Abstract
Background:
Progressive myoclonus epilepsies (PMEs) comprise a group of rare genetic disorders characterized by action myoclonus, epileptic seizures, and ataxia with progressive neurologic decline. Due to clinical and genetic heterogeneity of PMEs, it is difficult to decide which genes are affected. The aim of this study was to report an action myoclonus with or without renal failure syndrome (EPM4) family and summarize the clinical and genetic characteristics of all reported EPM4 patients.
Methods:
In the present study, targeted next-generation sequencing (NGS) was applied to screen causative genes in a Chinese PME family. The candidate variant was further confirmed by cosegregation analysis and further functional analysis, including the reverse transcription polymerase chain reaction and Western blot of the proband's muscle. Moreover, literature data on the clinical and mutational features of all reported EPM4 patients were reviewed.
Results:
The gene analysis revealed a novel homozygous splicing mutation (c.995-1G>A) of the SCARB2 gene in two brothers. Further functional analysis revealed that this mutation led to loss function of the SCARB2 protein. The classification of the candidate variant, according to the American College of Medical Genetics and Genomics standards and guidelines and functional analysis, was pathogenic. Therefore, these two brothers were finally diagnostically confirmed as EPM4.
Conclusions:
These present results suggest the potential for targeted NGS to conduct a more rapid and precise diagnosis for PME patients. A literature review revealed that mutations in the different functional domains of SCARB2 appear to be associated with the phenotype of EPM4.
Keywords: Progressive Myoclonus Epilepsies, Progressive Myoclonus Epilepsy with or without Renal Failure, SCARB2 Gene, Targeted Next-Generation Sequencing
摘要
背景:
进行性癫痫性肌阵挛(PMEs)是一组以肌阵挛、癫痫发作、共济失调及神经系统功能进行性减退为特点的罕见病。由于该病具有临床和基因异质性,明确疾病的致病基因有一定难度。 本文对一例EPM4家系进行了报道并总结了所有已报道EMP4患者的临床与基因突变特点。
方法:
本研究应用目标区域测序在一个家系中检测 PMEs致病基因并对候选基因变异位点进行家系共分离分析以及对先证者 肌肉细胞进行基因表达水平和蛋白表达水平分析。同时,回顾并总结所有已报道EPM4临床和突变特点的文献。
结果:
通过基因分析发现,先证者及其兄长均携带SCARB2基因c.995-1G>A剪切位点突变。进一步实验发现该突变导致SCARB2 蛋白功能缺失。根据美国医学遗传学和基因组学学院(ACMG)标准和指南,该候选变异位点为致病突变,该兄弟最终被确诊 EPM4。
结论:
靶向二代测序未来有望成为更加快速精准诊断PMEs的方法。通过回顾相关文献表明EPM4的表型与突变位点所在SCARB2 基因功能域有关。
INTRODUCTION
Progressive myoclonus epilepsies (PMEs) comprise a group of hereditary disorders characterized by action myoclonus, epileptic seizures, and ataxia with progressive neurologic decline.[1] Owing to clinical and molecular heterogeneity, PMEs present with various forms caused by different disease-causing genes. According to the Online Mendelian Inheritance in Man database and relevant literature, PMEs can be divided into 12 subtypes: Unverricht–Lundborg disease (EPM1A), EPM1B, Lafora body disease (EPM2A), Lafora body disease (EPM2B), EPM3, action myoclonus with or without renal failure syndrome (EPM4), PME-ataxia syndrome (EPM5), North Sea PME (EPM6), EPM7, EPM8, EPM9, and EPM10. The respective disease-causative genes are as follows: CSTB, PRICKLE1, MELF, NHLRC1, KCTD7, SCARB2, PRICKLE2, GOSR2, KCNC1, CERS1, LMNB2, and PRDM8.[1,2,3,4,5,6,7,8,9,10,11,12,13] However, there are other neurogenetic diseases mainly characterized by myoclonus, epileptic seizures, and ataxia, such as myoclonus epilepsy and ragged red fibers, neuronal ceroid lipofuscinoses, sialidosis, dentatorubral-pallidoluysian atrophy (DRPLA), and neuronopathic Gaucher disease, while a literature also regarded these diseases as PMEs.[1] In clinic, it is difficult to make an exact diagnosis among the various forms of PMEs due to homogeneous phenotypes. Moreover, other diseases, such as juvenile myoclonic epilepsy, inherited ataxia, and mitochondrial disease, also resemble or overlap PMEs in clinical features, which present challenges in the differential diagnosis. Therefore, the screening for the disease-related mutation of pathogenic genes is particularly valuable in the diagnosis of PMEs. Traditional Sanger sequencing lacks the efficiency to handle larger numbers of candidate genes associated with PMEs. As a powerful approach for genetic diagnostics in inherited Mendelian disorders, targeted next-generation sequencing (NGS) has increased the ability to rapidly and effectively sequence any genomic region of interest.[14,15]
In the present study, two brothers were confirmed as EPM4 with the application of a targeted NGS panel, which covers the causative genes of PMEs, juvenile myoclonic epilepsy,[16] inherited ataxia, and mitochondrial disease. The investigators found a novel splice mutation of scavenger receptor class B, member 2 (SCARB2), and further validated the candidate variant by functional research based on the patient's tissues. Furthermore, literature data were reviewed to summarize the clinical and mutational spectrum of all reported EPM4 patients worldwide.
METHODS
Ethical approval
The present study was approved by the Ethics Committee of the First Affiliated Hospital of Fujian Medical University. Written informed consent was obtained from all participants in the family and individuals in the control group.
Subject
A family from Fujian Province, China, who presented with a PME phenotype, was recruited in the present study. Detailed clinical data, including clinical presentations, physical examinations, laboratory tests, and neuroimaging and electroencephalography (EEG) results, were collected. Three hundred unrelated healthy individuals with no known history of neurogenetic disease were collected and assigned as the control group.
Genetic testing of dentatorubral-pallidoluysian atrophy
For the purpose of excluding DRPLA, the patient was tested for CAG trinucleotide repeats in the ATN1 gene of the proband by polymerase chain reaction (PCR), as previously described.[17]
Targeted next-generation sequencing and sequence analysis
Targeted NGS was performed on genomic DNA samples extracted from the proband and his elder brother's peripheral blood samples using a Blood Genomic Extraction Kit (Qiagen, Hilden, Germany). The sequences were performed by an NGS-based assay using the Illumina HiSeq2500 platform (Illumina, California, USA). The panel was prepared using a NimbleGen SeqCap EZ Choice kit (Roche, Basel, Switzerland), which included 927 disease-causative genes of the neurogenetic disease, containing progressive myoclonic epilepsy, hereditary ataxia, mitochondrial diseases, neuronal ceroid lipofuscinosis and other neurogenetic diseases. Targeted coding exons and intron-exon regions corresponded to 4.8 Mb of the genomic sequence.
First, the raw sequence reads were aligned to the human reference genome (UCSC hg 19) (http://hgdownload.cse.ucsc.edu/) using Burrows–Wheeler Aligner (Li and Durbin, 2009). Second, the gene-, region-, and filter-based levels of the variants were annotated using the ANNOVAR software (version Feb 11, 2013, GitHub, Philadelphia, USA). Then, the frequency of the variants was further determined using the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/), the 1000 Genomes Project (http://ftp.ncbi.nih.gov/), and the Exome Aggregation Consortium (http://exac.broadinstitute.org/). Finally, the filtered variants were classified according to the American College of Medical Genetics and Genomics (ACMG) standards and guidelines.
Sanger sequencing
Sanger sequencing was further performed to verify the candidate variants, and cosegregation analysis was performed among family members. The candidate regions were amplified by PCR and sequenced using the ABI PRISM 3730 gene analyzer (Applied Biosystems, California, USA).
Reverse transcriptase polymerase chain reaction
To evaluate whether the candidate variants influence the expression of mRNA, total RNA was isolated from frozen muscle treated with TRIzol Reagent (Life Technologies, California, USA), and single-strand cDNA was prepared using the PrimeScript® RTase Kit (Takara, Otsu, Japan). The candidate region of SCARB2 was amplified and sequenced using the following primers: Forward: 5'-TGACTATGAGAGTGTACAGG-3'; Reverse: 5'-TGGTCTTCCTGATTTGGGTG-3'.
Western blot
Protein was isolated from frozen muscle treated with RIPA and PMSF (Beyotime, Shanghai, China). Thirty microgram of total protein was resolved on 10% SDS-PAGE gels, and the proteins were transferred onto nitrocellulose (NC) membranes. Then, the NC membranes were blocked with 5% nonfat milk in Tris-buffered saline with Tween solution (0.01 mol/L of Tris HCl, 0.15 mol/L NaCl, and 0.1% Tween) and probed with the appropriate primary antibody (mouse anti-LIMP2 antibody, Santa Cruz Biotechnology, USA; mouse anti-glyceraldehyde phosphate dehydrogenase antibody, Beyotime, China). Next, the membranes were incubated with the appropriate peroxidase-labeled mouse anti-goat Ig and developed with enhanced chemiluminescent detection reagents (Beyotime, Shanghai, China).
RESULTS
Clinical features of the family
The proband is a 23-year-old male. He began to experience myoclonic jerks of the upper limbs and shoulders at the age of 21, especially when he felt nervous or fell asleep. Five episodes of generalized tonic–clonic seizures occurred 2 years later without any medication. On examination, his intelligence was normal. There was prominent action myoclonus that involved the upper limbs and shoulders. The other neurologic examination revealed cerebellar features, including dysarthria, a broad-based gait, abnormal heel–knee–tibia test, and finger–nose test. Mild generalized skeletal muscle atrophy without fasciculations and pes cavus was also observed.
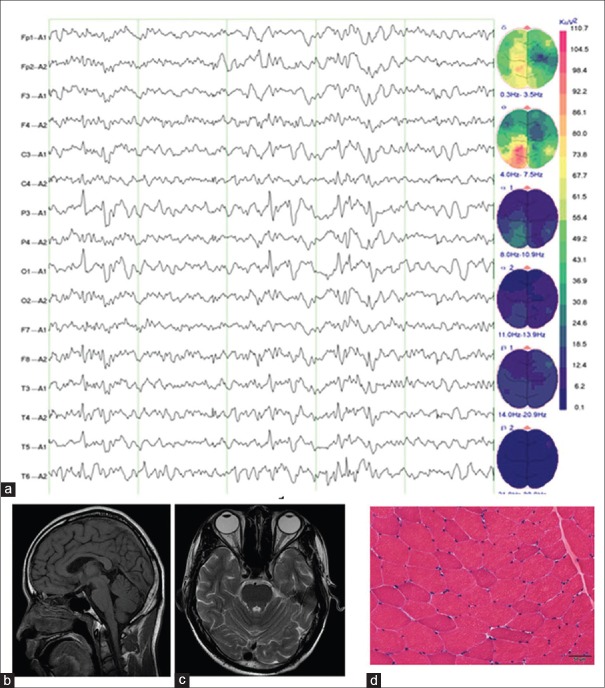
The routine blood biochemical test results, including normal serum blood urea nitrogen, creatinine, and urinalysis, were unremarkable. EEG revealed multifocal spike and wave complexes, especially in the left parietal lobe, occipital lobe, and temporal lobe [Figure 1a]. Brain MRI revealed mild cerebellar atrophy [Figure 1b and 1c]. Muscle biopsy presented with certain muscle atrophy and other less obvious signs, but there were no ragged red fibers [Figure 1d].
Figure 1.
The auxiliary examinations of the proband. (a) EEG revealed multifocal spike and wave complexes, especially in the left parietal lobe, occipital lobe, and temporal lobe. (b) Axial brain MRI revealed mild cerebellar atrophy. (c) Transverse MRI scan revealed mild cerebellar atrophy. (d) Muscle biopsy presented with certain muscle atrophy and other less obvious signs (H and E, original magnification ×100). MRI: Magnetic resonance imaging; EEG: Electroencephalography.
The elder brother of the proband was found to have similar symptoms at the age of 20 years, with myoclonus, ataxia, and generalized tonic-clonic seizures. He started to experience anxiety and myoclonic jerks of the upper limbs and presented with three generalized episodes of tonic-clonic seizures and loss of consciousness in 1 year. After the medication of sodium valproate and lamotrigine, the symptom of epilepsy did not occur. The neurologic examination revealed ataxia and muscle weakness. This patient did not undergo blood biochemical tests, brain MRI, and muscle biopsy.
Dentatorubral–pallidoluysian atrophy CAG trinucleotide repeats analysis
The PCR analysis demonstrated that the number of ATN1 CAG repeats was within the normal range in the proband.
Identification of variants by targeted next-generation sequencing analysis
Targeted NGS was performed in the two patients. The coverage of the fraction of the target base is presented in Supplementary Table 1. The mean coverage of the target bases was 92.1698 (II1) and 111.7527 (II2), respectively. Furthermore, the total SNP variants, including SNPs, noncoding region variants, synonymous mutations, and missense mutations, were 3499 (II1) and 3082 (II2), respectively, while the total initial map of insertion and deletion (INDEL) variants, including insertion and deletion, were 161 (II1) and 180 (II2), respectively. Through the further bioinformatic analysis of these two patients, it was found that these patients harbored a splice-site homozygous mutation in the SCARB2 gene (c.995-1G>A), which was an unreported splicing variant.
Supplementary Table 1.
The coverage of the fraction of target base of II1 and II2
| Items | II1 | II2 |
|---|---|---|
| Total read | 5,637,906 | 6,396,240 |
| Total mapped read | 5,575,859 | 6,311,962 |
| Unique mapped | 5,255,824 | 5,982,202 |
| No-mismatch mapped | 3,509,971 | 4,152,867 |
| Mismatch alignment bases rate | 0.3774 | 0.3507 |
| Reads on target regions | 4,037,140 | 4,883,548 |
| Fraction on target regions | 0.7161 | 0.7635 |
| Fraction on target regions covered by reads | 0.9982 | 0.9987 |
| Unique mapped reads on target regions | 3,832,984 | 4,658,405 |
| No-mismatch reads on target regions | 2,577,212 | 3,232,079 |
| Reads on target ± 150 regions | 4,652,214 | 5,360,719 |
| Fraction on target ± 150 regions | 0.8252 | 0.8381 |
| Fraction on target ± 150 regions covered by reads | 0.996 | 0.9961 |
| Unique mapped reads on target ± 150 regions | 4,407,479 | 5,100,383 |
| No-mismatch reads on target ± 150 regions | 2,937,340 | 3,524,865 |
| Reads on target ± 500 regions | 4,801,824 | 5,411,318 |
| Fraction on target ± 500 regions | 0.8517 | 0.846 |
| Fraction on target ± 500 regions covered by reads | 0.8957 | 0.8004 |
| Unique mapped reads on target ± 500 regions | 4,542,903 | 5,145,900 |
| No-mismatch reads on target ± 500 regions | 3,011,736 | 3,551,922 |
| Fraction of target bases covered | 0.9982 | 0.9987 |
| Fraction of target bases covered with 0~5X | 0.012 | 0.0071 |
| Fraction of target bases covered with 5~10X | 0.0171 | 0.0099 |
| Fraction of target bases covered with 10~15X | 0.0207 | 0.0122 |
| Fraction of target bases covered with 15~20X | 0.0257 | 0.0138 |
| Fraction of target bases covered with 20~25X | 0.0316 | 0.0157 |
| Fraction of target bases covered with 25~30X | 0.0355 | 0.0179 |
| Fraction of target bases covered with 30~35X | 0.0382 | 0.0222 |
| Fraction of target bases covered with 35~40X | 0.0417 | 0.0252 |
| Fraction of target bases covered with 40~45X | 0.0439 | 0.0287 |
| Fraction of target bases covered with 45~50X | 0.0452 | 0.0308 |
| Fraction of target bases covered with >50X | 0.6865 | 0.8152 |
| Mean Coverage of target bases | 92.1698 | 111.7527 |
Sanger sequencing and cosegregation analysis
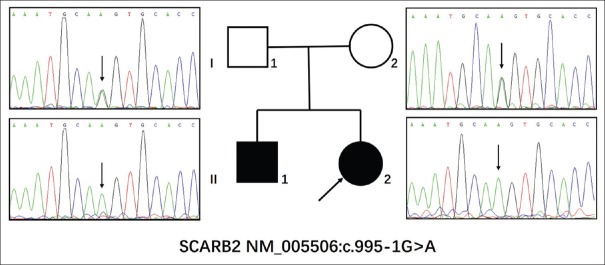
The SCARB2 c.995-1G>A homozygous mutation identified by targeted NGS was further confirmed in the two brothers by Sanger sequencing [Figure 2]. The homozygous mutation was also found in his elder brother, and SCARB2 c.995-1G>A was heterozygous in his unaffected parents [Figure 2]. Meanwhile, the mutation was not detected in the 300 unrelated controls. Therefore, the SCARB2 c.995-1G>A homozygous mutation cosegregated with the PME family.
Figure 2.
Sanger sequencing of the family with progressive myoclonus epilepsies. The two brothers were tested for the homozygous splice mutation (c.995-1G>A) of the SCARB2 gene. Then, their parents were tested for heterozygous mutations of the SCARB2 gene. The arrow indicated the homozygous splice mutation (c.995-1G>A).
SCARB2 gene and protein expression analysis
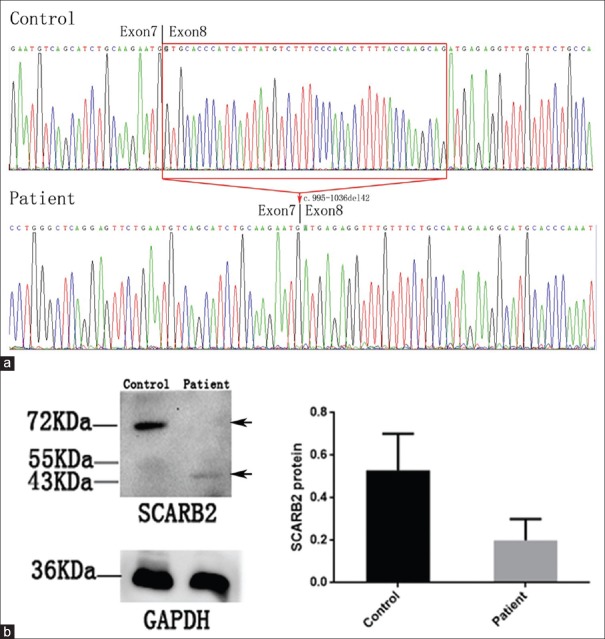
To evaluate the meaning of the SCARB2 c.995-1G>A homozygous mutation, the cDNA and protein levels of SCARB2 were analyzed by RT-PCR and Western blot, respectively. After Sanger sequencing to the cDNA of the SCARB2 gene, as presented in Figure 3a, a c.995-1036del42 mutation was observed in the proband. Furthermore, Western blot was performed to evaluate the expression and quality of the SCARB2 protein. As presented in Figure 3b, a truncated SCARB2 protein is observed, which is accordant with the c.995-1036del42 mutation in the cDNA of patients who harbored the SCARB2 c.995-1G>A homozygous mutation. Compared to the full length of the 72,000 of the SCARB2 protein in healthy controls, the expression of the truncated SCARB2 protein that weighted from 43,000 to 55,000 significantly decreased (t = 2.887, P = 0.0447) [Figure 3b].
Figure 3.
SCARB2 gene expression and protein expression analysis of the proband. (a) Sanger sequencing to the cDNA of the SCARB2 gene: a c. 995-1036del42 mutation was observed in the proband. (b) Western blot analysis of the protein obtained from the muscle of proband and controls. Compared to the full length of the 72,000 of SCARB2 protein in healthy controls, the expression of the truncated SCARB2 protein that weighted from 43,000 to 55,000 significantly decreased (t = 2.887, P = 0.0447). The arrow indicated a 43,000-protein band appeared in patients but a 72,000 band in control.
DISCUSSION
In the present study, targeted NGS technique was applied to screen disease-causative genes in a PME family. A novel SCARB2 splicing homozygous variant c.995-1G>A was identified in this family. According to the standards and guidelines of the ACMG, the variant was classified as a pathogenic variant. Further functional analysis confirmed that the c.995-1G>A variant can lead to the loss function of SCARB2 protein. Therefore, the two brothers were finally diagnostically confirmed with EPM4.
EPM4 is a rare form of PMEs and is an autosomal recessive inherited disorder caused by homozygous mutation and compound heterozygous mutation in the SCARB2 gene.[18,19] Due to the genetic heterogeneity and clinical variability of PMEs, it is often challenging to detect particular gene mutations by depending solely on phenotypes. In clinic, there is a need to establish a molecular diagnostic strategy for the screening of disease-causative genes in PME cases. Targeted NGS is a high-throughput and cost-effective method to screen genomic regions of interest. This approach has been applied for inherited Mendelian disorders.[14,15] Once the panel is established, it can be used for the same genomic region in different cases. In the present study, a panel that included 927 disease-causative genes of neurogenetic diseases was established and used for detecting the culprit genes of PMEs and other neurogenetic diseases. However, the disadvantage of targeted NGS is that the approach cannot accurately detect trinucleotide repeats and copy number variations. Hence, for PME cases, before applying targeted NGS, the CAG trinucleotide repeats of DRPLA needs to be initially screened.
To date, only few EPM4 patients have been recorded worldwide. The correlation of genotypes and phenotypes among EPM4 patients has seldom been summarized in previous literature. The studies conducted by the investigators identified both mutant alleles in the two brothers with EPM4, providing more information to further analyze the clinical features and mutational spectrum of all reported EPM4 patients [Table 1].[18,19,20,21,22,23,24,25,26,27,28,29,30] According to the literature review and the present study, the median age of onset is 20 years (range: 11–52 years) and the median age of death is 30.5 years (range: 23–59 years). All EPM4 patients begin with three typical manifestations: action myoclonus, generalized seizures, and ataxia. However, the median onset age of these three typical manifestations shows little statistical difference: action myoclonus presents at 21 years (range: 14–57 years), generalized seizure presents at 21.5 years (range: 16–63 years), and ataxia presents at 20.5 years (range: 14–58 years). With regard to renal failure, 11 patients suffered from this and developed EPM4 in adolescence (median: 17 years), while the other 17 patients had no renal dysfunction, but presented initial signs at a later age (median: 22 years) (P = 0.033). In addition, other malfunctions were also observed in previous studies: hearing loss occurred in two patients, cognitive decline occurred in two patients, and demyelinating polyneuropathy occurred in four patients.[22,23,24,25] In the present study, it was found that the proband and his brother presented with talipes cavus, which may suggest that they have acquired peripheral neuropathy.
Table 1.
Literature data on the clinical and mutational features of all reported EPM4
| Reference | Case | Mutation type | Exon/intron | Nucleotide mutation | Protein alteration | Location of mutation | Sex | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Balreira et al. Hum Mol Genet[18] | 1 | Homozygote | Exon 4 | c.533G>A | W178X | GBA binding domain | Female | |||
| 2 | Homozygote | Exon 4 | c.533G>A | W178X | GBA binding domain | Female | ||||
| Berkovic et al. Am J Hum Genet[19] | 3 | Homozygote | Intron 10 | c.1239+1G>T | N | CD36 like domain | Female | |||
| Berkovic et al. Am J Hum Genet[19] | 4 | Homozygote | Exon 4 | c.435_436insAG | W146SfsX16 | GBA binding domain | Female | |||
| Berkovic et al. Am J Hum Genet[19] | 5 | Compound heterozygote | Exon 3 | c.296 delA | N99IfsX34 | CD36 like domain | Male | |||
| Intron 5 | c.704+5G>A | N | GBA binding domain | |||||||
| Berkovic et al. Am J Hum Genet[19] | 6 | Homozygote | Exon 7 | c.862C>T | Q288X | GBA binding domain | – | |||
| Dardis et al. Mol Genet Metab[20] | 7 | Homozygote | Exon 8 | c.1087C>A | H363N | CD36 like domain | Female | |||
| Dibbens et al. Ann Neurol[21] | 8 | Homozygote | Intron 8 | c.1116-2A>C | N | CD36 like domain | Male | |||
| Dibbens et al. Ann Neurol[21] | 9 | Homozygote | Intron 5 | c.704+1G>C | N | GBA binding domain | Male | |||
| Dibbens et al. Ann Neurol[21] | 10 | Homozygote | Exon 11 | c.1258delG | E420RfsX5 | CD36 like domain | Female | |||
| Dibbens et al. Ann Neurol[21] | 11 | Homozygote | Exon 5 | c.666delCCTTA | Y222X | GBA binding domain | Female | |||
| Dibbens et al. Ann Neurol[21] | 12 | Compound heterozygote | Intron 3 | c.424-2A>C | N | CD36 like domain | Female | |||
| Exon 8 | c.1087C>A | H363N | CD36 like domain | |||||||
| Dibbens et al. Arch Neurol[22] | 13 | Compound heterozygote | Exon 7 | c.862C>T | Q288X | GBA binding domain | Male | |||
| Intron 9 | c.1187+3insT | N | CD36 like domain | |||||||
| Hopfner et al. BMC Neurol[23] | 14 | Homozygote | Exon 1 | c.111delC | I37MfsX7 | CD36 like domain | Male | |||
| 15 | Homozygote | Exon 1 | c.111delC | I37MfsX7 | CD36 like domain | Male | ||||
| 16 | Homozygote | Exon 1 | c.111delC | I37MfsX7 | CD36 like domain | Female | ||||
| Perandones et al. Mov Disord[24] | 17 | Homozygote | Intron 5 | c.704+1G>A | N | GBA binding domain | Female | |||
| Guerrero-López et al. Mov Disord[28] | 18 | Homozygote | Exon 8 | c.1015insT | F339FfsX9 | CD36 like domain | Female | |||
| Higashiyama et al. Mov Disord[29] | 19 | Homozygote | Exon 11 | c.1385_1390del6insATGCATGCACC | G462DfsX34 | TM domain | Female | |||
| 20 | Homozygote | Exon 11 | c.1385_1390del6insATGCATGCACC | G462DfsX34 | TM domain | Male | ||||
| Fu et al. Neuropathol Appl Neurobiol[25] | 21 | Homozygote | Exon 11 | c.1385_1390del6insATGCATGCACC | G462DfsX34 | TM domain | Male | |||
| Fu et al. Neuropathol Appl Neurobiol[25] | 22 | Homozygote | Exon 3 | c.361C>T | R121X | CD36 like domain | Female | |||
| Zeigler et al. J Neurol Sci[27] | 23 | Homozygote | Exon 11 | c.1270C>T | R424X | CD36 like domain | Male | |||
| 24 | Homozygote | Exon 11 | c.1270C>T | R424X | CD36 like domain | Female | ||||
| He et al. Clin Genet[26] | 25 | Homozygote | Exon 11 | c.1270C>T | R424X | CD36 like domain | Female | |||
| 26 | Homozygote | Exon 11 | c.1270C>T | R424X | CD36 like domain | Female | ||||
| This study | 27 | Homozygote | Exon 7 | c.995-1G>A | N | CD36 like domain | Male | |||
| 28 | Homozygote | Exon 7 | c.995-1G>A | N | CD36 like domain | Male | ||||
| Reference | Age of onset (years) | Age at death (years) | Action myoclonus (years) | Tonic clonic seizures (years) | Ataxia (years) | Renal failure (years) | Phenotype | Other specific phenotype | ||
| Balreira et al. Hum Mol Genet[18] | 15 | 23 | 15 | – | 18 | 18 | With renal failure | N | ||
| 17 | 26 | 15 | – | 17 | 21 | With renal failure | N | |||
| Berkovic et al. Am J Hum Genet[19] | 11 | – | – | – | – | – | With renal failure | N | ||
| Berkovic et al. Am J Hum Genet[19] | – | – | – | – | – | – | With renal failure | N | ||
| Berkovic et al. Am J Hum Genet[19] | 11 | – | – | – | – | – | With renal failure | N | ||
| Berkovic et al. Am J Hum Genet[19] | – | – | – | – | – | – | With renal failure | N | ||
| Dardis et al. Mol Genet Metab[20] | 26 | – | 26 | 27 | 26 | N | Without renal failure | N | ||
| Dibbens et al. Ann Neurol[21] | 14 | 29 | 14 | 17 | 17 | N | Without renal failure | N | ||
| Dibbens et al. Ann Neurol[21] | 15 | 27 | 15 | 16 | 16 | N | Without renal failure | N | ||
| Dibbens et al. Ann Neurol[21] | 23 | 33 | 23 | 23 | 24 | N | Without renal failure | N | ||
| Dibbens et al. Ann Neurol[21] | 25 | 40 | 25 | 28 | 31 | N | Without renal failure | N | ||
| Dibbens et al. Ann Neurol[21] | 26 | 32 | 26 | 26 | 27.5 | N | Without renal failure | N | ||
| Dibbens et al. Arch Neurol[22] | 16 | – | 16 | 20 | 20 | N | Without renal failure | Demyelinating polyneuropathy | ||
| Hopfner et al. BMC Neurol[23] | 14 | 31 | 14 | 20 | 14 | – | With renal failure | Demyelinating polyneuropathy | ||
| 20 | 38 | 26 | 32 | 20 | – | With renal failure | Hearing loss, demyelinating polyneuropathy | |||
| 20 | 34 | 20 | 20 | 20 | – | With renal failure | Demyelinating polyneuropathy | |||
| Perandones et al. Mov Disord[24] | 21 | – | 23 | 25 | 21 | 25 | With renal failure | Hearing loss | ||
| Guerrero-López et al. Mov Disord[28] | 22 | – | 22 | 22 | 30 | N | Without renal failure | N | ||
| Higashiyama et al. Mov Disord[29] | 43 | – | 43 | 58 | 58 | N | Without renal failure | N | ||
| 52 | – | 57 | 63 | 52 | N | Without renal failure | Acute ischemic stroke | |||
| Fu et al. Neuropathol Appl Neurobiol[25] | 45 | 59 | 48 | – | 51 | N | Without renal failure | Dementia | ||
| Fu et al. Neuropathol Appl Neurobiol[25] | 20 | 28 | 20 | 20 | 20 | N | Without renal failure | Cognitive decline | ||
| Zeigler et al. J Neurol Sci[27] | 17 | 30 | 17 | 17 | 17 | 29 | With renal failure | N | ||
| 17 | 27 | 17 | 17 | 17 | N | Without renal failure | ||||
| He et al. Clin Genet[26] | 21 | – | 21 | 25 | 22 | N | Without renal failure | N | ||
| 27 | – | 27 | N | 27 | N | Without renal failure | ||||
| This study | 21 | – | 21 | 21 | 21 | N | Without renal failure | Talipes cavus | ||
| 20 | – | 20 | 20 | 20 | N | Without renal failure | Talipes cavus | |||
–: Not mention in the literature; N: Do not have the symptom; GBA: Beta-glucocerebrosidase; TM: Transmembrane.
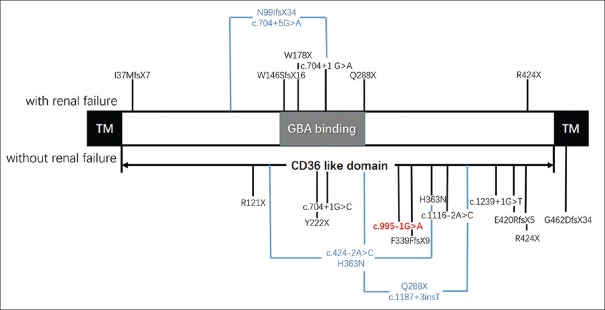
The SCARB2 gene encodes SCARB2 protein in humans and is also known as lysosomal integral membrane protein type-2 (LIMP2), which is a disease causative of EPM4, and is associated with Parkinson's disease.[18,19,31,32] LIMP2, which is a nonspecifically expressed transmembrane (TM) protein, is mainly located in lysosomes and endosomes.[33] While bounding with beta-glucocerebrosidase (GBA), LIMP2 can transfer GBA from the endoplasmic reticulum to the lysosome.[34,35,36,37,38,39] SCARB2 protein comprises two TM domains and one CD36-like domain, which contains a highly conserved coiled-coil domain (residue: 145-288 aa) that binds GBA [Figure 4].[35,36,37] Mutations in the SCARB2 gene result in decreasing and anomaly location of SCARB2 protein, affecting the combination of SCARB and GBA.
Figure 4.
The structure of the SCARB2 gene and the reported mutation of SCARB2. GBA: Beta-glucocerebrosidase; TM: Transmembrane.
The genotype and phenotype correlation of all reported EPM4 patients and the two brothers, including the 19 SCARB2 gene mutations in 28 EPM4 patients, is summarized in the present study [Table 1 and Figure 4].[18,19,20,21,22,23,24,25,26,27,28,29,30] Among these 19 mutations, 16 mutations (84.21%) were homozygous mutations, while only 3 mutations were compound heterozygous. The mutation types of SCARB2 were nonsense, frameshift, and splice-site mutations, which can be assumed to disrupt gene function, leading to the complete absence of the gene product through the lack of transcription or nonsense-mediated decay of the altered transcript. For the phenotype with renal failure, seven mutations were found in 11 patients, of which the four mutations located in the GBA-binding domain may directly disrupt SCARB2 bounding with GBA. Merely two mutations were not located in the GBA-binding domain. Although the I37MfsX7 homozygous mutation was located in this domain, it was a frameshift mutation that may produce a truncated protein with no GBA-binding domain. For the phenotype without renal failure, there were 11 homozygous mutations. Among these, eight mutations were located after the GBA-binding domain, which may not affect the combination of GBA and SCARB2, while two mutations were compound heterozygous mutations. Merely one mutation was located in the TM domain, which led to a late-onset phenotype.[25,29] These data possibly reveal that the functional domains of the SCARB2 gene are associated with the EPM4 phenotype. Interestingly, there was a marked variability of clinical features between these two patients in EPM4 families (family 17) with the same nucleotide position (R424X), and the variability was also found in family 14 (c.704+1G>A) though follow-up studies.[27,30] Hence, it could be speculated that the phenotypic difference may be due to genetic modifiers or environmental factors. Therefore, more data need to be acquired before a specific phenotype–genotype correlation could be determined.
In conclusion, the present study reported two EPM4 brothers with a novel splice mutation in the SCARB2 gene detected by targeted NGS analysis. A literature review revealed that pathogenic mutations of the SCARB2 gene in EPM4 patients are homozygous mutations rather than compound heterozygous mutations. The mutations in the different functional domains of SCARB2 appear to be associated with the phenotype of EPM4.
Supplementary information is linked to the online version of the paper on the Chinese Medical Journal website.
Financial support and sponsorship
This work was supported by the grants from the National Natural Science Foundation of China (No. U1505222, No. 81322017, No. 81500980, and No. 81571100) and the National Key Clinical Specialty Discipline Construction Program and Key Clinical Specialty Discipline Construction Program of Fujian.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
The authors sincerely thank the families that have contributed samples for the purposes of this study.
Footnotes
Edited by: Ning-Ning Wang
REFERENCES
- 1.Kälviäinen R. Progressive myoclonus epilepsies. Semin Neurol. 2015;35:293–9. doi: 10.1055/s-0035-1552620. doi: 10.1055/s-0035-1552620. [DOI] [PubMed] [Google Scholar]
- 2.Pennacchio LA, Lehesjoki AE, Stone NE, Willour VL, Virtaneva K, Miao J, et al. Mutations in the gene encoding cystatin B in progressive myoclonus epilepsy (EPM1) Science. 1996;271:1731–4. doi: 10.1126/science.271.5256.1731. doi: 101126/science27152561731. [DOI] [PubMed] [Google Scholar]
- 3.Gómez-Garre P, Sanz Y, Rodríguez De Córdoba SR, Serratosa JM. Mutational spectrum of the EPM2A gene in progressive myoclonus epilepsy of Lafora: High degree of allelic heterogeneity and prevalence of deletions. Eur J Hum Genet. 2000;8:946–54. doi: 10.1038/sj.ejhg.5200571. doi: 10.1038/sj.ejhg.5200571. [DOI] [PubMed] [Google Scholar]
- 4.Gómez-Abad C, Gómez-Garre P, Gutiérrez-Delicado E, Saygi S, Michelucci R, Tassinari CA, et al. Lafora disease due to EPM2B mutations: A clinical and genetic study. Neurology. 2005;64:982–6. doi: 10.1212/01.WNL.0000154519.10805.F7. doi: 10.1212/01.WNL.0000154519.10805.F7. [DOI] [PubMed] [Google Scholar]
- 5.Berkovic SF, Mazarib A, Walid S, Neufeld MY, Manelis J, Nevo Y, et al. A new clinical and molecular form of Unverricht-Lundborg disease localized by homozygosity mapping. Brain. 2005;128:652–8. doi: 10.1093/brain/awh377. doi: 10.1093/brain/awh377. [DOI] [PubMed] [Google Scholar]
- 6.Singh S, Suzuki T, Uchiyama A, Kumada S, Moriyama N, Hirose S, et al. Mutations in the NHLRC1 gene are the common cause for Lafora disease in the Japanese population. J Hum Genet. 2005;50:347–52. doi: 10.1007/s10038-005-0263-7. doi: 10.1007/s10038-005-0263-7. [DOI] [PubMed] [Google Scholar]
- 7.Ferlazzo E, Italiano D, An I, Calarese T, Laguitton V, Bramanti P, et al. Description of a family with a novel progressive myoclonus epilepsy and cognitive impairment. Mov Disord. 2009;24:1016–22. doi: 10.1002/mds.22489. doi: 10.1002/mds.22489. [DOI] [PubMed] [Google Scholar]
- 8.Corbett MA, Schwake M, Bahlo M, Dibbens LM, Lin M, Gandolfo LC, et al. A mutation in the Golgi qb-SNARE gene GOSR2 causes progressive myoclonus epilepsy with early ataxia. Am J Hum Genet. 2011;88:657–63. doi: 10.1016/j.ajhg.2011.04.011. doi: 10.1016/j.ajhg.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbull J, Girard JM, Lohi H, Chan EM, Wang P, Tiberia E, et al. Early-onset Lafora body disease. Brain. 2012;135:2684–98. doi: 10.1093/brain/aws205. doi: 10.1093/brain/aws205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salar S, Yeni N, Gündüz A, Güler A, Gökçay A, Velioğlu S, et al. Four novel and two recurrent NHLRC1 (EPM2B) and EPM2A gene mutations leading to Lafora disease in six Turkish families. Epilepsy Res. 2012;98:273–6. doi: 10.1016/j.eplepsyres.2011.09.020. doi: 10.1016/j.eplepsyres.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 11.Kousi M, Anttila V, Schulz A, Calafato S, Jakkula E, Riesch E, et al. Novel mutations consolidate KCTD7 as a progressive myoclonus epilepsy gene. J Med Genet. 2012;49:391–9. doi: 10.1136/jmedgenet-2012-100859. doi: 10.1136/jmedgenet-2012-100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damiano JA, Afawi Z, Bahlo M, Mauermann M, Misk A, Arsov T, et al. Mutation of the nuclear lamin gene LMNB2 in progressive myoclonus epilepsy with early ataxia. Hum Mol Genet. 2015;24:4483–90. doi: 10.1093/hmg/ddv171. doi: 10.1093/hmg/ddv171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muona M, Berkovic SF, Dibbens LM, Oliver KL, Maljevic S, Bayly MA, et al. A recurrent de novo mutation in KCNC1 causes progressive myoclonus epilepsy. Nat Genet. 2015;47:39–46. doi: 10.1038/ng.3144. doi: 10.1038/ng.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu ZJ, Li HF, Tan GH, Tao QQ, Ni W, Cheng XW, et al. Identify mutation in amyotrophic lateral sclerosis cases using HaloPlex target enrichment system. Neurobiol Aging. 2014;35:2881e11–2881e15. doi: 10.1016/j.neurobiolaging.2014.07.003. doi: 10.1016/j.neurobiolaging.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Li LX, Zhao SY, Liu ZJ, Ni W, Li HF, Xiao BG, et al. Improving molecular diagnosis of Chinese patients with Charcot-Marie-tooth by targeted next-generation sequencing and functional analysis. Oncotarget. 2016;7:27655–64. doi: 10.18632/oncotarget.8377. doi: 10.18632/oncotarget.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang XY, Yu JB, Yang D, Han CT, Lin WH. Late-onset juvenile myoclonic epilepsy or frontal lobe epilepsy with myoclonus. Chin Med J. 2016;129:2508–9. doi: 10.4103/0366-6999.191829. doi: 10.4103/03666999.191829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koide R, Ikeuchi T, Onodera O, Tanaka H, Igarashi S, Endo K, et al. Unstable expansion of CAG repeat in hereditary dentatorubral-pallidoluysian atrophy (DRPLA) Nat Genet. 1994;6:9–13. doi: 10.1038/ng0194-9. doi: 10.1038/ng0194-9. [DOI] [PubMed] [Google Scholar]
- 18.Balreira A, Gaspar P, Caiola D, Chaves J, Beirão I, Lima JL, et al. A nonsense mutation in the LIMP-2 gene associated with progressive myoclonic epilepsy and nephrotic syndrome. Hum Mol Genet. 2008;17:2238–43. doi: 10.1093/hmg/ddn124. doi: 10.1093/hmg/ddn124. [DOI] [PubMed] [Google Scholar]
- 19.Berkovic SF, Dibbens LM, Oshlack A, Silver JD, Katerelos M, Vears DF, et al. Array-based gene discovery with three unrelated subjects shows SCARB2/LIMP-2 deficiency causes myoclonus epilepsy and glomerulosclerosis. Am J Hum Genet. 2008;82:673–84. doi: 10.1016/j.ajhg.2007.12.019. doi: 10.1016/j.ajhg.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dardis A, Filocamo M, Grossi S, Ciana G, Franceschetti S, Dominissini S, et al. Biochemical and molecular findings in a patient with myoclonic epilepsy due to a mistarget of the beta-glucosidase enzyme. Mol Genet Metab. 2009;97:309–11. doi: 10.1016/j.ymgme.2009.04.011. doi: 10.1016/j.ymgme.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Dibbens LM, Michelucci R, Gambardella A, Andermann F, Rubboli G, Bayly MA, et al. SCARB2 mutations in progressive myoclonus epilepsy (PME) without renal failure. Ann Neurol. 2009;66:532–6. doi: 10.1002/ana.21765. doi: 10.1002/ana.21765. [DOI] [PubMed] [Google Scholar]
- 22.Dibbens LM, Karakis I, Bayly MA, Costello DJ, Cole AJ, Berkovic SF, et al. Mutation of SCARB2 in a patient with progressive myoclonus epilepsy and demyelinating peripheral neuropathy. Arch Neurol. 2011;68:812–3. doi: 10.1001/archneurol.2011.120. doi: 10.1001/archneurol.2011.120. [DOI] [PubMed] [Google Scholar]
- 23.Hopfner F, Schormair B, Knauf F, Berthele A, Tölle TR, Baron R, et al. Novel SCARB2 mutation in action myoclonus-renal failure syndrome and evaluation of SCARB2 mutations in isolated AMRF features. BMC Neurol. 2011;11:134. doi: 10.1186/1471-2377-11-134. doi: 10.1186/1471-2377-11- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perandones C, Micheli FE, Pellene LA, Bayly MA, Berkovic SF, Dibbens LM, et al. A case of severe hearing loss in action myoclonus renal failure syndrome resulting from mutation in SCARB2. Mov Disord. 2012;27:1200–1. doi: 10.1002/mds.25083. doi: 10.1002/mds.25083. [DOI] [PubMed] [Google Scholar]
- 25.Fu YJ, Aida I, Tada M, Tada M, Toyoshima Y, Takeda S, et al. Progressive myoclonus epilepsy: Extraneuronal brown pigment deposition and system neurodegeneration in the brains of japanese patients with novel SCARB2 mutations. Neuropathol Appl Neurobiol. 2014;40:551–63. doi: 10.1111/nan.12057. doi: 10.1111/nan.12057. [DOI] [PubMed] [Google Scholar]
- 26.He M, Tang BS, Li N, Mao X, Li J, Zhang JG, et al. Using a combination of whole-exome sequencing and homozygosity mapping to identify a novel mutation of SCARB2. Clin Genet. 2014;86:598–600. doi: 10.1111/cge.12338. doi: 10.1111/cge.12338. [DOI] [PubMed] [Google Scholar]
- 27.Zeigler M, Meiner V, Newman JP, Steiner-Birmanns B, Bargal R, Sury V, et al. A novel SCARB2 mutation in progressive myoclonus epilepsy indicated by reduced β-glucocerebrosidase activity. J Neurol Sci. 2014;339:210–3. doi: 10.1016/j.jns.2014.01.022. doi: 10.1016/j.jns.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 28.Guerrero-López R, García-Ruiz PJ, Giráldez BG, Durán-Herrera C, Querol-Pascual MR, Ramírez-Moreno JM, et al. A new SCARB2 mutation in a patient with progressive myoclonus ataxia without renal failure. Mov Disord. 2012;27:1826–7. doi: 10.1002/mds.25114. doi: 10.1002/mds.25114. [DOI] [PubMed] [Google Scholar]
- 29.Higashiyama Y, Doi H, Wakabayashi M, Tsurusaki Y, Miyake N, Saitsu H, et al. A novel SCARB2 mutation causing late-onset progressive myoclonus epilepsy. Mov Disord. 2013;28:552–3. doi: 10.1002/mds.25296. doi: 10.1002/mds.25296. [DOI] [PubMed] [Google Scholar]
- 30.Perandones C, Pellene LA, Micheli F. A new SCARB2 mutation in a patient with progressive myoclonus ataxia without renal failure. Mov Disord. 2014;29:158–9. doi: 10.1002/mds.25738. doi: 10.1002/mds.25738. [DOI] [PubMed] [Google Scholar]
- 31.Do CB, Tung JY, Dorfman E, Kiefer AK, Drabant EM, Francke U, et al. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson's disease. PLoS Genet. 2011;7:e1002141. doi: 10.1371/journal.pgen.1002141. doi: 10.1371/journal.pgen.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michelakakis H, Xiromerisiou G, Dardiotis E, Bozi M, Vassilatis D, Kountra PM, et al. Evidence of an association between the scavenger receptor class B member 2 gene and Parkinson's disease. Mov Disord. 2012;27:400–5. doi: 10.1002/mds.24886. doi: 10.1002/mds.24886. [DOI] [PubMed] [Google Scholar]
- 33.Kuronita T, Eskelinen EL, Fujita H, Saftig P, Himeno M, Tanaka Y. A role for the lysosomal membrane protein LGP85 in the biogenesis and maintenance of endosomal and lysosomal morphology. J Neurol Sci. 2002;115(Pt 21):4117–31. doi: 10.1242/jcs.00075. doi: 10.1126/science.271.5256.1731. [DOI] [PubMed] [Google Scholar]
- 34.Rothaug M, Zunke F, Mazzulli JR, Schweizer M, Altmeppen H, Lüllmann-Rauch R, et al. LIMP-2 expression is critical for β-glucocerebrosidase activity and α-synuclein clearance. Proc Natl Acad Sci U S A. 2014;111:15573–8. doi: 10.1073/pnas.1405700111. doi: 10.1073/pnas.1405700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reczek D, Schwake M, Schröder J, Hughes H, Blanz J, Jin X, et al. LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of beta-glucocerebrosidase. Cell. 2007;131:770–83. doi: 10.1016/j.cell.2007.10.018. doi: 10.1016/j.cell.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 36.Blanz J, Groth J, Zachos C, Wehling C, Saftig P, Schwake M, et al. Disease-causing mutations within the lysosomal integral membrane protein type 2 (LIMP-2) reveal the nature of binding to its ligand beta-glucocerebrosidase. Hum Mol Genet. 2010;19:563–72. doi: 10.1093/hmg/ddp523. doi: 10.1093/hmg/ddp523. [DOI] [PubMed] [Google Scholar]
- 37.Neculai D, Schwake M, Ravichandran M, Zunke F, Collins RF, Peters J, et al. Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36. Nature. 2013;504:172–6. doi: 10.1038/nature12684. doi: 10.1038/nature12684. [DOI] [PubMed] [Google Scholar]
- 38.Liou B, Haffey WD, Greis KD, Grabowski GA. The LIMP-2/SCARB2 binding motif on acid β-glucosidase: basic and applied implications for Gaucher disease and associated neurodegenerative diseases. J Biol Chem. 2014;289:30063–74. doi: 10.1074/jbc.M114.593616. doi:10.1074/jbc.M114.593616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malini E, Zampieri S, Deganuto M, Romanello M, Sechi A, Bembi B, et al. Role of LIMP-2 in the intracellular trafficking of β-glucosidase in different human cellular models. FASEB J. 2015;29:3839–52. doi: 10.1096/fj.15-271148. doi: 10.1096/fj.15-271148. [DOI] [PubMed] [Google Scholar]