Tsan J. T.; Funk W.; Wright W. E.; Hu J. S.; Kingston R. E.; Baer R. Preferred sequences for DNA recognition by the TAL1 helix–loop–helix proteins. Mol. Cell. Biol. 14:1256–1265; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kawaguchi Y.; Cooper B.; Gannon M.; Ray M.; MacDonald R. J.; Wright C. V. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 32:128–134; 2002. [DOI] [PubMed] [Google Scholar]
- 9. Kee B. L.; Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix–loop–helix transcription factor E12. J. Exp. Med. 188:699–713; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krapp A.; Knofler M.; Ledermann B.; Burki K.; Berney C.; Zoerkler N.; Hagenbuchle O.; Wellauer P. K. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 12:3752–3763; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ledent V.; Paquet O.; Vervoort M. Phylogenetic analysis of the human basic helix–loop–helix proteins. Genome Biol. 3:RESEARCH0030; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee J. C.; Smith S. B.; Watada H.; Lin J.; Scheel D.; Wang J.; Mirmira R. G.; German M. S. Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes 50:928–936; 2001. [DOI] [PubMed] [Google Scholar]
- 13. Lemercier C.; To R. Q.; Carrasco R. A.; Konieczny S. F. The basic helix–loop–helix transcription factor Mist1 functions as a transcriptional repressor of myoD. EMBO J. 17:1412–1422; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ludolph D. C.; Konieczny S. F. Transcription factor families: Muscling in on the myogenic program. FASEB J. 9:1595–1604; 1995. [DOI] [PubMed] [Google Scholar]
- 15. Luo X.; Shin D. M.; Wang S.; Konieczny S. F.; Muallem S. Aberrant localization of intracellular organelles, Ca2+ signaling, and exocytosis in Mist1 null mice. J. Biol. Chem. 280:12668–12675; 2005. [DOI] [PubMed] [Google Scholar]
- 16. Mak K. L.; Longcor L. C.; Johnson S. E.; Lemercier C.; To R. Q.; Konieczny S. F. Examination of mammalian basic helix–loop–helix transcription factors using a yeast one-hybrid system. DNA Cell Biol. 15:1–8; 1996. [DOI] [PubMed] [Google Scholar]
- 17. Massari M. E.; Murre C. Helix–loop–helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20:429–440; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mills J. C.; Roth K. A.; Cagan R. L.; Gordon J. I. DNA microarrays and beyond: completing the journey from tissue to cell. Nat. Cell Biol. 3:E175–178; 2001. [DOI] [PubMed] [Google Scholar]
- 19. Naya F. J.; Huang H. P.; Qiu Y.; Mutoh H.; DeMayo F. J.; Leiter A. B.; Tsai M. J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 11:2323–2334; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Obata J.; Yano M.; Mimura H.; Goto T.; Nakayama R.; Mibu Y.; Oka C.; Kawaichi M. p48 subunit of mouse PTF1 binds to RBP-Jkappa/CBF-1, the intracellular mediator of Notch signalling, and is expressed in the neural tube of early stage embryos. Genes Cells 6:345–360; 2001. [DOI] [PubMed] [Google Scholar]
- 21. Ono Y.; Fukuhara N.; Yoshie O. Transcriptional activity of TAL1 in T cell acute lymphoblastic leukemia (T-ALL) requires RBTN1 or -2 and induces TALLA1, a highly specific tumor marker of T-ALL. J. Biol. Chem. 272:4576–4581; 1997. [DOI] [PubMed] [Google Scholar]
- 22. Pin C. L.; Rukstalis J. M.; Johnson C.; Konieczny S. F. The bHLH transcription factor Mist1 is required to maintain exocrine pancreas cell organization and acinar cell identity. J. Cell Biol. 155:519–530; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roux E.; Strubin M.; Hagenbuchle O.; Wellauer P. K. The cell-specific transcription factor PTF1 contains two different subunits that interact with the DNA. Genes Dev. 3:1613–1624; 1989. [DOI] [PubMed] [Google Scholar]
- 24. Rukstalis J. M.; Kowalik A.; Zhu L.; Lidington D.; Pin C. L.; Konieczny S. F. Exocrine specific expression of Connexin32 is dependent on the basic helix–loop–helix transcription factor Mist1. J. Cell Sci. 116:3315–3325; 2003. [DOI] [PubMed] [Google Scholar]
- 25. Schwitzgebel V. M. Programming of the pancreas. Mol. Cell. Endocrinol. 185:99–108; 2001. [DOI] [PubMed] [Google Scholar]
- 26. Schwitzgebel V. M.; Scheel D. W.; Conners J. R.; Kalamaras J.; Lee J. E.; Anderson D. J.; Sussel L.; Johnson J. D.; German M. S. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development 127:3533–3542; 2000. [DOI] [PubMed] [Google Scholar]
- 27. Shen C. P.; Kadesch T. B-cell-specific DNA binding by an E47 homodimer. Mol. Cell. Biol. 15:4518–4524; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. St-Pierre B.; Flock G.; Zacksenhaus E.; Egan S. E. Stra13 homodimers repress transcription through class B E-box elements. J. Biol. Chem. 277:46544–46551; 2002. [DOI] [PubMed] [Google Scholar]
- 29. Williams J. A. Intracellular signaling mechanisms activated by cholecystokinin-regulating synthesis and secretion of digestive enzymes in pancreatic acinar cells. Annu. Rev. Physiol. 63:77–97; 2001. [DOI] [PubMed] [Google Scholar]
- 30. Wright W. E.; Binder M.; Funk W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol. Cell. Biol. 11:4104–4110; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yun K.; Wold B. Skeletal muscle determination and differentiation: Story of a core regulatory network and its context. Curr. Opin. Cell Biol. 8:877–889; 1996. [DOI] [PubMed] [Google Scholar]
- 32. Zhu L.; Tran T.; Rukstalis J. M.; Sun P.; Damsz B.; Konieczny S. F. Inhibition of Mist1 homodimer formation induces pancreatic acinar-to-ductal metaplasia. Mol. Cell. Biol. 24:2673–2681; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


