Abstract
Significance: Culture-independent methods have revealed the diverse and dynamic bacterial communities that colonize chronic wounds. Only recently have studies begun to examine fungal colonization and interactions with the bacterial component of the microbiome, their relationship with the host, and influence on wound outcomes.
Recent Advances: Studies using culture-independent sequencing methods reveal that fungi often go undetected in wounds. Candida spp. and Cladosporidium spp. are the most commonly identified fungi in wounds. The wound environment may promote multispecies biofilm formation between bacteria and fungi in wounds, with implications for pathogenicity, treatment, and outcomes.
Critical Issues: Identifying microorganisms that are problematic for healing will require a comprehensive understanding of all members of the polymicrobial wound community, including fungi and bacteria. Improved reference databases and bioinformatics tools for studying fungal communities will stimulate further research into the fungal microbiome.
Future Directions: Continued study of polymicrobial wound communities using culture-independent methods will further our understanding of the relationships between microbial bioburden, the host response, and impact on healing, complications, and patient outcomes. Future studies should encompass all types of microbiota, including fungi, and focus on potential multi-kingdom interactions that contribute to pathogenicity, biofilm formation, and poor outcomes.
Keywords: : fungi, microbiome, chronic wounds, biofilm

Lindsay Kalan, PhD
Scope and Significance
Microbiome analysis of chronic wounds has progressed significantly in recent years. Findings support the hypotheses that microbes contribute to impaired wound healing, and wound microbiomes encompass diverse species of bacteria. Only very recently have microbiome studies been published that include an analysis of both fungal and bacterial communities in chronic wounds, suggesting fungi are underappreciated agents of complications. This review will introduce the reader to the state-of-the art technology for studying fungi in the microbiome and discuss the current literature regarding fungi in chronic wounds. We will discuss the interactions of fungi and bacteria within biofilms and the relationship to clinical outcomes.
Translational Relevance
Application of next-generation sequencing has facilitated advanced insight into the polymicrobial nature of chronic wounds and eliminated biases associated with culture-dependent techniques. Pioneering studies have delineated the dynamics of bacterial communities in chronic wound microbiomes. Incorporating the fungal microbiome into bacteria-centric models will advance our understanding of the complex wound environment, permitting researchers to begin determining the mechanisms of microbial interactions, including vastly understudied fungal-bacterial interactions. This will lead to innovation of targeted treatment strategies aiming to intercept synergistic interactions that may lead to biofilm formation, enhanced virulence, and clinical complication.
Clinical Relevance
In recent years, the recognition of microbial biofilm and its relationship to impaired healing has resulted in the introduction of a “biofilm-based wound care” concept1 that aims to prevent biofilm formation in wound tissue. Advancing this concept further, is the increasing body of evidence that biofilms are polymicrobial communities more recalcitrant to intervention, than their single-species counterparts. This phenomenon is amplified in biofilms comprising bacteria and fungi, where fungi can provide a scaffold for bacterial attachment and offer additional protection.2,3 High-throughput sequencing allows rapid characterization of the microbiome, informing on the presence of bacterial and fungal species to guide antimicrobial therapy that targets both.
Discussion
Overview of microbiome research
Microbial communities assemble to form distinct and stable ecological systems within the human body, commonly referred to as the “microbiome.” With the advent of next-generation sequencing technologies comes the capability to study the microbiome with a new lens, highlighting that traditional culture-based approaches grossly underestimate the complexity and total microbial diversity of human-associated microbiota. As symbionts, the microbiome influences higher host biology in a manner we are only beginning to understand. In the context of the skin, when the barrier is breached, microbes from the healthy skin and the environment assemble in the wound tissue, in some cases disrupting the highly orchestrated phases of tissue repair. Discerning which microbial community types act to impede, promote, or have a neutral effect on wound healing would dramatically change clinical practice. As a result, the number of wound microbiome studies is continually increasing. This review will focus on study of the microbiome, and specifically the fungal microbiome of chronic, nonhealing wounds with an emphasis on diabetic foot ulcers (DFU).
One of the most popular approaches to characterizing the microbiome involves polymerase chain reaction (PCR) amplification and high-throughput DNA sequencing of the bacterial 16S ribosomal RNA (rRNA) gene, a highly conserved gene containing hypervariable regions that are informative for taxonomic identification.4 Classification is accomplished by clustering amplicon sequences into operational taxonomic units (OTUs) based on the percent similarity to each other (typically, ∼97% for species-level assignment) and choosing a representative sequence from each OTU. Taxonomy is determined by alignment of the representative sequence against a reference database of bacterial 16S rRNA genes. Bioinformatics software pipelines such as the Quantitative Insights Into Microbial Ecology (QIIME)5 and mothur6 provide a suite of tools to analyze data, including raw read processing, OTU picking, phylogenetic analysis, and determination of diversity metrics. Furthermore, the development of protocols for sample preparation, sequencing, and downstream software tools has enabled the noncomputer scientist to include microbiome research in their studies and push forward molecular diagnostics in the clinic.
However, a limitation to 16S rRNA gene amplicon methodology is that this approach is “bacteriocentric” and excludes other microorganisms such as fungi. Fungi are often overshadowed by their bacterial counterparts of the human microbiome, even though they are prominent members of some ecosystems (e.g., skin), and fungal infection is of serious concern.7,8 This is especially surprising, given that foot mycosis and onychomycosis occur at a high frequency in diabetic populations, with some surveys confirming fungal foot infections in over 75% of diabetic individuals tested.9,10 In fact, Candida infections can oftentimes be a first clinical symptom of prediabetic or diabetic status and should prompt further diagnostic measures on the part of the treating physician.11 Fungal foot infections, although often overlooked, are dangerous in that they create a portal of entry for secondary bacterial infection through fissures and splits.12 Toenail infections in particular, left untreated, increase the risk of developing a foot ulcer and subsequent infection-related complications.13
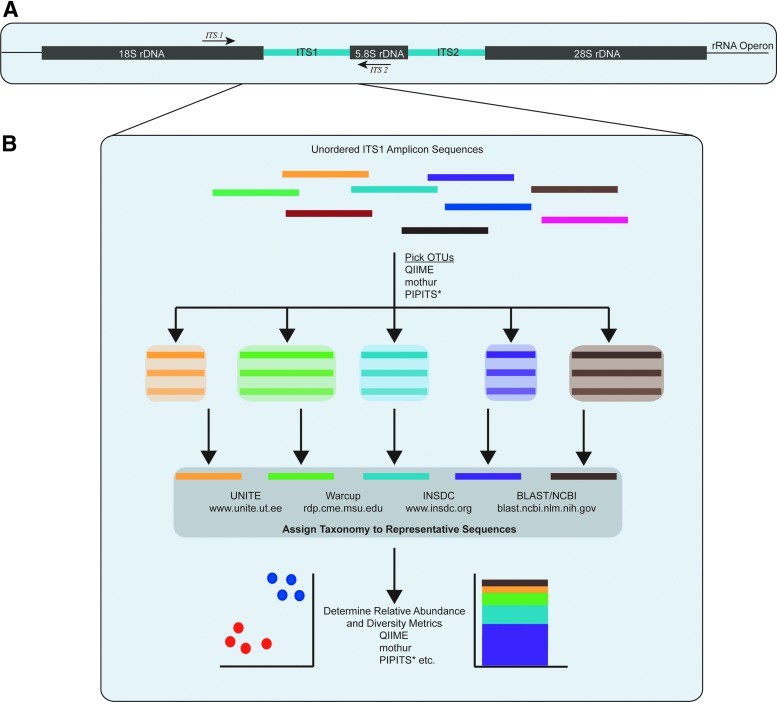
Collectively, fungi in the microbiome are referred to as the “mycobiome” and can be classified with culture-independent approaches by applying the same general workflow as the bacterial microbiome, or “bacteriome.” This is accomplished by sequencing the fungal rRNA gene operon (Fig. 1A). Target regions within the ribosomal DNA include the 18S small subunit (SSU) rDNA, 28S large subunit rDNA (LSU), or the internal transcribed spacer (ITS) region.14 Typically, the LSU and SSU are used for large-scale phylogenetic surveys because they are highly conserved, but have fewer hypervariable regions than the bacterial 16S rRNA gene, consequently limiting taxonomic resolution. The internal transcribed spacer (ITS) region contains two hypervariable regions, ITS1 and ITS2, with the 5.8S rRNA gene between them.15 This region has been adapted as the universal barcode region for fungi by the Consortium for the Barcode of Life because it has been shown to have superior species discrimination and PCR success rates across a broad range of fungi. Regardless of the barcode utilized, the available fungal databases represent a minor fraction (∼1.5%) of total fungal diversity on the planet, the vast majority of which are currently unknown.15,16 Software pipelines dedicated to fungal analysis are also available. PIPITS17 incorporates the RDP Classifier and curated UNITE database18 for taxonomic classification, after extracting the hypervariable ITS1 or ITS2 subregions from paired-end sequence reads with ITSx.14 PIPITS and other fungal-specific bioinformatics pipelines are reviewed in Gweon et al.17
Figure 1.
(A) Schematic of the fungal rRNA operon highlighting the ITS region and primer binding sites. (B) Bioinformatics workflow to process and analyze ITS amplicons from microbiome specimens. After polymerase chain reaction amplification, OTUs can be clustered with standardized pipelines and classified against fungal reference databases. The fungal ITS1 region can vary in size between different species, unlike amplification of hypervariable regions of the bacterial 16S rRNA gene that result in the same size amplicon. *PIPITS is a fungal-specific software pipeline. ITS, internal transcribed spacer; OTU, operational taxonomic units; QIIME, Quantitative Insights into Microbial Ecology; rRNA, ribosomal RNA.
The fungal microbiome of DFU
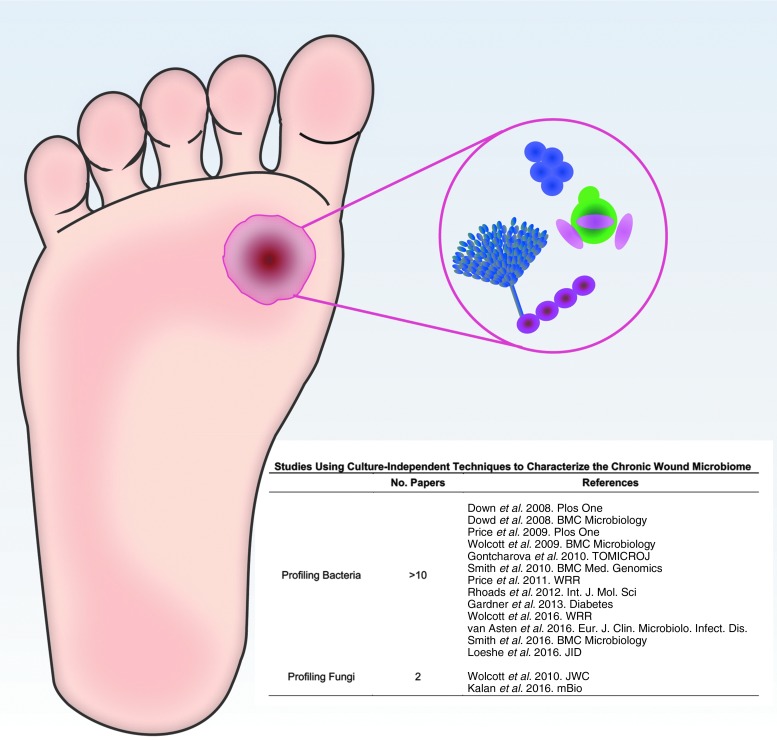
To date, the vast majority of research on chronic wound microbiomes has focused on the bacterial content (Fig. 2). Work by our group and others has demonstrated that the bacteriome in DFUs contain greater than 15 species on average and is associated with outcomes and clinical factors.19–21 Microbiome stability, measured as the change in overall community structure over time is significantly associated with slow healing wounds. That is, the more stable the DFU microbiome, the longer the healing time.20 However, each of these studies exclusively profiles bacteria and fails to integrate fungi into the prevailing model.
Figure 2.
The microbiome of DFU contains many species of both bacteria and fungi that are not always detected by culture-dependent techniques. Molecular-based approaches have primarily focused on bacteria as evidenced by the number of articles published since 2008. In comparison, only two studies have profiled fungi in chronic wound microbiomes with molecular approaches. DFU, diabetic foot ulcers.
Exclusion of fungi from DFU microbiome analysis is shortsighted because the healthy skin microbiome comprises bacteria and fungi. Moreover, the feet have been shown to exhibit the highest level of fungal diversity within the skin microbiome, with many species considered opportunistic pathogens.22,23 Culture-dependent and culture-independent studies focusing on fungi have consistently revealed that a significant portion of chronic wounds such as DFUs are colonized or infected with fungi. For example, Chellan et al. examined 518 diabetic lower leg wounds by directly inoculating fungal selective media with deep wound tissue sections collected after surgical debridement and saline cleansing. After observing growth for 4 weeks, fungi spanning 18 different species were detected in 27.2% of specimens. Approximately 6% of wounds solely contained fungi, while 21.4% of specimens had mixed bacterial and fungal flora.24
In 2011, the first study to employ culture-independent analysis of yeast and fungi in chronic wounds of mixed etiology found 23% of specimens tested positive for fungi. However, when classified by wound type, 40.8% of DFUs (n = 83) were fungal positive and in many specimens, the predicted fungal:bacterial ratio was predicted to be >50%.25 A more recent study exclusively examined DFU (n = 105) and found 28.6% of specimens were positive for fungi by microscopic examination, but only 20% were positive by culture, further supporting the premise that culture-dependent methods underestimate diversity and bioburden.26 Findings from these studies suggest that over a quarter of all DFUs contain fungi that are not detected or diagnosed by standard microbiology laboratory protocols.
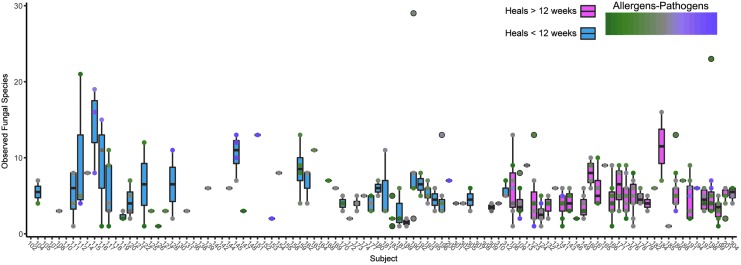
Recently, our group provided additional insights by employing PCR-based amplicon sequencing of the fungal ITS1 region to precisely define the prevalence and structure of fungal communities residing in DFUs over time (6 months), in an attempt to link these polymicrobial, inter-kingdom microbiomes to clinical outcomes.27 One of the most important findings was that fungi are highly diverse and prevalent members of DFU microbiomes (n = 100), occurring in 71.6% of specimens for at least one time point, and in 79% of wounds during the course of the 26-week study, far exceeding previous predictions. Figure 3 demonstrates that individual wound specimens contained >1 and up to 30 different fungal OTUs. While the “mycobiome” was highly heterogeneous both between patients (interpersonal variation) and over time (intrapersonal, temporal variation), we observed that patients administered systemic antibiotics had significantly higher fungal diversity in their wounds than those who were not. This suggests that using antibiotics solely targeting bacteria may create an environment favorable to fungal colonization and expansion. Conversely, wound deterioration was also marked by a significant increase in fungal diversity,27 suggesting an alternate scenario by which increased fungal colonization leads to a worsening wound environment, suspected bacterial infection, and subsequent increase in prescribed antibiotics. Notably, ∼30% of the ITS sequence reads were not classified beyond the Kingdom level, likely due to poorly populated fungal ITS reference databases.
Figure 3.
Number of observed fungal species detected in DFU. Individual subjects are labeled across the x-axis and the observed species along the y-axis for multiple time points per subject. Box plots are colored by healing time (<12 weeks, blue; >12 weeks, pink). Each point represents a single sample and the fill corresponds to the ratio of allergenic to pathogenic fungi (green, high allergens; purple, high pathogens; gray, equal allergens/pathogens).
Previous studies have reported that members of the genus Candida spp. are the most commonly isolated yeasts from DFUs.24,25 Over 75% of species isolated by Chellan et al. were Candida species (25% C. parapsilosis, 22.7% C. tropicalis, and 10.6% C. albicans). These same three species were each detected in greater than 30% of fungi-positive wounds by Dowd et al. with C. albicans found in 46% of fungi-positive wounds. We also observed that, of the specimens' culture positive for fungi, Candida spp. was the most commonly isolated. ITS1 sequencing confirmed that 22% of the specimens were positive for C. albicans, but this was not the most frequently detected fungal species. Cladosporium herbarum, a ubiquitous and saprophytic fungus, was present at multiple time points in 56% of subjects. Dowd et al. also reported C. herbarum in 9.32% of fungi-positive wounds of mixed etiology, sampled at a single point in time. While the C. herbarum species complex has a worldwide distribution associated with diverse substrates, isolates with clinical origins have been associated with pathology, including subcutaneous infection in humans and animals.28
We observed a clear distribution of wounds containing either a high proportion of pathogenic or allergenic fungi (Fig. 3). As an important agent linked to allergic rhinitis and respiratory disease, Cladosporium spp. along with other allergic fungi such as Aspergillus spp., Penicillium spp., Alternaria spp., Pleospora spp., and Fusarium spp. was detected with high frequency in DFUs. Pathogenic and opportunistic fungi, such as Candida spp., Trichosporon asahii, and Rhodotorula spp., were associated with poor outcomes, such as stalled open wounds after 6 months' treatment, or wounds resulting in an amputation. Specifically, polymicrobial pathogenic communities of fungi tend to reside in wounds with necrotic, nonviable tissue, in a significantly higher proportion compared to wounds absent for necrosis. However, with regular debridement and standardized care, not all wounds become or remain necrotic, and it is unclear if increased fungal bioburden contributes to, or is a result of, necrosis.
It is intriguing that like DFUs and other chronic wounds, chronic rhinitis is associated with polymicrobial colonization and biofilm formation,29,30 suggesting that the involvement of fungi detected in recalcitrant wounds is underestimated and warrants further investigation. Further supporting this idea, we found that allergenic fungi were positively associated with wounds of longer duration and surface area, but negatively associated with hemoglobin A1c and white blood cell levels. On the contrary, pathogens had a positive association with the latter two markers. This suggests that glucose control and the microbiome intersect, resulting in different immunological responses depending on the community structure of the fungal microbiome.
Multi-kingdom wound biofilms
In the environment, groups of microorganisms assemble into biofilms attached to surfaces for more efficient nutrient cycling and enhanced protection from external threats. These highly structured, sophisticated microbial communities can develop in wounds and require intervention to assist host defenses in overcoming biofilm-associated complications.31 Yet, despite the rapid increase in laboratory biofilm research and the development of biofilm-based wound care principles, the link between wound biofilm and clinical outcomes remains ambiguous. This is, in part, because biofilms in wound tissue are difficult to detect. Visual identification of biofilm in a wound is debatable, while the presence of the biofilm is not a strong predictor of healing rate. The biofilm lifecycle, and determination of subsequent biomarkers, is largely studied in vitro with a single species laboratory strain of bacteria. Because the wound microbiome comprises mixed bacterial-fungal communities, we postulate that this environment would support the interaction of bacteria and fungi to form multispecies biofilms. Interactions between some clinically important bacterial and fungal species have been described. Candida spp. have been reported to interact with diverse bacteria such as Staphylococcus aureus, Pseudomonas aeruginosa, Burkholderia cenocepacia, Streptococcus spp, Acinetobacter baumannii, Enterococcus faecalis, and E. coli (reviewed in Ref.32). Consistently, it has been reported that fungal hyphae can facilitate adhesion to surfaces and provide a substrate for bacteria to bind. This action can result in an increased resistance to antimicrobial penetration in biofilms, as is the case when S. aureus coats itself with secreted C. albicans cell wall polysaccharides enhancing aggregation and tolerance.3 Streptococcus gordonii also produces surface glycans that bind C. albicans; however, it is the production of the quorum-sensing molecule autoinducer-2 that is required for dense biofilm formation, pointing toward inter-kingdom chemical communication.
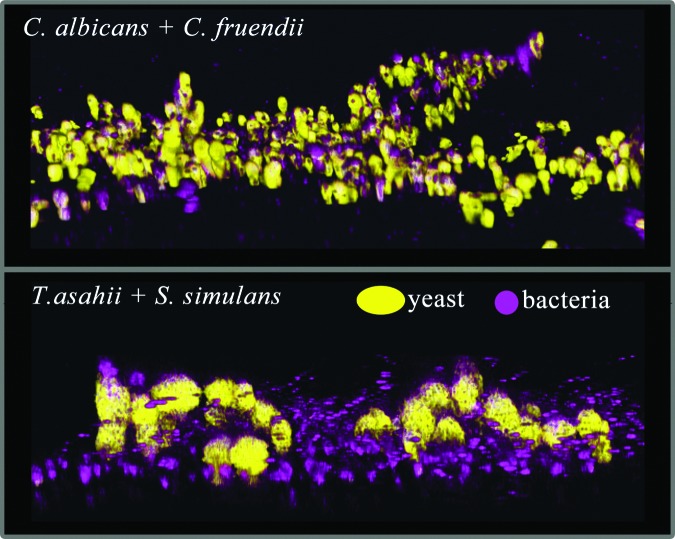
Kalan et al.27 used molecular-based computational analysis as a guide to predict the establishment of a biofilm in wounds with a stable bacteriome and mycobiome for a period of at least 4 weeks. Target organisms were then isolated directly from patient specimens with genera-specific cultivation media. Organisms that were co-isolated were tested for their ability to form mixed-species biofilms. This approach was tested on two wounds with disparate outcomes. The first wound healed within 12 weeks, while the second wound was open and led to an amputation after 26 weeks. From the healed wound, C. albicans and Citrobacter freundii were co-isolated and shown to assemble into a structured, three-dimensional biofilm in vitro. C. albicans provided the scaffold for C. freundii to intimately attach and proliferate (Fig. 4). Microscopic evidence of the interaction suggests that the fungi themselves are critical to the construction and establishment of the biofilm. Each individual fungal or bacterial species also possessed the capacity to form a biofilm, although less structured. This observation is important because C. albicans hyphae can invade epithelial cells by active penetration.33,34 This action facilitates bacterial epithelial entry as they act as passengers adhered to the hyphae. Work by Townsend et al. have demonstrated similar findings in a tripartite inter-kingdom biofilm model of C. albicans, Staphylococcus aureus, and P. aeruginosa. In a two-dimensional culture, C. albicans grew predominantly as hyphae, providing a scaffold for S. aureus and P. aeruginosa attachment. When this experiment was repeated with a hydrogel scaffold to create three-dimensional culture and mimic a wound surface, the phenotype shifted. Instead, C. albicans grew predominantly as yeast cells coaggregated with bacterial cells.35 This suggests that the microenvironment influences microbial gene expression to trigger different phenotypes.
Figure 4.
Inter-kingdom biofilms formed between fungi and bacteria isolated from the same DFU specimen. The fungi (yellow) form a three-dimensional structure that bacteria (purple) attach to. Candida albicans + Citrobacter fruendii (top) and Trichosporon asahii + Staphylococcus simulans (bottom).
T. asahii and Staphylococcus simulans were co-isolated from wound specimens collected after 22 weeks from a wound that ultimately resulted in an amputation. This fungal-bacterial pair also formed a mixed-species biofilm in vitro, with S. simulans adhering to clusters of T. asahii cells (Fig. 4) in a similar architecture to C. albicans + C. freundii biofilms. There is little information regarding the pathogenicity and host response of T. asahii biofilm and it is virtually nonexistent for co-infection with bacteria. Trichosporon species were the second most common fungal genus, after Candida spp., to be isolated by Chellan et al.24 Each of the T. asahii isolates was identified as resistant to the antifungal agent flucytosine. The Trichosporon genus was first described from a hair (Trichos) infection, with irregular nodules (sporon or spores) identified along the hair shaft of a man's beard in 1890. T. asahii is a known biofilm-forming organism36 with the potential to cause superficial and invasive infection, likely from their characteristic morphological diversity, including the capability to form mycelia, blastoconidia, and arthroconidia.37 Taken together, these data imply that T. asahii may be an important emerging pathogen for skin and soft tissue infection, which has the ability to go undetected in chronic wounds such as DFU. Indeed, compared to environmental isolates that grow mainly as hyphae, clinical isolates of T. ashaii have been reported to primarily grow as conidia with higher levels of glucuronoxylomannan antigen, a factor thought to be important for immune evasion and antiphagocytosis.38
Summary
As the microbiome field matures and expands into new environments, a consistent theme emerges: microbes do not live in isolation. Chronic wounds are no exception, hosting diverse microbiomes sensitive to the changing tissue microenvironment. Fungi are important members of the DFU microbiome; however the mechanisms driving their interaction with cohabitating bacteria remain unclear. Current treatments for wounds with suspected biofilm are primarily focused on targeting bacteria. There is now sufficient evidence to conclude that increased species diversity of biofilms is correlated to increased resistance to antimicrobials.3,39–41 Moreover, use of antibiotics targeting bacteria in mixed communities have been shown to increase fungal diversity in wound tissue27 and provide a niche for fungal expansion in mixed bacterial-fungal biofilms.
Fluconazole, an antifungal drug, has been shown to significantly reduce the mean healing time of DFUs when compared to the standard of care in a randomized control study.42 More recently, it was demonstrated that treatment of a tripartite bacterial-fungal biofilm with antibiotics alone resulted in a threefold increase of viable fungal cells, whereas only a combination treatment with antibiotics (ciprofloxacin and flucloxacillin) and fluconazole resulted in decreased viable cells of both bacteria and fungi.40 Current literature, while limited, suggest that wound care strategies could benefit from inclusion of antifungal drugs such as fluconazole, amphotericin B, or caspofungin43 concurrent with antibacterial therapy, or application of a broad-spectrum topical antimicrobial that targets both.
Abbreviations and Acronyms
- DFU
diabetic foot ulcers
- ITS
internal transcribed spacer
- LSU
large subunit rDNA
- OTUs
operational taxonomic units
- PCR
polymerase chain reaction
- QIIME
Quantitative Insights into Microbial Ecology
- rRNA
ribosomal RNA
- SSU
small subunit
Acknowledgments and Funding Sources
This work was supported by grants from the National Institutes of Health [NIAMS R00 AR060873 (EAG), NIAMS R01 AR066663 (EAG), and NINR R01 NR015639 (EAG)] and a grant from the Pennsylvania Department of Health (EAG). The content is solely the responsibility of the authors and does not necessarily represent the official view of the funding bodies.
Author Disclosure and Ghostwriting
The authors have no competing interests. The article was written solely by its authors.
About the Authors
Elizabeth Grice, PhD, is an Assistant Professor of Dermatology and Microbiology at the University of Pennsylvania Perelman School of Medicine. She received her doctorate in Human Genetics in 2006 from Johns Hopkins University, and performed postdoctoral work at the National Institutes of Health. Her research interests focus on genomic and metagenomic analyses of cutaneous host-microbe interactions, especially in nonhealing wounds. Lindsay Kalan, PhD, is a Postdoctoral Fellow in the Department of Dermatology at the University of Pennsylvania Perelman School of Medicine and is a member of EAG laboratory. She received her doctorate in 2012 from McMaster University where she studied the evolution of clinical antibiotic resistance. Her current research interests include application of metagenomic analyses to study microbial interactions in nonhealing wounds.
Take Home Messages.
Next-generation sequencing has revealed chronic wound microbiomes to be complex microbial communities. Culture-dependent methods do not capture the degree of both bacterial and fungal diversity in wound tissue.
Studies employing culture-dependent and culture-independent methods have concluded that fungi in DFU often go undetected, resulting in a vastly underestimated occurrence.
Fungal pathogens are highly associated with wound necrosis. Topical treatments targeting bacteria and fungi could lead to improved outcomes in these types of wounds.
Fungi have the ability to form intricate biofilms with gram-positive and gram-negative bacteria. The fungal partner can provide a structure for bacteria to attach and an additional protection from external threats such as antibiotics.
Continued study of the wound microbiome will further advance our understanding of the relationship between microbial bioburden, wound progression, host response, and individual characteristics.
References
- 1.Wolcott R. Economic aspects of biofilm-based wound care in diabetic foot ulcers. J Wound Care 2015;24:189–190, 192–194 [DOI] [PubMed] [Google Scholar]
- 2.Kean R, Rajendran R, Haggarty J, et al. . Candida albicans mycofilms support Staphylococcus aureus colonization and enhances miconazole resistance in dual-species interactions. Front Microbiol 2017;8:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kong EF, Tsui C, Kucharíková S, Andes D, Van Dijck P, Jabra-Rizk MA. Commensal protection of Staphylococcus aureus against antimicrobials by Candida albicans biofilm matrix. MBio 2016;7:e01365–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yarza P, Yilmaz P, Pruesse E, et al. . Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 2014;12:635–645 [DOI] [PubMed] [Google Scholar]
- 5.Caporaso JG, Kuczynski J, Stombaugh J, et al. . QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schloss PD, Westcott SL, Ryabin T, et al. . Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009;75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol 2014;14:405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui L, Morris A, Ghedin E. The human mycobiome in health and disease. Genome Med 2013;5:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckhard M, Lengler A, Liersch J, Bretzel RG, Mayser P. Fungal foot infections in patients with diabetes mellitus—results of two independent investigations. Mycoses 2007;50 Suppl 2(s2):14–19 [DOI] [PubMed] [Google Scholar]
- 10.Wijesuriya TM, Weerasekera MM, Kottahachchi J, et al. . Proportion of lower limb fungal foot infections in patients with type 2 diabetes at a tertiary care hospital in Sri Lanka. Indian J Endocrinol Metab 2014;18:63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wohlrab J, Wohlrab D, Meiss F. Skin diseases in diabetes mellitus. J Dtsch Dermatol Ges 2007;5:37–53 [DOI] [PubMed] [Google Scholar]
- 12.Roujeau JC, Sigurgeirsson B, Korting HC, Kerl H, Paul C. Chronic dermatomycoses of the foot as risk factors for acute bacterial cellulitis of the leg: a case-control study. Dermatology (Basel) 2004;209:301–307 [DOI] [PubMed] [Google Scholar]
- 13.Boyko EJ, Ahroni JH, Cohen V, Nelson KM, Heagerty PJ. Prediction of diabetic foot ulcer occurrence using commonly available clinical information: the Seattle Diabetic Foot Study. Dia Care 2006;29:1202–1207 [DOI] [PubMed] [Google Scholar]
- 14.Bengtsson-Palme J, Ryberg M, Hartmann M, et al. . Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Bunce M, ed. Methods Ecol Evol 2013;4:914–919 [Google Scholar]
- 15.Schoch CL, Seifert KA, Huhndorf S, et al. . Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci 2012;109:6241–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porras-Alfaro A, Liu KL, Kuske CR, Xie G. From genus to phylum: large-subunit and internal transcribed spacer rRNA operon regions show similar classification accuracies influenced by database composition. Appl Environ Microbiol 2014;80:829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gweon HS, Oliver A, Taylor J, et al. . PIPITS: an automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Bunce M, ed. Methods Ecol Evol 2015;6:973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kõljalg U, Nilsson RH, Abarenkov K, et al. . Toward a unified paradigm for sequence-based identification of fungi. Mol Ecol 2013;22:5271–5277 [DOI] [PubMed] [Google Scholar]
- 19.Gardner SE, Hillis SL, Heilmann K, Segre JA, Grice EA. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes 2013;62:923–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loesche M, Gardner SE, Kalan L, et al. . Temporal stability in chronic wound microbiota is associated with poor healing. J Invest Dermatol 2017;137:237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price LB, Liu CM, Melendez JH, et al. . Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. Ratner AJ, ed. PLoS One 2009;4:e6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh J, Byrd AL, Park M, NISC Comparative Sequencing Program, Kong HH, Segre JA. Temporal stability of the human skin microbiome. Cell 2016;165:854–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Findley K, Oh J, Yang J, et al. . Topographic diversity of fungal and bacterial communities in human skin. Nature 2013;498:367–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chellan G, Shivaprakash S, Karimassery Ramaiyar S, et al. . Spectrum and prevalence of fungi infecting deep tissues of lower-limb wounds in patients with type 2 diabetes. J Clin Microbiol 2010;48:2097–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dowd SE, Delton Hanson J, Rees E, et al. . Survey of fungi and yeast in polymicrobial infections in chronic wounds. J Wound Care 2011;20:40–47 [DOI] [PubMed] [Google Scholar]
- 26.Mehra BK, Singh AK, Gupta D, Narang R, Patil R. A clinicomicrobiological study on incidence of mycotic infections in diabetic foot ulcers. ijss-sncom. INJSS. 2017;4:50–54 [Google Scholar]
- 27.Kalan L, Loesche M, Hodkinson BP, et al. . Redefining the chronic-wound microbiome: fungal communities are prevalent, dynamic, and associated with delayed healing. MBio 2016;7:e01058–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandoval-Denis M, Gené J, Sutton DA, Wiederhold NP, Cano-Lira JF, Guarro J. New species of Cladosporium associated with human and animal infections. Persoonia 2016;36:281–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gelardi M, Passalacqua G, Fiorella ML, Quaranta N. Assessment of biofilm by nasal cytology in different forms of rhinitis and its functional correlations. Eur Ann Allergy Clin Immunol 2013;45:25–29 [PubMed] [Google Scholar]
- 30.Jung JH, Cha HE, Kang IG, Kim ST. Clinical characteristics of biofilms in patients with chronic rhinosinusitis: a prospective case-control study. Indian J Otolaryngol Head Neck Surg 2015;67:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swanson T, Angel D, Sussman G, et al. . Wound infection in clinical practice: principles of best practice. International Wound Infection Institute (IWII) Wound infection in clinical practice. Wounds International 2016
- 32.Allison DL, Willems HME, Jayatilake JAMS, Bruno VM, Peters BM, Shirtliff ME. Candida-bacteria interactions: their impact on human disease. Microbiol Spectr 2016;4:103–136 [DOI] [PubMed] [Google Scholar]
- 33.Schlecht LM, Peters BM, Krom BP, et al. . Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue. Microbiology (Reading, Engl) 2015;161(Pt 1):168–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong EF, Kucharíková S, Van Dijck P, Peters BM, Shirtliff ME, Jabra-Rizk MA. Clinical implications of oral candidiasis: host tissue damage and disseminated bacterial disease. Deepe GS, Jr., ed. Infect Immun 2015;83:604–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Townsend EM, Sherry L, Rajendran R, et al. . Development and characterisation of a novel three-dimensional inter-kingdom wound biofilm model. Biofouling 2016;32:1259–1270 [DOI] [PubMed] [Google Scholar]
- 36.Di Bonaventura G, Pompilio A, Picciani C, Iezzi M, D'Antonio D, Piccolomini R. Biofilm formation by the emerging fungal pathogen Trichosporon asahii: development, architecture, and antifungal resistance. Antimicrob Agents Chemother 2006;50:3269–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colombo AL, Padovan ACB, Chaves GM. Current knowledge of Trichosporon spp. and Trichosporonosis. Clin Microbiol Rev 2011;24:682–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karashima R, Yamakami Y, Yamagata E, Tokimatsu I, Hiramatsu K, Nasu M. Increased release of glucuronoxylomannan antigen and induced phenotypic changes in Trichosporon asahii by repeated passage in mice. J Med Microbiol 2002;51:423–432 [DOI] [PubMed] [Google Scholar]
- 39.Lemire JA, Kalan L, Gugala N, Bradu A, Turner RJ. Silver oxynitrate—an efficacious compound for the prevention and eradication of dual-species biofilms. Biofouling. 2017;33:460–469 [DOI] [PubMed] [Google Scholar]
- 40.Townsend EM, Sherry L, Kean R, et al. . Implications of antimicrobial combinations in complex wound biofilms containing fungi. Antimicrob Agents Chemother 2017;61:00672–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee KWK, Periasamy S, Mukherjee M, Xie C, Kjelleberg S, Rice SA. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J 2014;8:894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chellan G, Neethu K, Varma AK, et al. . Targeted treatment of invasive fungal infections accelerates healing of foot wounds in patients with Type 2 diabetes. Diabet Med 2012;29:e255–e262 [DOI] [PubMed] [Google Scholar]
- 43.Felton T, Troke PF, Hope WW. Tissue penetration of antifungal agents. Clin Microbiol Rev 2014;27:68–88 [DOI] [PMC free article] [PubMed] [Google Scholar]