Abstract
Background
The mammalian target of rapamycin (mTOR) pathway plays a significant role in osteogenic differentiation and bone maintenance. As the only known endogenous inhibitor of mTOR function, DEP domain containing mTOR interacting protein (DEPTOR) is potentially involved in stem cell differentiation, although the pathophysiological significance and its molecular mechanisms remain unclear. The present study aimed to elucidate the effects of DEPTOR on the progress of osteoporosis and investigate the underlying molecular mechanisms of osteogenic regulation.
Methods
An ovariectomy mouse model with decreased bone formation and osteogenic induction with bone marrow mesenchymal stem cells (BMSCs) were used to investigate the relationship between DEPTOR and osteogenic events. A loss-of-function investigation was then performed to explore the role of DEPTOR in the osteogenic differentiation of BMSCs both in vitro and in vivo. Finally, long noncoding RNA (lncRNA) and mRNA sequences were investigated to reveal the underlying mechanisms of DEPTOR in osteogenic regulation. RNA interference, western blotting, and chromatin immunoprecipitation assays were performed for further mechanistic determination.
Results
The results indicated that DEPTOR contributes to the progress of osteoporosis, and higher expression of Deptor was observed in osteoporotic bones. The expression of DEPTOR was reduced during the osteogenic differentiation of BMSCs, and knockdown of DEPTOR promoted BMSC osteogenesis in vitro and in vivo. lncRNA and mRNA sequences indicated that knockdown of DEPTOR upregulated the expression of maternally expressed 3 (nonprotein coding) (MEG3), which subsequently activated bone morphogenetic protein 4 (BMP4) signaling. Furthermore, DEPTOR could bind to a specific region (− 1000 bp ~ 0) of the MEG3 promoter to regulate its transcription, and inhibition of MEG3 reduced BMP4 activation triggered by DEPTOR knockdown.
Conclusions
Taken together, our study revealed a novel function of DEPTOR in osteogenic differentiation by inhibiting MEG3-mediated activation of BMP4 signaling, which suggested that DEPTOR could be a therapeutic target for bone loss diseases and skeletal tissue regeneration.
Electronic supplementary material
The online version of this article (10.1186/s13287-018-0935-9) contains supplementary material, which is available to authorized users.
Keywords: DEP domain containing mTOR interacting protein, Osteoporosis, Osteogenesis, Maternally expressed 3 (nonprotein coding), Bone morphogenetic protein 4, Bone engineering
Background
Osteoporosis is one of the most severe skeletal diseases in the elderly population and postmenopausal women [1]. It is characterized by decreased bone mineral density and destruction of the microarchitectural bone structure, resulting in higher susceptibility to bone fracture [2, 3]. Bone marrow mesenchymal stem cells (BMSCs) are multipotent progenitor cells that can be differentiated into osteoblastic, adipogenic, and chondrogenic lineages [4]. Accumulating research indicates that the aberrant lineage differentiation of endogenous BMSCs contributes to osteoporosis [5–7]. However, the molecular mechanisms by which BMSCs undergo committed osteogenic differentiation remain unclear, hindering the clinical treatment of osteoporosis.
As a serine/threonine kinase that responds to extracellular signaling, the mammalian target of rapamycin (mTOR) is a critical modulator of cell differentiation and tissue regeneration [8, 9]. Emerging studies have demonstrated that mTOR signaling plays a critical role in the osteogenic differentiation of MSCs [10–12]. mTORC1 is recognized as a common effector that mediates bone-related signaling, such as via WNT and bone morphogenetic protein (BMP) [9]. In addition, recent genetic research in mice indicated that impairment of mTORC1 function resulted in increased osteogenesis, while impairment of mTORC2 function led to reduced osteogenesis [12].
DEP domain containing mTOR interacting protein (DEPTOR) is a pivotal protein belonging to the mTORC1 and mTORC2 complexes [13, 14]. As an endogenous mTOR inhibitor, DEPTOR participates in multiple biological events, such as cell growth, apoptosis, autography, and cell differentiation [15]. DEPTOR could act as a stemness factor that plays an important role in the pluripotent maintenance of embryonic stem cells (ESCs). Knockdown of DEPTOR promoted ESC differentiation with a corresponding increase of mTORC1 activity [16]. Moreover, a recent study indicated that DEPTOR promoted adipogenic differentiation via activation of the AKT/PKB-PPAR-γ pathway [17]. Our previous research revealed that DEPTOR could function as a negative regulator in miR-375-mediated osteogenesis through feedback activation of IRS1/PI3K/AKT signaling [18].
Given the significance of DEPTOR in osteogenic/adipogenic regulation, the role and function of DEPTOR in bone metabolism and tissue engineering requires further study. In the present study, we demonstrated that DEPTOR participated in the progress of osteoporosis, and higher expression of Deptor was observed in osteoporotic bones. The expression of DEPTOR was reduced during the osteogenic differentiation of BMSCs, and knockdown of DEPTOR promoted BMSC osteogenesis, not only in vitro but also in vivo. Furthermore, long noncoding RNA (lncRNA) and mRNA sequencing indicated that reduction of DEPTOR upregulated the expression of maternally expressed 3 (nonprotein coding) (MEG3), which subsequently activated bone morphogenetic protein 4 (BMP4)/SMAD1/5/8 signaling, suggesting that DEPTOR could be a therapeutic target for bone loss diseases and skeletal tissue regeneration.
Methods
Cell culture and osteogenic induction
All materials were obtained from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise stated. Minimum essential medium alpha (α-MEM), fetal bovine serum (FBS), and 100× antibiotics were purchased from Gibco (Grand Island, NY, USA). Primary human bone marrow mesenchymal stem cells (hBMSCs) from three different healthy donors were obtained from ScienCell (7500; San Diego, CA, USA). Cells between passages 3 and 5 were utilized for the experiments and all of the in-vitro experiments were repeated in triplicate. hBMSCs were cultured in proliferation medium (PM) containing fresh α-MEM, 10% (v/v) FBS, and 1% (v/v) antibiotics at 37 °C in an incubator with an atmosphere consisting of 95% air and 5% CO2, with 100% relative humidity. For osteogenic induction, hBMSCs were cultured in osteogenic medium (OM), consisting of fresh α-MEM, 10% (v/v) FBS, 1% (v/v) antibiotics, 10 mmol·L− 1 β-glycerophosphate, 0.2 mmol·L− 1 l-ascorbic acid, and 100 nmol·L− 1 dexamethasone.
Lentivirus infection
Recombinant lentiviruses expressing short hairpin RNAs targeting DEPTOR (shDEPTOR #1, shDEPTOR #2) and the scrambled control (shNC) were purchased from GenePharma (Shanghai, China); the sequences are presented in Additional file 1: Table S1. hBMSCs were exposed with the viral supernatant at a multiplicity of infection of 100 together with polybrene (5 μg·ml− 1) for 24 h. Puromycin at 1 μg·ml− 1 was then added to the culture medium for at least 3 days to establish stable cell lines.
RNA sequencing
Total RNAs of each sample were extracted using TruSeq RNA sample preparation reagents (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions, with fragmentation for 4 min at 94 °C. The amplified fragmented cDNA ~ 300 bp in size were sequenced in paired-end mode using an Illumina HiSeq instrument, and two FASTQ files were generated for each sample. The alignment of the reads onto the reference genome and splice site identification were performed using Bowtie/TopHat, with mapping allowing up to two mismatches. The aligned reads were then assembled into transcripts using Cufflinks software, and then computed as normalized values termed fragments per kilobase of exon per million fragments mapped (FPKM). Statistical analysis of differentially expressed genes and transcript splice variants was performed using Cuffdiff, with a false discovery rate (FDR) of 5%.
Ovariectomy operation of mice
Eight-week-old female C57BL/6 mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA) and maintained in pathogen-free facilities on a 12-h light/dark cycle. Bilateral ovariectomy (OVX) or sham operation (N = 10 per group) was conducted as described previously [19]. The mice were sacrificed by CO2 asphyxiation 3 months after surgery. Femurs from the sham or OVX mice were dissected free of soft tissue, and analyzed using high-resolution Inveon microtomography (Siemens, Munich, Germany) after fixation in 4% paraformaldehyde. Images were acquired at an effective pixel size of 8.82 μm, a voltage of 80 kV, a current of 500 μA, and an exposure time of 1500 ms in each of the 360 rotational steps. Parameters of the trabecular region (1–2 mm distal to the proximal epiphysis), including bone volume/total volume (BV/TV), trabecular bone thickness (Tb.Th), and trabecular bone number (Tb.N), were measured using an Inveon Research Workplace (Siemens) according to the guidelines set by the American Society for Bone and Mineral Research (ASBMR). For histological analysis, the femurs were decalcified with 10% EDTA (pH 7.4), dehydrated, and then embedded in paraffin. Slices (5-μm thick) were prepared and stained with hematoxylin and eosin (H&E).
Immunofluorescence staining of mouse femurs
After fixation in 4% paraformaldehyde at 4 °C overnight, femurs of the sham or OVX mice were decalcified in 14% EDTA (pH 7.4) at room temperature, and then washed in phosphate-buffered saline (PBS) for 2 h. After soaking in 30% sucrose with constant agitation at 4 °C overnight, the femurs were embedded in 22-oxa-1,25-dihydroxyvitamin D3 optimum cutting temperature (OCT) compound, and cut into 30-μm thick sections. The primary antibody against Deptor (1:200 dilution; Proteintech, Chicago, IL, USA) was incubated with the slices overnight at 4 °C. Alexa Fluor-546-labeled-goat anti-rabbit IgG (1:1000 dilution; Invitrogen, Carlsbad, CA, USA) was utilized as the secondary antibody, and cell nuclei were counterstained with 2-(4-amidinophenyl)-1H-indole-6-carboxamidine (DAPI) (Invitrogen).
Isolation of primary mBMSCs
The ends of the femurs from sham and OVX mice were removed with scissors, and the bone marrow was harvested by inserting a syringe needle (27 gauge) into one end of the femur and flushing with α-MEM. After centrifuging at 1000 rpm for 5 min, the cells were collected and cultured in fresh α-MEM mixed with 20% (v/v) FBS and 1% (v/v) antibiotics. The medium was replaced every 2 days to remove the nonadherent cells until full confluency was obtained, and the mouse bone marrow mesenchymal stem cells (mBMSCs) at the first passage were used for the in-vitro experiments to analyze the expression of Deptor in osteoporotic conditions.
RNA interference
Short interfering (si)RNAs targeting MEG3 (si-MEG3 #1, si-MEG3 #2) and the scramble control (si-NC) were obtained from GenePharma, and the sequences are presented in Additional file 1: Table S1. Transfection of hBMSCs was conducted with Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. Cells were harvested 48 h after transfection for RNA and protein analyses. As for the osteogenic induction, transfection was reconducted every 5 days to guarantee the knockdown efficiency, and cells were collected 7 or 14 days after osteogenic differentiation.
Chemical administration
Recombinant human Noggin (6057-NG) was purchased from R&D Systems Inc. (Minneapolis, MN, USA). Noggin was reconstituted in sterile PBS containing 0.1% bovine serum albumin (BSA) and administrated at 500 ng·ml− 1 according to previous studies [20, 21].
Alkaline phosphatase staining and quantification
Cells were seeded in six-well plates, and cultured in PM or OM for 7 days. Alkaline phosphatase (ALP) staining was conducted using a nitro-blue tetrazolium chloride (NBT)/5-bromo-4-chloro-3-indolylphosphate toluidine (BCIP) staining kit (CoWin Biotech, Beijing, China) after fixation in 95% ethanol at room temperature. ALP activity was measured using an ALP assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s instructions, and normalized to the total protein contents as determined using the BCA method (Thermo Fisher Scientific, Rockford, IL, USA).
Alizarin red S staining and quantification
Cells were seeded in six-well plates, and cultured in PM or OM for 14 days. Alizarin red S (ARS) staining was performed with 1% ARS (pH 4.2) after fixation in 4% paraformaldehyde. To quantify the degree of mineralization, the stains were dissolved in 100 mmol·L− 1 cetylpyridinium chloride and the absorbance was measured at 562 nm.
von Kossa staining
Cells were seeded in six-well plates, and cultured in PM or OM for 21 days. The cells were fixed incubated with 5% silver nitrate for 30 min in a dark room after fixation in 4% paraformaldehyde, and then exposed to a 60-W UV lamp for 1 h. Unincorporated silver nitrate was dissolved with 5% sodium thiosulfate, and then washed with MilliQ water.
Heterotopic bone formation assay in vivo
Eight-week-old male BALB/c nude (nu/nu) mice were obtained from Jackson Laboratory and housed in pathogen-free facilities under a 12-h light and 12-h dark cycle. hBMSCs stably infected with shDEPTOR #1, shDEPTOR #2, and shNC mixed with beta-tricalcium phosphate particles (SynthoGraft; Bicon, Boston, MA, USA) were transplanted subcutaneously under the dorsal space of the nude mice (N = 6 per group). The mice were sacrificed by CO2 asphyxiation 8 weeks after transplantation. The specimens were decalcified with 10% EDTA (pH 7.4), dehydrated, and then embedded in paraffin. Slices (5-μm thick) were prepared and stained with H&E and Masson’s trichrome. Immunohistochemical (IHC) staining using antibodies against Ocn (Osteocalcin, 1:200 dilution; Abcam, Cambridge, UK) and Deptor (1:100 dilution; Proteintech) was performed with a diaminobenzidine (DAB) Staining kit (ZSGB-BIO, Beijing, China) under the manufacturer’s instructions.
RNA extraction, reverse transcription, and quantitative real-time PCR
Total cellular RNAs were extracted using the TRIzol reagent (Invitrogen). Reverse transcription was conducted with a PrimeScript RT Reagent Kit (Takara Bio Inc., Shiga, Japan) under the manufacturer’s instructions. Quantitative real-time PCR (qPCR) was performed with a SYBR Green Master Mix (Roche Applied Science, Mannheim, Germany). A 7500 Real-Time PCR Detection System (Applied Biosystems, Foster City, CA, USA) was used to detect gene expression, and the thermal settings used were as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. The primers are presented in Additional file 1: Table S1, and the expression of GAPDH or Gapdh was utilized for normalization. The data were analyzed using the 2–ΔΔCt relative expression method.
Western blotting analysis
Western blotting analysis was conducted as reported previously [18]. Briefly, cells were lysed on ice with radioimmunoprecipitation assay (RIPA) buffer and a protease inhibitor cocktail (Roche Applied Science). The lysates were then centrifuged at 12000 rpm for 30 min to remove the debris. Proteins were separated by 10% SDS-PAGE and transferred to 0.45-μm polyvinylidene fluoride (PVDF) membranes. Primary antibodies against runt related transcription factor 2 (RUNX2), DEPTOR, BMP4, p-SMAD1/5/8, SMAD1 (Cell Signaling Technology, Beverly, MA, USA), OCN, and GAPDH (Abcam) were diluted 1:1000 and incubated with the membranes at 4 °C overnight. Secondary antibodies against rabbit or mouse (Cell Signaling Technology) were diluted 1:10,000 and incubated with the membranes at room temperature for 1 h. An ECL kit (CoWin Biotech) was utilized to visualize the immunoreactive proteins, and the band intensities were analyzed using ImageJ software (https://imagej.nih.gov/ij/) for quantitative calculation.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed using an EZ-Magna ChIP assay kit (Merck Millipore, Darmstadt, Germany) according to the manufacturer’s instructions. hBMSCs were seeded in 10-cm dishes, and cells were cross-linked with 1% formaldehyde after they reached 100% confluency. The cell lysate was then sonicated into DNA fragments, and the DNA–protein complexes were isolated using antibodies against DEPTOR (Merck Millipore) and isotype IgG (Cell Signaling Technology) at 4 °C overnight with constant rotation. Unbound substances were removed using elution buffer. After reversing the cross-link, the DNA was purified using spin columns, and used for quantitative ChIP-qPCR with a 7500 Real-Time PCR Detection System (Applied Biosystems). The primers specific for the MEG3 promoter are presented in Additional file 1: Table S1. The data were calculated using the 2–ΔΔCt relative expression method. Relative enrichment was then analyzed as the amount of amplified DNA normalized to the input and relative to values obtained from immunoprecipitation with isotype IgG.
Statistical analysis
Statistical analysis was performed using SPSS Statistics 20.0 software (IBM, Armonk, NY, USA). All values were expressed as the mean ± SE from three independent experiments. Student’s t test was performed to analyze the differences between two groups, and for multiple comparisons a one-way analysis of variance (ANOVA) followed by Tukey’s test was conducted. P < 0.05 was considered statistically significant.
Results
High DEPTOR levels are associated with reduced bone formation in vivo
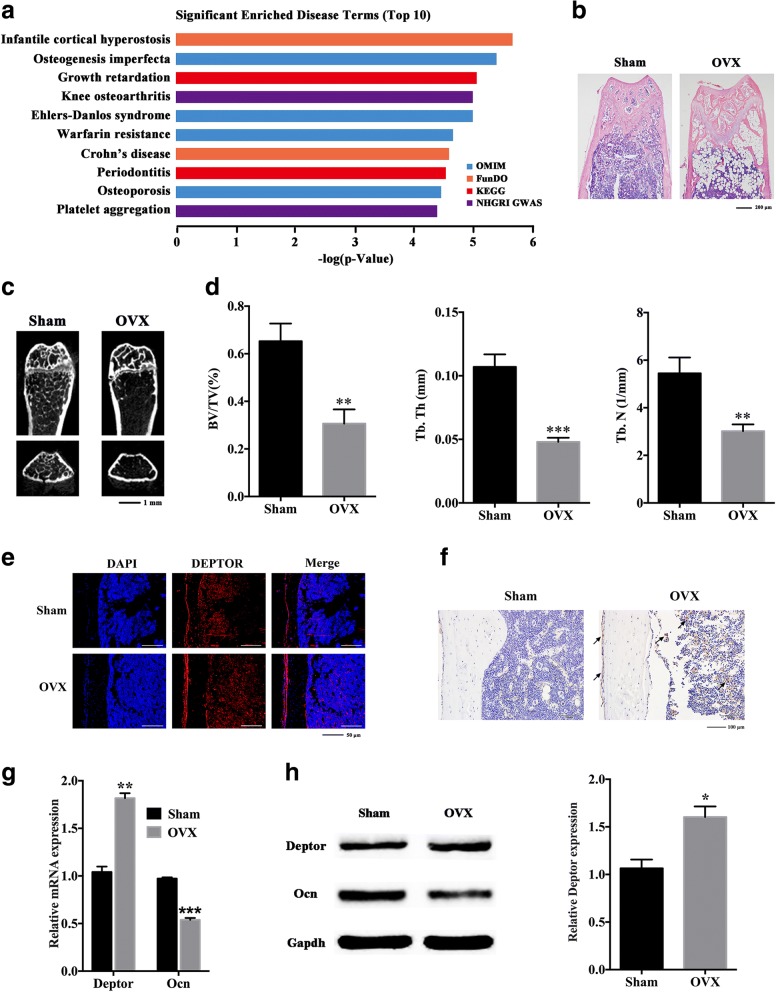
To explore the role of DEPTOR in biological events, we established its global expression profile in BMSCs with DEPTOR knockdown, where NC was utilized as the scramble control. Lentiviruses expressing DEPTOR shRNA were utilized to knockdown DEPTOR in hBMSCs, and two shRNA sequences were generated in case of off-target effects. More than 80% of hBMSCs were GFP positive as shown in Additional file 2: Figure S1a. qRT-PCR and western blotting analysis indicated that the expression of DEPTOR was decreased significantly in the DEPTOR knockdown groups (Additional file 2: Figure S1b, c). According to the disease enrichment analysis, DEPTOR has a significant relationship with bone-associated diseases, such as infantile cortical hyperostosis, osteogenesis imperfecta, periodontitis, and osteoporosis (Fig. 1a). An OVX mouse model was constructed to detect the relationship between Deptor expression and bone formation in vivo. H&E staining, μCT images, and analyses revealed that the trabecular bone of OVX mice was dramatically reduced compared with that in the sham group (Fig. 1b–d). Immunofluorescence analyses of femur sections indicated Deptor expression both in the cortical bone and bone marrow in OVX mice in contrast to that in the sham group (Fig. 1e). Moreover, IHC analyses of Deptor indicated more positive staining both in the cortical bone and bone marrow of OVX mice compared with that in the sham mice, suggesting that the expression of Deptor was elevated in osteoporotic conditions (Fig. 1f). mBMSCs were then isolated from both sham and OVX mice. qRT-PCR and western blotting analysis demonstrated that the expression of Deptor was increased in mBMSCs from OVX mice compared with those from the sham group, while the expression of Ocn, an osteogenic marker, was significantly decreased in mBMSCs from OVX mice (Fig. 1g, h).
Fig. 1.
High DEPTOR levels associated with reduced bone formation in vivo. a Significantly enriched disease terms (top 10) associated with DEPTOR knockdown. b H&E staining of femurs at distal metaphysis growth plate area in Sham and OVX mice (N = 6). Scale bars: 200 μm. c μCT images of femurs at distal metaphysis growth plate area in Sham and OVX mice (N = 6). Scale bars: 1 mm. d Bone volume to total volume (BV/TV), trabecular bone thickness (Tb.Th), and trabecular bone number (Tb.N) reduced in OVX mice (N = 6). e Deptor expression of Sham and OVX mice evaluated by histology and immunofluorescence (N = 6). Scale bars: 50 μm. f Deptor expression of Sham and OVX mice evaluated by IHC (N = 6). Arrows indicate Deptor-positive areas. Scale bars: 100 μm. g Relative mRNA expression of Deptor and Ocn by qRT-PCR in BMSCs isolated from Sham and OVX mice. Gapdh used for normalization. h Left: western blot analysis of Deptor and Ocn protein levels in BMSCs isolated from Sham and OVX mice. Gapdh used as internal control. Right: quantification of band intensities. Data presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 (n = 3 independent experiments). See Additional file 2: Figure S1. DAPI 2-(4-amidinophenyl)-1H-indole-6-carboxamidine, DEPTOR DEP domain containing mTOR interacting protein, GAPDH glyceraldehyde 3-phosphate dehydrogenase, Ocn osteocalcin, OVX ovariectomy
DEPTOR is downregulated during the osteogenic differentiation of hBMSCs
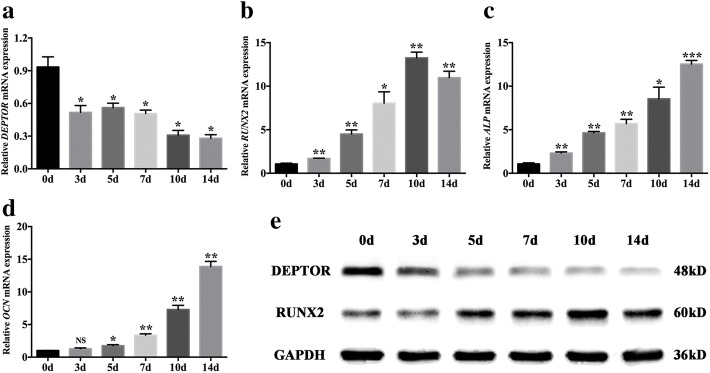
The expression profile of DEPTOR was determined in the osteogenic differentiation of hBMSCs to further explore its effects in vitro. As the qRT-PCR analyses indicated, downregulation of DEPTOR was observed during osteogenic differentiation, accompanied by upregulation of the osteogenic markers RUNX2, ALP, and OCN (Fig. 2a–d). Moreover, western blotting analysis demonstrated a similar decrease in the level of DEPTOR, while the level of RUNX2 increased during the osteogenic differentiation of hBMSCs (Fig. 2e).
Fig. 2.
DEPTOR is downregulated in osteogenic differentiation of hBMSCs. a Relative expression of DEPTOR during osteogenic differentiation of hBMSCs determined by qRT-PCR. GAPDH used for normalization. b–d Relative mRNA levels of osteogenic markers (b) RUNX2, (c) ALP, and (d) OCN during osteogenic differentiation of hBMSCs, determined by qRT-PCR. GAPDH used for normalization. e Western blot analysis of DEPTOR and RUNX2 protein levels during osteogenic differentiation of hBMSCs. GAPDH used as internal control. Data presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, NS not significant (n = 3 independent experiments). ALP alkaline phosphatase, d day, DEPTOR DEP domain containing mTOR interacting protein, GAPDH glyceraldehyde 3-phosphate dehydrogenase, Ocn osteocalcin, RUNX2 runt related transcription factor 2
Knockdown of DEPTOR enhances osteogenic differentiation of hBMSCs in vitro
Lentiviruses were utilized to stably knockdown DEPTOR in hBMSCs to further investigate the effects of DEPTOR on osteogenic differentiation. Seven days after culturing the cells in PM or OM, ALP staining and quantification showed that the ALP activity was remarkably increased in the DEPTOR knockdown groups (Fig. 3a, b). The extracellular mineralization, as revealed by ARS staining and AZR quantification, and von Kossa staining, on day 14 and 21, respectively, was also enhanced in the DEPTOR knockdown groups (Fig. 3a, c). Consistently, qRT-PCR indicated that the expression levels of RUNX2, ALP, Osterix (OSX), and OCN were dramatically increased after DEPTOR knockdown in PM or OM (Fig. 3d–g). Moreover, western blotting analysis demonstrated that the protein levels of RUNX2 and OCN were elevated in the DEPTOR knockdown groups after osteogenic induction on day 7 and 14, respectively (Fig. 3h).
Fig. 3.
Knockdown of DEPTOR promotes osteogenic differentiation of hBMSCs in vitro. hBMSCs transfected with lentivirus expressing DEPTOR shRNAs (shDEPTOR #1, shDEPTOR #2) or scrambled control (shNC), and cultured in proliferation medium (PM) or osteogenic medium (OM). a ALP staining on day 7, ARS staining on day 14, and VK staining on day 21 after proliferative culture or osteogenic induction. b, c ALP activity on day 7 and AZR mineralization assay on day 14 after proliferative culture or osteogenic induction. d, e Relative mRNA levels of ALP and RUNX2 measured by qRT-PCR on day 7 of proliferative culture or osteogenic induction. f, g Relative mRNA levels of OSX and OCN measured by qRT-PCR on day 14 of proliferative culture or osteogenic induction. GAPDH used for normalization. h Left: western blot analysis of RUNX2 and OCN protein levels on day 14 after osteogenic induction. GAPDH used as internal control. Right: quantification of band intensities. Data presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, NS not significant (n = 3 independent experiments). See Additional file 3: Figure S2. DEPTOR DEP domain containing mTOR interacting protein, ALP alkaline phosphatase, ARS/AZR Alizarin red S, VK von Kossa, GAPDH glyceraldehyde 3-phosphate dehydrogenase, Ocn osteocalcin, OSX osterix, RUNX2 runt related transcription factor 2
Knockdown of DEPTOR promotes bone formation in vivo
Eight weeks after implantation, H&E staining showed that more osteoid tissues were formed in the DEPTOR knockdown group compared with the control group (Additional file 3: Figure S2a). Masson’s trichrome staining displayed more collagen deposition (blue color) after DEPTOR knockdown (Additional file 3: Figure S2b). Moreover, IHC staining against OCN demonstrated a higher number of brown granules around the nuclei in the DEPTOR knockdown groups compared with that in the control group (Additional file 3: Figure S2c).
Knockdown of DEPTOR elevates MEG3 expression to regulate osteogenesis
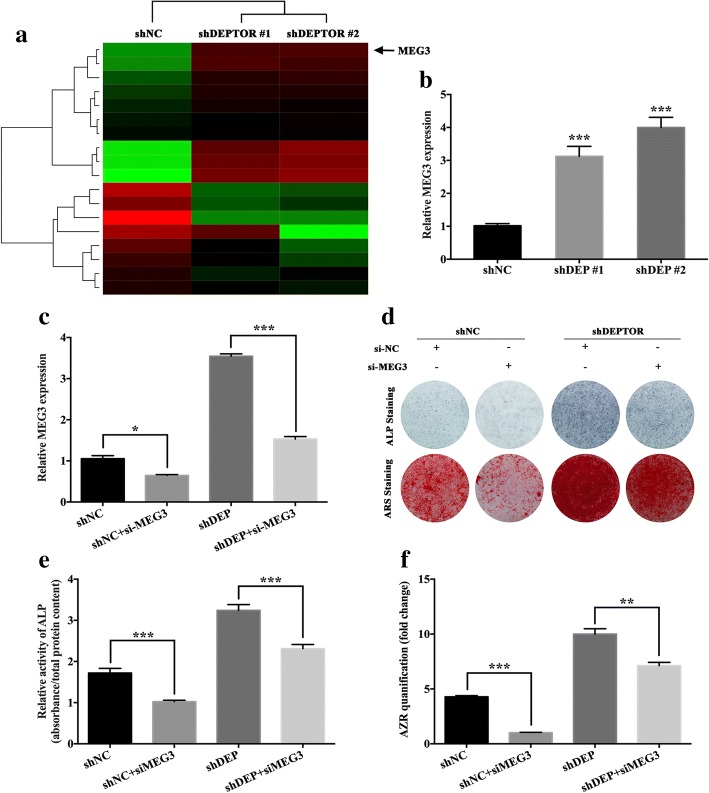
To determine the underlying molecular mechanisms of DEPTOR in the regulation of hBMSC osteogenesis, the expression profiles of both lncRNAs and mRNAs were constructed in the DEPTOR knockdown groups compared with the control group. Seventy-eight lncRNAs were differentially expressed according to the threshold values of fold change ≥ 2.0 and P < 0.05. Among the differentially expressed lncRNAs, MEG3 showed significant upregulation, and qRT-PCR validated that MEG3 was increased in the DEPTOR knockdown cells (Fig. 4a, b). MEG3 was demonstrated to promote osteogenic differentiation of BMSCs in recent studies [22, 23]. We further transfected MEG3 siRNAs into hBMSCs, and the knockdown efficiency was verified using qRT-PCR (Additional file 4: Figure S3a). After culturing the cells in OM for 7 days, ALP staining and quantification indicated that knockdown of MEG3 downregulated ALP activity (Additional file 4: Figure S3b, c). ARS staining and AZR quantification on day 14 for OM culture was also reduced in the MEG3 knockdown groups (Additional file 4: Figure S3b, d). Moreover, to clarify whether DEPTOR regulated hBMSC osteogenesis through MEG3, we transfected the MEG3 siRNA into hBMSCs with DEPTOR knockdown, and the knockdown efficiency of MEG3 was estimated using qRT-PCR (Fig. 4c). Seven days after osteogenic induction, ALP staining and quantification revealed that depletion of MEG3 significantly reduced the ALP activity triggered by DEPTOR knockdown (Fig. 4d, e). ARS staining and AZR quantification on day 14 indicated a similar tendency (Fig. 4d, f).
Fig. 4.
Knockdown of DEPTOR elevates MEG3 expression to regulate osteogenesis. a Hierarchical clustering indicated MEG3 was differentially expressed (P < 0.05) in DEPTOR knockdown hBMSCs. b Relative MEG3 expression measured by qRT-PCR in DEPTOR knockdown hBMSCs. GAPDH used for normalization. c–f MEG3 siRNA (si-MEG3) transfected into hBMSCs with DEPTOR knockdown. NC used as negative control. c Relative MEG3 expression determined by qRT-PCR. GAPDH used for normalization. d ALP staining on day 7 and ARS staining on day 14 after osteogenic induction. e, f ALP activity on day 7 and AZR mineralization assay on day 14 after osteogenic induction. Data presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 (n = 3 independent experiments). See Additional file 4: Figure S3. ALP alkaline phosphatase, ARS/AZR Alizarin red S, DEPTOR DEP domain containing mTOR interacting protein, MEG3 maternally expressed 3 (nonprotein coding), shDEPTOR #1/shDEPTOR #2 lentivirus expressing shRNAs targeting DEPTOR, shNC scrambled control, sh short hairpin, si short interfering
BMP4 signaling is involved in the regulation of hBMSC osteogenesis during DEPTOR knockdown
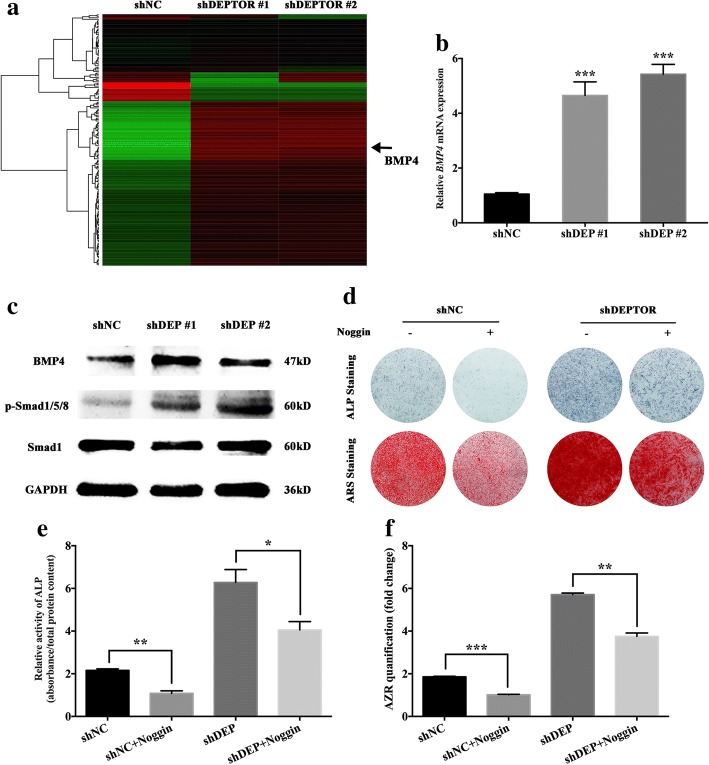
We identified 1107 mRNAs that were differentially expressed between DEPTOR knockdown and control cells, according to the thresholds values of fold change ≥ 2.0 and P < 0.05. Among the differentially expressed mRNAs, genes within the BMP signaling pathway were dramatically upregulated in DEPTOR siRNA cells, in which the upstream BMP4 gene showed a remarkable increase (Fig. 5a). qRT-PCR and western blotting analysis indicated that knockdown of DEPTOR increased the expression of BMP4, leading to more phosphorylated SMAD1/5/8, while the total SMAD1 remained barely changed (Fig. 5b, c). Next, we treated DEPTOR knockdown hBMSCs with the BMP antagonist Noggin. ALP staining and quantification after osteogenic induction for 7 days indicated that delivery of Noggin dramatically attenuated the ALP activity in the DEPTOR knockdown hBMSCs (Fig. 5d, e). Similar outcomes were observed for ARS staining and AZR quantification on day 14 after osteogenic differentiation (Fig. 5d, f).
Fig. 5.
BMP4 signaling involved in regulation of hBMSC osteogenesis after DEPTOR knockdown. a Hierarchical clustering indicated BMP4 was differentially expressed (P < 0.05) in DEPTOR knockdown hBMSCs. b Relative mRNA expression of BMP4 measured by qRT-PCR in DEPTOR knockdown hBMSCs. GAPDH used for normalization. c Western blot analysis of BMP4 and its downstream Smad expression in DEPTOR knockdown hBMSCs. GAPDH utilized as internal control. d–f BMP antagonist Noggin (500 ng·mL− 1) administered to DEPTOR knockdown and NC hBMSCs. d ALP staining on day 7 and ARS staining on day 14 after osteogenic induction. e, f ALP activity on day 7 and AZR quantification on day 14 after osteogenic induction. Data presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 (n = 3 independent experiments). ALP alkaline phosphatase, ARS/AZR Alizarin red S, BMP4 bone morphogenetic protein 4, DEPTOR DEP domain containing mTOR interacting protein, GAPDH glyceraldehyde 3-phosphate dehydrogenase, shDEPTOR #1/shDEPTOR #2 lentivirus expressing shRNAs targeting DEPTOR, shNC scrambled control, sh short hairpin
Knockdown of DEPTOR activates BMP4 signaling through MEG3
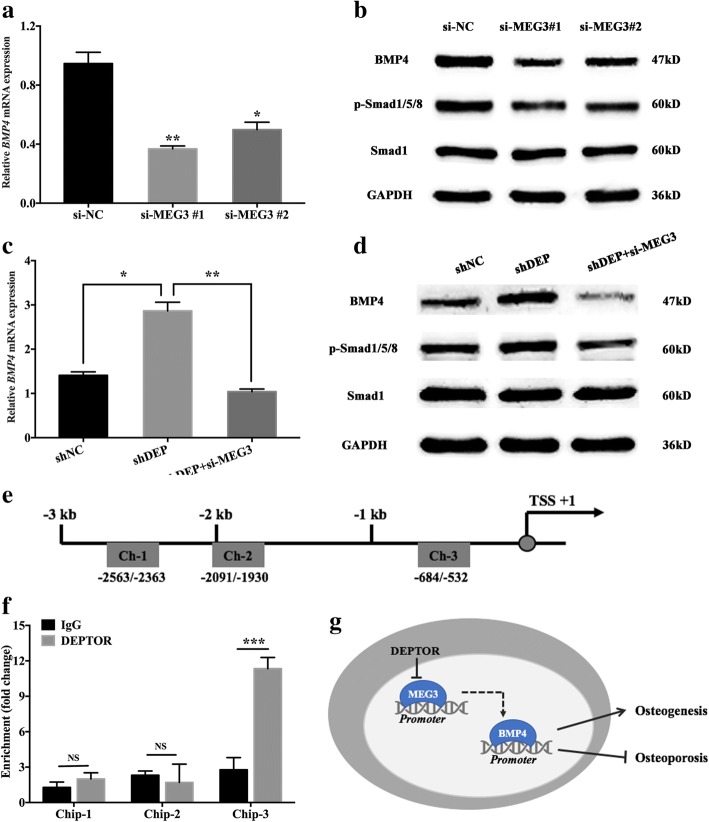
Previous research demonstrated that MEG3 could modulate BMP4 transcription through directly interacting with the core element of the BMP4 promoter [22]. In the present study, we detected BMP4 expression in hBMSCs transfected with MEG3 siRNAs. As the qRT-PCR analysis indicated, the mRNA level of BMP4 was reduced in the MEG3 knockdown groups (Fig. 6a). Moreover, western blotting analysis indicated that knockdown of MEG3 in hBMSCs inhibited BMP4 signaling, which was evidenced by the decrease in the BMP4 level, and the subsequent attenuation of phosphorylated SMAD1/5/8 (Fig. 6b). To further clarify whether DEPTOR modulates BMP4 signaling through MEG3, we established DEPTOR and MEG3 double knockdown cells. qRT-PCR indicated that depletion of MEG3 reduced the BMP4 expression in the DEPTOR knockdown group (Fig. 6c) compared with that in the control group. Moreover, western blotting analysis showed that BMP4 and its downstream phosphorylated SMAD1/5/8 were also decreased in the DEPTOR and MEG3 double knockdown cells compared with that in the DEPTOR knockdown cells (Fig. 6d). DEPTOR might function as a transcriptional regulator; therefore, we further investigated whether DEPTOR was directly responsible for MEG3 transcription [24]. Sequence analysis of the MEG3 promoter, comprising − 3000 bp upstream from the transcription start site (TSS), demonstrated three potential binding sites for DEPTOR (Fig. 6e). ChIP-qPCR showed that DEPTOR could bind to the specific region of the MEG3 promoter, with dramatic enrichment in the third region (− 1000 bp ~ 0) (Fig. 6f).
Fig. 6.
Knockdown of DEPTOR activates BMP4 signaling through MEG3. a Relative mRNA expression of BMP4 in MEG3 knockdown (si-MEG3 #1, si-MEG3 #2) and control vector (si-NC) groups. GAPDH used for normalization. b Western blot analysis of BMP4 and its downstream Smad expression in MEG3 knockdown and NC groups. GAPDH utilized as internal control. c, d MEG3 siRNA (si-MEG3) introduced into DEPTOR knockdown hBMSCs. c Relative mRNA expression of BMP4 measured using qRT-PCR. GAPDH used for normalization. d Western blot analysis of BMP4 and its downstream Smad expression. GAPDH utilized as internal control. e MEG3 promoter and location of primers. Positions marked relative to the transcription start site (TSS). f ChIP-qPCR showing interaction between DEPTOR and MEG3 promoter in hBMSCs. IgG used for normalization. Data presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, NS not significant (n = 3 independent experiments). g Proposed mechanism of DEPTOR modulation of osteogenesis via regulation MEG3-mediated BMP4 activation. BMP4 bone morphogenetic protein 4, Chip chromatin immunoprecipitation, DEPTOR DEP domain containing mTOR interacting protein, GAPDH glyceraldehyde 3-phosphate dehydrogenase, MEG3 maternally expressed 3 (nonprotein coding), shNC scrambled control, sh short hairpin, si short interfering
Discussion
Recent studies indicated that the abnormal lineage commitment of BMSCs is closely related to the bone loss and fat accumulation in bone marrow during osteoporosis [5]. BMSCs derived from elderly people have a decreased tendency to differentiate into osteoblasts, accompanied with an increased tendency toward the adipogenic lineage [25, 26]. As part of the Fob3a quantitative trait locus (QTL), a gene that was linked to obesity/leanness, Deptor, was involved in adipogenic regulation [27]. Our disease enrichment analysis revealed that DEPTOR might be associated with osteoporosis. In the present study, we used OVX mice as an osteoporotic model, which has been commonly recognized as a model of BMSCs with low osteogenic capability [19, 21, 28]. Elevated expression of Deptor was observed in both trabecular bones and BMSCs of osteoporotic mice, indicating that DEPTOR might play an important role during the progress of osteoporosis.
The osteoblastic and adipocytic lineages have been recognized as alternatives during cell differentiation [6, 7]. DEPTOR has been established as a positive regulator of adipogenesis. The expression of Deptor in white adipose tissue (WAT) correlated with the degree of obesity, and its overexpression accumulated more WAT in mice [17], while its effects in the osteogenic differentiation of BMSCs remain elusive. We demonstrated that DEPTOR expression was consistently decreased as BMSCs committed to the osteoblastic lineage. Moreover, knockdown of DEPTOR via lentivirus transfection enhanced the osteogenic differentiation of BMSCs both in vitro and in vivo, indicating DEPTOR as a novel regulator of osteogenesis.
The negative effects of DEPTOR in regulating osteogenesis seem counterintuitive, considering that DEPTOR was reported as an endogenous inhibitor of mTOR, which has controversial effects on the osteogenic differentiation of BMSCs [11, 12]. Functioning in two distinct multiprotein complexes, mTORC1 mainly phosphorylates S6 K1 and 4E-BP1 to control protein synthesis, whereas mTORC2 phosphorylates AKT at Ser 473 to modulate cell survival [9, 29]. It has been reported that deletion of S6 K1 enhanced resistance to age-related bone loss, while ablation of Rictor reduced the osteogenic differentiation of BMSCs [30, 31]. A recent study using Cre-mediated deletion of Rptor (RapKO) or Rictor (RicKO) revealed different roles of mTORC1 and mTORC2 in the osteogenic differentiation of MSCs. With impaired mTORC1 signaling, RapKO MSCs displayed an increased capacity for osteogenesis, while RicKO MSCs demonstrated a reduced capacity for osteogenesis in the presence of limited mTORC2 signaling [12]. This can be accounted for the confounding result that suppression of mTOR signaling with rapamycin had both stimulatory and inhibitory osteogenic effects [32, 33]. As the research showed, rapamycin primarily inhibited mTORC1 activity to promote osteogenesis, while prolonged exposure disrupted mTORC2 function to inhibit osteogenesis [34, 35]. Our previous study demonstrated that knockdown of DEPTOR activated S6 K1, which subsequently enhanced the negative feedback loop on IRS1/PI3K to inhibit AKT signaling, whereas the activity of mTOR remained inactivated as the phosphorylated mTOR was reduced by DEPTOR knockdown [18]. The insufficient activation of mTOR prompted further investigation of how DEPTOR regulates osteogenic differentiation.
Recent studies indicated that, independent of mTOR inhibitors, DEPTOR could function as a transcriptional regulator, and binds to specific promoter regions of genes to regulate endoplasmic reticulum homeostasis, such as PSEN2, CKAP4, and KEAP1 [24]. Recently, lncRNAs were observed to play important roles in the differentiation of MSCs, and high-throughput gene sequencing revealed that more than 1000 differentially expressed lncRNAs were identified during the osteogenic differentiation of BMSCs [36, 37]. We established an RNA-seq analysis to determine the expression profiles of both lncRNAs and mRNAs with DEPTOR knockdown in BMSCs, and 78 lncRNAs and 1107 mRNAs were differentially expressed. Among the differentially expressed lncRNAs, MEG3 has been implicated in the osteogenic differentiation of MSCs [22, 23, 38]. As a maternally expressed imprinted gene representing an lncRNA, MEG3 is widely recognized as a tumor suppressor that is expressed at low levels in some cancers and correlates with prognosis [39, 40]. Previous research demonstrated that MEG3 promoted osteogenic differentiation of MSCs from patients with multiple myeloma, and our recent study displayed a similar promotion of osteogenesis on hASCs with MEG3 overexpression [22, 23]. However, another study of postmenopausal osteoporosis revealed that MEG3 inhibited the osteogenic differentiation of BMSCs [38]. In the present study, we found that inhibition of DEPTOR remarkably upregulated the expression of MEG3, and further investigation indicated that DEPTOR could directly bind to a specific region (− 1000 bp ~ 0) of the MEG3 promoter to regulate its transcription. Moreover, inhibition of MEG3 reduced the enhancement of osteogenic differentiation triggered by DEPTOR knockdown, suggesting that DEPTOR regulated BMSC osteogenesis through MEG3.
lncRNAs participate in gene expression by regulating the transcriptional activity of neighboring coding regions, and can enhance gene expression over several Mbs [41, 42]. Previous research has shown that MEG3 can dissociate the transcriptionally inhibitory factor SOX2 from the BMP4 promoter to enhance BMP4 expression [22]. Here we revealed that knockdown of DEPTOR increased BMP4 expression, which subsequently phosphorylated Smad1/5/8 to activate Smad signaling. Furthermore, when treated with Noggin, a BMP4 antagonist, BMSCs with DEPTOR knockdown exhibited reduced capacity toward osteogenesis. As a member of the transforming growth factor beta (TGF-β) family, BMP4 was identified as a regulator of bone and cartilage formation [43]. Emerging evidence indicates that BMP signaling plays a pivotal role in osteogenic differentiation and bone formation [44, 45]. Given the relationship between MEG3 and BMP4, we speculated that knockdown of DEPTOR could activate BMP4 signaling through MEG3. As the results suggested, depletion of MEG3 reduced the activation of BMP4 signaling triggered by DEPTOR knockdown.
Conclusions
In this study, we determined the functions of DEPTOR in the osteogenic differentiation of BMSCs using a cellular biology approach and an osteoporotic animal approach that represents a BMSC model with low osteogenic capability. Our findings increased our understanding of the regulatory mechanisms of the osteogenic differentiation of BMSCs, and might contribute to bone tissue engineering and the treatment of osteoporotic diseases.
Additional files
Table S1. Sequences of RNA and DNA oligonucleotides. (DOCX 17 kb)
Figure S1. Transduction efficiency of lentivirus expressing DEPTOR shRNAs (shDEPTOR #1, shDEPTOR #2) or scrambled control (shNC) in hBMSCs. (TIF 1443 kb)
Figure S2. Knockdown of DEPTOR promotes bone formation in vivo. (TIF 1782 kb)
Figure S3. Knockdown of MEG3 reduces osteogenic differentiation of hBMSCs. (TIF 1035 kb)
Acknowledgments
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers 81570953, 81402235, 81700938), the Tason Stomatological Development Foundation (grant numbers 2015), and the Project for Culturing Leading Talents in Scientific and Technological Innovation of Beijing (grant numbers Z171100001117169).
Availability of data and materials
All data generated or analyzed during this study are included in this published article. The datasets supporting the conclusions of this article are available in the SRA repository with accession number SRP132791 (https://www.ncbi.nlm.nih.gov/sra/SRP132791).
Abbreviations
- ALP
Alkaline phosphatase
- ANOVA
Analysis of variance
- ARS/AZR
Alizarin red S
- ASBMR
American Society for Bone and Mineral Research
- BMSC
Bone marrow mesenchymal stem cell
- BV/TV
Bone volume/total volume
- DEPTOR
DEP domain containing mTOR interacting protein
- ESC
Embryonic stem cell
- FBS
Fetal bovine serum
- FDR
False discovery rate
- FPKM
Fragments per kilobase of exon per million fragments mapped
- H&E
Hematoxylin and eosin
- hBMSC
Human bone marrow mesenchymal stem cell
- IHC
Immunohistochemical
- mBMSC
Mouse bone marrow mesenchymal stem cell
- mTOR
Mammalian target of rapamycin
- OCT
Optimum cutting temperature
- OM
Osteogenic medium
- OSX
Osterix
- OVX
Ovariectomy
- PM
Proliferation medium
- qRT-PCR
Quantitative real-time reverse transcription PCR
- QTL
Quantitative trait locus
- Tb.N
Trabecular bone number
- Tb.Th
Trabecular bone thickness
- TGF-β
Transforming growth factor beta
- TSS
Transcription start site
- WAT
White adipose tissue
- α-MEM
Minimum essential medium alpha
Authors’ contributions
SC, LfJ, and SZ conducted the study, and collected and analyzed the data. SC, LfJ, and YfZ drafted the manuscript. SC, YfZ, and YsZ revised the manuscript. All authors approved the final version of the manuscript for submission.
Ethics approval and consent to participate
All applicable institutional and national guidelines for the care and use of animals were followed, and all animal experiments were approved by the Ethics Committee of Peking University Health Science Center (approval accession: LA2014223).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13287-018-0935-9) contains supplementary material, which is available to authorized users.
Contributor Information
Si Chen, Email: sysuchensi@126.com.
Lingfei Jia, Email: jialingfei1984@sina.com.
Shan Zhang, Email: amile_amy@sina.com.
Yunfei Zheng, Phone: +86-10-82195775, Email: yunfei_zheng@bjmu.edu.cn.
Yongsheng Zhou, Phone: +86-10-82195370, Email: kqzhouysh@hsc.pku.edu.cn.
References
- 1.Watts NB, GLOW investigators Insights from the global longitudinal study of osteoporosis in women (GLOW) Nat Rev Endocrinol. 2014;10:412–422. doi: 10.1038/nrendo.2014.55. [DOI] [PubMed] [Google Scholar]
- 2.Farr JN, Khosla S. Skeletal changes through the lifespan—from growth to senescence. Nat Rev Endocrinol. 2015;11:513–521. doi: 10.1038/nrendo.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cauley JA. Osteoporosis: fracture epidemiology update 2016. Curr Opin Rheumatol. 2017;29:150–156. doi: 10.1097/BOR.0000000000000365. [DOI] [PubMed] [Google Scholar]
- 4.Grayson WL, Bunnell BA, Martin E, Frazier T, Hung BP, Gimble JM. Stromal cells and stem cells in clinical bone regeneration. Nat Rev Endocrinol. 2015;11:140–150. doi: 10.1038/nrendo.2014.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verma S, Rajaratnam JH, Denton J, Hoyland JA, Byers RJ. Adipocytic proportion of bone marrow is inversely related to bone formation in osteoporosis. J Clin Pathol. 2002;55:693–698. doi: 10.1136/jcp.55.9.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jing H, Liao L, An Y, Su X, Liu S, Shuai Y, et al. Suppression of EZH2 prevents the shift of osteoporotic MSC fate to adipocyte and enhances bone formation during osteoporosis. Mol Ther. 2016;24:217–229. doi: 10.1038/mt.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li CJ, Cheng P, Liang MK, Chen YS, Lu Q, Wang JY, et al. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J Clin Invest. 2015;125:1509–1522. doi: 10.1172/JCI77716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu JS, Cui W. Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signaling in pluripotency and cell fate determination. Development. 2016;143:3050–3060. doi: 10.1242/dev.137075. [DOI] [PubMed] [Google Scholar]
- 9.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Long F. mTOR signaling in skeletal development and disease. Bone Res. 2018;6:1. doi: 10.1038/s41413-017-0004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sen B, Xie Z, Case N, Thompson WR, Uzer G, Styner M, et al. mTORC2 regulates mechanically induced cytoskeletal reorganization and lineage selection in marrow-derived mesenchymal stem cells. J Bone Miner Res. 2014;29:78–89. doi: 10.1002/jbmr.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin SK, Fitter S, Dutta AK, Matthews MP, Walkley CR, Hall MN, et al. Brief report: the differential roles of mTORC1 and mTORC2 in mesenchymal stem cell differentiation. Stem Cells. 2015;33:1359–1365. doi: 10.1002/stem.1931. [DOI] [PubMed] [Google Scholar]
- 13.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dibble CC, Cantley LC. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015;25:545–555. doi: 10.1016/j.tcb.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Catena V, Fanciulli M. Deptor: not only a mTOR inhibitor. J Exp Clin Cancer Res. 2017;36:12. doi: 10.1186/s13046-016-0484-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agrawal P, Reynolds J, Chew S, Lamba DA, Hughes RE. DEPTOR is a stemness factor that regulates pluripotency of embryonic stem cells. J Biol Chem. 2014;289:31818–31826. doi: 10.1074/jbc.M114.565838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laplante M, Horvat S, Festuccia WT, Birsoy K, Prevorsek Z, Efeyan A, et al. DEPTOR cell-autonomously promotes adipogenesis, and its overexpression is associated with obesity. Cell Metab. 2012;16:202–212. doi: 10.1016/j.cmet.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S, Zheng Y, Zhang S, Jia L, Zhou Y. Promotion effects of miR-375 on the osteogenic differentiation of human adipose-derived mesenchymal stem cells. Stem Cell Reports. 2017;8:773–786. doi: 10.1016/j.stemcr.2017.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv L, Ge W, Liu Y, Lai G, Liu H, Li W, et al. Lysine-specific demethylase 1 inhibitor rescues the osteogenic ability of mesenchymal stem cells under osteoporotic conditions by modulating H3K4 methylation. Bone Res. 2016;4:16037. doi: 10.1038/boneres.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiba S, Lee YM, Zhou W, Freed CR. Noggin enhances dopamine neuron production from human embryonic stem cells and improves behavioral outcome after transplantation into parkinsonian rats. Stem Cells. 2008;26:2810–2820. doi: 10.1634/stemcells.2008-0085. [DOI] [PubMed] [Google Scholar]
- 21.Jin C, Zhang P, Zhang M, Zhang X, Lv L, Liu H, et al. Inhibition of SLC7A11 by sulfasalazine enhances osteogenic differentiation of mesenchymal stem cells by modulating BMP2/4 expression and suppresses bone loss in ovariectomized mice. J Bone Miner Res. 2017;32:508–521. doi: 10.1002/jbmr.3009. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W, Ge X, Yang S, Huang M, Zhuang W, Chen P, et al. Upregulation of lncRNA MEG3 promotes osteogenic differentiation of mesenchymal stem cells from multiple myeloma patients by targeting BMP4 transcription. Stem Cells. 2015;33:1985–1997. doi: 10.1002/stem.1989. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Jin C, Chen S, Zheng Y, Huang Y, Jia L, et al. Long non-coding RNA MEG3 inhibits adipogenesis and promotes osteogenesis of human adipose-derived mesenchymal stem cells via miR-140-5p. Mol Cell Biochem. 2017;433:51–60. doi: 10.1007/s11010-017-3015-z. [DOI] [PubMed] [Google Scholar]
- 24.Catena V, Bruno T, De Nicola F, Goeman F, Pallocca M, Lezzi S, et al. Deptor transcriptionally regulates endoplasmic reticulum homeostasis in multiple myeloma cells. Oncotarget. 2016;7:70546–70558. doi: 10.18632/oncotarget.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moerman EJ, Teng K, Lipschitz DA, Lecka-Czernik B. Aging activates adipogenic and suppresses osteogenic programs in mesenchymal marrow stroma/stem cells: the role of PPAR-gamma2 transcription factor and TGF-beta/BMP signaling pathways. Aging Cell. 2004;3:379–389. doi: 10.1111/j.1474-9728.2004.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen W, Chen J, Gantz M, Punyanitya M, Heymsfield SB, Gallagher D, et al. MRI-measured pelvic bone marrow adipose tissue is inversely related to DXA-measured bone mineral in younger and older adults. Eur J Clin Nutr. 2012;66:983–988. doi: 10.1038/ejcn.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stylianou IM, Clinton M, Keightley PD, Pritchard C, Tymowska-Lalanne Z, Bunger L, et al. Microarray gene expression analysis of the Fob3b obesity QTL identifies positional candidate gene Sqle and perturbed cholesterol and glycolysis pathway. Physiol Genomics. 2005;20:224–232. doi: 10.1152/physiolgenomics.00183.2004. [DOI] [PubMed] [Google Scholar]
- 28.Qi M, Zhang L, Ma Y, Shuai Y, Li L, Luo K, et al. Autography maintains the function of bone marrow mesenchymal stem cells to prevent estrogen deficiency-induced osteoporosis. Theranostics. 2017;7:4498–4516. doi: 10.7150/thno.17949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 30.Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J, Holguin N, Shi Y, Silva MJ, Long F. mTORC2 signaling promotes skeletal growth and bone formation in mice. J Bone Miner Res. 2015;30:369–378. doi: 10.1002/jbmr.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isomoto S, Hattori K, Ohgushi H, Nakajima H, Tanaka Y, Takakura Y. Rapamycin as an inhibitor of osteogenic differentiation in bone marrow-derived mesenchymal stem cells. J Orthop Sci. 2007;12:83–88. doi: 10.1007/s00776-006-1079-9. [DOI] [PubMed] [Google Scholar]
- 33.Lee KW, Yook JY, Son MY, Kim MJ, Koo DB, Han YM, et al. Rapamycin promotes the osteoblastic differentiation of human embryonic stem cells by blocking the mTOR pathway and stimulating the BMP/Smad pathway. Stem Cells Dev. 2010;19:557–568. doi: 10.1089/scd.2009.0147. [DOI] [PubMed] [Google Scholar]
- 34.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly by Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 35.Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song WQ, Gu WQ, Qian YB, Ma X, Mao YJ, Liu WJ. Identification of long non-coding RNA involved in osteogenic differentiation from mesenchymal stem cells using RNA-Seq data. Genet Mol Res. 2015;14:18268–18279. doi: 10.4238/2015.December.23.14. [DOI] [PubMed] [Google Scholar]
- 37.Zhang W, Dong R, Diao S, Du J, Fan Z, Wang F. Differential long noncoding RNA/mRNA expression profiling and functional network analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cell Res Ther. 2017;8:30. doi: 10.1186/s13287-017-0485-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, Li Y, Zhang Y, Ma L, Lin L, Meng J, et al. LncRNA MEG3 inhibits osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis by targeting miR-133a-3p. Biomed Pharmacother. 2017;89:1178–1186. doi: 10.1016/j.biopha.2017.02.090. [DOI] [PubMed] [Google Scholar]
- 39.Mitra R, Chen X, Greenawalt EJ, Maulik U, Jiang W, Zhao Z, et al. Decoding critical long non-coding RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat Commun. 2017;8:1604. doi: 10.1038/s41467-017-01781-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48:R45–R53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;43:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Z, Liu X, Liu L, Deng H, Zhang J, Xu Q, et al. Regulation of lncRNA expression. Cell Mol Biol Lett. 2014;19:561–575. doi: 10.2478/s11658-014-0212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuji K, Cox K, Bandyopadhyay A, Harfe BD, Tabin CJ, Rosen V. BMP4 is dispensable for skeletogenesis and fracture-healing in the limb. J Bone Joint Surg Am. 2008;90(Suppl 1):14–18. doi: 10.2106/JBJS.G.01109. [DOI] [PubMed] [Google Scholar]
- 44.Rahman MS, Akhtar N, Jamil HM, Banik RS, Asaduzzaman SM. TGF-β/BMP signaling and other mulecular events: regulation of osteoblastogenesis and bone formation. Bone Res. 2015;3:11–30. doi: 10.1038/boneres.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salazar VS, Gamer LW, Rosen V. BMP signaling in skeletal development, disease, and repair. Nat Rev Endocrinol. 2016;12:203–221. doi: 10.1038/nrendo.2016.12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Sequences of RNA and DNA oligonucleotides. (DOCX 17 kb)
Figure S1. Transduction efficiency of lentivirus expressing DEPTOR shRNAs (shDEPTOR #1, shDEPTOR #2) or scrambled control (shNC) in hBMSCs. (TIF 1443 kb)
Figure S2. Knockdown of DEPTOR promotes bone formation in vivo. (TIF 1782 kb)
Figure S3. Knockdown of MEG3 reduces osteogenic differentiation of hBMSCs. (TIF 1035 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article. The datasets supporting the conclusions of this article are available in the SRA repository with accession number SRP132791 (https://www.ncbi.nlm.nih.gov/sra/SRP132791).