Abstract
Technological advances have led to the identification of biomarkers and development of novel target‐based therapies. While some novel therapies have improved patient outcomes, the prevalence and diversity of biomarkers and targets in patient populations, especially patients with cancer, has created a challenge for the design and performance of clinical trials. To address this challenge we propose that prospective cohort surveillance of patients may be a solution to promote clinical trial matching for patients in need.
CHALLENGES FACING THE DESIGN AND PERFORMANCE OF PRECISION‐BASED CLINICAL TRIALS
A greater understanding of the molecular biology and complexity of cancer has led to the discovery of new biomarkers that may predict response to novel target‐based therapies. Target‐based therapies add a new dimension of precision care by treating cancer patients who are known to express specific biomarkers predictive of increased likelihood of response, thereby creating hope and optimism for improved patient outcome. Furthermore, patients with disease originating from the same tissue can actually be further characterized into subcohorts based on the differential presence or expression of unique prognostic or predictive biomarkers.
Identifying patients within specific biomarker‐defined subcohorts is a major challenge in performing biomarker target‐based clinical trials. Most often, at the time a patient is in need of a clinical trial their biomarker status is not known. In addition, depending on the prevalence of a given marker, many patients may need to be screened in order to find sufficient numbers of patients who are eligible for a given trial. The current system of screening patients for trials and enrolling them in biomarker‐driven trials is often not adequate to complete the trial in a timely manner and creates unmet expectations for patients seeking trial enrollment—often when only a very few are biomarker‐eligible. In addition, the present system further increases the time and cost of conducting clinical trials. A new paradigm for clinical trial design and conducting clinical trials must be developed in order to deliver target‐based therapies.
One approach to address these challenges is to consent patients to observational studies and create patient and tumor registries that can be accessed to prescreen patient populations to identify those who are phenotypically and genotypically eligible for target‐based clinical trials. This type of approach has the potential of quickly determining the prevalence of biomarkers and targets across different patient populations, designing trials based on prevalence of targets, and efficiently matching patients to clinical trials.
CREATING NETWORKS FOR DATA SHARING AND COLLABORATIVE LEARNING TO ACCELERATE DEVELOPMENT OF NEW THERAPIES
In recent years, alliances of multiple stakeholders involved in the discovery, development, and delivery of new therapies have formed networks that pursue a common mission, including finding approaches for getting new therapies to patients faster and generating evidence of value.1 Collaborations between multiple stakeholders, including academic research centers, healthcare systems, pharma, and patient advocacy groups have emerged to create a “precompetitive space” to support data and tissue procurement. Data sharing is an important element of these networks, exemplified by ORIEN,2 TAPUR,3 GENIE,4 WIN,5 and APOLLO.6 These and other networks that support data sharing and collaboration represent new models to advance personalized cancer therapy trials. Alliances formed by patient advocacy groups, such as the Multiple Myeloma Research Foundation, have organized multiple stakeholders, including healthcare systems and pharmaceutical companies, to design and implement target‐based clinical trials for myeloma patients with an emphasis on improving patient access to clinical trials.7
One network that has integrated prospective patient cohort surveillance as an approach to improve design and performance of personalized cancer therapy trials is ORIEN (Oncology Research Information Exchange Network; http://www.oriencancer.org). ORIEN is comprised of multiple cancer centers that have agreed to use the same Institutional Review Board (IRB)‐approved protocol and consent (Total Cancer Care Protocol, TCC) to follow patients throughout their lifetime. Patients consent to donate medical records and tissue specimens for molecular profiling. Most patients consent to the protocol at the time of diagnosis, but may be consented at any time in the course of their disease. Patients understand that deidentified data will be shared with researchers from both academia and the pharmaceutical industry for the purpose of collaborative research and matching patients to clinical trials. Patients may withdraw from the study at any time. In the process of matching patients to clinical trials, all clinical trials available, both investigator‐initiated and pharma sponsored, are considered.
Supplementary Text 1 is available that further details the TCC Protocol. The clinical data collection is harmonized through data standards and common data dictionaries among all participating centers. The acquisition, processing, storage, and release of human tissue are also uniform across all sites, as they all follow the same policies and procedures.
A hybrid information network system, consisting of both a federated data management system and a centralized data warehouse, has been designed to facilitate the distribution of data from multiple sources and to accommodate the stakeholder's data use requirements.8 Stakeholders include academic researchers, clinicians, administrators, pharmaceutical companies, and patients themselves. A major focus of the harmonized clinical data is to address the needs of patients with disease progression who will likely benefit from the opportunity to participate in clinical trials of novel target‐based therapies or therapeutic regimens. Importantly, as part of the TCC consent, patients allow themselves to be contacted and notified if they become eligible for a clinical trial. The goal of this approach is to anticipate clinical trial options for patients and, when needed, to determine the “best” trial options for each patient. Figure 1 illustrates the concept of prospective cohort surveillance by forming in silico communities of patients consented to TCC and how these data can be used to enrich patient populations for specific target‐based clinical trials. The ORIEN approach is summarized here:
Consent patients to the TCC protocol across multiple clinical centers.
Collect longitudinal clinical data over the course of the patient's care.
Profile patient tumor‐derived samples using standardized molecular tests.
Store patient information in a data warehouse.
Screen patient data using inclusion/exclusion criteria of clinical trials.
Assign patients to in silico communities based on similar phenotypic and genotypic characteristics.
Notify the patient's physician of study options available.
Figure 1.
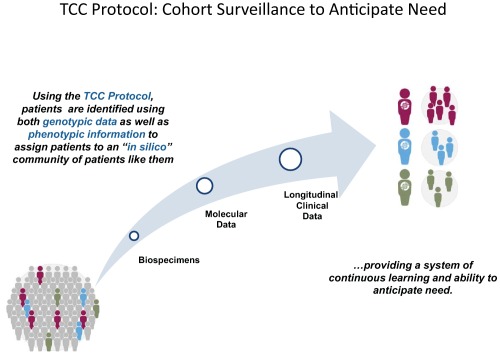
TCC protocol: Cohort surveillance to anticipate need.
The ORIEN system is a self‐governed and federated alliance of cancer centers. Each center participating in ORIEN may, or may not, choose to share data for any given project. The protocol allows for patient data to be submitted to the coordinating center (M2Gen) for data aggregation and trial matching. M2Gen (http://www.M2Gen.com) is a for‐profit enterprise owned primarily by the H. Lee Moffitt Cancer Center. The implementation and funding of the TCC protocol at each of the ORIEN member institutions is a combination of self‐funding and support from M2Gen.
Supplementary Text 2 is available, which includes a partial list of publications that have utilized the TCC protocol for new biomarker discovery, clinical trial matching, and population studies examining treatment effectiveness and risk. These publications also describe quality assurance and control methods that were used to examine clinical and molecular data derived from the TCC protocol.
A FEASIBILITY STUDY USING THE TCC PROTOCOL TO ENRICH THE PATIENT POPULATION FOR A TARGET‐BASED CLINICAL TRIAL
Recently, a study was published to examine the feasibility of using the TCC Protocol to enrich for patients to be tested for CD‐30‐positive immunohistochemical testing of solid tumors prior to patient enrollment into a brentuximab vedotin target‐based clinical trial sponsored by Takeda Pharmaceutical.9 The TCC Data Warehouse includes data from multiple sources, including cancer registry data, electronic medical record data, tissue data, consent data, and molecular data. An analytic tool allows investigators to identify groups of patients based on a set of parameters, including patient inclusion/exclusion criteria and molecular signatures that define the patient population to be screened. At the time of study, 8,307 patients with gene expression data across four tumor types of interest were stratified from low to high gene expression using a global Z‐score analysis. Banked tumor tissue samples were used to determine CD30 protein expression by semiquantitative immunohistochemistry. Statistical comparisons of Z‐ and H‐scores demonstrated that patients with a high level of CD30 gene expression significantly enriched for companion diagnostic immunohistochemical detection of CD30‐positive protein expression in breast, lung, skin, and ovarian cancer. This study demonstrated that patient surveillance that includes molecular analysis may improve clinical trial design and screening efficiency for enrolling patients into biomarker‐based clinical trials.
ADDRESSING CHALLENGES IN USING PROSPECTIVE COHORT SURVEILLANCE TO IMPROVE CLINICAL TRIAL DESIGN AND PATIENT ENROLLMENT
There are numerous challenges in developing a network of multiple stakeholders to follow patients throughout their lifetime and using the derived data to predict patient need, especially the need for a clinical trial. Rodon et al. described in broad terms the challenges of an international consortium (Worldwide Innovative Network, WIN) conducting personalized cancer therapy trials.10 Table 1 enumerates some of the challenges faced by ORIEN in developing prospective patient cohorts in anticipation of the need for assignment of patients to precision‐based clinical trials. Perhaps the greatest challenge is the creation of data standards that will allow for comparison of information generated from multiple sources, and the various networks being formed to study patient outcomes. Development of integrated data systems that promote data sharing in real time, and collaborative learning among all stakeholders, is required to improve clinical trial design and matching patients to target‐based clinical trials. Ultimately, design of the system to enrich patient populations expressing treatment targets should include patient consent for performance of an array of laboratory analyses, comprehensive and timely data aggregation, and interoperable data analytics to design patient cohorts required for matching patients to target‐based clinical trials.
Table 1.
Challenges associated with forming prospective patient cohorts to improve precision based clinical trials
| Challenges | Potential solutions |
|---|---|
| 1. Defining a patient's phenotype relies on accurate and standardized interpretation of unstructured data from diverse health records. Health records are primarily unstructured, making difficult the identification and transference of data that is necessary to match patients to clinical trials. | 1. Development of common data dictionaries and automated natural language processing (NLP) technologies are needed. |
| 2. Patient data must be secure, yet shared to achieve collaborative learning. Access and use of the data donated by patients requires a secure environment with sound governance to assure the data are being used in the patients' best interest. | 2. Use of honest brokers, limited datasets and deidentification allow compliance with HIPAA and FISMA while not overly restricting data aggregation and analysis. It will be important that regulations not be overly restrictive, making it difficult to access and aggregate data for analysis. |
| 3. Sound scientific oversight and measurements of quality of the data need to be in place to identify and recommend clinical trials for patients in need. | 3. Data quality standards must be established and automated so that data from disparate sources can be integrated for analysis and decision making. |
| 4. Communicating with patients who have consented to cohort surveillance and providing information to them in a meaningful and constructive way. | 4. Patients who consent to donate data and biospecimens may request results and reports of studies performed and this must be communicated in an understandable format with access to counseling to explain findings. |
| 5. Recognizing that patients' cancers genotypically evolve and are heterogeneous, how do we determine the current genomic state of a patient at the time of clinical trial enrollment? | 5. Performance of longitudinal assays, including liquid biopsies, may address the genotypic evolution of a patient's disease. Creating in silico communities based on genotype and phenotype with longitudinal clinical follow‐up will become the basis for deep learning and pattern recognition to predict events. |
| 6. With advances in technology how do we integrate new “‐omic” analysis into the repertoire of studies of patients to better understand the disease? | 6. Integrating new “‐omic” technologies will enhance systems analysis but will also require prospective validation of new assays to determine the value of each. |
| 7. Creation of a national (global) infrastructure to share data from all networks involved in collecting and studying patient data will enable all stakeholders to access and learn from the data to better meet patient needs, including need for clinical trials. | 7. Development of a “network of networks” will require the development of data standards and tools that promote interoperability. |
FUNDING
Research described is partially funded by Takeda Pharmaceutical Company.
CONFLICT OF INTEREST
William Dalton is employed by M2Gen as Executive Chair of the board. Jeffrey Ecsedy is employed by Takeda Pharmaceutical Company.
Supporting information
Supplementary Text 1‐4: The Total Cancer Care Protocol, Publications using the TCC Protocol describing quality assessment and control methods of data generated, Governance and funding of ORIEN, Challenges associated with implementing the TCC Protocol, and gaining patient input through formation of the Patient Advisory Council
Figure S1: Total Cancer Care Protocol: A Partnership with Patients
Figure S2: ORIEN Governance Structure: A Self‐Governed Alliance of Cancer Centers
References
- 1. Fiore, L.D. et al Data sharing, clinical trials, and biomarkers for precision oncology: challenges, opportunities, and programs at the Department of Veteran Affairs. Clin. Pharmacol. Ther. 101, 586–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. ORIEN Network . <http://http:oriencancer.org/>. Accessed 27 December 2017.
- 3.The Targeted Agent and Profiling Utilization Registry (TAPUR) Study. <http://www.tapur.org/> Accessed 27 December 2017.
- 4. AACR Project Genomics Evidence Neoplasia Information Exchange (GENIE) . <http://www.aacr.org/Research/Research/Pages/aacr-project-genie.aspx#.Wk0bblWnGUk./>. Accessed 27 December 2017.
- 5. Worldwide Innovative Networking in Personalized Cancer Medicine . <http://www.winconsortium.org/>. Accessed 27 December 2017.
- 6. The Applied Proteogenomics OrganizationaL Learning and Outcomes (APOLLO) network. <https://proteomics.cancer.gov/programs/apollo-network/>. Accessed 27 December 2017. [DOI] [PubMed]
- 7. The Multiple Myeloma Research Consortium . <https://www.themmrf.org/research-partners/clinic/about-the-mmrc/>. Accessed 3 January 2018.
- 8. Dalton, W.S. , Sullivan, D.M. , Yeatman, T.J. & Fenstermacher, D.A. The 2010 Health Care Reform Act: a potential opportunity to advance cancer research by taking cancer personally. Clin. Cancer Res. 16, 5987–5996 (2010). [DOI] [PubMed] [Google Scholar]
- 9. Li, B . et al Use of the Total Cancer Care System to enrich screening for CD30‐positive solid tumors for patient enrollment into a brentuximab vedotin clinical trial: a pilot study to evaluate feasibility. JMIR Res. Protoc. 6, e45 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rodon, J . et al Challenges in initiating and conducting personalized cancer therapy trials: perspectives from WINTHER, a Worldwide Innovative Network (WIN) Consortium trial. Ann. Oncol. 26, 1791–1798 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text 1‐4: The Total Cancer Care Protocol, Publications using the TCC Protocol describing quality assessment and control methods of data generated, Governance and funding of ORIEN, Challenges associated with implementing the TCC Protocol, and gaining patient input through formation of the Patient Advisory Council
Figure S1: Total Cancer Care Protocol: A Partnership with Patients
Figure S2: ORIEN Governance Structure: A Self‐Governed Alliance of Cancer Centers


