Abstract
Previously considered as a component of transcriptional noise, long noncoding RNAs (lncRNAs) were neglected as a therapeutic target, however, recently increasing evidence has shown that lncRNAs can participate in numerous biological processes involved in genetic regulation including epigenetic, transcriptional, and post-transcriptional regulation. In this review, we discuss the fundamental functions of lncRNAs at different regulatory levels and their roles in metabolic balance. Typical examples are introduced to illustrate their diverse molecular mechanisms. The comprehensive investigation and identification of key lncRNAs will not only contribute to insights into diseases, such as breast cancer and type II diabetes, but also provide promising therapeutic targets for related diseases.
Keywords: Disease, Epigenetics, LncRNAs, Post-transcription, Transcription
INTRODUCTION
The Human Genome Project revealed that there are only approximately 20,000 protein coding genes in humans, which is much less than previously estimated (1, 2), suggesting that the noncoding genome can influence a significant portion of cellular functionality. While not all noncoding genes play an active role in cells, long noncoding RNAs (lncRNAs) have a significant function (2). LncRNAs are a general class of non-coding RNAs (> 200 nucleotides in length), which have been shown to participate in many steps of gene transcription, including at the epigenetic and genetic level, but lack the ability to encode proteins. LncRNAs exist in the nucleus, cytoplasm, or both, and therefore their functions are closely related to their localization (3, 4). In recent years, the application of deep RNA sequencing (RNA-Seq) and ribosome profiling has made it easier to analyze transcriptomes, discover numerous new lncRNAs and annotate them (5–8). To date, 548,640 lncRNA transcripts and 354,855 lncRNA genes have been found in seventeen species, including human and mouse, and these are listed in the NONCODE database (http://www.noncode.org/index.php).
Both lncRNAs and their genes have similar chromatin states, meaning that lncRNAs may be able to function as a gene in cells (4, 9). However, it has been demonstrated that some lncRNAs contains a small open reading frames (ORF), that can encode for a peptide. Therefore, the definition of lncRNAs may change in the future (8, 10–12).
Compared with mRNA, the relative expression levels of lncRNAs are lower, but lncRNA expression is more specific than mRNA in different cell types, tissues, developmental stages and even diseases. They interact with mRNAs, proteins and DNA elements in many forms (4, 13–18). Therefore, lncRNAs have more intricate and multiple roles in regulating biological processes. They relieve the pressure that miRNAs exert on their target genes by acting as a sponge, compete with miRNAs for the same targets, and even become precursors of some miRNAs (19–21). During the past few years, many studies have revealed the crucial roles of lncRNAs in gene control and potential molecular mechanisms. These mechanisms may facilitate our understanding of the functions of lncRNAs and provide us with a complex and precise view of gene regulation.
EPIGENETIC REGULATION
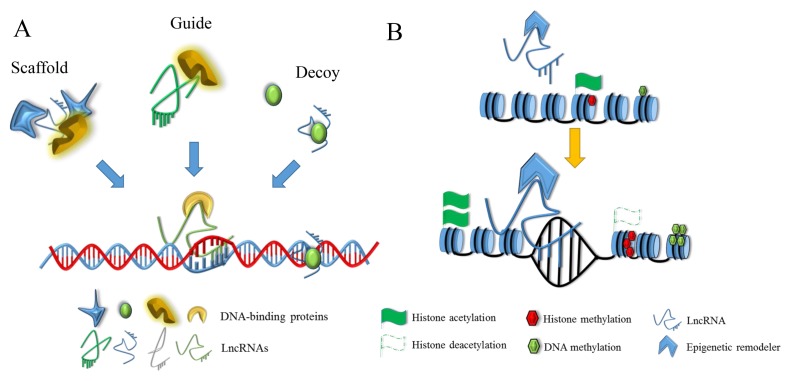
As a multifunctional regulator, lncRNAs may act as scaffolds and guides to recruit or directly modify the basic epigenetic modification elements, such as DNA, histones, and non-histones (Fig. 1) (22–25). LncRNAs can lead chromatinmodifying complexes to their genomic targets as guides or just deceive them as decoys (Fig. 1A, Table 1) (26–29). However, how do they recognize their target sites to govern gene expression?
Fig. 1.
The regulation of lncRNAs in epigenetics. (A) LncRNA may recruit protein complexes as scaffold, deceive chromatin-remodeling components as decoy, and direct remodelers as guide. (B) LncRNA guides epigenetic modifiers to change the chromatin structure, histone methylation or acetylation level, and DNA methylation level.
Table 1.
Characterized lncRNAs with potential roles in epigenetic regulation and peptide-mediated regulation
| LncRNAs | Target | Functions | References |
|---|---|---|---|
| MEG3 | PRC2 | Targets the cis or trans of PRC2 to mediate H3K27 methylation and gene silencing for dosage compensation, imprinting, and developmental gene expression | 29 |
| LRP1-AS | HMGB2 | Modulate the activity of non-histone chromatin modifier HMGB2 to decrease the expression of LRP1 | 31 |
| H19 | DNMT3B | Prevent DNMT3B from DNA methylation through attenuating SAHH hydrolysis to SAH | 32 |
| Kcnq1ot1 and Airn | G9a | Targets H3K9 methylase G9a for imprinting | 33, 34 |
| Xist | PCR1 | Recruit epigenetic complexes to change the status of histones and DNA, and then inactive X chromatin | 35, 36 |
| MLN | SERCA | Interact with SERCA and impede Ca2+ uptake into the SR | 12 |
| SPAR | mTORC1 | Bind to v-ATPase and blunts mTORC1 activation by amino acids | 39 |
| HOXB-AS3 peptide | PKM, miR-18 | Inhibit tumorigenesis by blocking PKM splicing, PKM2 formation, miR-18a processing, and subsequent metabolic reprogramming in colon cancer (CRC) cells | 40 |
In recent years, immunoprecipitation-coupled high-throughput sequencing (ChIRP-Seq) revealed the principles of RNA-Chromatin interactions and found that the occupancy sites of RNA are focal, specific, and numerous in the genome (30). For example, researchers found that a lncRNA, maternally expressed gene 3 (MEG3), was enriched in chromatin, and it can modulate the activity of transforming growth factor-β (TGFβ) by binding to distal regulatory elements, such as GA-rich DNA motifs, suggesting that lncRNAs may recognize their target sites through combining with specific DNA sequence motifs (29).
In addition to interacting with histone modifiers, lncRNAs also interplay with non-histone chromatin modifiers, such as LPR1-AS. As natural antisense transcript of low-density lipoprotein receptor-related protein 1 (LPR1), LRP1-AS can modulate the activity of non-histone chromatin modifier high-mobility group protein B2 (HMGB2) to decrease the expression of LRP1 (31).
Besides combining with DNA, histones, and non-histones, lncRNAs can also affect genome methylation. For instance, H19 knockdown activated a combination of U-rich elements (URE) with S-adenosylhomocysteine hydrolase (SAHH), leading to increased DNA methyltransferase 3 beta (DNMT3B)-mediated methylation. Furthermore, genome-wide methylation profiling also indicated that the interaction of H19 and SAHH changed the methylation of numerous gene loci, suggesting that DNA methylation might be regulated by lncRNA (32).
Genomic imprinting is an example of epigenetic regulation. As two representative monoallelic, parental-specific noncoding transcripts, Kcnq1ot1 and Airn have been demonstrated to induce silencing of imprinted neighboring genes called Kcnq1 and Igf2r by recruiting histone H3 lysine 9 methylase G9a, respectively (33, 34). However, X chromosome dosage compensation is another example to illustrate the biological function of lncRNAs. X-inactive specific transcript (Xist), a large noncoding transcript with several tandem repeats, is transcribed exclusively from the Xist gene on the X inactivation center of X chromatin and is necessary for X chromosome inactivation (35). Specifically, Xist can recruit epigenetic complexes, such as PRC1, PRC2, and DNA methyltransferases, to change the status of histones and DNA to inactive X chromatin (36).
Therefore, chemical modification, such as the methylation and acetylation of histones and DNA, influences gene expression by changing the structure of chromatin (Fig. 1B). LncRNAs partner with epigenetic modifiers as scaffolds, guides and decoys to change the accessibility of the DNA sequence. RNA-protein and DNA-RNA-protein complexes are the basic form of lncRNAs during this process. The secondary structure of lncRNAs, the structural characteristics of proteins, and the condition of chromatin may be crucial for their combination.
PEPTIDE-MEDIATED REGULATION OF lncRNA
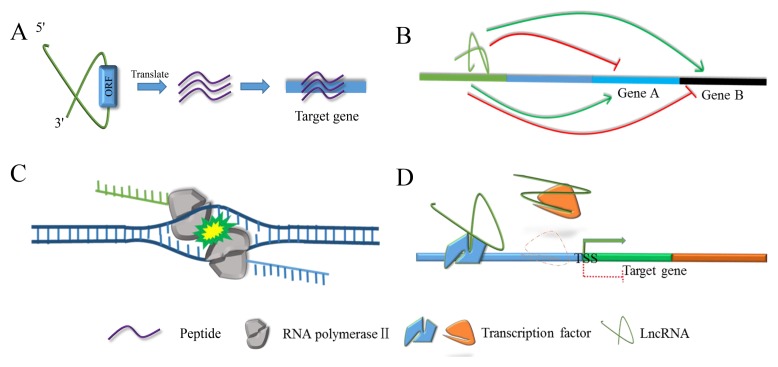
In the Introduction, we mentioned that some lncRNAs could encode peptides, which are translated from an ORF (Fig. 2A, Table 1). In general, the sequence of this type of peptide is rarely conserved between different species, and considered to have no function (37, 38). However, in recent years, the role of these peptides, which are translated from the ORF of lncRNAs, has been reported, such as myoregulin (MLN), small regulatory polypeptide of amino acid response (SPAR), and the HOXB cluster antisense RNA 3 (HOXB-AS3) peptide. As a peptide, which is encoded by a skeletal muscle-specific lncRNA LINC00948, MLN can directly interact with sarcoplasmic reticulum Ca2+-ATPase (SERCA) and impede Ca2+ uptake into the sarcoplasmic reticulum (SR), resulting in decreased Ca2+ handling in skeletal muscle and exercise performance (12). Coincidently, a similar functional mechanism of LINC00948 was showed in LINC00961. SPAR, a polypeptide encoded by lncRNA LINC00961, directly binds to v-ATPase and blunts mammalian target of rapamycin complex 1 (mTORC1) activation by amino acids (39). Furthermore, the HOXB-AS3 peptide, not HOXB-AS3 lncRNA, inhibits tumorigenesis by blocking PKM splicing, PKM2 formation, miR-18a processing, and subsequent metabolic reprogramming in colon cancer (CRC) cells, suggesting that lncRNAs can plays a role in cell through the peptide encoded by its own ORF (40). However, this type of research has predominantly focused on the function of rather than the effect of their related lncRNAs on biological processes. Taking HOXB-AS3 lncRNA as an example, although the HOXB-AS3 peptide, not HOXB-AS3 lncRNA, has been reported as playing a role in CRC, HOXB-AS3 lncRNA could also regulate the cell cycle progression of OCI-AML3 cells in Npm1 mutated acute myeloid leukemia, suggesting that it is possible that there is an unknown interaction between lncRNA and peptides that we need to further investigate (41).
Fig. 2.
The regulation of lncRNAs in transcription. (A) LncRNA can encode a peptide from its own ORF, and then play a role in biological process by these peptides. (B) LncRNA and its DNA locus in genome play different roles to their target genes. (C) Co-transcriptional collision of two converging polymerases during transcription processes of lncRNA and mRNA. (D) LncRNA combines with transcription factor as guide or decoy to promote or suppress transcription of downstream genes.
TRANSCRIPTIONAL REGULATION
LncRNAs can fulfil their roles during transcription (Table 2). The lncRNA Khps1, as a transcript, could recruit histone acetyltransferase p300/CBP to the sphingosine kinase 1 (SPHK1) promoter so that the transcriptional factor E2F1 could more easily combine with its binding sites and activate transcription of SPHK1 (42). However, there is a type of lncRNA called enhancer-associated RNAs (eRNAs), which are transcribed from enhancers, and can participate in the transcriptional process, such as Lockd, Haunt, and LEENE. In general, eRNAs likely facilitate enhancer interactions and thereby activate target genes. For example, as a DNA element, the lncRNA Lockd had no effect on the transcription of Cyclin dependent kinase inhibitor 1B (Cdkn1b), but it has been reported that the lncRNA Lockd could significantly reduce the transcription of Cdkn1b because of an enhancer-like element on its locus (43). Coincidently, an enhancer-associated lncRNA that enhances endothelial nitric oxide synthase (eNOS) expression (LEENE) has been reported the LEENE-associated enhancer formed a proximity association with the eNOS locus, and then facilitated the recruitment of RNA Pol II to the eNOS promoter to enhance eNOS nascent RNA transcription in endothelial cells (ECs) (44). In contrast with Khps1 and Lockd, the lncRNA HOXA upstream noncoding transcript (Haunt) was transcribed from approximately 40 kb upstream of the HOXA cluster and there was a potential enhancer of homeobox A (HOXA) in its DNA locus. Both Haunt and its DNA locus are responsible for the expression of HOXA, but interestingly, Haunt and its DNA locus performed exactly the opposite function during the expression of HOXA (Fig. 2B) (45).
Table 2.
Characterized lncRNAs with potential roles in transcriptional and post-transcriptional regulation
| LncRNAs | Target | Functions | References |
|---|---|---|---|
| Khps1 | SPHK1 | Promote E2F1 to combine with binding sites of SPHK1 | 42 |
| Lockd | Cdkn1b | As an enhancer-like element in regulating Cdkn1b on its locus | 43 |
| LEENE | eNOS | Enhance eNOS nascent RNA transcription through facilitating the recruitment of RNA Pol II to the eNOS promoter in endothelial cells | 44 |
| Haunt | HOXA | Responsible for the expression of HOXA | 45 |
| Airn | Igf2r | Silence the transcription of Igf2r by disturbing the recruitment of RNA polymerase II to the promoter of Igf2r | 50 |
| Blnc1 | EBF2 | Combine with the transcription factor EBF2 to form ribonucleoprotein complex that carry out this function | 51 |
| PANDA | NF-YA | p53 inducible and titrates away NF-YA to favor survival over cell death during DNA damage | 52 |
| lnc-DC | STAT3 | Combine with STAT3 to prevent the dephosphorylation of its tyrosine-705 by SHP1 | 53 |
| Uc.283+A | pri-miR-195 | Interact with stem region of the pri-miR-195 transcript and inhibit the processing of pri-miR-195 finally | 58 |
| LncND | miR-143-3p | Sponge with adsorbed miR-143-3p and enhance the Notch signaling pathway as a sponge during primate brain expansion | 21 |
| Sirt1 AS | Sirt1 | Interact with 3′UTR of Sirt1 mRNA to form RNA-RNA duplex, mask the binding sites of miR-34a, and enhance the stability of Sirt1 mRNA | 62 |
| OIP5-AS1 | GAK | Suppress GAK protein abundance and then inhibit cell division | 64 |
| 1/2sbsRNAs | Staufen1 | Regulate C2C12 cell myogenesis through triggering staufen1-mediated mRNA decay | 65 |
| H19 | KRSP | Strengthen the mRNA stability of myogenin, and then to boost the maturation of miRNAs | 67 |
| LincRNA-p21 | CTNNB1 JUNB | Interact with the translational repressors Rck to prevent the translation of CTNNB1 and JUNB | 55 |
| LncMyoD | IMP2 | Perturb the translation of some proliferation relative genes by competitive binding to the structure domain of IMP2 | 68 |
| LUNAR1 | Notch | A Notch-regulated pro-oncogenic lncRNA that is essential for T cell acute lymphoblastic leukemia growth | 69 |
| NBR2 | AMPK | Combine with AMPK and elevates its activity, and then form a positive feed-forward loop to alter kinase signaling pathway | 70 |
| LINK-A | HIF1α | Recruit BRK and LRRK2 to phosphorylate HIF1α at Tyr 565 and Ser 797, and then enhance the stabilization of HIF1α | 71 |
In addition to the above-mentioned mechanism, the transcriptional process of RNA can also interfere with the transcription of other genes. Antisense lncRNAs (AS lncRNAs), transcribed from the strand which is opposite to the previously annotated transcripts, may disturb transcription by co-transcriptional collision of two converging polymerases, such as Antisense Igf2r RNA noncoding (Airn) (46–49). Airn can silence the transcription of Igf2r by disturbing the recruitment of RNA polymerase II to the overlap section (Fig. 2C) (50).
Furthermore, some lncRNAs can fulfill their roles through their own transcription. Overexpression or knockdown of an inducible Brown fat lncRNA1 (Blnc1) could upregulate or downregulate the expression of thermogenesis genes, during brown adipose tissue development and thermogenesis, respectively (51). Further research provided compelling evidence that Blnc1 was positively regulated by a ribonucleoprotein complex, which was composed of Blnc1 and transcription factor called EBF2, suggesting a novel feedback regulatory loop during this process (Fig. 2D). Moreover, lncRNAs can also act as decoys in the interaction between transcription factors and DNA elements. For example, the promoter lncRNA PANDA restricts the expression of pro-apoptotic genes by combining with the transcription factor NF-YA to decrease its occupancy at target genes, thereby preventing p53-mediated apoptosis (Fig. 2D) (52).
In addition, lncRNAs may influence the phosphorylation and nuclear translocation of transcription factors to enhance or attenuate downstream gene expression. The tyrosine phosphatase SHP1 can downregulate the phosphorylation level of STAT3, and prevent its nuclear translocation. Based on this mechanism, Wang et al. found that lnc-DC can prevent the dephosphorylation of STAT3 on tyrosine-705 by SHP1 (53). During these processes, lncRNAs play their roles through various mediators such as transcripts and DNA elements, and even participate in the transcription of sense and antisense transcripts.
POST-TRANSCRIPTIONAL REGULATION
The maturation of pre-mRNAs to mature RNA plays a critical role in proteins coding. In these steps, there are different molecular mechanisms involved in the processes of splicing, stability, decay, and translation. Recent studies have shown that lncRNAs could be involved in these processes (54–56). Moreover, lncRNAs can also interact with protein kinases to further affect cytoplasmic signal transduction (Table 2).
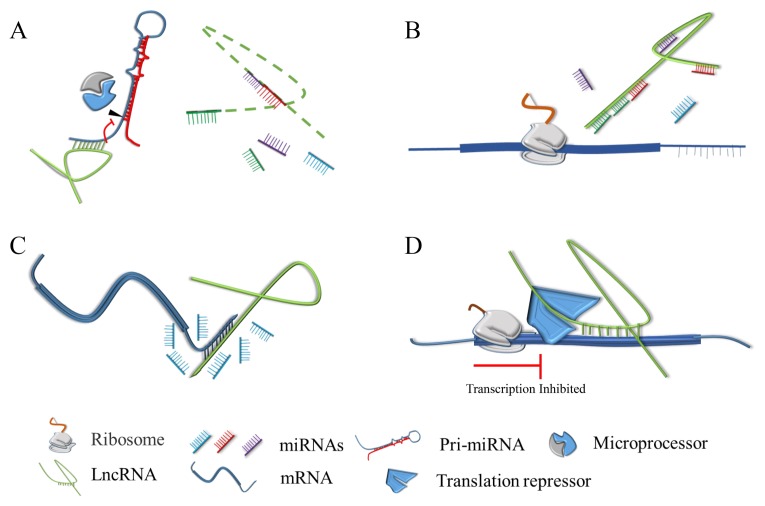
The Drosha and DGCR8 complexes are necessary for microRNA maturation (57). In 2014, an ultraconserved lncRNA, Uc.283+A, was shown to interact with the stem region of the pri-miR-195 transcript, and downregulate mature miR-195 levels (58). In addition, Uc.283+A can inhibit pri-miR-195 processing by Drosha through directing RNA-RNA interactions and impairing the binding of DGCR8, suggesting that lncRNAs could affect the formation of miRNAs (Fig. 3A). As a member of noncoding RNAs, miRNAs usually modulate mRNA stability or protein translation by targeting their seed sequence to the 3′ untranslated region (UTR) of mRNAs (59–61). As a sponge of miRNAs, it has been shown that LncND has a dozen miRNA response elements (MREs) for miR-143-3p, so that LncND could sponge this miRNA and enhance the Notch signaling pathway during the primate brain expansion (Fig. 3B) (21).
Fig. 3.
The regulation of lncRNAs in post-transcription. (A) LncRNA combines with pri-miRNA to inhibit its maturation or as the precursor of some miRNAs to regulate their maturation. (B) LncRNA absorbs miRNAs as a sponge or decoy to regulate target genes of miRNA (C). LncRNA competes with miRNA for same site to prevent the combination of genes and miRNAs. (D) LncRNA interacts with the coding regions of mRNA, and then combines with translation repressor to inhibit translation of target mRNA.
In addition to the above interaction, endogenous competition between miRNA and lncRNA has also been reported (62). This research identified a new AS lncRNA named Sirtuin 1 (Sirt1) AS lncRNA, which is transcribed from the antisense strand and is a tail-to-tail orientation of the sirt1 gene (63). Sirt1 is a target of miR-34a, and experimental results have demonstrated that Sirt1 AS lncRNA could cooperate with the 3′ UTR of Sirt1 mRNA to form an RNA-RNA duplex, and then mask the binding sites of miR-34a, finally enhancing the stability of Sirt1 mRNA (Fig. 3C) (62). In contrast with mechanism of Sirt1 AS lncRNA, OIP5-AS1 lncRNA could negatively affect mRNA stability with G-associated kinase (GAK) in HeLa cells. OIP5-AS1 lncRNA was shown to interact with GAK mRNA, and elevated OIP5-AS1 could suppress GAK protein abundance and then inhibit cell division (64). Coincidently, half–STAU1 (staufen double-stranded RNA-binding protein 1)–binding site RNAs (1/2sbsRNAs) was shown to regulate C2C12 cell myogenesis through decreasing target mRNA stability, suggesting that AS lncRNAs have dual roles for gene regulation (65). Another example of lncRNAs affecting mRNA stability is the relationship between H19 and K homology (KH)-type splicing regulatory protein (KSRP), which can negatively regulate target genes by promoting the decay of labile mRNA and favoring the maturation of select miRNAs from precursors (66). Giovarelli et al. found that H19 could directly interact with KSRP as a scaffold, and demonstrated that the disassociation of H19 from KSRP could strengthen the mRNA stability of myogenin, and then recruit Drosha and Dicer complexes to boost the maturation of selected miRNAs (67).
Furthermore, the translation of mRNAs is also under the control of lncRNAs. A prior study has demonstrated that LincRNA-p21 could interact with the translational repressor Rck to prevent the translation of Catenin beta-1 (CTNNB1) and jun B proto-oncogene (JUNB) (Fig. 3D) (55). Coincidentally, another lncRNA, LncMyoD, could perturb the translation of some genes involved in proliferation, such as N-Ras and c-Myc through competition for binding to the structure domain of IGF2-mRNA-binding protein 2 (IMP2), which is beneficial to the translation of proliferation genes. Furthermore, owing to its binding sites, LncMyoD could also prevent other genes from combining with IMP2 during the myogenesis period (68).
However, lncRNAs may be a downstream target of signaling pathways, such as mRNA. Leukemia-induced noncoding RNA, LUNAR1, was demonstrated to be under the control of the Notch signaling pathway (69). Conversely, lncRNAs also play crucial roles in different types of cytoplasmic signal transduction to regulate cellular metabolism. LncRNA NBR2 (neighbor of BRCA1 gene 2) is induced by the LKB1-AMPK pathway under energy stress, but NBR2 combines with adenosine 5′-monophosphate (AMP)-activated protein kinase (AMPK) conversely and elevates its activity. Therefore, they form a positive feed-forward loop to alter kinase signaling pathways (70). Another example is the activation of HIF1α signaling by lncRNA LINK-A (long intergenic non-coding RNA for kinase activation) under normoxic conditions. LINK-A recruits BRK and LRRK2 to phosphorylate HIF1α at Tyr 565 and Ser 797, and then it enhances the stabilization of HIF1α under normoxic conditions and facilitates the interplay between HIF1α and p300 on HB-EGF stimulation. The expression of target genes can be regulated in this way (71). These cases show that lncRNAs are not only regulated by signaling pathways, but can also be involved in cytoplasmic signal transduction. Therefore, the further study of the complex roles of lncRNAs in gene expression regulation is required.
METABOLISM BALANCE AND DISEASES
As a diverse class of regulators, lncRNAs play critical roles in affecting gene expression to maintain health, and ameliorate or aggravate pathological conditions. lncRNAs are also key regulators in the etiology of several disease states. At present, most studies of lncRNAs have focused on cancer. Furthermore, metabolic balance can also be controlled by lncRNAs (72, 73). The liver, skeletal muscle, and adipose tissue are major metabolic tissues, and the balance of glucose metabolism and lipid metabolism mainly depends on their proper function. Dysfunction of metabolic tissues could lead to whole-body diseases such as type 2 diabetes mellitus (T2D), non-alcoholic fatty liver disease (NAFLD), insulin resistance and obesity and so on (Table 3).
Table 3.
Characterized lncRNAs with potential roles in disease
| LncRNAs | Target | Disease | References |
|---|---|---|---|
| LncLSTR | TDP-43 | Fatty liver | 74 |
| Dum | Dppa2 | Muscle atrophy | 75, 76 |
| PU.1 AS | PU.1 | Type 2 diabetes mellitus | 51, 79, 80, 81 |
| Blnc1, Lnc-BATE1 | Ucp1 | ||
| HOTAIR | PRC2 | Breast cancer | 89, 90 |
| NKILA | NF-κB | ||
| lncTCF7 | Wnt | Liver cancer | 91 |
| SChLAP1 | SWI/SNF complex | Prostate cancer | 92, 93 |
| CTBP1-AS | CTBP1 |
The liver, a central metabolic organ, plays an important role in lipid metabolism. Depletion of the liver-specific triglyceride regulator (LncLSTR), which is beneficial for systemic lipid homeostasis, could impair the negative regulation of TDP-43 on the promotor of Cyp8b1, and then boost the lipoprotein lipase activation and clearance of plasma triglyceride (74).
As the largest metabolic organ in the body, skeletal muscle has a very important function in metabolic homeostasis. The atrophy and hypertrophy of skeletal muscle affects whole-body energy homeostasis. For instance, the Developmental pluripotency-associated 2 Upstream binding Muscle lncRNA (Dum) is linked with myogenic differentiation and muscle regeneration (75). The activation of Dum can strengthen the DNA methylation of Developmental pluripotency-associated 2 (Dppa2) by recruiting multiple methyltransferases to its promotor CpG sites, and then inhibits the transcription of Dppa2, which can regulate Oct4 to suppress muscle cell differentiation (75, 76).
Metabolic homeostasis in adipose tissue is important for health. The prevalence of obesity has led researchers to search for more detailed and accurate mechanisms underlying adipogenesis. In fact, hundreds of lncRNAs have been shown to be involved in the regulatory network of adipogenesis (77, 78), such as PU.1 AS lncRNA, which can promote the differentiation of preadipocytes by suppressing the translation of PU.1 mRNA in mouse and porcine models (79, 80). However, brown and beige adipocytes are considered to provide an ideal pathway to fat loss, suggesting that related lncRNAs, which can regulate the adipogenesis of brown and beige adipocytes, may play a role in the treatment of obesity. Both brown fat lncRNA 1 (Blnc1) and BAT-selective lncRNA (Lnc-BATE1) have been shown to promote thermogenesis gene expression, impair lipid accumulation, and improve energy homeostasis (51, 81). Therefore, lncRNAs may be a powerful weapon to against obesity and obesity induced metabolic diseases.
Furthermore, the endocrine system, immunity, hematopoiesis and cardiac development are also under the control of lncRNAs (82–84). Therefore, many previous studies have concentrated on the therapeutic role of lncRNAs, especially in cancer (24, 85–88). During the process of breast cancer metastasis, the expression of a biomarker, HOTAIR, is significantly increased. HOTAIR, a metastasis-associated lincRNA, has been shown to increase cancer invasiveness and metastasis by altering the histone H3K27 methylation of PRC2, suggesting that downregulation or disassociation of HOTAIR and PRC2 might be a prospective therapeutic target for breast cancer metastases (89). In another example, NF-κB Interacting lncRNA (NKILA) represses the breast cancer metastasis and cancer associated inflammation by inhibiting nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling (90). In contrast with NKILA, lncTCF7 promotes liver cancer stem cell self-renewal and tumor propagation by activating Wnt signaling (91). Additionally, another two lncRNAs, CTBP1-AS and SChLAP1, have been validated to promote prostate cancer through different molecular pathway (92, 93). From the above examples, we can deduce that lncRNAs are two-sided regulators of cancer progression. On the one hand, the aberrant expression of lncRNAs is closely linked with many types of cancer, and on the other hand their function in cancer could provide us with prospective therapeutic targets.
PROSPECT
LncRNAs positively or negatively regulate the expression of key genes to affect biological processes through various molecular mechanisms. Further studies will reveal additional characteristics of lncRNAs. For instance, some lncRNAs may have the same or different functional domains, which allow them to combine with more epigenetic modifiers to regulate gene expression. Although lncRNAs have poor conservation, the common features between lncRNAs and their interacting proteins, DNA, mRNAs, or miRNAs also deserve to be further investigated and classified. In addition to their role as regulators, lncRNAs are also under the control of some transcription factors and signaling pathways, and even can encode some peptide to regulate biological processes, and if we pay more attention to the role of intrinsic RNA rather than that of peptides, and this approach could lead to promising results. Furthermore, localization of lncRNAs would restrict their functions, so research on the mechanisms of lncRNA transposition could provide another perspective. These transcripts could be potential biomarkers in predicting the development of cancers or other diseases, and they represent a promising therapeutic target. The physiologic roles of the majority of lncRNAs are diverse and remain elusive, so there is a lot to discover. An enormous, complex and accurate gene regulatory network awaits further exploration.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Basic Research Programs of China (2015CB943102), the National Natural Science Foundation (31572366), and National Natural Science Foundation (21402162).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Ponting CP, Belgard TG. Transcribed dark matter: meaning or myth? Hum Mol Genet. 2010;19:R162–R168. doi: 10.1093/hmg/ddq362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 3.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 4.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM. Long non-coding RNAs as a source of new peptides. Elife. 2014;3:e03523. doi: 10.7554/eLife.03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banfai B, Jia H, Khatun J, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome Profiling Provides Evidence that Large Noncoding RNAs Do Not Encode Proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson DM, Anderson KM, Chang CL, et al. A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pang KC, Dinger ME, Mercer TR, et al. Genome-Wide Identification of Long Noncoding RNAs in CD8(+) T Cells. J Immunol. 2009;182:7738–7748. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- 14.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batista PJ, Chang HY. Long Noncoding RNAs: Cellular Address Codes in Development and Disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flynn RA, Chang HY. Long Noncoding RNAs in Cell-Fate Programming and Reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gloss BS, Dinger ME. The specificity of long noncoding RNA expression. Biochim Biophys Acta. 20161859:16–22. doi: 10.1016/j.bbagrm.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han XR, Yang F, Cao HQ, Liang ZC. Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 2015;29:3054–3064. doi: 10.1096/fj.14-259952. [DOI] [PubMed] [Google Scholar]
- 21.Rani N, Nowakowski TJ, Zhou HJ, et al. A Primate lncRNA Mediates Notch Signaling during Neuronal Development by Sequestering miRNA. Neuron. 2016;90:1174–1188. doi: 10.1016/j.neuron.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han P, Hang CT, Yang J, Chang CP. Chromatin remodeling in cardiovascular development and physiology. Circ Res. 2011;108:378–396. doi: 10.1161/CIRCRESAHA.110.224287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang CP, Han P. Epigenetic and lncRNA regulation of cardiac pathophysiology. Biochim Biophys Acta. 20161863:1767–1771. doi: 10.1016/j.bbamcr.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome Long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6:539–543. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–U158. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan JY, Xing Y, Wen XY, et al. Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis. Genome Biol. 2015;16 doi: 10.1186/s13059-015-0705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mondal T, Subhash S, Vaid R, et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6 doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamanaka Y, Faghihi MA, Magistri M, Alvarez-Garcia O, Lotz M, Wahlestedt C. Antisense RNA Controls LRP1 Sense Transcript Expression through Interaction with a Chromatin-Associated Protein, HMGB2. Cell Rep. 2015;11:967–976. doi: 10.1016/j.celrep.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou JC, Yang LH, Zhong TY, et al. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat Commun. 2015;6:10221. doi: 10.1038/ncomms10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandey RR, Mondal T, Mohammad F, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 34.Nagano T, Mitchell JA, Sanz LA, et al. The Air Noncoding RNA Epigenetically Silences Transcription by Targeting G9a to Chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 35.Pontier DB, Gribnau J. Xist regulation and function eXplored. Hum Genet. 2011;130:223–236. doi: 10.1007/s00439-011-1008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galupa R, Heard E. X-chromosome inactivation: new insights into cis and trans regulation. Curr Opin Genet Dev. 2015;31:57–66. doi: 10.1016/j.gde.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Chew GL, Pauli A, Rinn JL, Regev A, Schier AF, Valen E. Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development. 2013;140:2828–2834. doi: 10.1242/dev.098343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bazzini AA, Johnstone TG, Christiano R, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33:981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rion N, Ruegg MA. LncRNA-encoded peptides: More than translational noise? Cell Res. 2017;27:604–605. doi: 10.1038/cr.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang JZ, Chen M, Chen, et al. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol Cell. 2017;68:171–184 e176. doi: 10.1016/j.molcel.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 41.Papaioannou D, Petri A, Thrue CA, et al. HOXB-AS3 Regulates Cell Cycle Progression and Interacts with the Drosophila Splicing Human Behavior (DSHB) Complex in NPM1-Mutated Acute Myeloid Leukemia. Blood. 2016;128:1514. [Google Scholar]
- 42.Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, et al. LncRNA Khps1 Regulates Expression of the Proto-oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure. Mol Cell. 2015;60:626–636. doi: 10.1016/j.molcel.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 43.Paralkar VR, Taborda CC, Huang P, et al. Unlinking an lncRNA from Its Associated cis Element. Mol Cell. 2016;62:104–110. doi: 10.1016/j.molcel.2016.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miao Y, Ajami NE, Huang TS, et al. Enhancer-associated long non-coding RNA LEENE regulates endothelial nitric oxide synthase and endothelial function. Nat Commun. 2018;9:292. doi: 10.1038/s41467-017-02113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin YF, Yan PX, Lu JL, et al. Opposing Roles for the lncRNA Haunt and Its Genomic Locus in Regulating HOXA Gene Activation during Embryonic Stem Cell Differentiation. Cell Stem Cell. 2015;16:504–516. doi: 10.1016/j.stem.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 46.Shearwin KE, Callen BP, Egan JB. Transcriptional interference - a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmer AC, Ahlgren-Berg A, Egan JB, Dodd IB, Shearwin KE. Potent Transcriptional Interference by Pausing of RNA Polymerases over a Downstream Promoter. Mol Cell. 2009;34:545–555. doi: 10.1016/j.molcel.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hobson DJ, Wei W, Steinmetz LM, Svejstrup JQ. RNA Polymerase II Collision Interrupts Convergent Transcription. Mol Cell. 2012;48:365–374. doi: 10.1016/j.molcel.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pelechano V, Steinmetz LM. NON-CODING RNA Gene regulation by antisense transcription. Nat Rev Genet. 2013;14:880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- 50.Latos PA, Pauler FM, Koerner MV, et al. Airn Transcriptional Overlap, But Not Its lncRNA Products, Induces Imprinted Igf2r Silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 51.Zhao XY, Li SM, Wang GX, Yu Q, Lin JD. A Long Noncoding RNA Transcriptional Regulatory Circuit Drives Thermogenic Adipocyte Differentiation. Mol Cell. 2014;55:372–382. doi: 10.1016/j.molcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hung T, Wang YL, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang P, Xue YQ, Han YM, et al. The STAT3-Binding Long Noncoding RNA lnc-DC Controls Human Dendritic Cell Differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 54.Tripathi V, Ellis JD, Shen Z, et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon JH, Abdelmohsen K, Srikantan S, et al. LincRNA-p21 Suppresses Target mRNA Translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional Gene Regulation by Long Noncoding RNA. J Mol Biol. 2013;425:3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han JJ, Lee Y, Yeom KH, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 58.Liz J, Portela A, Soler M, et al. Regulation of pri-miRNA Processing by a Long Noncoding RNA Transcribed from an Ultraconserved Region. Mol Cell. 2014;55:138–147. doi: 10.1016/j.molcel.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 59.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shukla GC, Singh J, Barik S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 62.Wang GQ, Wang Y, Xiong Y, et al. Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci Rep. 2016;6:21865. doi: 10.1038/srep21865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Pang WJ, Wei N, et al. Identification, stability and expression of Sirt1 antisense long non-coding RNA. Gene. 2014;539:117–124. doi: 10.1016/j.gene.2014.01.037. [DOI] [PubMed] [Google Scholar]
- 64.Kim J, Noh JH, Lee SK, et al. LncRNA OIP5-AS1/cyrano suppresses GAK expression to control mitosis. Oncotarget. 2017;8:49409–49420. doi: 10.18632/oncotarget.17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang JS, Gong CG, Maquat LE. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 2013;27:793–804. doi: 10.1101/gad.212639.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gherzi R, Chen CY, Ramos A, Briata P. KSRP controls pleiotropic cellular functions. Semin Cell Dev Biol. 2014;34:2–8. doi: 10.1016/j.semcdb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 67.Giovarelli M, Bucci G, Ramos A, et al. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc Natl Acad Sci U S A. 2014;111:E5023–E5028. doi: 10.1073/pnas.1415098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gong CG, Li ZZ, Ramanujan K, et al. A Long Non-coding RNA, LncMyoD, Regulates Skeletal Muscle Differentiation by Blocking IMP2-Mediated mRNA Translation. Dev Cell. 2015;34:181–191. doi: 10.1016/j.devcel.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 69.Trimarchi T, Bilal E, Ntziachristos P, et al. Genome-wide Mapping and Characterization of Notch-Regulated Long Noncoding RNAs in Acute Leukemia. Cell. 2014;158:593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu XW, Xiao ZD, Han L, et al. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol. 2016;18:431–442. doi: 10.1038/ncb3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin AF, Li CL, Xing Z, et al. The LINK-A lncRNA activates normoxic HIF1 alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kornfeld JW, Bruning JC. Regulation of metabolism by long, non-coding RNAs. Front Genet. 2014;5:57. doi: 10.3389/fgene.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao XY, Lin JD. Long Noncoding RNAs: A New Regulatory Code in Metabolic Control. Trends Biochem Sci. 2015;40:586–596. doi: 10.1016/j.tibs.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li P, Ruan XB, Yang L, et al. A Liver-Enriched Long Non-Coding RNA, lncLSTR, Regulates Systemic Lipid Metabolism in Mice. Cell Metab. 2015;21:455–467. doi: 10.1016/j.cmet.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lang KC, Lin IH, Teng HF, et al. Simultaneous overexpression of Oct4 and Nanog abrogates terminal myogenesis. Am J Physiol Cell Physiol. 2009;297:C43–C54. doi: 10.1152/ajpcell.00468.2008. [DOI] [PubMed] [Google Scholar]
- 76.Wang LJ, Zhao Y, Bao XC, et al. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015;25:335–350. doi: 10.1038/cr.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun L, Goff LA, Trapnell C, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wei SJ, Du M, Jiang ZH, Hausman GJ, Zhang LF, Dodson MV. Long noncoding RNAs in regulating adipogenesis: new RNAs shed lights on obesity. Cell Mol Life Sci. 2016;73:2079–2087. doi: 10.1007/s00018-016-2169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pang WJ, Lin LG, Xiong Y, et al. Knockdown of PU.1 AS lncRNA Inhibits Adipogenesis Through Enhancing PU.1 mRNA Translation. J Cell Biochem. 2013;114:2500–2512. doi: 10.1002/jcb.24595. [DOI] [PubMed] [Google Scholar]
- 80.Wei N, Wang Y, Xu RX, et al. PU.1 antisense lncRNA against its mRNA translation promotes adipogenesis in porcine preadipocytes. Anim Genet. 2015;46:133–140. doi: 10.1111/age.12275. [DOI] [PubMed] [Google Scholar]
- 81.Alvarez-Dominguez JR, Bai ZQ, Xu D, et al. De Novo Reconstruction of Adipose Tissue Transcriptomes Reveals Long Non-coding RNA Regulators of Brown Adipocyte Development. Cell Metab. 2015;21:764–776. doi: 10.1016/j.cmet.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Turner M, Galloway A, Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat Immunol. 2014;15:484–491. doi: 10.1038/ni.2887. [DOI] [PubMed] [Google Scholar]
- 83.Knoll M, Lodish HF, Sun L. Long non-coding RNAs as regulators of the endocrine system. Nat Rev Endocrinol. 2015;11:151–160. doi: 10.1038/nrendo.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Satpathy AT, Chang HY. Long Noncoding RNA in Hematopoiesis and Immunity. Immunity. 2015;42:792–804. doi: 10.1016/j.immuni.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 85.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martin L, Chang HY. Uncovering the role of genomic “dark matter” in human disease. J Clin Invest. 2012;122:1589–1595. doi: 10.1172/JCI60020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu BD, Sun LJ, Liu Q, et al. A Cytoplasmic NF-kappa B Interacting Long Noncoding RNA Blocks I kappa B Phosphorylation and Suppresses Breast Cancer Metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 91.Wang YY, He L, Du Y, et al. The Long Noncoding RNA IncTCF7 Promotes Self-Renewal of Human Liver Cancer Stem Cells through Activation of Wnt Signaling. Cell Stem Cell. 2015;16:413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 92.Prensner JR, Iyer MK, Sahu A, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takayama K, Horie-Inoue K, Katayama S, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013;32:1665–1680. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]