Abstract
Rab7 facilitates vesicular transport and delivery from early endosomes to late endosomes as well as from late endosomes to lysosomes. The role of Rab7 in vesicular transport is dependent on its interactions with effector proteins, among them Rab-interacting lysosomal protein (RILP), which aids in the recruitment of active Rab7 (GTP-bound) onto dynein–dynactin motor complexes to facilitate late endosomal transport on the cytoskeleton. Here we detail a novel bead-based flow cytometry assay to measure Rab7 interaction with the Rab-interacting lysosomal protein (RILP) effector protein and demonstrate its utility for quantitative assessment and studying drug–target interactions. The specific binding of GTP-bound Rab7 to RILP is readily demonstrated and shown to be dose-dependent and saturable enabling Kd and Bmax determinations. Furthermore, binding is nearly instantaneous and temperature-dependent. In a novel application of the assay method, a competitive small molecule inhibitor of Rab7 nucleotide binding (CID 1067700 or ML282) is shown to inhibit the Rab7–RILP interaction. Thus, the assay is able to distinguish that the small molecule, rather than incurring the active conformation, instead ‘locks’ the GTPase in the inactive conformation. Together, this work demonstrates the utility of using a flow cytometry assay to quantitatively characterize protein–protein interactions involving small GTPases and which has been adapted to high-throughput screening. Further, the method provides a platform for testing small molecule effects on protein–protein interactions, which can be relevant to drug discovery and development.
Keywords: Ras superfamily, Rab, Protein-protein interaction, Guanine nucleotide binding, GTP hydrolysis, GTPase effector, Quantitative flow cytometry, Glutathione-S-transferase (GST) assay, Drug discovery, HTS–High-throughput screening, Structure–activity relationship (SAR), G-Trap assay
1 Introduction
1.1 Rab7 and Its Effector Protein RILP Are Disease Relevant Proteins
Rab7 is a ubiquitously expressed Ras-superfamily GTPase, which functions in the endocytic pathway of mammalian cells to regulate vesicular traffic from early to late endosomes and then to lysosomes while playing a role in lysosome biogenesis [1–4]. Rab7 function is pivotal for lysosome-mediated degradation of endocytosed signaling receptors, defective cell surface proteins, and excess internalized lipid [4, 5]. Under conditions of nutrient starvation, Rab7 mediates fusion of autophagic vacuoles with lysosomes to release key metabolites for cell survival [6, 7]. The homeostatic balance of these processes is crucial to cell growth and differentiation, with misregulation often resulting in human disease.
Rab7 is associated with inherited and acquired neurologic diseases, cancer, and infectious diseases. Typical examples include Charcot–Marie–Tooth type 2B (CMT2B) disease, which causes axonal degeneration and is linked to four Rab7 missense mutants [8], other neurodegenerative diseases and retinal degeneration associated with reduced Rab7 function in autophagy [9–13], and genetic lipid storage disorders such as Batten and Niemann–Pick Type C diseases that are in part caused by Rab7 inactivation due to the accumulation of cholesterol in late endosomes [14, 15]. Disease-causing pathogens also often hijack the endocytic machinery where Rab7 is pivotal to gain access to the cell interior for infection, replication, and long-term survival [16–18]. These representative examples of Rab7-associated diseases illustrate how alteration of the GTPase regulatory cycle, effector protein interactions, as well as vesicular trafficking and signal transduction processes can give rise to disease and thus, collectively calls for the need to explore diverse intervention approaches.
The 45 kDa Rab-interacting lysosomal protein (RILP) is one of several Rab7 effector proteins [19–23]. Recruitment of RILP by active (GTP-bound) Rab7 regulates endolysosomal morphogenesis and microtubule minus-end directed transport through the recruitment of the dynein–dynactin motor complex [23–25]. Recruitment of RILP is also vital for processes such as phagosome maturation and fusion with late endosomes and lysosomes [26, 27].
Biochemically, Rab7 interaction with RILP has been assessed primarily using glutathione beads and GST-RILP for pull-down assays followed by protein electrophoresis and immunodetection, which often do not provide quantitative information [28, 29]. The method described here uses glutathione flow cytometry beads and GST-RILP, followed by detection using a flow cytometer. Such, use of bead-based flow cytometry to measure protein–protein interactions is not only quantitative, but also provides an enabling environment for the application of high-throughput screening (HTS) approaches. For HTS, the bead-based method can be adapted to screening a protein of interest against protein libraries to identify potential interacting partners or screening libraries of small molecules that may modulate protein–protein interactions. Applying the latter case to Rab7, for instance, can help in identifying small molecules with the potential to regulate Rab7 activity that may be relevant to Rab7-associated diseases.
1.2 Rationale for a Bead-Based Flow Cytometry Assay for Quantitative Measurement of Rab7-Effector Protein Interactions
Use of the described bead-based flow cytometry approach to quantitatively characterize Rab7 interaction with RILP, as a model effector protein, is a viable alternative to the conventional glutathione-S-transferase (GST) pull-down method that is at best semiquantitative. We demonstrate that RILP interacts with active Rab7 only when bound to GTP, demonstrating specificity and that this specific binding is dose dependent and saturable, enabling Bmax and Kd determination. Furthermore, the interaction is shown for the first time to be time and temperature dependent. When the small molecule 2-(benzoylcarbamothioylamino)-5, 5-dimethyl-4, 7-dihydrothieno[2,3-c]pyran-3-carboxylic acid (PubChem: CID 1067700 or ML282) was incorporated in the assay as a modulator of Rab7 activity, we obtained the novel result that this small molecule, previously characterized as a competitive inhibitor of Rab7 nucleotide binding [30], freezes Rab7 in a GDP-like conformation that does not bind RILP. The method has utility for quantitatively assaying effects of small molecules on Rab7 nucleotide bound status in cells, by using cell lysates in lieu of purified Rab7 in the assay. The experimental procedure is also readily adapted to investigating individual protein–protein interactions or identifying novel partners using HTS approaches [31, 32].
2 Materials
2.1 Reagent Sources
Reagents used in this report were obtained from various sources. Sephadex G-25, glutathione (GSH) Sepharose 4B, and Superdex (dextran/cross-linked agarose beads designed for peptide separation; 13 μm particle size with an exclusion limit of 7 kDa) are available from GE Healthcare (Piscataway, NJ). BODIPY (4,4-difluoro-4-bora-3a,4a-diaza-s-indacene or dipyrromethene boron difluoride) nucleotide analogs (BODIPY-GTP; 2′-(or 3′)-O-[N-(2-aminoethyl)urethane] G-35778 and BODIPY-GDP; 2′-(or-3′)-O-[N-(2-aminoethyl)urethane], G-22360) were from Invitrogen (merged with Life Technologies, Grand Island, NY). ProBond™ system for His-tagged protein purification was from Life Technologies (Grand Island, NY). GSH Sepharose 4B (GE Healthcare Bio-Sciences, Uppsala, Sweden). GFP-Rab7 was cloned into the pE-Sumo T7, amp vector from LifeSensors (Malvern, PA), which has a 6×His Tag for optimal bacterial expression of soluble protein and ease of purification [33]. Amicon Ultra Centrifugal Filters (30K MWCO) were obtained from Millipore Ireland Ltd. Protein concentrations were determined using a Bradford Colorimetric Assay from Bio-Rad (Hercules, CA) Unlabeled nucleotides (GDP, GTP, and GTP-γ-S) and buffer components were from Sigma (St. Louis, MO, USA). 2-(benzoylcarbamothioylamino)-5,5-dimethyl-4, 7-dihydrothieno[2,3-c]pyran-3-carboxylic acid (PubChem: CID 1067700 or ML282) was from ChemDiv Inc. (San Diego, CA, USA) and is also available from Sigma-Aldrich and Cayman Chemical. Where applicable, a Becton Dickinson FACScan flow cytometer with a 488-nm excitation Argon (Ar) laser and standard detection optics was used for all assays [34]. Concentrations of BODIPY-linked nucleotides were based on absorbance measurements and an extinction coefficient value of 80,000 M−1 cm−1 at 488 nm. The major materials used in each relevant experimental procedure are further captured below.
2.2 Synthesis of GSH Beads for Flow Cytometry Assays
Superdex dextran/cross-linked agarose beads (13 μm, 7 kDa exclusion limit) in buffered suspension.
Reduced GSH solution.
Water soluble bis-epoxide, ethanol, sodium hydroxide, and 4-butanediol diglycidol ether.
15 ml coarse sintered glass filter.
Sodium phosphate and EDTA solutions.
Appropriately supplemented HEPES (4-(2-hydroxyethyl)-1- piperazineethanesulfonic acid) based buffers as explained in Subheading 3.
2.3 Expression and Purification of GST, GST-RILP, GST-Rab7, His-Rab7, and His-SUMO-GFP-Rab7 in Bacterial Cells
Competent E. coli BL21 (DE3) cells.
LB agar plates and liquid broth supplemented with 100 μg/ml ampicillin.
pGEX vector for GST expression.
Plasmids encoding GST-Rab7 (pGEX Rab7), GST-RILP (pGEX RBD-RILP), His-Rab7 (pRSET Rab7), and His-SUMO-GFP-Rab7 (pE-Sumo T7 amp GFP-Rab7).
Isopropyl-beta-d-1-thiogalactopyranoside (IPTG).
Affinity purification resins (nickel resin—ProBond™ and GSH Sepharose 4B).
Appropriately supplemented phosphate-based buffers and other necessary chemicals.
2.4 Immobilization of GST, GST-Rab7, and GST-RILP on GSH Beads
13 μm GSH-coated beads.
Purified GST, GST-Rab7, and GST-RILP.
Appropriately supplemented HEPES-based buffer as explained in Subheading 3.
2.5 Measurement of Specific Interaction Between Rab7 and RILP Effector Protein
13 μm GSH coated beads.
Purified GST, GST-RILP, and His-Rab7.
Nucleotides: BODIPY-GTP dissolved in water.
Appropriately supplemented HEPES-based buffer as explained in Subheading 3.
2.6 Measurement of Time-Dependent Rab7 Interaction with RILP
13 μm GSH-coated beads.
Purified GST, GST-RILP, His-Rab7, and GFP-Rab7.
Nucleotides: Unlabeled GDP, BODIPY-GTP, BODIPY-GDP, and GTP-γ-S in water.
Appropriately supplemented HEPES and phosphate-based buffers as explained in Subheading 3.
2.7 Measurement of Temperature- Dependent Rab7 Interaction with RILP
13 μm GSH-coated beads.
Purified GST-RILP and His-Rab7.
Nucleotides: BODIPY-GTP in water.
Appropriately supplemented HEPES-based buffer as explained in Subheading 3.
2.8 Measurement of the Kinetics of BODIPY-GTP Displacement from Rab7 by Competitive Guanine Nucleotide- Binding Inhibitor (CID 1067700 or ML282)
13 μm GSH-coated beads.
Purified GST-Rab7.
DMSO solvent.
Small molecule (PubChem: CID 1067700 or ML282) dissolved in DMSO.
Nucleotides: Unlabeled GDP and BODIPY-GTP in water.
HEPES-based buffers with appropriate supplementation as explained in Subheading 3.
2.9 Measurement of Rab7 Interaction with RILP in the Presence of a Competitive Guanine Nucleotide- Binding Inhibitor (CID 1067700 or ML282)
13 μm GSH-coated beads.
Purified GST-RILP and GFP-Rab7.
Nucleotide: GTP-γ-S dissolved in water.
Small molecule: CID 1067700 or ML282.
HEPES-based buffers supplemented appropriately as explained in Subheading 3.
3 Methods
Subheading 3.1 provides an overview of a flow cytometric Rab7- effector binding assay with application to other Ras-superfamily GTPases. Subheadings 3.2–3.6 provide more detailed descriptions of methods and representative results for Rab7-effector protein binding measurements using RILP as a representative example; including determination of Kd and Bmax, kinetic measurements, temperature dependence of binding, and the effect of a small molecule inhibitor of Rab7 nucleotide binding on effector interaction. Subheading 3.7 provides information on data analysis methods.
3.1 Experimental Design of a Bead-Based Flow Cytometry Assay and Its Application to Measurement of Protein–Protein Interactions
In biochemical assays, elucidation and characterization of protein–protein interactions often involves time-consuming preparation of cell lysates containing candidate proteins and then confirming evidence of interaction with antibody-based immunoblotting methods. The caveats of these approaches tend to be: (1) it is difficult to obtain quantitative information, (2) other proteins in the cell lysates can influence the assay outcome, and (3) assays are often subject to the quality and performance of the individual antibodies used for detection.
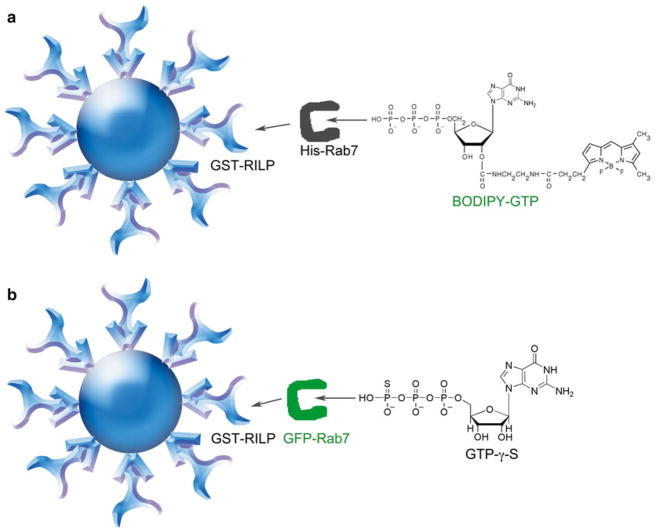
Sklar and colleagues pioneered the use of a bead-based flow cytometry approach to quantitatively measure protein–protein interactions associated with G-protein coupled receptors, initially taking advantage of streptavidin–biotin interactions [31, 32]. Further development of methodologies based on the use of GSH-beads and GST-fusion proteins led to the award of US patent 7,785,900 [35]. Nevertheless, traditional GST pull down and other nonquantitative or semiquantitative methods still remain in wide use [36, 37]. A bead-based flow cytometry assay for characterizing Rab GTPase and effector protein interactions can be set up in two ways using recombinant GST-fused RILP immobilized on GSH beads (Fig. 1a, b). In the first way, Rab7–RILP interaction is measured indirectly based on an increase in bead-associated fluorescence reflecting the binding of a Rab7-BODIPY-GTP complex to RILP (Fig. 1a). In the second way, Rab7–RILP interaction is measured directly based on the binding of GFP-Rab7 in the presence of nonhydrolyzable GTP-γ-S with GFP fluorescence measured by the flow cytometer (Fig. 1b). By applying these two approaches, the specificity of the protein–protein interaction between Rab7 and GSH bead-immobilized RILP is readily established.
Fig. 1.
GSH bead-based flow cytometry assay configurations for quantitative measurements of GTPase-effector protein binding. (a) Assay design for detecting Rab7 and Rab-interacting lysosomal protein (RILP) effector protein interaction based on detection of bound fluorescent BODIPY-GTP. GST-RILP (Rab binding domain of RILP only) is immobilized on 13 μm Superdex beads coated with GSH and incubated with purified His-tagged Rab7 complexed to fluorescent BODIPY-GTP. Flow cytometry detection is based on bead-associated fluorescence when His-Rab7-BODIPY-GTP binds to RILP. (b) Assay design for detecting Rab7 and Rab-interacting lysosomal protein (RILP) protein interaction based on detection of GFP-tagged Rab7. GST-RILP (Rab-binding domain of RILP only) is immobilized on 13 μm Superdex beads coated with GSH and incubated with GFP- tagged Rab7 complexed to nonhydrolyzable GTP-γ-S. Flow cytometry detection is based on bead-associated fluorescence when GFP-Rab7-GTP-γ-S binds to RILP
3.2 Reagent Preparation for Quantitative Protein–Protein Interaction Measurements
3.2.1 Synthesis of GSH Beads for Flow Cytometry Assays (see also Note 1)
Superdex dextran/cross-linked agarose beads are extruded from a column, activated with a water-soluble bis-epoxide, and then coupled to reduced GSH purged with nitrogen or argon gas. One milliliter of 50 % slurry of beads in 20 % ethanol is reduced to a wet cake using a 15 ml coarse sintered glass filter, and washed three times with 15 ml water to remove the ethanol. The wet cake is transferred to a small screw-cap tube, and the filter is rinsed with 0.3 ml of water, which is added to the tube. The beads are suspended by vortexing, then 60 μl of 10 M NaOH and 300 μl of 1,4-butanediyl diglycidyl ether (formerly butanediol diglycidyl ether) are added, and the suspension is rocked gently at 40 °C for 4 h.
The epoxy-activated beads are rinsed four times on a coarse sintered-glass filter. For the preparation of high site density beads, 600 μl of 100 mM GSH in 100 mM sodium phosphate (pH is 7.5), 1 mM EDTA, is added to the epoxy-activated beads. The beads are kept in suspension for 16 h at 40 °C, then rinsed twice with 0.01 % dodecyl maltoside. Beads with about 40-fold less GSH derivatization are prepared using 20 mM GSH and reacted for 2 h. The remaining active sites are blocked with 1 % 2-mercaptoethanol for 2 h. The beads are then rinsed twice followed by storage in 30 mM HEPES, pH 7.5, 100 mM KCl, 20 mM NaCl, 1 mM MgCl2 with 0.01 % dodecyl maltoside and 0.02 % NaN3 at 4 °C as a 50 % slurry, which corresponds to ~2.5 × 105 beads per microliter, or 25 assays of 104 beads each per microliter. High site density GSH derivatized beads have been successfully used for flow cytometry-based assays of GTPases and HTS of activators and inhibitors [38] (see also Note 2).
3.2.2 Expression and Purification of Recombinant Proteins
Competent E. coli (BL21) are used for transformation and expression of GST-Rab7, GST-Rab7-binding domain (RBD) of RILP (prepared as previously described by Dan Cimino [39]). Competent BL21(DE3)pLysS E. coli are used for transformation and expression of His-Rab7 or His-SUMO-GFP-Rab7. Transformants in all cases are selected on Luria–Broth (LB) agar plates containing 100 μg/ml ampicillin. Individual drug-resistant colonies (~20) are used to inoculate 100 ml of LB broth liquid cultures that are grown at 37 °C to a bacterial density of 0.5–0.7 absorbance units measured at 595 nm. Protein expression was induced by transfer to room temperature and addition of 0.2 mM isopropyl-beta-d-1-thiogalactopyranoside (IPTG) for 16–18 h to maximize yield of properly folded active fusion protein. (For pGEX vectors, IPTG induces gene expression directly under the control of a Ptac (hybrid trp/ lac) promoter; for pRSET vectors, IPTG enables gene expression under the control of a T7 promoter via activation of LacZ-dependent T7 polymerase expression encoded by a lamba lysogen carried by the bacterial strain).
Purification of GST-Rab7 and GST-RILP is then performed according to standard published procedures [34, 38]. In brief, bacterial cells are snap frozen and thawed and lysed using a micro-tip sonicator (Misonix Inc., Newtown, CT, U.S.A.). Cell lysates are centrifuged at 8,000 × g for 10 min to pellet the cellular debris. GST proteins are purified by batch purification method per GSH Sepharose 4B manufacturer’s instructions (GE Healthcare) and eluted with 10 mM GSH. Eluted protein is concentrated using Amicon® Ultra centrifugal filters (30K MWCO) and washed with three times with 10 volumes of HEPES buffer (30 mM HEPES, pH 7.5, 20 mM NaCl, and 100 mM KCl supplemented with 1 mM EDTA, 1 mM dithiothreitol (DTT)) to simultaneously remove excess GSH. Protein concentrations are determined using a BCA assay. Single-use aliquots are snap frozen and stored at −80 °C until used for experiments.
Purification of His-Rab7 or His-SUMO-GFP-Rab7 is performed by suspending the harvested bacterial cell pellets in cold native binding buffer for purification of His-tagged Rab7 proteins using the native protein purification method (manufacturer’s instructions for protein isolation via nickel affinity resin, Life Technologies). Bacterial cells are lysed using lysozyme and a microtip sonicator (Misonix Inc., Newtown, CT, U.S.A.), and cell debris removed by centrifugation as earlier. The supernatant is mixed with freshly prepared ProBond™ nickel chelating resin and bound at room temperature for 45 min using gentle agitation to keep the resin suspended in the lysate solution. Resin with bound protein is settled using low speed centrifugation (800×g) followed by multiple washes before eluting the nickel bound His-tagged protein with imidazole-based native elution buffer. Imidazole is removed by dialysis and protein is concentrated using Amicon® Ultra centrifugal filters (10K MWCO) followed by quantification using BCA protein assay. Single-use aliquots stored at −80 °C are used in the experiments.
3.2.3 Immobilization of GST, GST-Rab7, and GST-RILP on GSH Beads for Flow Cytometry
Purified GST, GST-Rab7 protein, or GST-RILP at 1 μM is incubated in 96-well plates or microfuge tubes at 4 °C overnight with 105 GSH beads in a total volume of 100 μl of HEPES buffer supplemented with 1 mM EDTA and freshly prepared 1 mM dithiothreitol (DTT). Beads in 96-well plates were kept suspended by rotation at ~300 rpm, while beads in microfuge tubes were kept suspended by slow end-over-end rotation. At the maximal concentration of protein, approximately 5 × 106 GST-Rab7 molecules are bound to each bead. This represents a concentration of ~8 nM bead-bound protein [38]. Unbound protein is removed by centrifugation twice at 800 × g followed by resuspension of washed beads in fresh buffer (30 mM HEPES, pH 7.5, 20 mM NaCl and 100 mM KCl, 1 mM EDTA, 1 mM DTT, 0.1 % BSA).
3.2.4 Preparation of Nucleotide-Bound, Active Rab7
The assay requires exchanging endogenous nucleotide by preincubating purified Rab7 with excess BODIPY-GTP in the presence of ethylenediaminetetraacetic acid (EDTA) as metal cation chelator. Addition of exogenous magnesium cations to ‘lock’ bound BODIPY-GTP onto Rab7 prior to testing on immobilized RILP is also an important step in ensuring quantitative measurements. Increasing concentrations of His-Rab7 (purified on nickel beads as described under Subheading 3.2.2) are incubated with a fixed concentration of BODIPY-GTP (500 nM) in a buffer-promoting nucleotide exchange (30 mM HEPES, pH 7.5, 20 mM NaCl and 100 mM KCl, 5 mM EDTA, and 0.1 % BSA) for 20 min at room temperature. Bound nucleotide is ‘locked-on’ the GTPase with the addition of 20 mM MgCl2 (final) and then the solutions are transferred to ice. The nucleotide must be kept cold to avoid GTP-hydrolysis and consequent conversion of the GTPase to the inactive conformation. Interaction measurements between Rab7 and RILP are initiated by mixing BODIPY-GTP-bound Rab7 with thoroughly washed GST-RILP immobilized on GSH beads. BODIPY-GTP-bound Rab7 mixed with GSH beads without bound protein and GST-coated GSH beads serve as negative controls. The mixtures are allowed to incubate for 15 min at room temperature before making fluorescence measurements on a FACScan flow cytometer using the fluorescein filter set (FL1). For GTPases with rapid hydrolysis rates, use non-hydrolyzable GTP-γ-S in lieu of GTP. His-SUMO-GFP-Rab7 is loaded with GTP-γ-S as detailed in 3.4.1.
3.2.5 Flow Cytometry Measurement of Bead-Associated Fluorescence
Recorded parameters of each event are based on light scattering and fluorescent properties. On the FACScan flow cytometer, GSH beads are first identified based on forward and side scattering. Fluorescence measurements from 1,000 events (beads) are averaged to mean channel fluorescence (MCF). Tubes suitable (Product No. 352008, BD Bioscience, San Jose, California, U.S.A.) for flow cytometry are used and reactions are diluted at least tenfold in HPSM buffer (30 mM HEPES, pH 7.5, 20 mM NaCl, 100 mM KCl, 1 mM DTT, 1 mM EDTA, 0.1 % BSA, and 20 mM MgCl2). This dilution step is necessary to ensure discrimination between bead-associated fluorescence and background fluorescence of soluble proteins as well as to ensure sufficient sample volume for the measurement.
3.3 Quantitative Measurement of the Specific Interaction Between Rab7 and RILP
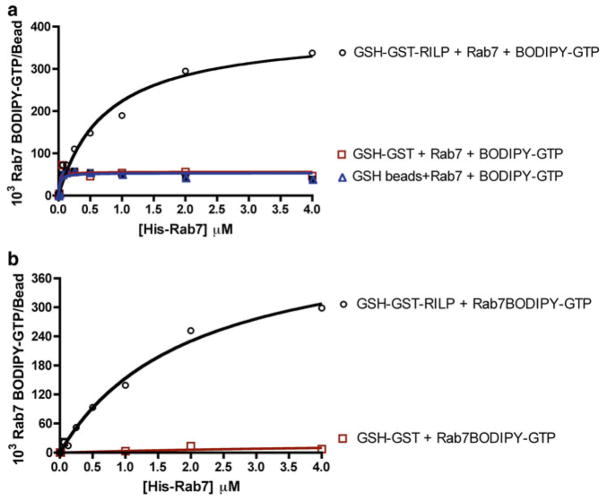
GST-RILP-coated beads (10E5 beads/100 μl buffer) are incubated with increasing concentrations of His-Rab7 prebound to BODIPY-GTP for 30 min with mild agitation at 4 °C as described in 3.2.4 and the bead-associated fluorescence is compared to that obtained with GST-coated or naked GSH beads. Rab7 is able to specifically bind to GSH beads coated with GST-RILP in a dose-dependent saturable manner with minimal interaction with either immobilized GST or naked GSH beads (Fig. 2a, b). The specificity of the observed interaction agrees with studies that have used techniques other than flow cytometry to show that RILP is a protein–protein interaction partner of Rab7 [28, 29, 40]. The Rab7–RILP interaction measured using the described bead-based flow cytometry approach, yielded a Bmax value of (506.20±28.66)×103 Rab7-BODIPY-GTP bound molecules per bead and equilibrium dissociation constant (Kd) of 1.87 ± 0.25 μM, calculated assuming one binding site per immobilized RILP for every Rab7 molecule. These results show that the interaction of Rab7 with RILP can be quantitatively measured by flow cytometry using small amounts of pure protein.
Fig. 2.
Quantitative measurements of GTPase-effector protein binding. (a) Flow cytometry-based measurement of total Rab7 binding to RILP by detecting fluorescent BODIPY-GTP shows binding is saturable, quantitative, and specific. Rab7 binding to GSH beads coated with GST alone or without any protein coating was minimal. (b) Specific binding of Rab7-RILP with unwanted nonspecific background binding subtracted
3.4 Measurement of the Time-Dependent Rab7 and RILP Interaction
3.4.1 Methods for Long-Term and Rapid Kinetic Measurements Using Flow Cytometry
For long-term kinetic measurements, purified GST or GST-RILP is incubated with 13 μm GSH beads as already described. One micromolar His-Rab7 ‘locked-on’ with 500 nM BODIPY-GTP or BODIPY-GDP is then incubated with bead immobilized GST or GST-RILP for increasing time intervals (0–150) min. Mean channel fluorescence (MCF) is then obtained on the FACScan flow cytometer by diluting the bead mixture 10-fold in HPSM buffer (see Section 3.2.6) based on BODIPY fluorescence.
Early time point protein–protein association kinetic measurement is used to assay Rab7 interaction with RILP. GST-RILP is first incubated with 13 μm GSH beads overnight as already described. Nucleotide-bound Rab7 is then prepared by incubating 1 μM GFP-Rab7 purified on nickel beads with 1 μM GTP-γ-S or 1 μM unlabeled GDP in a nucleotide exchange buffer (30 mM HEPES, pH 7.5, 20 mM NaCl and 100 mM KCl, 5 mM EDTA, 0.1 % BSA, and 1 mM DTT) at room temperature for 20 min. Bound nucleotide is ‘locked-on’ with 20 mM MgCl2 (final) and then put on ice. Real-time interaction between Rab7 and RILP is then initiated by mixing thoroughly washed GSH mobilized GST-RILP with nucleotide ‘locked-on’ GFP-Rab7. Mean channel fluorescence (MCF) is then obtained in HPSM buffer (30 mM HEPES pH 7.5, 20 mM NaCl, 100 mM KCl, 1 mM DTT, 1 mM EDTA and 0.1 % BSA, 20 mM MgCl2) on the FACScan flow cytometer based on GFP fluorescence.
3.4.2 Representative Kinetic Measurements of Rab7–RILP Interaction
Traditional GST pull-down assay procedures are not suited for kinetic measurements. In contrast, the described bead-based flow cytometry method for protein–protein interaction measurement provides a robust and economical way of quantitatively measuring the time dependence of the interaction with only minor modification of the procedures.
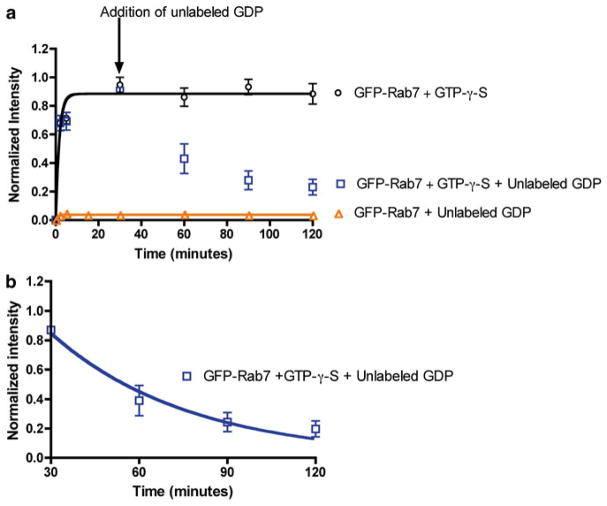
To measure the time-dependent interaction of Rab7 with RILP, the binding of purified GFP-Rab7 to immobilized RILP is measured by monitoring bead-associated changes in GFP fluorescence. Concentrations of GST-RILP, GFP-Rab7, unlabeled GDP, and GTP-γ-S are held fixed in this case. GFP-Rab7 interaction with GSH bead-immobilized GST-RILP is found to occur within sample mixing time and with almost instantaneous kinetics both in long term (Fig. 3a), and in early time point assessments of binding kinetics (Fig. 3b)—seen as a rapid and saturable rise in bead- associated fluorescence. Further tests of the specificity and validity of flow cytometry-based Rab7-RILP binding measurement using unlabeled GDP show no binding to RILP by GFP-Rab7 prebound to unlabeled GDP (Fig. 3a), confirming that Rab7 has to be in the GTP-bound state for interaction with the RILP effector protein to take place as previously observed [28, 29, 40]. Addition of unlabeled GDP is also able to competitively displace GTP-γ-S from the Rab7 nucleotide-binding pocket and cause the dissociation of GFP-Rab7 from the GST-RILP beads (Fig. 3a). When the rate of GFP-Rab7-GDP dissociation (Fig. 3b) is fitted to a single-phase exponential decay function using PRISM software, a calculated value of 0.020 ± 0.004 min−1 is deduced. Taken together, bead- based flow cytometry measurements are highly specific with only Rab7 in the GTP-bound nucleotide being able to bind RILP and further illuminate that Rab7 binding to RILP is very fast. Moreover, the use of GFP-Rab7 does not compromise RILP binding and is a useful tool for defining the equilibrium and kinetic parameters of the Rab7-RILP protein–protein interaction.
Fig. 3.
Quantitative measurements of Rab7 GTPase-RILP effector binding is rapid and nucleotide specific. (a) Flow cytometry-based measurement of the long-term kinetics of Rab7 binding to RILP by detecting fluorescent GFP-Rab7 shows binding is rapid and dependent on Rab7 being GTP bound. GFP-Rab7 prebound to nonhydrolyzable GTP-γ-S is nearly instantaneous and stable over 120 min. Addition of GDP results in displacement of GTP-γ-S from Rab7 and dissociation of GFP-Rab7GDP complex detected as a loss of bead-associated fluorescence. There is no binding of GFP-Rab7 in the GDP-bound state to RILP. (b) Data from panel (a) were replotted starting at the 30 min time point to allow determination of the dissociation rate of GFP-Rab7-GDP from RILP. Data was fitted to single-phase exponential decay function using PRISM software yielding a dissociation rate of 0.020 ± 0.003 min−1
3.5 Measurement of the Temperature Dependence of Rab7 and RILP Interaction
3.5.1 Methods for Measuring Temperature-Dependent Protein–Protein Interaction
For the measurement of protein–protein interaction at varying temperatures (4–37 °C), GST-RILP (1 μM) is incubated with 13 μm GSH beads overnight at 4 °C in HPSM buffer (see Section 3.2.6). The following day, increasing concentrations of His-Rab7 are incubated with fixed concentrations of 500 nM BODIPY-GTP in a nucleotide exchange buffer (30 mM HEPES, pH 7.5, 20 mM NaCl and 100 mM KCl, 5 mM EDTA, 0.1 % BSA, and 1 mM DTT) for 20 min before adding 20 mM MgCl2 (final) to ‘lock’ the bound nucleotide. Bead-immobilized GST-RILP is washed thoroughly with the same buffer system and then in cubated with nucleotide-bound Rab7 for 15 min at temperatures of 4, 22, and 37 °C. Fluorescence measurements are then obtained on Flow Cytometer using HPSM buffer as outlined in Section 3.4.1.
For temperature-shift experiments, purified 1 μM GST-RILP is first bound to beads in HPSM buffer. BODIPY-GTP-bound Rab7 is prepared as described in 3.2.4. To initiate the protein–protein interaction, BODIPY-GTP-bound Rab7 and thoroughly washed bead-immobilized GST-RILP are first mixed together on ice and then progressively shifted to 4 °C, 22 °C, and finally to 37 °C. On the FACScan flow cytometer, fluorescence measurements or MCF are made of Rab7-BODIPY-GTP bound to the RILP beads as a function of time following each temperature shift. All measurements are obtained in HPSM buffer as outlined in Section 3.4.1.
3.5.2 Representative Temperature-Dependent Rab7 and RILP Interaction Measurements
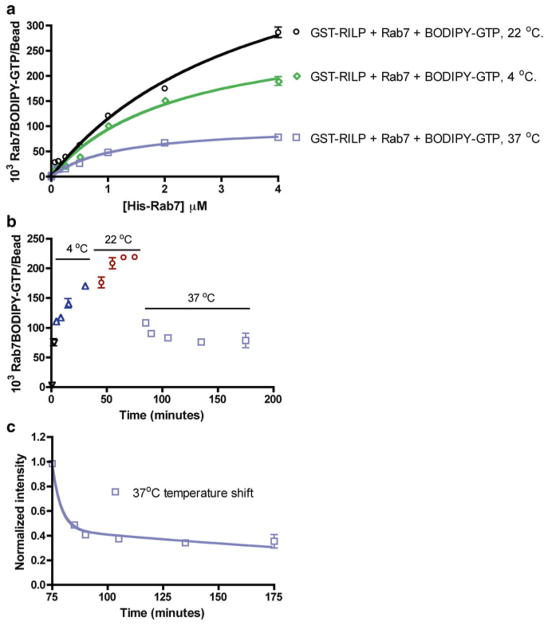
Temperature is an important determinant of protein–protein interactions because of its ability to affect the rate of protein conformation changes and molecular collisions. Typically, the rate of protein–protein interaction is favored at elevated temperatures due to enhanced frequency of molecular collisions at elevated temperatures. The bead-based flow cytometry assay was used for testing the impact of varying temperatures at 4 °C, 22 °C, and 37 °C on Rab7 and RILP interactions. The experiment is carried out with increasing concentrations of His-Rab7 against fixed concentrations of bead-immobilized RILP and BODIPY-GTP. His-Rab7 binding to RILP increases as the temperature is raised from 4 °C to 22 °C (Fig. 4a). Not surprisingly, when the assay is conducted at 37 °C, the fluorescence intensities—used as the measure of BODIPY-GTP-His-Rab7 bound to the RILP beads—are substantially lower across all concentrations of His-Rab7. In a kinetic temperature- shift experiment, His-Rab7, BODIPY-GTP, and bead-immobilized RILP are first mixed on ice and then progressively shifted to 4 °C, 22 °C and finally to 37 °C. Fluorescence measurements are made of Rab7-BODIPY-GTP bound to the RILP beads as a function of time following each temperature shift. A steady increase in binding is observed for temperature shifts from 0 °C through 22 °C, but again a sudden drop in fluorescence is observed as the reaction mixture is shifted from 22 °C to 37 °C (Fig. 4b). Estimation of the rate of temperature-induced fluorescence loss is made by fitting the 37 °C time points (Fig. 4c) to a two-phase exponential decay function. The rate constants of 0.0436 ± 0.053 and 0.014 ± 0.003 min−1 for the fast and slow phases of fluorescence loss, respectively, are deduced, where the rate constant value calculated for the fast phase of fluorescence loss is statistically close to that measured for the dissociation of GFP-Rab7-GDP from RILP (Fig. 3a). The sudden drop in fluorescence at 37 °C is most likely due to Rab7-mediated hydrolysis of BODIPY-GTP to BODIPY-GDP resulting in a Rab7 conformational change which leads to Rab7 dissociation from RILP. The lack of fluorescent GTP-γ-S availability from commercial vendors precluded further testing of this significant observation though it remains of interest. The ability to rapidly measure nucleotide hydrolysis in real time helps mechanistic analyses and is a simple alternative to radioisotope-based filter-binding measurements of nucleotide hydrolysis.
Fig. 4.
Flow cytometry-based measurements show Rab7 GTPase-RILP effector binding is temperature dependent and sensitive to nucleotide hydrolysis. (a–c) Flow cytometry-based measurement of His-Rab7 binding to RILP by detecting fluorescent BODIPY-GTP. (a) Dose-dependent His-Rab7 binding is temperature dependent and negatively affected by GTP hydrolysis at higher temperature (b and c). (b) A kinetic temperature-shift experiment shows His-Rab7 binding to RILP increases steadily at 4 and 22 °C, but decreases rapidly upon shift to 37 °C, likely due to GTP hydrolysis and dissociation of Rab7 from RILP. (c) Data in (b) were replotted starting at the 75 min time point to allow determination of the dissociation rate. Data were fitted to a two-phase exponential decay function using PRISM software yielding a dissociation rate of 0.014 ± 0.003 min−1 for the slow phase and 0.0436 ± 0.053 min-1 for the fast phase. The rate constant value deduced for the fast phase is statistically close to that measured for the dissociation of GFP-Rab7-GDP from RILP in Fig. 2a, supporting the conclusion that 37 °C stimulates Rab7 GTPase hydrolysis of BODIPY-GTP and consequent dissociation from RILP
3.6 Measurement of the Effect of a Novel Guanine Nucleotide-Binding Inhibitor on Rab7 and RILP Interaction
3.6.1 Measuring the Kinetics of Direct BODIPY-GTP Nucleotide Displacement by a Competitive Guanine Nucleotide-Binding Inhibitor (CID 1067700)
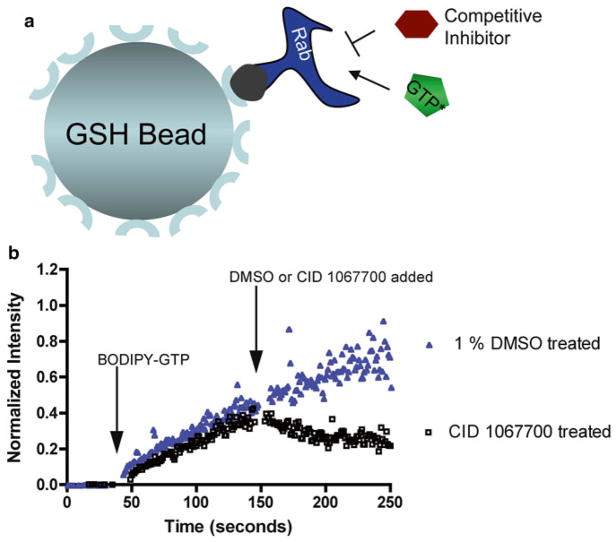
As a general note, direct fluorescent nucleotide displacement measurements are performed in HEPES buffer at nonequilibrium nucleotide-binding conditions. In brief, 1 μM GST-Rab7 is immobilized on GSH-beads as described in 3.2.3. Following GSH-bead immobilization of GST-Rab7, fluorescence baseline measurements are first obtained on the FACScan flow cytometer by measuring the fluorescence of thoroughly washed GSH bead-immobilized GST-Rab7 (2×103 GSH beads) before adding BODIPY-GTP (100 nM: Kd concentration for BODIPY-GTP binding to Rab7). After ~150 s, competition is initiated in situ against binding/loading BODIPY-GTP using either DMSO (1 % final) or CID 1067700 (10 μM final). As a negative control for all measurements, GST-Rab7 is prebound to GDP (500 μM) prior to adding BODIPY-GTP at the above concentration and the low level nonspecific bead-associated fluorescence is subtracted from fluorescence obtained from the DMSO or CID 1067700 treated samples to ensure that only specific nucleotide binding is finally considered.
3.6.2 Assessment of Rab7 Interaction with RILP in the Presence of Competitive Guanine Nucleotide-Binding Inhibitor (CID 1067700) Under Equilibrium Binding Conditions
GST-RILP at 1 μM is first immobilized on 13 μM GSH beads as described in Section 3.2.3. GFP-Rab7 (1 μM) purified on nickel beads is incubated with increasing concentrations of GTP-γ-S (1 % DMSO final treated), unlabeled GDP or CID 1067700 (dissolved in DMSO) hereby referred to as ligands in nucleotide exchange buffer for 20 min at room temperature. Bound nucleotide is ‘locked-on’ with 20 mM MgCl2 (final) and then all sample sets are put on ice. The samples are then incubated with thoroughly washed GSH mobilized GST-RILP for 15 min at room temperature before obtaining fluorescence measurements in an HPSM buffer as outlined in Section 3.4.1. Mean channel fluorescence (MCF) as a measure of bead-associated GFP fluorescence is determined on a FACScan flow cytometer.
3.6.3 Measurement of the Effect of a Competitive Guanine Nucleotide-Binding Inhibitor on the Interaction Between Rab7 and RILP
We previously identified a first-in-class competitive guanine nucleotide-binding inhibitor (PubChem: CID 1067700 or ML282) with activity against Rab7 in vitro [30] and in cell-based assays [41]. The importance of the compound is recognized through the award of a US patent [42]. In the published work, GST-Rab7 was immobilized on GSH beads and BODIPY- nucleotide binding to Rab7 was measured as an increase in bead- associated fluorescence by flow cytometry and used to mechanistically characterize the small molecule competitor, as illustrated in cartoon form (Fig. 5a). The same assay setup is also useful to measure nucleotide-binding kinetics and changes induced by small molecule addition. For example, addition of CID 1067700 in such an assay results in a rapid decrease in bead-associated fluorescence as a function of time (Fig. 5b). This is due to displacement of BODIPY-GTP from Rab7 and disruption of further fluorescent nucleotide loading. One question that could not be resolved by the flow cytometry-based nucleotide-binding assay is whether or not CID 1067700 binding to Rab7 promotes the active conformation of Rab7 (normally induced by GTP) or instead retains Rab7 in the inactive conformation (normally induced by GDP).
Fig. 5.
GSH bead-based flow cytometry assays for quantitative measurements of Rab7 guanine nucleotide binding and dissociation kinetics. (a) Assay design for detecting nucleotide binding and dissociation kinetics on Rab7 based on detection of bound fluorescent BODIPY-GTP. GST-Rab7 is immobilized on 13 μm Superdex beads coated with GSH and detection is based on fluorescent BODIPY-GTP binding. (b) BODIPY-GTP (100 nM final) was added to GST-Rab7 immobilized on GSH beads suspended in 300 μl of buffer (first arrow). The ligand was allowed to bind for 100 s and then DMSO (1 % final) or CID 1067700 (10 μM final) was added at 150 s (second arrow). While the addition of a competitive guanine nucleotide-binding inhibitor (CID 1067700) causes dissociation of BODIPY-GTP, addition of DMSO has no effect on BODIPY-GTP-binding kinetics
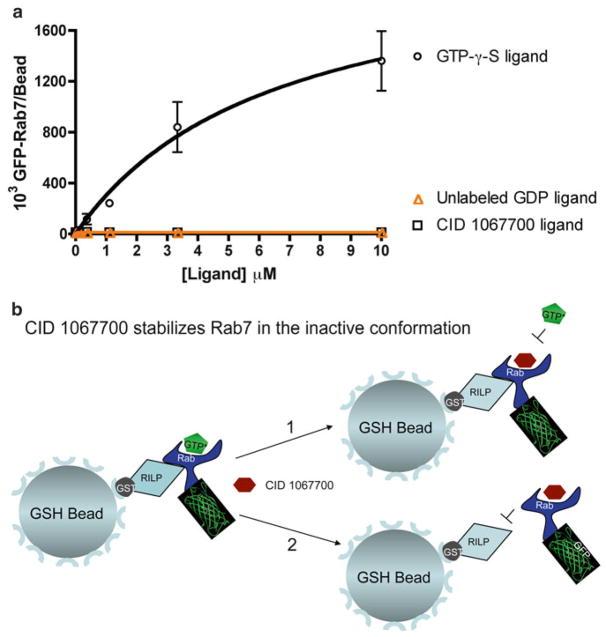
The RILP-binding assay detailed here is able to discriminate Rab7 in the active conformation from the inactive conformation and hence is ideally suited for testing the impact of CID 1067700 on Rab7 conformational status. We monitored the interaction of bead-immobilized RILP with GFP-Rab7 in the presence of increasing concentrations of: (1) GTP-γ-S, (2) CID 1067700, or (3) GDP. CID1067700 and nucleotide stocks for this experiment are prepared in DMSO and diluted 1000x in the assay. In the presence of CID 1067700 alone, Rab7 is unable to adopt the ‘active’ like conformation and fails to bind to the RILP effector protein, analogous to the GDP negative control (Fig. 6a). In contrast, GFP-Rab7 is able to bind RILP in a saturable manner in the presence of GTP-γ-S. From this, one can conclude that the presence of CID 1067700 most likely freezes Rab7 in an inactive conformational state that does not support interaction with RILP (Fig. 6b). Together, this evidence supports the conclusion that CID 1067700 is a competitive inhibitor of Rab7 nucleotide binding, which does not induce Rab7 to adopt the active conformation, and thus also precludes interaction with downstream effector proteins. Such information is critical in the assessment of small molecule GTPase inhibitor suitability for in vivo utility.
Fig. 6.
GSH bead-based flow cytometry assay establishes that a competitive guanine nucleotide-binding inhibitor (CID 1067700) retains Rab7 in an inactive conformation. (a) Flow cytometry-based measurement of Rab7 binding to RILP by detecting fluorescent GFP-Rab7 in the presence of CID 1067700. GFP-Rab7 increasingly binds to RILP at increasing concentrations of nonhydrolyzable GTP-γ-S but fails to bind to RILP with increasing concentrations of GDP or CID 1067700. Rab7 does not adopt ‘active’ like conformation in the presence of CID 1067700 alone. (b) Graphical display of the two distinct possible scenarios that can result from competitive inhibitor (CID 1067700) binding to Rab7. In scenario 1, binding of the competitive inhibitor dissociates GTP from the nucleotide-binding pocket, but keeps Rab7 in the active conformation, which would still allow binding to the RILP effector. In scenario 2, binding of the competitive inhibitor to Rab7 causes the GTPase to assume or remain in the inactive conformation, which does not favor interaction with RILP. The data we have presented support scenario 2 and suggest that the guanine nucleotide-binding inhibitor should functionally inhibit Rab7 in cell-based assays
3.6.4 Broader applications: Probing Other GTPases and Performing High Throughput Screens
The described effector binding assay can be extended to the study of other Ras-related GTPases andthe analyses of active GTPase status in cell lysates (see Note 3). Using multiplexing approaches requiring differentially labeled bead sets the assay can be adapted for high throughput screening assays (see Note 4). Advantages of the assay over conventional methods are summarized in Notes 5–6.
3.7 Data Analyses
All data processing and analyses presented in this report employed GraphPad Prism software (GraphPad Software). For kinetic experiments, raw data acquired were first processed using IDLE query software (obtained from University of New Mexico Center for Molecular Discovery, UNM CMD) before further analysis using GraphPad Prism. All experiments are representative of at least three independent trials.
Acknowledgments
This work was generously supported by National Science Foundation (MCB0956027) and the National Institutes of Health (R21NS7740241) to AWN and (P30CA1181000, U54MH074425, and U54MH084690) to LAS. DS was supported as a visiting MARC scholar (T34 GM008395, PI Zavala, CSUN) and as a summer intern (ASERT IRACDA K12 GM088021, PI Wandinger-Ness). We thank Ms. Janet Kelly for administrative support. We also acknowledge Elsa Romero and Patricia Jim for technical support. Small molecule screening was performed in the NMMLSC/ UNMCMD and flow cytometry assays were conducted in the Flow Cytometry Shared Resource Center supported by the University of New Mexico Cancer Center (P30 CA11810).
Footnotes
GSH Bead Synthesis and Alternatives. GSH prepared in phosphate buffer can reduce the pH of the solution. It is crucial to readjust the pH to 7.5 prior to incubation with the epoxy-activated Superdex beads for the coupling reaction to be effective. As an alternative, polystyrene beads (6–8 μm) precoated with GSH may be purchased (GSHP-60-5) from Spherotech (Lake Forest, IL). However, it should be noted that the polystyrene beads exhibit higher nonspecific binding than GSH cross-linked to dextran/cross-linked agarose beads as described here. Therefore, the substitution of GSH-polystyrene beads must be carefully optimized using relevant controls. For example, an irrelevant GST-fusion protein is best used as a negative control instead of beads coated with GST, which gives higher than expected background signal. Additionally, when using beads of a smaller diameter, the conditions for saturating with GST-tagged proteins and maximal binding of ligand must be optimized accordingly [34, 38], see also Note 2.
Calculating Bead-Binding Sites and Assuring Saturation Binding of GST-Fusion Proteins to GSH Beads. Utilizing the previously determined kinetic and equilibrium-binding constants for GST fused to green fluorescent protein (GST–GFP) binding to GSH beads is useful in establishing the optimal stoichiometric mixtures of GSH beads and individual GST-fusion proteins so as to achieve saturating site occupancies [34, 38]. The saturable site occupancy values for GST–GFP are a measure of the amount of bead-associated GSH. Based on a Kd for GST–GFP binding of approximately 80 nM, the optimal concentration of GST-fusion protein required to fully saturate the GSH sites on the beads is taken as 10 × Kd, giving 91 % saturation.
Extrapolation to Different GTPases and Cell-Based Assays. The effector binding assay described here can be extrapolated to different families of Ras-related GTPases and has utility for measuring GTPase activation status in cells. Examples of such applications include: determination of EC50 values of Rab and Rho GTPase-targeted small molecules in cell-based assays [41, 43]; for monitoring GTPase cascades in Sin nombre hantavirus infection [39], and responses of cancer patient samples to treatment with drugs that target GTPase activation [44]. Based on these examples, the assay has utility for monitoring host GTPase responses to viral, fungal, and bacterial pathogens and for studying changes in GTPase activation in response to specific disease processes, growth factors, cytokines, toxins, among other extracellular stimuli.
In lieu of using BODIPY nucleotides or GFP-tagged GTPases for detection, effector binding can also be efficaciously measured using antibodies to detect bound GTPases (e.g., using a fluorescently conjugated primary antibody directed against the GTPase of interest or using an nonfluorescent primary antibody followed by a fluorescent secondary antibody). Key negative controls include use of a control GST-effector protein to which the GTPase of interest is not expected to bind and omission of the primary antibody.
Adapting the Assay to Multiplex and HTS Applications. For multiplex analyses, 4 μm diameter, polystyrene GSH-beads with 1.2×106 GSH sites per bead and labeled with differing intensities of red fluorescent dye have been successfully used [45]. Up to seven different bead sets with varying emission magnitudes at 665 ± 10 nm can be uniformly excited at 635 nm and readily discriminated by flow cytometry. To capitalize on such beads for multiplex, HTS applications, GST-fusion proteins are individually coated onto different bead sets and the red fluorescence intensity serves as a ‘zipcode’ in the multiplex measurement for identifying sets of bound GST-proteins. Briefly, bead sets (at a concentration of 1.4 × 105 beads/μl and total volume of 240–250 μl) are first blocked with 0.1 % bovine serum albumin in 30 mM HEPES pH 7.5, 100 mM KCl, 20 mM NaCl containing 0.01 % (v/v) NP-40, and 1 mM EDTA for 30 min at room temperature. GST-fusion protein binding is conducted overnight at 4 °C with 1 μM GST-fusion protein in 100 μl of buffer (30 mM HEPES pH 7.5, 100 mM KCl, 20 mM NaCl containing 0.01 % (v/v) NP-40, and 1 mM EDTA). Protein-coupled beads are washed two times with 30 mM HEPES pH 7.5, 100 mM KCl, 20 mM NaCl, 0.01 % (v/v) NP-40, 1 mM EDTA buffer supplemented with 0.1 % BSA and 1 mM DTT. Bead sets are subsequently pooled for multiplex analyses and individual assays are conducted in 384-well plates. The described assay configuration has been used for HTS, multiplex measurement of small molecule interference with nucleotide binding to families of bead-immobilized GST-GTPases in compound library screens [45, 46], as well as for characterization of small molecules and their chemical optimization through structure–activity analyses [41, 43, 47–50]. The effector-binding assay described here is also suited for multiplex analyses and has been shown to have utility for quantitative, spatiotemporal resolution of GTPase cascades that are activated in response to hantavirus infection of cells [39] and growth factor stimulation [39, 41]. Further potential applications include screens for small molecule inhibitors of GTPase-effector protein interactions and parallel analyses of GTPase responses to diverse stimuli.
Highlights of Comparative Advantages of Bead-Based Flow Cytometry Measurements. The bead-based flow cytometry technique we have presented offers a more robust approach for quantitatively assaying equilibrium binding and kinetic parameters in comparison to some of the methods commonly used to achieve the same [36, 37, 51–53]. The assays are simple to set up on account of the following realities: (1) it is easy to generate GST tagged protein, (2) tagging of interacting partners with flow cytometry-suited fluorophores does not require elaborate work, and (3) in the case of nucleotide-binding proteins, both labeled and unlabeled nucleotides can easily be obtained commercially. With respect to addressing specificity questions, a bead-based flow cytometry assay is also not susceptible to challenges inherent in cell-based assays such as difficulties in telling if the interaction between two proteins of interest is a direct process or occurs via a protein complex involving intermediaries. Elucidation of this question more often tends to involve time-consuming experiments that can lead to false-positive results. Moreover, it is also easy to assay the impact of other confounding factors on the interaction between two proteins. One can assess whether the interaction between two proteins is influenced by kinetics of protein–protein association or thermodynamics of the system, solution viscosity, pH, and/or salt concentrations [54].
Conclusions. Much as our current report centered primarily on Rab7 interaction with RILP as model system for method and protocol illustration, the design of the GSH bead-based flow cytometry assay can also be extended to some of the well- characterized Rab7 regulatory protein partners such as hVps39 [55], TBC1D15 [55, 56], ORPIL [25, 57], Rabring7 [58, 59], and an α-subunit of the proteasome–XAPC7 [60, 61]. Rab7 interaction with regulatory proteins is an important physiological process and controls important physiological processes that can result into human disease states when mis-regulated. Taken together, our findings present GSH bead-based flow cytometry as a simpler method for quantitatively measuring Rab7 interactions with guanine nucleotides and with regulatory proteins. Based on our evidence with CID 1067700, bead-based flow cytometry can also be used for HTS of small molecule modulators of Rab7 protein–protein interactions that may be pertinent for identifying activator and inhibitor small molecules that may have relevance in the long-term development of new therapeutics for Rab7-associated diseases.
References
- 1.Feng Y, Press B, Wandinger-Ness A. Rab 7: an important regulator of late endocytic membrane traffic. J Cell Biol. 1995;131:1435–1452. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meresse S, Gorvel JP, Chavrier P. The rab7 GTPase resides on a vesicular compartment connected to lysosomes. J Cell Sci. 1995;108:3349–3358. doi: 10.1242/jcs.108.11.3349. [DOI] [PubMed] [Google Scholar]
- 3.Press B, Feng Y, Hoflack B, et al. Mutant Rab7 causes the accumulation of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J Cell Biol. 1998;140:1075–1089. doi: 10.1083/jcb.140.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bucci C, Thomsen P, Nicoziani P, et al. Rab7: a key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saxena S, Bucci C, Weis J, et al. The small GTPase Rab7 controls the endosomal trafficking and neuritogenic signaling of the nerve growth factor receptor TrkA. J Neurosci. 2005;25:10930–10940. doi: 10.1523/JNEUROSCI.2029-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutierrez MG, Munafó DB, Berón W, et al. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 7.Jager S, Bucci C, Tanida I, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 8.Spinosa MR, Progida C, De Luca A, et al. Functional characterization of Rab7 mutant proteins associated with Charcot-Marie-Tooth type 2B disease. J Neurosci. 2008;28:1640–1648. doi: 10.1523/JNEUROSCI.3677-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castino R, Lazzeri G, Lenzi P, et al. Suppression of autophagy precipitates neuronal cell death following low doses of methamphetamine. J Neurochem. 2008;106:1426–1439. doi: 10.1111/j.1471-4159.2008.05488.x. [DOI] [PubMed] [Google Scholar]
- 10.Bains M, Zaegel V, Mize-Berge J, et al. IGF-I stimulates Rab7–RILP interaction during neuronal autophagy. Neurosci Lett. 2011;488:112–117. doi: 10.1016/j.neulet.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan CC, Epstein D, Hiesinger PR. Intracellular trafficking in Drosophila visual system development: a basis for pattern formation through simple mechanisms. Dev Neurobiol. 2011;71:1227–1245. doi: 10.1002/dneu.20940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Midorikawa R, Yamamoto-Hino M, Awano W, et al. Autophagy-dependent rhodopsin degradation prevents retinal degeneration in Drosophila. J Neurosci. 2010;30:10703–10719. doi: 10.1523/JNEUROSCI.2061-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takacs-Vellai K, Bayci A, Vellai T. Autophagy in neuronal cell loss: a road to death. Bioessays. 2006;28:1126–1131. doi: 10.1002/bies.20489. [DOI] [PubMed] [Google Scholar]
- 14.Choudhury A, Dominguez M, Puri V, et al. Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J Clin Invest. 2002;109:1541–1550. doi: 10.1172/JCI15420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haskell RE, Carr CJ, Pearce DA, et al. Batten disease: evaluation of CLN3 mutations on protein localization and function. Hum Mol Genet. 2000;9:735–744. doi: 10.1093/hmg/9.5.735. [DOI] [PubMed] [Google Scholar]
- 16.Seabra MC, Mules EH, Hume AN. Rab GTPases, intracellular traffic and disease. Trends Mol Med. 2002;8:23–30. doi: 10.1016/s1471-4914(01)02227-4. [DOI] [PubMed] [Google Scholar]
- 17.Zhang M, Chen L, Wang S, et al. Rab7: roles in membrane trafficking and disease. Biosci Rep. 2009;29:193–209. doi: 10.1042/BSR20090032. [DOI] [PubMed] [Google Scholar]
- 18.Vonderheit A, Helenius A. Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol. 2005;3:e233. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agola JO, Jim PA, Ward HH, et al. Rab GTPases as regulators of endocytosis, targets of disease and therapeutic opportunities. Clin Genet. 2011;80:305–318. doi: 10.1111/j.1399-0004.2011.01724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bucci C, De Gregorio L, Bruni CB. Expression analysis and chromosomal assignment of PRA1 and RILP genes. Biochem Biophys Res Commun. 2001;286:815–819. doi: 10.1006/bbrc.2001.5466. [DOI] [PubMed] [Google Scholar]
- 21.Cantalupo G, Alifano P, Roberti V, et al. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cogli L, Piro F, Bucci C. Rab7 and the CMT2B disease. Biochem Soc Trans. 2009;37:1027–1031. doi: 10.1042/BST0371027. [DOI] [PubMed] [Google Scholar]
- 23.Jordens I, Fernandez-Borja M, Marsman M, et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- 24.Johansson M, Lehto M, Tanhuanpaa K, et al. The oxysterol-binding protein homologue ORP1L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol Biol Cell. 2005;16:5480–5492. doi: 10.1091/mbc.E05-03-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansson M, Rocha N, Zwart W, et al. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison RE, Brumell JH, Khandani A, et al. Salmonella impairs RILP recruitment to Rab7 during maturation of invasion vacuoles. Mol Biol Cell. 2004;15:3146–3154. doi: 10.1091/mbc.E04-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marsman M, Jordens I, Kuijl C, et al. Dynein-mediated vesicle transport controls intracellular Salmonella replication. Mol Biol Cell. 2004;15:2954–2964. doi: 10.1091/mbc.E03-08-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun J, Deghmane AE, Bucci C, et al. Detection of activated Rab7 GTPase with an immobilized RILP probe. Methods Mol Biol. 2009;531:57–69. doi: 10.1007/978-1-59745-396-7_5. [DOI] [PubMed] [Google Scholar]
- 29.Peralta ER, Martin BC, Edinger AL. Differential effects of TBC1D15 and mammalian Vps39 on Rab7 activation state, lysosomal morphology, and growth factor dependence. J Biol Chem. 2010;285:16814. doi: 10.1074/jbc.M110.111633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agola JO, Hong L, Surviladze Z, et al. A competitive nucleotide binding inhibitor: in vitro characterization of Rab7 GTPase inhibition. ACS Chem Biol. 2012;7:1095–1108. doi: 10.1021/cb3001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simons PC, Shi M, Foutz T, et al. Ligand-receptor-G-protein molecular assemblies on beads for mechanistic studies and screening by flow cytometry. Mol Pharmacol. 2003;64:1227–1238. doi: 10.1124/mol.64.5.1227. [DOI] [PubMed] [Google Scholar]
- 32.Waller A, Simons PC, Biggs SM, et al. Techniques: GPCR assembly, pharmacology and screening by flow cytometry. Trends Pharmacol Sci. 2004;25:663–669. doi: 10.1016/j.tips.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 33.Butt TR, Edavettal SC, Hall JP, et al. Sumo fusion technology for difficult-to-express proteins. Protein Expr Purif. 2005;43:1–9. doi: 10.1016/j.pep.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tessema M, Simons PC, Cimino DF, et al. Glutathione-S-transferase-green fluorescent protein fusion protein reveals slow dissociation from high site density beads and measures free GSH. Cytometry A. 2006;69:326–334. doi: 10.1002/cyto.a.20259. [DOI] [PubMed] [Google Scholar]
- 35.Simons PC, Sklar LA, Prossnitz ER, et al. Glutathione beads and GST fusion proteins. STCUNM; Albuquerque, NM: Sanford- Burnham Medical Research Institute; La Jolla, CA, USA: 2010. [Google Scholar]
- 36.Phizicky EM, Fields S. Protein-protein interactions: methods for detection and analysis. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen TN, Goodrich JA. Protein- protein interaction assays: eliminating false positive interactions. Nat Methods. 2006;3:135–139. doi: 10.1038/nmeth0206-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz SL, Tessema M, Buranda T, et al. Flow cytometry for real-time measurement of guanine nucleotide binding and exchange by Ras-like GTPases. Anal Biochem. 2008;381:258–266. doi: 10.1016/j.ab.2008.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buranda T, BasuRay S, Swanson S, et al. Rapid parallel flow cytometry assays of active GTPases using effector beads. Anal Biochem. 2013;442:149–157. doi: 10.1016/j.ab.2013.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosales KR, Peralta ER, Guenther GG, et al. Rab7 activation by growth factor withdrawal contributes to the induction of apoptosis. Mol Biol Cell. 2009;20:2831–2840. doi: 10.1091/mbc.E08-09-0911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong L, Guo Y, BasuRay S, et al. A Pan- GTPase inhibitor as a molecular probe. PLoS One. doi: 10.1371/journal.pone.0134317. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wandinger-Ness A, Sklar LA, Agola JO, et al. Rab7 GTPase inhibitors and related methods of treatment. STCUNM; Albuquerque, NM: University of Kansas; Lawrence, KS, USA: 2014. [Google Scholar]
- 43.Oprea TI, Sklar LA, Agola JO, et al. Novel activities of select NSAID R-enantiomers against Rac1 and Cdc42 GTPases. PLoS One. doi: 10.1371/journal.pone.0142182. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo Y, Kenney SR, Cook L, et al. Novel mechanism of therapeutic benefit through ketorolac usage in ovarian cancer patients. J Clin Oncol. under review. [Google Scholar]
- 45.Surviladze Z, Waller A, Wu Y, et al. Identification of a small GTPase inhibitor using a high-throughput flow cytometry bead-based multiplex assay. J Biomol Screen. 2010;15:10–20. doi: 10.1177/1087057109352240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Surviladze Z, Young SM, Sklar LA. High-throughput flow cytometry bead-based multiplex assay for identifi cation of Rho GTPase inhibitors. Methods Mol Biol. 2012;827:253–270. doi: 10.1007/978-1-61779-442-1_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Surviladze Z, Ursu O, Miscioscia F, et al. Three small molecule pan activator families of Ras-related GTPases. Probe reports from the NIH Molecular Libraries Program. 2010 [PubMed] [Google Scholar]
- 48.Surviladze Z, Waller A, Strouse JJ, et al. A potent and selective inhibitor of Cdc42 GTPase. Probe reports from the NIH Molecular Libraries Program. 2010 [PubMed] [Google Scholar]
- 49.Hong L, Simons P, Waller A, et al. A small molecule pan-inhibitor of Ras-superfamily GTPases with high efficacy towards Rab7. Probe reports from the NIH Molecular Libraries Program. 2010 [PubMed] [Google Scholar]
- 50.Hong L, Surviladze Z, Ursu O, et al. Characterization of a Cdc42 protein inhibitor and its use as a molecular probe. J Biol Chem. 2013;288:8531–8543. doi: 10.1074/jbc.M112.435941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He L, Olson DP, Wu X, et al. A flow cytometric method to detect protein-protein interaction in living cells by directly visualizing donor fluorophore quenching during CFP → YFP fluorescence resonance energy transfer (FRET) Cytometry A. 2003;55:71–85. doi: 10.1002/cyto.a.10073. [DOI] [PubMed] [Google Scholar]
- 52.Dye BT, Schell K, Miller DJ, et al. Detecting protein-protein interaction in live yeast by flow cytometry. Cytometry A. 2005;63:77–86. doi: 10.1002/cyto.a.20112. [DOI] [PubMed] [Google Scholar]
- 53.Chen J, Carter MB, Edwards BS, et al. High throughput flow cytometry based yeast two-hybrid array approach for large-scale analysis of protein-protein interactions. Cytometry A. 2012;81:90–98. doi: 10.1002/cyto.a.21144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schreiber G. Kinetic studies of protein- protein interactions. Curr Opin Struct Biol. 2002;12:41–47. doi: 10.1016/s0959-440x(02)00287-7. [DOI] [PubMed] [Google Scholar]
- 55.Flinn RJ, Yan Y, Goswami S, et al. The late endosome is essential for mTORC1 signaling. Mol Biol Cell. 2010;21:833–841. doi: 10.1091/mbc.E09-09-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang XM, Walsh B, Mitchell CA, et al. TBC domain family, member 15 is a novel mammalian Rab GTPase-activating protein with substrate preference for Rab7. Biochem Biophys Res Commun. 2005;335:154–161. doi: 10.1016/j.bbrc.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 57.Rocha N, Kuijl C, van der Kant R, et al. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J Cell Biol. 2009;185:1209–1225. doi: 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mizuno K, Kitamura A, Sasaki T. Rabring7, a novel Rab7 target protein with a RING finger motif. Mol Biol Cell. 2003;14:3741–3752. doi: 10.1091/mbc.E02-08-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mizuno K, Sakane A, Sasaki T. Rabring7: a target protein for rab7 small g protein. Methods Enzymol. 2005;403:687–696. doi: 10.1016/S0076-6879(05)03059-4. [DOI] [PubMed] [Google Scholar]
- 60.Dong J, Chen W, Welford A, et al. The proteasome alpha-subunit XAPC7 interacts specifically with Rab7 and late endosomes. J Biol Chem. 2004;279:21334–21342. doi: 10.1074/jbc.M401022200. [DOI] [PubMed] [Google Scholar]
- 61.Mukherjee S, Dong J, Heincelman C, et al. Functional analyses and interaction of the XAPC7 proteasome subunit with Rab7. Methods Enzymol. 2005;403:650–663. doi: 10.1016/S0076-6879(05)03056-9. [DOI] [PubMed] [Google Scholar]