Abstract
Ahead of display, a non‐original layer was observed on the surface of a fragment of a wall painting by Ambrogio Lorenzetti (active 1319, died 1348/9). FTIR analysis suggested proteinaceous content. Mass spectrometry was used to better characterise this layer and revealed two protein components: sheep and cow glue and chicken and duck egg white. Analysis of post‐translational modifications detected several photo‐oxidation products, which suggest that the egg experienced prolonged exposure to UV light and was likely applied long before the glue layer. Additionally, glycation products detected may indicate naturally occurring glycoprotein degradation or reaction with a carbohydrate material such as starch, identified by ATR‐FTIR in a cross‐section of a sample taken from the painting. Palaeoproteomics is shown to provide detailed characterisation of organic layers associated with mural paintings and therefore aids reconstruction of the conservation history of these objects.
Keywords: conservation science, cultural heritage, mass spectrometry, post-translational modifications, proteomics
High‐throughput mass spectrometry (MS) allows highly efficient and reproducible sequencing of modern and ancient proteins.1 Protein‐based materials have been used extensively in cultural heritage objects, both by artists, as paint binders for example, or for conservation as consolidants or adhesives. Their identification can provide insight into artistic technique or history of the object and can be helpful in defining display and storage conditions, as well as conservation protocols. Proteomics, or palaeoproteomics when applied to ancient samples, has already been successfully used for studying proteins in samples from works of art, providing accurate information about the nature of the material, its biological source, and degradation status.2, 3, 4, 5, 6, 7, 8, 9, 10, 11 Herein we present the first attempt, to our knowledge, to investigate in detail multiple post‐translational modifications (PTMs) of unknown proteinaceous layers applied to a wall painting.
Ambrogio Lorenzetti's A Group of Four Poor Clares (1336‐40, National Gallery, London, NG1147; Figure 1) is a fragment of a wall painting, made using the a fresco technique with areas of secco, in the Chapter House in San Francesco, Siena (Italy). The work was discovered under whitewash shortly before 1855 and the fragment was removed and remounted in Siena before 1878.12 It was then purchased by the National Gallery and has been in storage or on display in a controlled museum environment ever since.
Figure 1.

The wall painting “A Group of Four Poor Clares” by Ambrogio Lorenzetti (active 1319, died 1348/9), National Gallery, London. Sampling areas for proteomic analysis are indicated. Copyright The National Gallery, London.
During preparation for display of the piece, conservators observed a water‐soluble layer applied on the surface. Analysis by Fourier transform infrared spectroscopy (FTIR) identified the layer to be protein‐based (see the Supporting Information). The source of the protein should not be the original fresco itself, where pigments are dispersed in water, without proteinaceous binder, and applied on fresh plaster. The wall painting fragment also contains elements of secco, an additional paint layer applied to the dry plaster containing both pigment and an organic binding material, typically whole egg, or, less frequently, casein, animal glue, gums, or oils.13, 14 However, the cross‐section samples taken from the fragment (Supporting Information, Figures S1, S2) do not show any secco layers in the areas examined. It was therefore of interest to understand the source and function of this proteinaceous material. Since FTIR is not able to confidently discriminate between different protein sources, MS‐based proteomics techniques were chosen to achieve an accurate identification of the biological species and protein source of the material. Additionally, the characterisation of protein damage through PTMs was used to try to clarify the history of the artwork, important for effective conservation strategies.
Two samples of the unknown layer, OS6 and OS7 (Figure 1), were removed. Protein residues were extracted in a guanidinium solution, and digested with Lys‐C and trypsin. The tryptic peptides were analysed by nanoflow liquid chromatography tandem mass spectrometry (nLC‐MS/MS) by means of a single injection per sample. Protein identification and additional data analysis was performed with the MaxQuant software.15 The resulting proteins and their modifications were compared with data from control samples prepared using the same workflow. Modern dried chicken (Gallus gallus) egg white and pigmented bovine (Bos taurus) glue mock‐ups were analysed to control for PTMs generated in vivo or during sample preparation, while skeletal remains of relatively similar age, but not exposed to visible or UV light,16 were used as controls for age‐induced degradation (see the Supporting Information for the full methodology).
In both samples, endogenous proteins, absent in the laboratory blanks, allowed for identification of two sources of protein: egg white and collagens (Supporting Information, Table S3). All major egg white proteins were identified, including ovalbumin, ovotransferrin, ovomucoid, and ovoglobulins. No egg yolk proteins were detected. Based on peptide‐spectra matches against publicly available protein databases, most egg proteins contained peptides characteristic for chicken (Gallus gallus). Additionally, ovotransferrin from domestic mallard, that is, duck (Anas platyrhynchos), was confidently sequenced from both samples (Supporting Information, Figures S3–4), covering several amino acids polymorphic in chicken. It could be that peptides uniquely assigned to duck could belong to other, phylogenetically closely related, wild bird species whose protein sequences are not publicly available yet. However, the identification of domestic duck egg white proteins seems the most plausible. The more limited number of duck‐specific sequences identified, compared to the chicken‐specific ones, could be explained either through a deliberate use of different proportions of eggs from the two species during the layer preparation, or through sporadic/accidental incorporation of duck egg white. Since egg white becomes brittle when dry, it was rarely used as secco binder, but more likely as varnish (glair).17, 18 Therefore, this layer was likely applied to saturate the colours and to act as a protective coating. During conservation, the surface material was found to be soluble in water, a property of dry glue but not of dry egg white.19 As a mixture of the two proteins is unlikely in this context, we suggest the presence of two layers, an earlier one of egg white and a subsequent one of animal glue.
Animal glue was indicated by the presence of sheep‐specific (Ovis aries) collagen (COL1A1, COL1A2, COL3A1) peptides in both samples. In OS6, two bovine COL1A2 peptides, most probably from cow (Bos taurus) given the context, were also detected. While animal glue can be used in secco, the thickness (Supporting Information, Figure S1 c), general absence of pigment particles, and presence over most of the painting, suggests the glue was applied as part of later conservation treatments. This hypothesis is supported by FTIR analysis (Supporting Information, Figure S2), which shows a proteinaceous material on the surface as well as inside cracks in the cross‐section, suggesting its application to an already aged painting. A previous study of another Lorenzetti wall painting also found evidence of a glue layer applied during restoration.20
The identification of collagen from different species may simply be due to the use of a multi‐species derived glue. Alternatively, glue from various animal sources may have been used at different times. For example, techniques to remove wall paintings from their original support typically employ several layers of cloth glued onto the surface of the wall painting. The glued cloths, together with the paint layers and sometimes underlying plaster, are then removed. Once the fragment is on a new support, the facing cloths are detached by dissolving, and consequently removing, the glue with hot water.14 Residual glue from this type of process has been identified on other wall painting fragments,21 so it is possible that the presence of Bos collagen in a single sample on the edge of the painting relates to a different treatment than the sheep‐specific peptides present in higher abundance.
To understand the differences between these egg and glue layers, the molecular damage patterns of the peptides were investigated. First, deamidation of asparagine (Asn) and glutamine (Gln) residues were examined. Asn and Gln naturally deamidate over time.22 The glue peptides were significantly more deamidated than those of the egg. On average, 91.8 % Asn (SD 2.0 %) and 48.4 % Gln (SD 3.6 %) collagen residues were deamidated versus only 8.9 % Asn (SD 1.7 %) and 5.7 % Gln (SD 1.1 %) for the egg proteins (Supporting Information, Figure S6). However, glue preparation involves prolonged boiling of animal connective tissue that would significantly increase the rate of deamidation of Asn. This reaction is relatively slow in intact collagen but more rapid in gelatin.23 The low level of intact Asn in the collagen supports this interpretation. This protein damage is therefore not representative of the overall history of the wall painting.
Additional PTMs were then investigated with focus on photo (UV) damage. Proteins are known to be the main targets of photo‐oxidation owing to endogenous chromophoric amino acids.24, 25, 26 Photo‐oxidative damage of proteins occurs via two major pathways: direct photo‐oxidation through excitement by UV light (Type 1),24, 25, 26, 27 and indirect oxidation by singlet state oxygen 1O2 (Type 2), resulting from energy transfer from excited sensitiser(s) to the molecular oxygen, which can then oxidise chromophoric amino acids tryptophan (Trp), histidine (His), tyrosine (Tyr), cysteine (Cys), methionine (Met), and phenylalanine (Phe).25, 26, 27, 28 Photosensitisers can be both endogenous (flavins, porphyrins, vitamins) and exogenous (aromatic compounds, dyes, and pigments).26
Several photo‐oxidative PTMs were detected and compared to mock‐up egg white, pigmented glue samples, and previously published circa 350‐year‐old bones16 (Supporting Information). The analysis of OS6 and OS7 shows that half of all proteinogenic amino acids, including all chromophoric residues, underwent oxidation (Supporting Information, Table S5). However, mass shift information is often not sufficient to distinguish between chemical and Type 1/Type 2 photo‐oxidation. In contrast to most oxidised amino acids, which show a stochastic distribution of oxidative modifications with mainly mono‐ and little di‐oxidation, His di‐oxidation was four times more abundant than mono‐oxidation in OS6 and OS7, whereas in the controls His was mainly or equivalently mono‐oxidised. This indicates that His is mostly di‐oxidised in a single step via intermediate peroxidic species (Figure 2). The unstable peroxidic intermediate then undergoes rearrangement to a more stable tautomeric form that can further break down to aspartic24, 28 and potentially hydroxyglutamic acids, whose corresponding mass shifts are observed four times more frequently in OS6 and OS7, compared to the bone negative controls, and at least twice as much as pure egg white. The comparatively low levels of multiply photo‐oxidised His observed in all controls indicate that sample preparation had little effect in the introduction of these PTMs (Figure 2; Supporting Information, Table S6).
Figure 2.
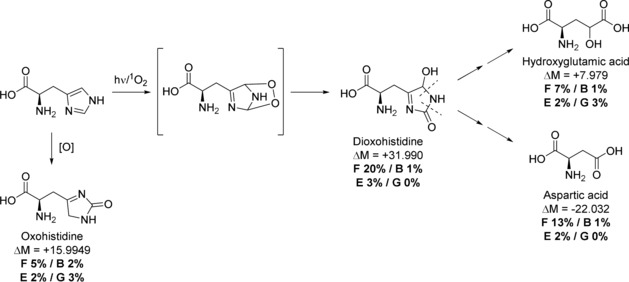
Photo‐oxidation of histidine via peroxidic state. ΔM=mass shift, F%=occurrence in wall painting, B%=bone references, E%=egg white control, G%=glue+pigment controls.
Oxidation products of Trp were also found in relatively high amounts in OS6 and OS7. It is known that Trp has a relatively high photo‐oxidation reaction rate compared to the other chromophoric amino acids. It is an efficient photosensitiser24 and has been used to determine UV damage in archaeological samples.29 Trp can undergo mono‐, di‐, and tri‐oxidation, as well as formation of kynurenine and hydroxykynurenine. The wall painting samples contain more mono‐ and di‐oxidative Trp PTMs than the controls (an average of 32 % di‐oxidised Trp compared to 18 % in the pure egg white, 9 % in the bone samples, and 0 % in the glue mock‐ups; Supporting Information, Table S7). OS7 showed higher relative levels of oxidation products than OS6, which could be related to the presence of pigment particles in this area (Figure 1) acting as photosensitisers.30 In contrast to other publications,29, 31 we observed no to little increase in the amount of kynurenine or hydroxykynurenine in the UV‐exposed painting samples (average of 15 % and 3 %, respectively) compared to the controls (ranging 0–21 % and 0–5 %). Based on these observations, it cannot be determined whether these PTMs occur during sample preparation or in vivo, especially in the more Trp‐rich egg white. It seems that all of the oxidative Trp PTMs observed can also form during aging processes, through sample preparation, or via biological pathways.
Interestingly, we also found a mass‐shift indicative of a His Cα−Cβ bond cleavage (Supporting Information, Figure S8, Table S8), which results in the formation of glycine (Gly). Current literature32, 33, 34 describes only photolytic Cα−Cβ bond cleavage. However, this PTM is also observed in the non‐irradiated bone controls, indicating that this modification might also be due to aging rather than exclusively being UV‐mediated. It is observed at least twice as much in OS6 and OS7 when compared to the egg and glue controls (Supporting Information, Table S8). So far, no mechanism other than photolytic radical‐based Cα−Cβ bond cleavage is reported in literature. The occurrence of this PTM in ancient samples may thus be of future interest for researching degradation in palaeoproteomics.
Owing to the higher number of detected egg white proteins (11), compared to the few in the glue (3), and the absence of Trp in collagen,35 it is problematic to compare photo‐oxidative PTMs between the two layers. However, the fact that the egg white peptides contain relatively high amounts of photo‐oxidative damage, especially of His, indicates that it was likely exposed to light for a significant period of time before the addition of the glue, which would protect it to some extent from damage.17 This means the egg white layer might have been applied before the whitewash, thus illustrating how the study of PTMs can aid in understanding the conservation history of works of art.
Unrelated to UV damage, PTMs on OS6 and OS7 revealed a high prevalence of mass shifts on lysine (Lys) and arginine (Arg) side chains corresponding to PTMs described in the literature as advanced glycation end products (AGEs, Figure 3), caused by carbohydrate excess36 and ascribed to diagenesis in archaeological bones.37, 38 The degradation of carbohydrates can produce glyoxal and methylglyoxal in vitro, which react with Lys and Arg residues to form carboxymethyl or carboxyethyl Lys and (methyl‐) imidazolone Arg.39 Combined, AGEs were at least four times more abundant in the painting samples than in the controls. In a semi‐quantitative analysis (Supporting Information, Table S9), we found at least 1 glyoxal or methylglyoxal modification for 102 of all 608 Lys residues in both OS6 and OS7 (17 %). Compared to the bone and egg white controls, we saw over three‐fold more AGE‐linked Lys residues in the painting samples. The presence of natural glycosylation in collagen and egg white proteins35, 40 is a potential cause for the high prevalence of AGEs. However, the ratios in OS6 and OS7 are much higher than in the modern egg white and aged bone samples. A carbohydrate‐based material, such as starch, was detected in the surface layer of the painting using attenuated total reflection (ATR)‐FTIR and may have been an addition to the glue. It is therefore more likely that protein glycation was further enhanced in OS6 and OS7 by the presence of starch.
Figure 3.
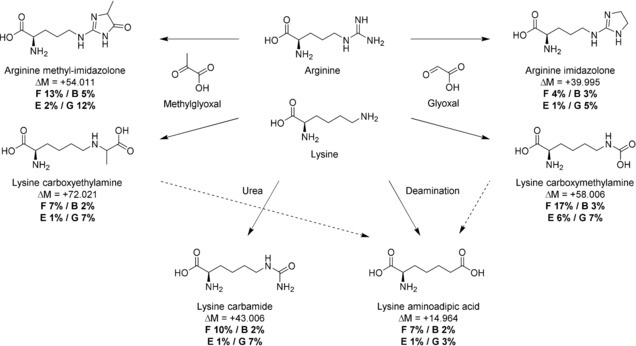
Advanced glycation end products from arginine and lysine. ΔM=mass shift, F%=occurrence in wall painting, B%=bone references, E%=egg white control, G%=glue+pigment controls (combined). F% almost exclusively represents numbers for egg peptides and are therefore not directly comparable with the G%.
Surprisingly, the AGE‐levels in collagens of the modern glue mock‐ups (“G” in Figure 3) were sometimes as high as the levels in OS6 and OS7. These glycations were possibly caused by the addition of pigment or formed during heating as part of the glue making process, since they do not occur as frequently in the ancient bones. Along with AGEs, Lys carbamylation (potentially caused by urea release during His or Arg decay) and Lys to aminoadipic acid conversion (a marker for protein degradation41 described in some archaeological studies42) were also observed.
In conclusion, mass spectrometry based palaeoproteomic analysis enabled the identification of the proteinaceous materials applied on the surface of Ambrogio Lorenzetti's A Group of Four Poor Clares, whereas profiling of PTMs helped to reconstruct its conservation history. Proteins from chicken and duck egg white, as well as sheep and cow glue were identified as surface coatings, probably applied during conservation treatments. Furthermore, the evidence of protein UV damage and the solubility of the surface layer suggest the application of egg for a period of time before the glue layer. Finally, glycation was discovered as a likely result of possible reaction between proteins and starch on the painting. This work shows that palaeoproteomics has a strong potential to improve our understanding of the history and function of the materials identified in cultural heritage objects and to inform proposed conservation treatments.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
M.M. is supported by the University of Copenhagen (KU2016 UCPH Excellence Programme) and by the Danish National Research Foundation award PROTEIOS to Matthew Collins (who we also thank for comments on the manuscript). P.R., D.S. and F.D.G. are supported by the Marie Skłodowska‐Curie European Training Network (ETN) “TEMPERA” coordinated by E.C., a project funded by the European Union's EU Framework Programme for Research and Innovation Horizon 2020 (Grant Agreement number 722606). C.G. is supported by the Marie Skłodowska‐Curie Individual Fellowship “EGYPTOMICS”, a project funded by the European Union's EU Framework Programme for Research and Innovation Horizon 2020 (Grant Agreement number 750270). Work at the Novo Nordisk Foundation Center for Protein Research is funded in part by a generous donation from the Novo Nordisk Foundation (Grant number NNF14CC0001).
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository43 with the dataset identifiers PXD008486 and PXD009298.
M. Mackie, P. Rüther, D. Samodova, F. Di Gianvincenzo, C. Granzotto, D. Lyon, D. A. Peggie, H. Howard, L. Harrison, L. J. Jensen, J. V. Olsen, E. Cappellini, Angew. Chem. Int. Ed. 2018, 57, 7369.
References
- 1. Cappellini E., Collins M. J., Gilbert M. T. P., Science 2014, 343, 1320–1322. [DOI] [PubMed] [Google Scholar]
- 2. Hynek R., Kuckova S., Hradilova J., Rapid Commun. Mass Spectrom. 2004, 18, 1896–1900. [DOI] [PubMed] [Google Scholar]
- 3. Tokarski C., Martin E., Rolando C., Cren-Olivé C., Anal. Chem. 2006, 78, 1494–1502. [DOI] [PubMed] [Google Scholar]
- 4. Leo G., Cartechini L., Pucci P., Sgamellotti A., Marino G., Birolo L., Anal. Bioanal. Chem. 2009, 395, 2269–2280. [DOI] [PubMed] [Google Scholar]
- 5. Dallongeville S., Koperska M., Garnier N., Reille-Taillefert G., Rolando C., Tokarski C., Anal. Chem. 2011, 83, 9431–9437. [DOI] [PubMed] [Google Scholar]
- 6. Fremout W., Kuckova S., Crhova M., Sanyova J., Saverwyns S., Hynek R., Kodicek M., Vandenabeele P., Moens L., Rapid Commun. Mass Spectrom. 2011, 25, 1631–1640. [DOI] [PubMed] [Google Scholar]
- 7. Leo G., Bonaduce I., Andreotti A., Marino G., Pucci P., Colombini M. P., Birolo L., Anal. Chem. 2011, 83, 2056–2064. [DOI] [PubMed] [Google Scholar]
- 8. Kuckova S., Sandu I. C. A., Crhova M., Hynek R., Fogas I., Schafer S., J. Cult. Herit. 2013, 14, 31–37. [Google Scholar]
- 9. van der Werf I. D., Calvano C. D., Laviano R., Simonetti A., Sabbatini L., Microchem. J. 2013, 106, 87–94. [Google Scholar]
- 10. Mazurek J., Svoboda M., Maish J., Kawahara K., Fukakusa S., Taniguchi Y., e-PreservationScience 2014, 11, 76–83. [Google Scholar]
- 11. Lluveras-Tenorio A., Vinciguerra R., Galano E., Blaensdorf C., Emmerling E., Perla Colombini M., Birolo L., Bonaduce I., PLoS One 2017, 12, e0172990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gordon D., The Italian Paintings Before 1400, National Gallery Company, London, 2011. [Google Scholar]
- 13. Cennini C., The Craftsman's Handbook (Il Libro Dell′arte), Dover, New York, 1954. [Google Scholar]
- 14. Mora P., Mora L., Philippot P., The Conservation of Wall Paintings, Butterworth & Co., Sevenoaks, 1984. [Google Scholar]
- 15. Cox J., Mann M., Nat. Biotechnol. 2008, 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- 16. Sawafuji R., Cappellini E., Nagaoka T., Fotakis A. K., Jersie-Christensen R. R., Olsen J. V., Hirata K., Ueda S., R. Soc. Open Sci. 2017, 4, 161004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Imbrogno J., Nayak A., Sorci M., Belfort G., Angew. Chem. Int. Ed. 2014, 53, 7014–7017; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 7134–7137. [Google Scholar]
- 18. Woudhuysen-Keller R., Wouldhuysen-Keller P., Bulletin: The Hamilton Kerr Institute 1994, 2, 90–141. [Google Scholar]
- 19. Mills J. S., White R., The Organic Chemistry of Museum Objects, Butterworth-Heinemann, Oxford, 1999. [Google Scholar]
- 20. Benetti F., Perra G., Damiani D., Atrei A., Marchettini N., Int. J. Mass Spectrom. 2015, 392, 111–117. [Google Scholar]
- 21. Bonaduce I., Colombini M. P., Rapid Commun. Mass Spectrom. 2003, 17, 2523–2527. [DOI] [PubMed] [Google Scholar]
- 22. Robinson N. E., Robinson A. B., Proc. Natl. Acad. Sci. USA 2001, 98, 944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Duin A. C. T., Collins M. J., Org. Geochem. 1998, 29, 1227–1232. [Google Scholar]
- 24. Davies M. J., Photochem. Photobiol. Sci. 2004, 3, 17–25. [DOI] [PubMed] [Google Scholar]
- 25. Kerwin B. A., R. L. Remmele, Jr. , J. Pharm. Sci. 2007, 96, 1468–1479. [DOI] [PubMed] [Google Scholar]
- 26. Pattison D. I., Rahmanto A. S., Davies M. J., Photochem. Photobiol. Sci. 2012, 11, 38–53. [DOI] [PubMed] [Google Scholar]
- 27. Davies M. J., Truscott R. J. W., J. Photochem. Photobiol. B 2001, 63, 114–125. [DOI] [PubMed] [Google Scholar]
- 28. Agon V. V., Bubb W. A., Wright A., Hawkins C. L., Davies M. J., Free Radical Biol. Med. 2006, 40, 698–710. [DOI] [PubMed] [Google Scholar]
- 29. Solazzo C., Clerens S., Plowman J. E., Wilson J., Peacock E. E., Dyer J. M., J. Cult. Herit. 2015, 16, 896–903. [Google Scholar]
- 30. Duce C., Ghezzi L., Onor M., Bonaduce I., Colombini M. P., Tine′ M. R., Bramanti E., Anal. Bioanal. Chem. 2012, 402, 2183–2193. [DOI] [PubMed] [Google Scholar]
- 31. Grosvenor A. J., Morton J. D., Dyer J. M., Amino Acids 2010, 39, 285–296. [DOI] [PubMed] [Google Scholar]
- 32. Schöneich C., J. Pharm. Pharmacol. 2017, https://doi.org/10.1111/jphp.12688. [DOI] [PubMed] [Google Scholar]
- 33. Haywood J., Mozziconacci O., Allegre K. M., Kerwin B. A., Schöneich C., Mol. Pharm. 2013, 10, 1146–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lucas B., Barat M., Fayeton J. A., Perot M., Jouvet C., Gregoire G., Brøndsted Nielsen S., J. Chem. Phys. 2008, 128, 164302. [DOI] [PubMed] [Google Scholar]
- 35. Karpowicz A., Stud. Conserv. 1981, 26, 153–160. [Google Scholar]
- 36. Chen S. J., Aikawa C., Yoshida R., Matsui T., Physiol. Rep. 2015, 3, e12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cleland T. P., Schroeter E. R., Schweitzer M. H., Proc. R. Soc. B 2015, 282, 20150015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cleland T. P., Schroeter E. R., Feranec R. S., Vashishth D., Proc. R. Soc. B 2016, 283, 20160593. [Google Scholar]
- 39. Chetyrkin S., Mathis M., Pedchenko V., Sanchez O. A., McDonald W. H., Hachey D. L., Madu H., Stec D., Hudson B., Voziyan P., Biochemistry 2011, 50, 6102–6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Terajima M., Perdivara I., Sricholpech M., Deguchi Y., Pleshko N., Tomer K. B., Yamauchi M., J. Biol. Chem. 2014, 289, 22636–22647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sell D. R., Strauch C. M., Shen W., Monnier V. M., Biochem. J. 2007, 404, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cappellini E., Jensen L. J., Szklarczyk D., Ginolhac A., da Fonseca R. A. R., Stafford T. W., Holen S. R., Collins M. J., Orlando L., Willerslev E., et al., J. Proteome Res. 2012, 11, 917–926. [DOI] [PubMed] [Google Scholar]
- 43. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., et al., Nucleic Acids Res. 2013, 41, D1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


