Abstract
Background
The gut microbiome has an important role in infant health and immune development and may be affected by early-life exposures. Maternal diet may influence the infant gut microbiome through vertical transfer of maternal microbes to infants during vaginal delivery and breastfeeding. We aimed to examine the association of maternal diet during pregnancy with the infant gut microbiome 6 weeks post-delivery in mother-infant dyads enrolled in the New Hampshire Birth Cohort Study. Infant stool samples were collected from 145 infants, and maternal prenatal diet was assessed using a food frequency questionnaire. We used targeted sequencing of the 16S rRNA V4-V5 hypervariable region to characterize infant gut microbiota. To account for differences in baseline and trajectories of infant gut microbial profiles, we stratified analyses by delivery mode.
Results
We identified three infant gut microbiome clusters, characterized by increased abundance of Bifidobacterium, Streptococcus and Clostridium, and Bacteroides, respectively, overall and in the vaginally delivered infant stratum. In the analyses stratified to infants born vaginally and adjusted for other potential confounders, maternal fruit intake was associated with infant gut microbial community structure (PERMANOVA, p < 0.05). In multinomial logistic regression analyses, increased fruit intake was associated with an increased odds of belonging to the high Streptococcus/Clostridium group among infants born vaginally (OR (95% CI) = 2.73 (1.36, 5.46)). In infants delivered by Cesarean section, we identified three clusters that differed slightly from vaginally delivered infants, which were characterized by a high abundance of Bifidobacterium, high Clostridium and low Streptococcus and Ruminococcus genera, and high abundance of the family Enterobacteriaceae. Maternal dairy intake was associated with an increased odds of infants belonging to the high Clostridium cluster in infants born by Cesarean section (OR (95% CI) = 2.36 (1.05, 5.30)). Linear models suggested additional associations between maternal diet and infant intestinal microbes in both delivery mode strata.
Conclusions
Our data indicate that maternal diet influences the infant gut microbiome and that these effects differ by delivery mode.
Electronic supplementary material
The online version of this article (10.1186/s40168-018-0490-8) contains supplementary material, which is available to authorized users.
Keywords: Infant gut microbiome, Infant gut microbiome clusters, Maternal prenatal diet, Delivery mode, 16S rRNA gene, Fruit
Background
Studies have examined the maternal contribution to infant health, including the effect of maternal diet during pregnancy and lactation. Prenatal diet influences the risk of infant and child allergy. For instance, the Mediterranean diet during pregnancy has been associated with a reduced risk of both persistent and atopic wheeze and atopy in children at 6.5 years old [1, 2]. High meat consumption during pregnancy is associated with an increased risk of wheeze in the first year of life, while maternal dairy intake is associated with a reduced risk of infantile wheeze [2]. Little is known about the actual mechanisms by which maternal diet affects children’s health; we hypothesize that maternal diet impacts the development of the gut microbiome in infancy and subsequently influences child health outcomes. It is established that diet is an important driver of the gut microbiome. Short-term diets comprised solely of either plant or animal foods have been shown to alter the human gut microbiome [3]. In addition, studies in humans and humanized gnotobiotic mice show that diets with reduced carbohydrates [4], or high in polysaccharides, alter gut microbiome composition [5].
To date, there has been limited investigation of the relation of maternal diet with the developing infant gut microbiome. One study observed that maternal high-fat diet during pregnancy was associated with meconium microbial composition but had limited maternal diet data [6]. One mechanism the infant gut microbiome may be affected by maternal diet is via vertical transfer of maternal microbes to infants at delivery. There is a well-established relation of the infant gut microbiome with delivery mode (Cesarean section versus vaginally born infants) [7, 8] which may persist through adulthood [9]. Alternatively, maternal diet may affect fetal development and subsequent host response to microbial populations at and after birth. Additionally, differences in breast milk composition due to maternal diet may contribute to the infant gut microbiome in breastfed infants. We examined the relationship of maternal diet during pregnancy with the infant stool microbiome in 6-week-old infants and identified associations within delivery mode groups.
Results
Study population
Characteristics of the study population are summarized in Table 1 (n = 145). Maternal age ranged from 22 to 44 years, with an average of 31.9 years. More than 70% of mothers in the population had at least a college degree, more than 90% were married, and most were first-time mothers. Smoking during pregnancy was rare at 4.8%, and the population was slightly overweight with an average pre-pregnancy BMI of 25.6. Our population had more male infants than female infants (57.2% males), and an average birth weight and gestational age of 3427 g and 39.3 weeks, respectively. Most infants were born vaginally (66.9%) and were exclusively breastfed (70.3%) at 6 weeks of age. Daycare attendance and infant antibiotic exposure at or before 4 months of age were rare. Measures and distributions of maternal prenatal diet are shown in Table 1. Most subject characteristics and maternal dietary factors were not different between delivery mode groups (Table 1).
Table 1.
Subject characteristics by delivery mode (n = 145)
| n (%) or mean [range] | p value1 | |||
|---|---|---|---|---|
| All (n = 145) | Vaginal (n = 97) | Cesarean (n = 48) | ||
| Maternal characteristics | ||||
| Maternal age | 31.9 [22–44] | 31.8 [22–44] | 32.2 [23–42] | 0.65 |
| Education level | 0.14 | |||
| Less than 11th grade | 1 (0.7) | 1 (1.0) | 0 (0.0) | |
| High school graduate or equivalent | 15 (10.3) | 7 (7.2) | 8 (16.7) | |
| Junior college graduate or some college | 25 (17.2) | 16 (16.5) | 9 (18.8) | |
| College graduate | 54 (37.2) | 34 (35.1) | 20 (41.7) | |
| Any post-graduate schooling | 50 (34.5) | 39 (40.2) | 11 (22.9) | |
| Relationship status | 0.29 | |||
| Married | 133 (91.7) | 91 (93.8) | 42 (87.5) | |
| Separated or divorced | 3 (2.1) | 1 (1.0) | 2 (4.2) | |
| Single and never married | 9 (6.2) | 5 (5.2) | 4 (8.3) | |
| Smoking | 0.43 | |||
| No | 138 (95.2) | 91 (93.8) | 47 (97.9) | |
| Yes | 7 (4.8) | 6 (6.2) | 1 (2.1) | |
| Parity | 0.8 [0–4] | 0.9 [0–4] | 0.7 [0–2] | 0.07 |
| Maternal BMI | 25.6 [17.4–47.8] | 26.0 [17.4–47.8] | 24.7 [18.4–42.1] | 0.22 |
| Gestational diabetes2 | 0.58 | |||
| No | 121 (83.4) | 84 (86.6) | 37 (77.1) | |
| Yes | 16 (11.0) | 10 (10.3) | 6 (12.5) | |
| Dietary factors (servings/day)3 | ||||
| aMED score | 3.67 [0–7] | 3.8 [0–7] | 3.5 [0–7] | 0.32 |
| Dairy | 3.6 [0.0–8.5] | 3.7 [0.0–8.5] | 3.5 [0.8–6.8] | 0.40 |
| Fruit | 2.3 [0.0–5.9] | 2.3 [0.2–5.9] | 2.2 [0.0–5.6] | 0.52 |
| Vegetables | 3.3 [0.0–9.6] | 3.3 [0.5–9.6] | 3.2 [0.0–7.6] | 0.78 |
| Whole grains | 1.0 [0.0–4.1] | 1.2 [0.1–4.1] | 0.9 [0.0–3.1] | 0.03 |
| Fish and seafood | 0.2 [0.0–1.0] | 0.2 [0.0–1.0] | 0.2 [0.0–0.6] | 0.56 |
| Nuts, legumes, and soy | 0.9 [0.0–4.5] | 0.9 [0.0–3.1] | 0.9 [0.0–4.5] | 1.00 |
| Red and processed meat | 0.8 [0.0–2.3] | 0.7 [0.0–2.0] | 0.8 [0.0–2.3] | 0.21 |
| Polyunsaturated fat (g/day) | 13.9 [8.67–23.7] | 13.6 [8.7–21.0] | 14.5 [9.2–23.7] | 0.12 |
| EPA (g/day) | 0.1 [0.0–0.5] | 0.1 [0.0–0.5] | 0.1 [0.0–0.5] | 0.28 |
| DHA (g/day) | 0.1 [0.0–0.6] | 0.1 [0.0–0.6] | 0.1 [0.0–0.5] | 0.79 |
| Monounsaturated:saturated fatty acid ratio | 1.1 [0.7–2.3] | 1.1 [0.7–2.1] | 1.2 [0.7–2.3] | 0.35 |
| Infant characteristics | ||||
| Sex | 0.38 | |||
| Female | 62 (42.8) | 44 (45.4) | 18 (37.5) | |
| Male | 83 (57.2) | 53 (54.6) | 30 (62.5) | |
| Birth weight (g)4 | 3427.4 [1960–4710] | 3466.3 [2030–4565] | 3348.9 [1960–4710] | 0.18 |
| Gestational age (weeks) | 39.3 [30.4–43.4] | 39.5 [30.4–42.1] | 39.1 [33.4–43.4] | 0.21 |
| Feeding method | 0.04 | |||
| Exclusively breastfed | 102 (70.3) | 74 (76.3) | 28 (58.3) | |
| Combination | 37 (25.5) | 21 (21.7) | 16 (33.3) | |
| Exclusively formula fed | 6 (4.1) | 2 (2.1) | 4 (8.3) | |
| Antibiotics by 4 months old5 | 0.66 | |||
| No | 135 (93.1) | 91 (93.8) | 44 (91.7) | |
| Yes | 5 (3.5) | 3 (3.1) | 2 (4.2) | |
1p value determined by two-sided Welch’s t test or Fisher’s exact test
2Missing data on eight subjects
3All dietary factors are standardized to a 2000 cal/day diet, excluding aMED score and monounsaturated:saturated fatty acid ratio
4Missing data on three subjects
5Missing data on five subjects
Microbial community composition and maternal dietary factors
We identified 11,029,093 bacterial sequences in 145 6-week infant stool samples passing quality control. The most abundant taxonomic group was Enterobacteriaceae comprising 20.0% of the 6-week infant stool microbiome, followed by Bifidobacterium (18.4%), Bacteroides (10.4%), and Streptococcus (8.10%) (Additional file 1: Table S1). Open-reference OTU picking identified a total of 4260 OTUs in our population of 6-week infant stool samples, with 2850 OTUs corresponding to Greengenes IDs and 1410 de novo OTUs. Our main analyses focused on vaginally delivered infants (n = 97). Adjusting for infant feeding method, maternal BMI, parity, and batch, maternal fruit consumption was associated with infant stool microbiome composition (generalized UniFrac distance PERMANOVA, p = 0.028; Table 2; Fig. 1a). In a sensitivity analysis, the effect of maternal fruit consumption on the infant gut microbiome in babies born vaginally persisted when restricted to infants who were exclusively breastfed (generalized UniFrac distance PERMANOVA, p = 0.022; Table 2). Excluding infants who may have received antibiotics, or those delivered prematurely (Additional file 1: Table S10, Figure S7a), did not qualitatively change the results.
Table 2.
Relation of microbial community composition in vaginally delivered 6-week old infants with maternal diet
| p value1 | ||
|---|---|---|
| Dietary factor | All (n = 97)2 | Exclusively breastfed (n = 74)3 |
| aMED score | 0.19 | 0.26 |
| Dairy | 0.60 | 0.91 |
| Fruit | 0.028 | 0.022 |
| Vegetables | 0.56 | 0.92 |
| Whole grains | 0.47 | 0.25 |
| Fish and seafood | 0.33 | 0.21 |
| Nuts, legumes, and soy | 0.52 | 0.71 |
| Red and processed meat | 0.86 | 0.69 |
| Polyunsaturated fat | 0.76 | 0.84 |
| EPA | 0.29 | 0.45 |
| DHA | 0.40 | 0.50 |
| MUFA:SFA ratio | 0.66 | 0.43 |
1All p values are determined by PERMANOVA
2p values are adjusted for infant feeding method, maternal BMI, parity, and batch
3p values are adjusted for maternal BMI, parity, and batch
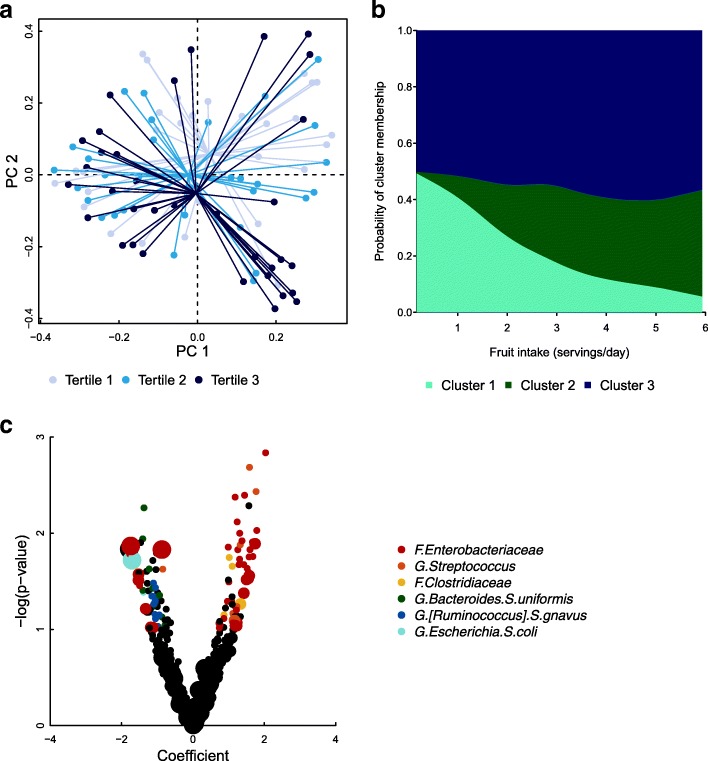
Fig. 1.
Associations between maternal diet and infant gut microbial communities in infants delivered vaginally. Plots include only infants delivered vaginally (n = 97). a Principal coordinate plot of generalized UniFrac distances (PERMANOVA p = 0.028 for maternal fruit intake as a continuous variable), colored by maternal fruit intake tertiles. Each point represents an individual, and lines indicate the distance from tertile centroid. b Predicted probability plot of infant stool cluster membership by maternal fruit intake during pregnancy from multinomial logistic regression models adjusted for infant feeding method, maternal BMI, parity, and batch. Cluster 1 is the reference group. c Linear model associations between maternal aMED score and relative abundance of infant stool OTUs. The size of each point indicates the log-ratio transformed relative abundance (LRTA) of each OTU (LRTA ≥ 30, LRTA ≥ 20, or LRTA < 20). Points are colored by taxonomy represented heavily in top results, for p < 0.10. F., G., and S. in taxonomy labels indicate that the level of taxonomy is family, genus, or species, respectively
Infant gut microbiome clusters
We identified three distinct clusters of the infant gut microbiome in vaginally delivered infants (n = 97), with cluster 1 characterized by a high abundance of the genus Bifidobacterium, cluster 2 by the genera Streptococcus and Clostridium, and cluster 3 by the genus Bacteroides (Additional file 1: Figure S1, Table S1). To assess the relationship between maternal dietary factors and infant gut cluster membership, we used multinomial logistic regression with cluster 1 as the reference group. The odds of belonging to cluster 2 are 2.73 times greater for each additional maternal serving of fruit per day (95% CI 1.36, 5.46; Table 3, Fig. 1b). These results were consistent when infants delivered prematurely were not included in the analyses (Additional file 1: Table S9, Figure S6a).
Table 3.
Infant gut microbiome cluster is influenced by maternal diet
| OR (95% confidence interval)1, 2 | ||
|---|---|---|
| Dietary factor | Cluster 2 | Cluster 3 |
| aMED score | 1.31 (0.90, 1.89) | 0.96 (0.73, 1.27) |
| Dairy | 0.87 (0.56, 1.37) | 0.84 (0.59, 1.18) |
| Fruit | 2.73 (1.36, 5.46)3 | 1.61 (0.97, 2.68)3 |
| Vegetables | 0.88 (0.60, 1.28) | 0.82 (0.60, 1.11) |
| Whole grains | 0.94 (0.36, 2.41) | 1.10 (0.57, 2.13) |
| Fish and seafood | 0.67 (0.01, 32.26) | 0.50 (0.02, 12.57) |
| Nuts, legumes, and soy | 0.49 (0.20, 1.20) | 0.48 (0.24, 0.96) |
| Red and processed meat | 4.39 (0.82, 23.47) | 2.35 (0.62, 8.95) |
| Polyunsaturated fat | 0.84 (0.65, 1.09) | 0.87 (0.72, 1.06) |
| EPA | 0.24 (0.00, 72.57) | 4.13 (0.08, 217.72) |
| DHA | 0.15 (0.00, 62.82) | 2.66 (0.04, 158.91) |
| MUFA:SFA ratio | 1.64 (0.15, 17.73) | 1.78 (0.26, 12.28) |
1Models include vaginally delivered infants (n = 97)
2Cluster 1 is the reference group
3Corresponds to Fig. 1b
Operational taxonomic units in the infant gut and maternal dietary factors
To investigate the microbial taxa that may drive differences observed above, we measured associations between each maternal dietary factor and the relative abundance of individual infant stool operational taxonomic units (OTUs) with linear models adjusted for feeding method, maternal BMI, parity, and batch. While the microbiome-wide significance of maternal dietary factors after adjusting p values using the false discovery rate (FDR) method was not observed, OTUs with the same taxonomic assignment tended to appear together in the most significant results, supporting a true association of maternal dietary factors and infant gut microbiota. Maternal aMED score was positively associated with OTUs classified to the family Enterobacteriaceae, the genus Streptococcus, and the family Clostridiaceae, and negatively with OTUs classified to the species Bacteroides uniformis, the Enterobacteriaceae family, and the species Escherichia coli and [Ruminococcus] gnavus (Fig. 1c; Additional file 1: Table S2a). Maternal dairy intake was positively associated with OTUs classified to the species Clostridium neonatale and C. butyricum and the genus Staphylococcus, and negatively related to OTUs classified to the Lachnospiraceae family (Additional file 1: Table S2b). OTUs classified to the genus Streptococcus including the species Streptococcus agalactiae were positively associated with maternal fish and seafood intake, while OTUs classified to the species Bacteroides uniformis were negatively associated (Additional file 1: Table S2c). We observed a negative association between fruit intake and OTUs of the genus Bifidobacterium and a positive association with OTUs in the Clostridiaceae family in the infant gut (Additional file 1: Table S2d). We observed additional associations between these and other maternal dietary factors and infant stool OTUs, which are presented in the supplementary tables.
Associations in infants delivered by Cesarean section
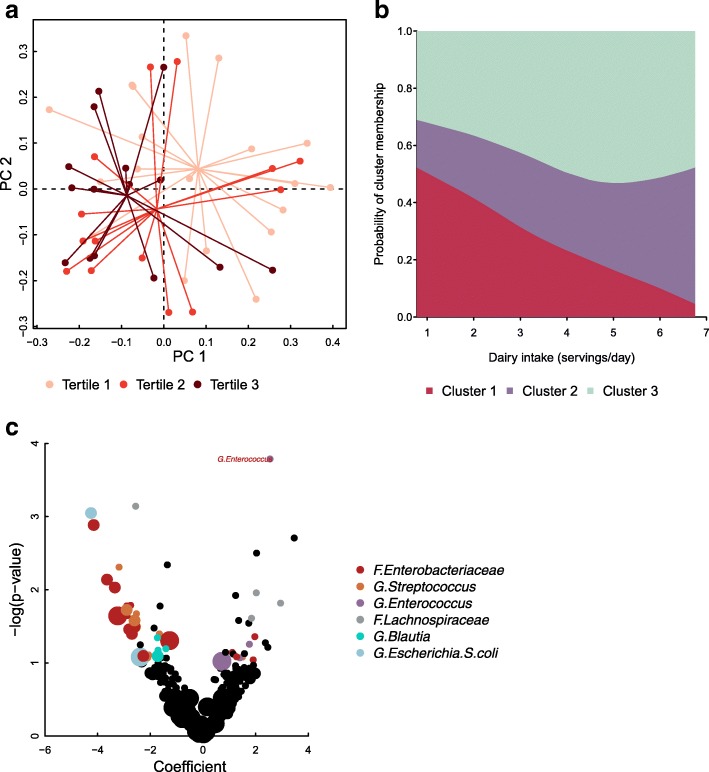
We separately assessed the relation of maternal diet with the infant gut microbiome in infants delivered by Cesarean section (n = 48). In an adjusted analysis, maternal dairy intake was associated with infant stool microbiome composition (generalized UniFrac distance PERMANOVA, p = 0.034; Fig. 2a; Additional file 1: Table S3). Additionally, we identified clusters of infant gut microbiota in infants delivered by Cesarean section that differed slightly from those identified in vaginally delivered infants; cluster 1 was characterized by a high abundance of the genus Bifidobacterium, cluster 2 by high Clostridium and low Streptococcus and Ruminococcus genera, and cluster 3 by high abundances of the family Enterobacteriaceae, the genus Ruminococcus, and the family Lachnospiraceae (Additional file 1: Table S1, Figure S1). The odds of belonging to the cluster 2 were 2.36 times greater for each additional maternal serving of dairy per day (95% CI 1.05, 5.30; Fig. 2b; Additional file 1: Table S4). Linear models indicated a positive association of maternal dairy intake with OTUs classified to the family Enterobacteriaceae and the species Escherichia coli and negative association with the OTUs that belong to the genera Bifidobacterium, Pseudomonas, and Bacteroides (Additional file 1: Table S5b). One OTU classified to the genus Corynebacterium was negatively associated with maternal dairy intake (p = 0.0044), and one each OTU classified to the species Acinetobacter rhizosphaerae (p = 0.0040) and to the family Ruminococcaceae (p = 0.0061), respectively, were also positively associated with maternal dairy intake, but we did not observe multiple related OTUs with similar associations. OTUs classified to the genus Enterococcus (one q value < 0.10) and the Lachnospiraceae family were positively related with and OTUs classified to the family Enterobacteriaceae, the species Escherichia coli and the genera Streptococcus and Blautia, were negatively related with the maternal aMED score (Additional file 1: Table S5a). As in vaginally delivered infants, maternal fish and seafood intake was positively associated with OTUs in the genus Streptococcus in infants delivered by Cesarean section (Additional file 1: Table S5c). In contrast to infants born vaginally, maternal dietary fish and seafood were positively associated with OTUs in the genus Bacteroides including the species B. uniformis and negatively associated with OTUs in the species Clostridium neonatale and other OTUs of the Streptococcus genus (Additional file 1: Table S5c). Additionally, maternal red and processed meat was positively associated with OTUs in the genus Bifidobacterium among others including the species Escherichia coli and the genus Enterococcus (Additional file 1: Table S5e). Results were qualitatively the same when infants born before 37 weeks gestation were excluded (see Additional file 1: Supplementary Information).
Fig. 2.
Associations between maternal diet and infant gut microbial communities in infants delivered vaginally. Plots include only infants delivered by Cesarean section (n = 48). a Principal coordinate plot of generalized UniFrac distances (PERMANOVA p = 0.034 for maternal dairy as a continuous variable), colored by maternal dairy intake tertiles. Each point represents an individual, and lines indicate the distance from tertile centroid. b Predicted probability plot of infant stool cluster membership by maternal dairy intake during pregnancy from multinomial logistic regression models adjusted for infant feeding method, maternal BMI, parity, and batch. Cluster 1 is the reference group. c Linear model associations between maternal aMED score and relative abundance of infant stool OTUs. The size of each point indicates the log-ratio transformed relative abundance (LRTA) of each OTU (LRTA ≥ 30, LRTA ≥ 20, or LRTA < 20). Red text indicates q < 0.10. Points are colored by taxonomy represented heavily in top results, for p < 0.10. F., G., and S. in taxonomy labels indicate that the level of taxonomy is family, genus, or species
Discussion
We observed differences in both overall infant gut microbial community structure and specific microbes in relation to maternal dietary factors, often in a delivery mode-dependent pattern. Some effects of maternal diet were more apparent in exclusively breastfed infants; however, we were unable to compare to exclusively formula-fed infants since most infants received some breast milk.
We found maternal fish and seafood consumption to be positively related to OTUs in the genus Streptococcus in the infant gut, which included the species Streptococcus agalactiae, or Group B Streptococcus. Streptococcus agalactiae is known to infect farmed fish populations [10], and fishmongers have presented with infections of Streptococcus iniae related to fish handling [11]. Fish consumption is generally recommended as healthful due to its DHA and EPA content despite the exposure to heavy metals and other contaminants that may be detrimental to health [12]. For instance, greater fish consumption is associated with child development outcomes including decreased risk of asthma and improved cognition [13, 14]. The decrease in OTUs classified to the species Clostridium neonatale with increased maternal fish and seafood intake in infants born by Cesarean section is a possibly beneficial change to infant intestinal microbial communities. We observed consistent associations between maternal DHA and EPA intake and infant gut microbiota. The associations of other maternal dietary factors with specific infant gut microbes were also somewhat unexpected, namely for red and processed meat and for fruit consumption. While fruit is considered healthful and red and processed meat consumption is recommended to be limited, OTUs in the genus Bifidobacterium, generally recognized as a beneficial microbe, were decreased with increasing maternal fruit consumption in vaginally born infants yet increased with higher maternal red and processed meat consumption in those born by Cesarean section. The results from infant gut microbiome cluster analyses are consistent with this observation. For example, it is surprising that higher maternal fruit intake is associated with an increased probability of an infant delivered vaginally belonging to cluster 2, in which the genera Bifidobacterium and Bacteroides are underrepresented and the family Clostridiaceae is overrepresented compared to the other two clusters.
The effect of maternal dairy intake on infant gut microbiota was primarily observed in infants delivered by Cesarean section. Infants delivered by Cesarean section are at increased risk of dairy allergies compared to those born vaginally [15, 16] and show decreased colonization of milk-digesting bacteria [7, 17] including the genus Lactobacillus. One recent study suggests that both the microbial and fatty acid composition of breast milk may differ by delivery mode [18], so the differential effect of maternal dairy intake by delivery mode on the infant gut microbiome may be attributable to the differences in breast milk microbiota or lipids in relation to maternal diet by delivery mode. Our observation of increased colonization with the species Acinetobacter rhizosphaerae, a microbe that colonizes and promotes growth in plants [19], with maternal dairy in the Cesarean group is interesting in conjunction with the positive associations found between maternal dairy and the species Clostridium neonatale and Staphylococcus genus in vaginally delivered infants. Species of the genus Acinetobacter are increasingly involved in nosocomial infections [20], while the species Clostridium neonatale was initially isolated from infants affected by necrotizing enterocolitis [21], and the pathogenic potential of species in the genus Staphylococcus is well-established [22, 23]. Thus, it will be important to investigate whether maternal dairy consumption fosters the growth of potentially pathogenic microbes in the infant gut, particularly for infants delivered operatively.
In several cases, a maternal dietary factor was associated with a microbe in both delivery groups but in opposite directions. Since Cesarean delivery results in differential microbial community structure of the infant gut [24], and the gut microbiome is a complex system, such differences are plausible; experimental studies will be required to confirm this observation. However, the associations between maternal diet and infant gut microbial communities did not always differ by delivery mode. For example, the maternal aMED score was consistently positively and negatively associated with the Enterobacteriaceae family, and additionally negatively associated with the species Escherichia coli, in both delivery mode strata.
Our study sample was drawn from Northern New England, which has a relatively homogenous population; this could limit the generalizability of the study. However, by limiting unmeasured confounding due to differences in human microbiomes between populations that may result in erroneous associations with dietary differences, our study also may have greater internal validity. Maternal diet was assessed by self-report between 24 and 28 weeks of gestation, and it is possible that dietary patterns change near the end of pregnancy or during lactation. However, diets are often stable, and a mid-pregnancy food frequency questionnaire (FFQ)-assessment is likely to be sufficiently representative of late pregnancy and lactation diet in a high proportion of individuals [25, 26]. Nonetheless, the effects we observe may be due in part to maternal diet during lactation. Further, we cannot conclude if the effects of maternal diet occur only in breastfed babies, or also in those mostly fed formula, or a combination of formula and breast milk. It is also possible that associations between maternal exposures and infant microbiota at 6 weeks of life are influenced by factors not investigated in this study.
Future studies examining the relationship between maternal diet and components of breast milk including microbial and nutritional profiles, as well as the potential influence of maternal diet on the fetal microbiome, may offer insight into the mechanisms by which maternal diet influences the infant gut microbiome. Determining the impact of changes in the gut microbiome of infants due to maternal diet on infant health and development is an opportunity to refine dietary recommendations for pregnant and lactating women to support infant health.
Conclusions
In conclusion, we identified three clusters of infant gut microbial communities and observed effects of multiple maternal prenatal dietary factors on the infant gut microbiome that often varied by delivery mode.
Methods
Study population
Subjects for the present study were from the New Hampshire Birth Cohort Study (NHBCS) who provided infant stool samples at 6-week postpartum. As described previously [27, 28], eligible participants for the NHBCS are pregnant women obtaining prenatal care in New Hampshire, US clinics, who reported relying on a private, unregulated well as their home water source. Participants between 18 and 45 years old were recruited between 24 and 28 weeks of gestation. The Center for the Protection of Human Subjects at Dartmouth provided institutional review board approval. All methods were carried out in accordance with the guidelines. Written informed consent was obtained for participation from all subjects for themselves and their children. Between 31 March 2011 when the first potential subject with the opportunity to collect a 6-week infant stool sample from was screened for eligibility and 30 June 2015 when the data were frozen for this analysis, 6569 women were screened for eligibility and 1334 were eligible. Nine hundred seventy-six subjects were enrolled and 361 provided 6-week infant stool samples, 187 of which had 16S rRNA gene sequencing data available (Additional file 1: Figure S2).
Telephone interviews conducted every 4 months ascertained the infant’s feeding method, including the timing of breastfeeding cessation and the start date of each new formula, as applicable, as well as medication use such as antibiotics. Infants reported to have never been introduced to formula and to be breastfed were considered to be exclusively breastfed, while those who were ever breastfed and ever formula-fed were considered to be fed with a combination of breast milk and formula regardless of whether or not the infant was still breastfed at the time of stool collection. Those who were never breastfed and only received formula were considered exclusively formula-fed. The delivery mode was drawn from maternal delivery records, and maternal pre-pregnancy height, weight, and parity were determined via a self-administered questionnaire upon study entry and used to compute pre-pregnancy BMI.
Maternal dietary data and Mediterranean diet score
Maternal diet during pregnancy was assessed during gestational weeks 24–28 with a validated food frequency questionnaire (FFQ) [25]. We computed the alternative Mediterranean diet (aMED) score based on FFQ-derived measures of daily food intake, following the method of Tobias et al. 2012 [29], excluding alcohol consumption to adapt the score to a pregnant cohort. Daily consumption of vegetables, fruits, legumes, soy, and nuts, whole grains, fish and seafood, and monounsaturated to saturated fat ratio (MUFA:SFA) above the median is considered healthful and confers 1 point to the aMED score. Red and processed meat intake is considered less healthful, and consumption below the median confers 1 point to the aMED score. This results in possible aMED score values from 0 to 7, with a higher score indicating greater adherence to a Mediterranean dietary pattern. We additionally considered each of those dietary components and maternal dairy, polyunsaturated fatty acids (PUFA), and omega-3 fatty acids eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) intakes separately.
Sample collection and DNA extraction
Infant stool samples were collected at the 6-week maternal postpartum follow-up appointment and were aliquoted and frozen at − 80 °C within 24 h. Following established methods reviewed by Wu et al. [30], we used the Zymo DNA extraction kit (Zymo Research) to extract microbial DNA from thawed samples and quantified the DNA using OD260/280 nanodrop.
Targeted 16S rRNA gene sequencing
The V4-V5 hypervariable region of the bacterial 16S rRNA gene was sequenced at the Marine Biological Laboratory (MBL) in Woods Hole, MA, using established methods [31, 32]. As described previously [24], 16S rDNA V4-V5 amplicons were generated from purified genomic DNA samples using fusion primers. The use of forward primers containing one of eight five-nucleotide barcodes between the Illumina-specific bridge and sequencing primer regions and the 16S-specific region and a single reverse primer containing 1 of 12 Illumina indices enables 96 samples per lane multiplexing. Amplifications were done in triplicate with one negative control for internal quality control at the MBL. We used qPCR (Kapa Biosystems) to quantify the amplicon pool, and one Illumina MiSeq 500 cycle paired-end run to sequence each pool of 96 libraries. We demultiplexed and divided datasets using Illumina MiSeq Reporter and a custom Python script.
Microbiome profiling
To identify microbial population structure profiles, we used full-length amplicon sequences for the rRNA gene V4-V5 hypervariable regions. We merged the forward and reverse reads based on sequence overlap [33], removed the primer sequences, and discarded the sequences containing any ambiguous nucleotides. We used vsearch [34] to remove chimeras both de novo and in comparison to the RDP classifier training reference.
Data processing and statistical analysis
We identified open reference operational taxonomic units (OTUs) using QIIME version 1.9.1 [35] with the UCLUST algorithm and 97% similarity [36]. We used PyNAST alignment [37] with Greengenes core reference [38, 39] to build OTU tables and assign taxonomy [39, 40]. Phylogenetic trees were constructed using the FastTree method [41]. Subsequent analyses were performed using R version 3.2.2 [42]. We computed generalized UniFrac distances between pairs of samples using OTU tables and the midpoint rooted phylogenetic tree. Generalized UniFrac unifies weighted and unweighted UniFrac to detect differences in moderately abundant lineages instead of those exclusively in highly abundant or rare lineages [43, 44]. Of 187 infant stool samples with available sequencing data, we dropped 18 for missing maternal FFQ data, 14 for missing infant feeding method, and 7 for missing maternal BMI. We dropped three samples of low quality, with read counts < 5000, yielding a final sample size of 145 (Additional file 1: Figure S2), with 97 vaginally delivered infants and 48 delivered by Cesarean section.
Primary analyses were of vaginally delivered infants. We tested the contribution of maternal dietary measures (as continuous variables) to the infant microbiome at 6 weeks using the adonis function in the R package vegan [45] with 10,000 permutations. This is a nonparametric permutational multivariate analysis of variance (PERMANOVA) test that partitions a distance matrix by sources of variation and can accept both categorical and continuous variables. In preliminary analyses of all infants, delivery mode, feeding method, as previously shown in our cohort [24], and parity were significantly related to infant stool microbial community composition at 6 weeks of age. We performed crude and adjusted analyses for each dietary food group variable individually, normalized by maternal calorie consumption, and maternal aMED score and MUFA:SFA ratio in vaginally delivered infants. All dietary variables were modeled as continuous variables. Adjusted analyses included both variables known to be related to the infant gut microbiome by previous studies and variables significantly related to infant gut microbiota in our population to control confounding. Accordingly, we adjusted for feeding method (exclusively breastfed, combination fed, exclusively formula fed), maternal BMI (continuous), parity (continuous), and batch and performed a sensitivity analysis in exclusively breastfed infants. We considered p < 0.05 to be significant. To visualize the results, we performed principal coordinates analysis (PCoA) on generalized UniFrac distances using the cmdscale function in R [42] and plotted the samples by the first two coordinates, colored by each dietary variable tertile to aid in visualization.
Inspired by the work identifying adult enterotypes by Arumugam et al. [46] and Wu et al. [47], we identified clusters of the infant gut microbiome at 6 weeks of age using the partitioning around medoids (PAM) method of clustering on generalized UniFrac distances. We identified clusters in all infants (n = 145) and within infants delivered vaginally (n = 97) or by Cesarean section (n = 48) for within delivery group analyses. We used the elbow method to determine the optimal number of clusters, which is based on the within-cluster sum of squares. To assess the relationship between each maternal dietary factor and infant gut microbiome cluster membership, we used multinomial logistic regression in the R package nnet adjusting all models for the same covariates as in previous analyses. The outcome in each model was the infant gut microbiome cluster membership; cluster 1 was used as the reference in all models.
To more precisely identify which microbial taxa contribute to the observed differences in infant stool microbial community composition by maternal dietary factors, we used a series of linear models to test the relationships between each maternal dietary factor and each OTU. We computed the relative abundance for each OTU by dividing the read count in each cell by the total number of reads in the sample and filtered out OTUs with zero or near-zero variance using the caret package in R. This results in 624 OTUs. We used a zero replacement procedure and log-ratio transformation to account for the zero-inflated and compositional nature of the data [48] before running linear models. We adjusted p values for multiple comparisons using the Benjamini and Hochberg method to control the false discovery rate [49]. In reporting the results, we use the lowest level of assigned taxonomy available, which is most commonly the genus, but ranges from the family to the species level.
All analyses were analogously performed in the Cesarean section group to assess if an effect of maternal diet on the infant gut microbiome exists in the case of surgical delivery. We were unable to adjust for antibiotic usage due to a paucity of exposure and imprecise data on timing of exposure. For sensitivity, we repeated analyses (1) excluding infants with possible antibiotic exposure (nvaginal = 3, nCesarean = 2) and those with missing antibiotic exposure information (nvaginal = 3, nCesarean = 2) and (2) excluding all infants delivered before 37 weeks gestation (n = 8), reported in the supplemental information. We also report unadjusted estimates for analyses and analyses further adjusted for infant age at sample collection in the supplemental information.
Additional file
Contains supplemental tables (Tables S1–S16) and figures (Figures S1–S7). (PDF 3429 kb)
Acknowledgements
We thank the participants of the New Hampshire Birth Cohort Study and to the study staff, without whom this research would not be possible.
Funding
This study was by supported by funding from the National Institutes of Health (grants NIGMS P20GM104416, NIEHS P01ES022832, NLM K01LM011985, NLM R01LM012723 and NIDCR R01DE022772), the US Environmental Protection Agency (RD83544201), and the Rosaline Borison Memorial Fund. None of the funding bodies had a role in the design of the study or the collection, analysis, and interpretation of data and in writing of the manuscript.
Availability of data and materials
The 16S rRNA gene sequencing datasets used in this study are stored in the National Center for Biotechnology Information (NCBI) Sequence Read Archive: http://www.ncbi.nlm.nih.gov/sra under accession number PRJNA296814.
Abbreviations
- aMED
Alternative Mediterranean diet
- DHA
Docosahexaenoic acid
- EPA
Eicosapentaenoic acid
- FDR
False discovery rate
- FFQ
Food frequency questionnaire
- MBL
Marine Biological Laboratory
- MUFA:SFA
Monounsaturated to saturated fat ratio
- NHBCS
New Hampshire Birth Cohort Study
- OTU
Operational taxonomic unit
- PAM
Partitioning around medoids
- PCoA
Principal coordinates analysis
- PERMANOVA
Permutational multivariate analysis of variance
- PUFA
Polyunsaturated fatty acids
Authors’ contributions
MRK and JCM designed the research. SNL performed the statistical analyses and wrote the paper. SNL, AGH, BCC, MRK, and JCM interpreted the results. JAE calculated the maternal intake of food groups and the Mediterranean diet score from the food frequency questionnaires. HGM provided technical and methodological support from the Marine Biological Laboratory. AGH has primary responsibility for the final content. All authors reviewed the manuscript, offered critical feedback, and approved the final version.
Ethics approval and consent to participate
The Center for the Protection of Human Subjects at Dartmouth provided institutional review board approval. All methods were carried out in accordance with the guidelines. Written informed consent was obtained for participation from all subjects for themselves and their children.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s40168-018-0490-8) contains supplementary material, which is available to authorized users.
Contributor Information
Sara N. Lundgren, Email: Sara.N.Lundgren.GR@dartmouth.edu
Juliette C. Madan, Email: Juliette.C.Madan@hitchcock.org
Jennifer A. Emond, Email: Jennifer.A.Emond@dartmouth.edu
Hilary G. Morrison, Email: hmorrison1981@gmail.com
Brock C. Christensen, Email: Brock.C.Christensen@dartmouth.edu
Margaret R. Karagas, Email: Margaret.R.Karagas@dartmouth.edu
Anne G. Hoen, Phone: 603-653-6087, Email: Anne.G.Hoen@dartmouth.edu
References
- 1.Chatzi L, Torrent M, Romieu I, Garcia-Esteban R, Ferrer C, Vioque J, Kogevinas M, Sunyer J. Mediterranean diet in pregnancy is protective for wheeze and atopy in childhood. Thorax. 2008;63:507–513. doi: 10.1136/thx.2007.081745. [DOI] [PubMed] [Google Scholar]
- 2.Chatzi L, Garcia R, Roumeliotaki T, Basterrechea M, Begiristain H, Iniguez C, Vioque J, Kogevinas M, Sunyer J, group Is, group Rs: Mediterranean diet adherence during pregnancy and risk of wheeze and eczema in the first year of life: INMA (Spain) and RHEA (Greece) mother-child cohort studies. Br J Nutr 2013, 110:2058–2068. [DOI] [PubMed]
- 3.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. 2007;73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu DM, Antony KM, Ma J, Prince AL, Showalter L, Moller M, Aagaard KM. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 2016;8:77. doi: 10.1186/s13073-016-0330-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, Sears MR, Becker AB, Scott JA, Kozyrskyj AL, Investigators CS. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. 2013;185:385–394. doi: 10.1503/cmaj.121189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goedert JJ, Hua X, Yu G, Shi J. Diversity and composition of the adult fecal microbiome associated with history of cesarean birth or appendectomy: analysis of the American Gut Project. EBioMedicine. 2014;1:167–172. doi: 10.1016/j.ebiom.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun J, Fang W, Ke B, He D, Liang Y, Ning D, Tan H, Peng H, Wang Y, Ma Y, et al. Inapparent Streptococcus agalactiae infection in adult/commercial tilapia. Sci Rep. 2016;6:26319. doi: 10.1038/srep26319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinstein MR, Litt M, Kertesz DA, Wyper P, Rose D, Coulter M, McGeer A, Facklam R, Ostach C, Willey BM, et al. Invasive infections due to a fish pathogen, Streptococcus iniae. S iniae Study Group. N Engl J Med. 1997;337:589–594. doi: 10.1056/NEJM199708283370902. [DOI] [PubMed] [Google Scholar]
- 12.Mozaffarian D, Rimm EB. Fish intake, contaminants, and human health: evaluating the risks and the benefits. JAMA. 2006;296:1885–1899. doi: 10.1001/jama.296.15.1885. [DOI] [PubMed] [Google Scholar]
- 13.Oken E, Radesky JS, Wright RO, Bellinger DC, Amarasiriwardena CJ, Kleinman KP, Hu H, Gillman MW. Maternal fish intake during pregnancy, blood mercury levels, and child cognition at age 3 years in a US cohort. Am J Epidemiol. 2008;167:1171–1181. doi: 10.1093/aje/kwn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salam MT, Li YF, Langholz B, Gilliland FD. Maternal fish consumption during pregnancy and risk of early childhood asthma. J Asthma. 2005;42:513–518. doi: 10.1081/JAS-200067619. [DOI] [PubMed] [Google Scholar]
- 15.Metsala J, Lundqvist A, Kaila M, Gissler M, Klaukka T, Virtanen SM. Maternal and perinatal characteristics and the risk of cow’s milk allergy in infants up to 2 years of age: a case-control study nested in the Finnish population. Am J Epidemiol. 2010;171:1310–1316. doi: 10.1093/aje/kwq074. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez-Valverde F, Gil F, Martinez D, Fernandez B, Aznal E, Oscoz M, Olivera JE. The impact of caesarean delivery and type of feeding on cow’s milk allergy in infants and subsequent development of allergic march in childhood. Allergy. 2009;64:884–889. doi: 10.1111/j.1398-9995.2008.01931.x. [DOI] [PubMed] [Google Scholar]
- 17.Gronlund MM, Lehtonen OP, Eerola E, Kero P. Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J Pediatr Gastroenterol Nutr. 1999;28:19–25. doi: 10.1097/00005176-199901000-00007. [DOI] [PubMed] [Google Scholar]
- 18.Kumar H, du Toit E, Kulkarni A, Aakko J, Linderborg KM, Zhang Y, Nicol MP, Isolauri E, Yang B, Collado MC, Salminen S. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front Microbiol. 2016;7:1619. doi: 10.3389/fmicb.2016.01619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulati A, Vyas P, Rahi P, Kasana RC. Plant growth-promoting and rhizosphere-competent Acinetobacter rhizosphaerae strain BIHB 723 from the cold deserts of the Himalayas. Curr Microbiol. 2009;58:371–377. doi: 10.1007/s00284-008-9339-x. [DOI] [PubMed] [Google Scholar]
- 20.Hanlon GW. The emergence of multidrug resistant Acinetobacter species: a major concern in the hospital setting. Lett Appl Microbiol. 2005;41:375–378. doi: 10.1111/j.1472-765X.2005.01791.x. [DOI] [PubMed] [Google Scholar]
- 21.Benamar S, Cassir N, La Scola B. Genome sequence of a Clostridium neonatale strain isolated in a Canadian neonatal intensive care unit. Genome Announc. 2016;4:e01431–15. [DOI] [PMC free article] [PubMed]
- 22.Ericson JE, Popoola VO, Smith PB, Benjamin DK, Fowler VG, Benjamin DK, Jr, Clark RH, Milstone AM. Burden of invasive Staphylococcus aureus infections in hospitalized infants. JAMA Pediatr. 2015;169:1105–1111. doi: 10.1001/jamapediatrics.2015.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt BK, Kirpalani HM, Corey M, Low DE, Philip AG, Ford-Jones EL. Coagulase-negative staphylococci as true pathogens in newborn infants: a cohort study. Pediatr Infect Dis J. 1987;6:1026–1031. doi: 10.1097/00006454-198706110-00004. [DOI] [PubMed] [Google Scholar]
- 24.Madan JC, Hoen AG, Lundgren SN, et al. Association of cesarean delivery and formula supplementation with the intestinal microbiome of 6-week-old infants. JAMA Pediatr. 2016;170:212–9. [DOI] [PMC free article] [PubMed]
- 25.Willett WC, Sampson L, Stampfer MJ, Rosner B, Bain C, Witschi J, Hennekens CH, Speizer FE. Reproducibility and validity of a semiquantitative food frequency questionnaire. Am J Epidemiol. 1985;122:51–65. doi: 10.1093/oxfordjournals.aje.a114086. [DOI] [PubMed] [Google Scholar]
- 26.Crozier SR, Robinson SM, Godfrey KM, Cooper C, Inskip HM. Women’s dietary patterns change little from before to during pregnancy. J Nutr. 2009;139:1956–1963. doi: 10.3945/jn.109.109579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert-Diamond D, Cottingham KL, Gruber JF, Punshon T, Sayarath V, Gandolfi AJ, Baker ER, Jackson BP, Folt CL, Karagas MR. Rice consumption contributes to arsenic exposure in US women. Proc Natl Acad Sci U S A. 2011;108:20656–20660. doi: 10.1073/pnas.1109127108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farzan SF, Korrick S, Li Z, Enelow R, Gandolfi AJ, Madan J, Nadeau K, Karagas MR. In utero arsenic exposure and infant infection in a United States cohort: a prospective study. Environ Res. 2013;126:24–30. doi: 10.1016/j.envres.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tobias DK, Zhang C, Chavarro J, Bowers K, Rich-Edwards J, Rosner B, Mozaffarian D, Hu FB. Prepregnancy adherence to dietary patterns and lower risk of gestational diabetes mellitus. Am J Clin Nutr. 2012;96:289–295. doi: 10.3945/ajcn.111.028266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu GD, Lewis JD, Hoffmann C, Chen YY, Knight R, Bittinger K, Hwang J, Chen J, Berkowsky R, Nessel L, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. doi: 10.1186/1471-2180-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huse SM, Young VB, Morrison HG, Antonopoulos DA, Kwon J, Dalal S, Arrieta R, Hubert NA, Shen L, Vineis JH, et al. Comparison of brush and biopsy sampling methods of the ileal pouch for assessment of mucosa-associated microbiota of human subjects. Microbiome. 2014;2:5. doi: 10.1186/2049-2618-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newton RJ, McLellan SL, Dila DK, Vineis JH, Morrison HG, Eren AM, Sogin ML. Sewage reflects the microbiomes of human populations. MBio. 2015;6:e02574. doi: 10.1128/mBio.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eren AM, Vineis JH, Morrison HG, Sogin ML. A filtering method to generate high quality short reads using illumina paired-end technology. PLoS One. 2013;8:e66643. doi: 10.1371/journal.pone.0066643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rognes T, Mahé F, Flouri T, McDonald D: Vsearch: VSEARCH 1.4. 0. Zenodo, European Organization for Nuclear Research, Geneva 2015.
- 35.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 37.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Werner JJ, Koren O, Hugenholtz P, DeSantis TZ, Walters WA, Caporaso JG, Angenent LT, Knight R, Ley RE. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J. 2012;6:94–103. doi: 10.1038/ismej.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R Core Team. R: a language and environment for statistical computing [online]. Vienna: R Foundation for Statistical Computing; 2015.
- 43.Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, Collman RG, Bushman FD, Li H. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. 2012;28:2106–2113. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J. Package ‘GUniFrac’. 2012. [Google Scholar]
- 45.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara R, Simpson GL, Solymos P, Stevens MHH, Wagner H. Package ‘vegan’. Community ecology package, version. 2015:2.2–1.
- 46.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aitchison J. The statistical analysis of compositional data. London; New York: Chapman and Hall; 1986. [Google Scholar]
- 49.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contains supplemental tables (Tables S1–S16) and figures (Figures S1–S7). (PDF 3429 kb)
Data Availability Statement
The 16S rRNA gene sequencing datasets used in this study are stored in the National Center for Biotechnology Information (NCBI) Sequence Read Archive: http://www.ncbi.nlm.nih.gov/sra under accession number PRJNA296814.