Abstract
Importance
Although a variety of well characterized diseases such as sarcoidosis and granulomatosis with polyangiitis (GPA) affect the lacrimal gland, many patients with dacryoadenitis are labelled as having nonspecific orbital inflammation (NSOI) on the basis of histology and systemic disease evaluation. The ability to subdivide these patients should facilitate selection of effective therapies.
Objective
The a priori hypothesis was that gene expression profiles would complement clinical and histopathologic evaluations in identifying well-characterized diseases and in subdividing NSOI into clinically-relevant groups.
Design
Gene expression levels in biopsies of inflamed and control lacrimal glands were measured with microarrays. Stained sections of the same biopsies were used for evaluation of histopathology.
Setting
Tissues were obtained from oculoplastic surgeons at 7 centers representing 5 countries. Gene expression analysis was done at the Oregon Health & Science University.
Participants
48 subjects: 4 thyroid eye disease, 7 sarcoidosis, 3 GPA, 28 NSOI, and 6 healthy controls.
Main Outcome(s) and Measure(s)
The primary outcome was subdivision of samples based on gene expression of our published list of ~40 differentially expressed transcripts in blood, lacrimal gland, and orbital adipose tissue from patients with sarcoidosis. Stained sections were evaluated for no, mild, moderate, or marked inflammation, granulomas, nodules, or fibrosis by two independent ocular pathologists masked to the clinical diagnosis.
Results
The mclust algorithm segregated the samples into 4 subsets with differences illustrated by a heat map and multidimensional scaling plots. Most of the sarcoidosis samples are in subset 1, which had the highest granuloma score. Interestingly, 3 NSOI samples in subset 1 had no apparent granulomas. 32% of the NSOI samples could not be distinguished from samples of healthy controls, while other examples of NSOI tended to group with gene expression resembling TED or GPA. The 4 subsets could also be partially differentiated by their fibrosis, inflammation, and granuloma scores.
Conclusions and Relevance
Gene expression profiling discloses clear heterogeneity among patients with lacrimal inflammatory disease. Comparison of the expression profiles suggests that a subset of patients with nonspecific dacryoadenitis might have a limited form of sarcoidosis, while other patients with NSOI cannot be distinguished from healthy controls.
INTRODUCTION
Dacryoadenitis has a broad differential diagnosis that includes sarcoidosis, nonspecific orbital inflammation (NSOI), Sjögren’s syndrome, and less commonly, entities such as granulomatosis with polyangiitis (GPA), thyroid eye disease (TED), or IgG4-related disease1–5. Other considerations in the differential diagnosis include infections, tumors including lymphoma, and lymphoid hyperplasia. Among these diagnoses, nonspecific inflammation is the most common biopsy diagnosis2.
Although histopathology is certainly a component of the gold standard for diagnosing diseases of the lacrimal gland, the approach has limitations. For example, GPA is a medium-sized vessel vasculitis, but vessels of that size are rarely present within a lacrimal specimen. Although the pathology of sarcoidosis includes granuloma, which are readily appreciated by histology, a sampling error might exclude granulomas from the tissue which is microscopically examined. The presence of IgG4 positive plasma cells is emerging as an important factor in lacrimal inflammation4, but the specificity of these cells has been questioned6. Clearly there is opportunity to improve the diagnostic yield from lacrimal gland biopsy. Further, an unresolved issue is whether nonspecific inflammation represents a single diagnostic entity or a variety of different inflammations.
Implementation of molecular techniques has the potential to increase the accuracy and specificity of diagnosing the different forms of dacryoadenitis. For example, gene expression profiling can distinguish different causes of synovitis7, esophagitis8, or myocarditis.9 We previously employed gene expression profiling to characterize different cause of uveitis10,11 and of orbital adipose inflammation12–15. In another report, we analyzed gene expression in lacrimal gland, orbital adipose tissue, and blood from subjects affected by sarcoidosis16. In that study, we selected a set of 40 mRNA transcripts that were differentially expressed in all three diseased tissues compared to the same tissues from healthy controls16.
We now have compared gene expression profiles of lacrimal gland from subjects with TED, GPA, sarcoidosis, or nonspecific orbital inflammation to profiles from normal controls. Based on the 40 transcripts identified from our prior sarcoidosis study, we used the mclust clustering algorithm17 to test the hypothesis that nonspecific lacrimal gland inflammation is a heterogeneous collection of diseases which sometimes resembles sarcoidosis.
METHODS
Tissue
We collected 49 lacrimal gland biopsies from 48 subjects (one lacrimal gland from a patient with sarcoidosis was sampled from two separate locations) from 7 international centers: Emory University, Kaohsiung Veteran’s General Hospital, King Khaled Eye Specialist Hospital, Medical College of Wisconsin, Ophthalmic Surgeons and Consultants (Ohio), Oregon Health & Science University, and University of British Columbia. All tissue had been formalin fixed and paraffin embedded. All tissues had been reviewed by an ophthalmic pathologist from the contributing center and then further reviewed by two additional pathologists (DJW and HEG) who collaborated in the preparation of this report. These centers and the pathologists have previously used a similar method of tissue collection to analyze gene expression from a variety of orbital diseases including TED13, GPA12, sarcoidosis16, and nonspecific orbital inflammation14. Control tissue from healthy individuals was obtained at the time of cosmetic surgery or blepharoplasty. In some instances, surgeons removed portions of normal lacrimal gland if the gland prolapsed during the course of orbital surgery. The study was approved by the IRB at Oregon Health & Science University as well as the IRB at each participating center.
Pathology Review
The two reviewing pathologists were tasked with confirming the diagnosis from the institution where the biopsy was obtained. The pathologists also independently scored each tissue on the basis of granulomas, lymphoid nodules, fibrosis, and inflammation. In each case, an ordinal scale from zero to three was used. The scores reflected none, mild, moderate or marked change for each given descriptor. The two pathologists’ scores were in agreement two-thirds of the time. The scores were averaged for analyses.
RNA extraction and microarray
All tissue was sent to Oregon Health & Science University, Portland, Oregon, for RNA extraction and microarray as previously described12,13,16. In brief, cDNA was synthesized from purified RNA, amplified, labeled, and hybridized to Affymetrix U 133 plus 2.0 arrays, which include probe sets for about 45,000 transcripts. Further, we have reported on the RNA quality and the correlation between our array data and quantitative PCR18.
Statistical methods
Affymetrix cel files were imported into R statistical language19 and expression levels were calculated by the robust multi-array analysis20. Mclust17 was used to cluster the normalized gene expression values of selected probe sets. A clustering analysis is a type of unsupervised machine learning that groups samples into clusters such that those within a cluster are more closely related to one another than those assigned to different clusters based on some dissimilarity measure21. Most of the conventional clustering algorithms such as hierarchical clustering or k-means clustering require the number of clusters pre-specified, which can be a subjective decision and disadvantage in applications22. In contrast, mclust assumes that samples are collected from a number of Gaussian distributions (or normal distributions). First, It fits a range of different Gaussian mixture models, and then it chooses an optimal number of Gaussian distributions or clusters based on the Bayesian information criterion23. Therefore, mclust can provide a more objective number of clusters than the conventional clustering algorithms.
Heat maps and multidimensional scaling (MDS) plots were employed to visualize the cluster analysis results. All computations were done using affy, limma and mclust packages of R statistical language.
Selection of discriminating probe sets
In our previous study investigating gene expression differences due to sarcoidosis in lacrimal gland, orbital tissue, or blood, we employed linear models and empirical Bayes methods24 while adjusting for potential confounding effects of sex and age. The linear models were fitted to each tissue type separately and race was not included in the models due to too many missing values. We reported 159 Affymetrix probe sets indicating differential expression common to all three of these tissues (at least 1.5-fold change compared to healthy controls with an adjusted false discovery rate (FDR) p-value < .05)16. Among them, 45 probe sets had FDR p-values < 0.005 and were used for the analyses shown in Figures 1 and 2. Because of redundancy in the probe sets, the 45 probe sets represent 40 different genes.
Figure 1.
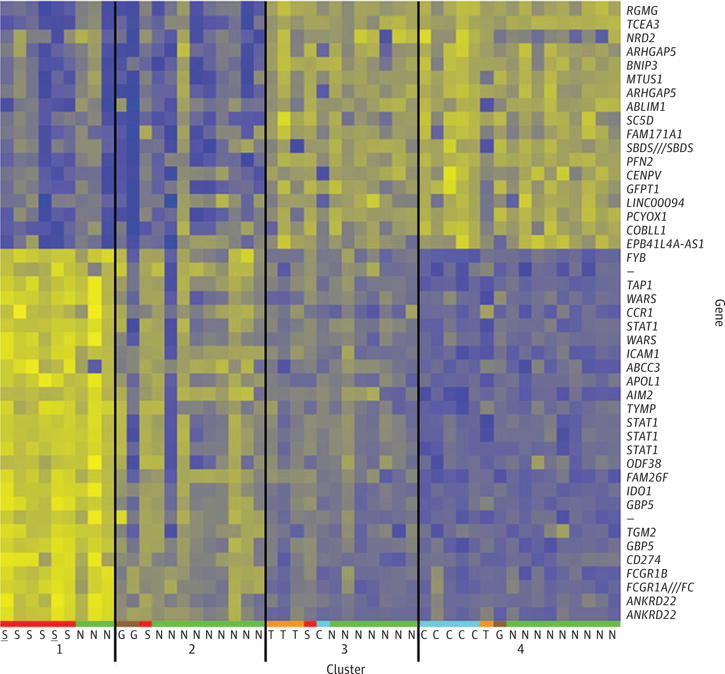
Heat map showing relative expression levels in 4 clusters based on the core probe sets. Yellow, high expression; blue, low expression; S, sarcoidosis; G, GPA; T, TED; N, NSOI; C, control. The number of clusters and allocation of specimens were determined by the mclust algorithm. The underlined “S”s in cluster 1 indicate two specimens from the same subject.
Figure 2.
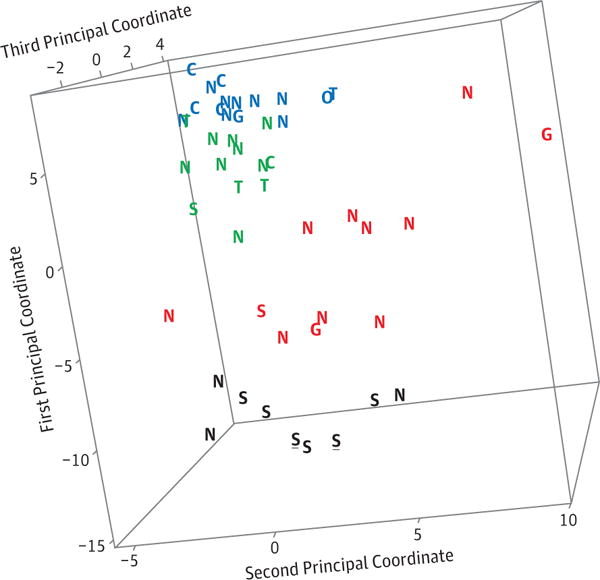
The 3-dimensional MDS plot illustrates the clustering of the 4 clusters determined by mclust analysis using 45 probe sets previously shown to be differential expressed in subjects with sarcoidosis. The four colors indicate the 4 clusters. A rotating view of this plot is seen in Video 1. S, sarcoidosis; G, GPA; T, TED; N, NSOI; C, control. The underlined black “S”s indicate two specimens from the same subject.
RESULTS
We analyzed 49 lacrimal biopsies including 7 from subjects with sarcoidosis, 3 from subjects with GPA, 4 from subjects with TED, 6 from healthy controls, and 28 from subjects with NSOI. The mean age and sex for each of these five groups is shown in Table 1. Information on race is also provided in this table. Although the groups do differ in some comparisons on the basis of age, sex or race, the differences are not statistically significant. Because of the numbers of specimens in each of our disease groups, we focused our initial analysis on 45 core probe sets that we previously reported to discriminate between gene expression profiles of lacrimal gland, orbital tissue, and blood from subjects with sarcoidosis when compared to profiles from healthy controls16. Application of the mclust algorithm with the selected probe sets used unsupervised machine learning to divide the subjects into an optimal number of clusters with different gene expression patterns based on the Bayesian information criterion23. Inclusion of additional probe sets that we previously found to be differentially expressed in orbital tissues from subjects with these diseases decreased the consistency of clustering as indicated by the mclust partition with the disease diagnosis.
Table 1.
Subject demographics
| n | Age (St. Dev.) |
Sex
|
Race
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| F | M | Asian | African-American | Hawaiian, Pacific Islander | White | Unknown | |||
| Control | 6 | 51.7 (25.4) | 4 | 2 | 0 | 1 | 0 | 3 | 2 |
| GPA | 3 | 32.4 (17.4) | 3 | 0 | 2 | 1 | 0 | 0 | 0 |
| NSOI | 28 | 40.6 (18.7) | 18 | 10 | 1 | 7 | 1 | 8 | 11 |
| Sarcoidosis | 7 | 32.9 (12.4) | 5 | 2 | 0 | 4 | 0 | 1 | 2 |
| TED | 4 | 57.1 (8.8) | 2 | 2 | 0 | 0 | 0 | 3 | 1 |
|
| |||||||||
| Total | 48 | 32 | 16 | 3 | 13 | 1 | 15 | 16 | |
The mclust results based on the core probe sets are shown in the heat map (Figure 1) and the multidimensional scaling plot (Figure 2). These probe sets discriminate among the controls and the 3 known diseases: sarcoidosis, TED, and GPA. As seen in both the heat map and the MDS plot, 6 of the 8 sarcoidosis samples, two of the three GPA samples, three of the four TED samples, and 5 of the 6 control samples cluster together. These same genes subdivide the NSOI samples into the 4 clusters Three of the 28 NSOI samples have a gene expression pattern resembling sarcoidosis even though granulomas were not detected in the biopsies by either pathologist. Clinical information was available on two of the 3 subjects with NSOI whose gene expression profile resembled sarcoidosis. Both subjects were observed without therapy and neither had chest computerized tomography imaging. Other NSOI samples group with other diagnoses including GPA, TED, or controls. In both Figures 1 and 2, underlining is used to identify the two samples from different portions of the lacrimal gland from the same individual. The pattern seen in the heat map and the proximity of the two data points shown in the MDS plot indicate that the reproducibility of the method is good.
We next analyzed the lacrimal biopsies based on the pathologist’s scoring for fibrosis, inflammation, lymphoid nodules or granulomas. In Figure 3A, the biopsies have been grouped based on the gene expression cluster. Using Chi square analysis, the clusters differ for each of these four pathology scores (Fibrosis: p-value = 0.014; Inflammation: p-value = 0.001; Granuloma: p-value = 0.001; Nodules: p-value = 0.002). We then analyzed how the four pathology scores correlated with the histologic diagnosis. As shown visually in Figure 3B and as confirmed by Chi square analysis, lymphoid nodules do not discriminate among the various diagnoses (p=0.75). The quantification of fibrosis tends to be a useful discriminator, although the differences did not reach statistical significance (p=0.09). On the other hand, either the detection of granulomas or the extent of inflammation had a non-random distribution among the diagnoses as determined by Chi square (p<0.001 for either variable), although the inflammation scores showed considerable overlap.
Figure 3.
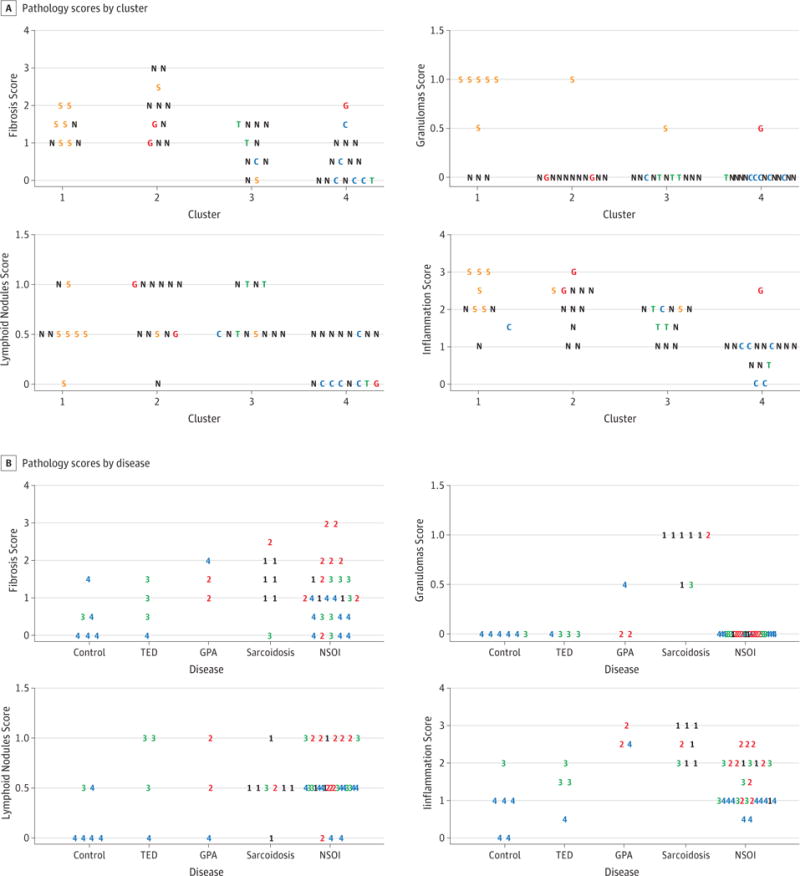
The tissues were scored for fibrosis, granulomas, lymphoid nodules, and inflammation. A. The scores are grouped by cluster with the letters and colors indicating the disease group. S – sarcoidosis, T – TED, G – GPA, N – NSOI, C – control. B. The scores are grouped by disease with the numbers and colors indicating the clusters as shown in figure 1.
An important question to address is whether results could be attributed to an effect from medication. Data on prednisone usage was known for 25 subjects in this study. For the six subjects receiving prednisone, the dosages were moderately high and ranged from 20 to 60 mg/day (mean 39.2 mg/day). With only 6 subjects receiving prednisone, statistical conclusions about the impact of the prednisone should be tentative. However, the MDS plot shown as Figure 4 highlights those on prednisone therapy. Both of the sarcoidosis subjects who did not cluster with the other sarcoidosis samples were receiving prednisone at the time of the biopsy. The subject with GPA receiving prednisone appears to be shifted to the right as well. Corticosteroids are known to have major effects on gene expression23, and it is plausible therefore to speculate that their use affected some results.
Figure 4.
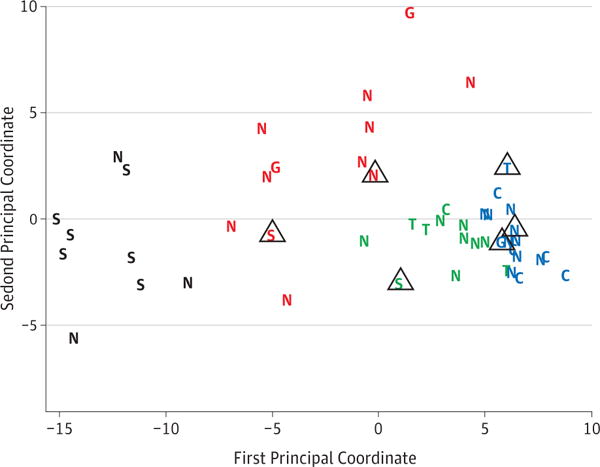
The effect of prednisone on gene expression. Subjects who were known to be taking prednisone at the time of the biopsy have been marked with a triangle. The shift to the right of two subjects with sarcoidosis and one subject with GPA suggests that prednisone might have affected gene expression.
Finally, potential uses of gene expression profiling are to be able to predict prognosis or to identify therapy that has a high likelihood of success. Although the size of the database and the duration of follow up data are not adequate to address these issues thoroughly, one clinical vignette might be instructive. One subject who was included in the study had a history of well documented Graves’ disease more than a decade prior to the onset of orbital disease. Her orbital imaging showed bilateral but asymmetric inflammation that involved the inferior and superior rectus muscles, as well as orbital fat, the lacrimal glands and the cavernous sinus. The radiologist felt that the findings were atypical for thyroid orbitopathy due to the lacrimal and sinus involvement. The pathologist read the biopsy as nonspecific inflammation. Her gene expression profile also supported this conclusion with a pattern of gene expression that fell into cluster 1 rather than with the thyroid eye disease samples. The patient responded to rituximab therapy, an approach which was not effective for thyroid eye disease in a randomized controlled trial25.
DISCUSSION
Our observations based on gene expression indicate clearly that nonspecific inflammation within the lacrimal gland is a heterogeneous collection of diseases. The data suggest that a minority of patients with nonspecific lacrimal gland inflammation might have a localized form of sarcoidosis, even if granulomas are not present in the biopsy section examined. Other examples of NSOI resemble either GPA or TED in terms of the pattern of gene expression. More detailed clinical information and more extensive follow up information would help determine if these subjects followed different clinical courses that correlated with the pattern of gene expression.
Our study found some degree of fibrosis, inflammation and even lymphoid nodules among healthy controls. This corresponds well with what has been reported in minor salivary gland and lacrimal gland biopsies26. Some degree of fibrosis and lymphoid nodularity is frequently detected in this tissue from individuals who have no known abnormality regarding function of these glands. The clustering on the basis of gene expression divided the biopsies into groups that correlated with the pathology scores more consistently than dividing the tissue on the basis of a diagnosis such as TED, GPA, or sarcoidosis. Either grouping, i.e. by gene expression or by diagnosis, reveals that the four pathology scores (fibrosis, inflammation, granulomas, lymphoid nodules) do not reliably subdivide the tissues based on the overlap shown in Figures 3A and 3B.
Our study has a variety of limitations. Centers might vary in clinical approach such as the duration of symptoms acceptable prior to biopsy. These centers might also vary as to pharmacologic treatment prior to biopsy. The length of time that the biopsy was in fixative could influence the integrity of the RNA. Our data on clinical course are somewhat limited. Finally, we show data that suggest that oral corticosteroid affects gene expression, but we do not have adequate data to assess other medications. We also emphasize that our observations on corticosteroids are based on a small number of subjects taking a relatively high dosage of prednisone. In a prior study on orbital adipose tissue that focused on a different set of transcripts, we could not show that corticosteroids affected gene expression13. Additional study of this issue is warranted.
Despite these limitations, this is arguably the largest study to date on gene expression in lacrimal inflammation; and the first to show definitively the heterogeneity of nonspecific inflammation in that gland. Prior studies on gene expression in the lacrimal gland27 or in the lacrimal accessory gland28 were limited to normal tissues. Our analysis supports the conclusion that some patients with nonspecific lacrimal gland inflammation might have a limited form of sarcoidosis, GPA, or even TED. Furthermore, the analysis demonstrates how patterns of gene expression can provide data that complements the information gleaned by light microscopy.
Supplementary Material
Video 1: A rotating view of the 3-dimensional MDS plot shown in Figure 2.
Key Point.
Question
Is nonspecific orbital inflammation (NSOI) affecting the lacrimal gland a single disease or multiple different diseases and is it a limited form of other diseases such as sarcoidosis or granulomatosis with polyangiitis?
Key Points/Finding
A cohort study comparing the expression of 40 genes from biopsies of patients with lacrimal NSOI with controls indicates that NSOI is a heterogeneous collection of diseases and suggests that it is often a limited form of known lacrimal inflammations.
Meaning
The pattern of gene expression can subdivide NSOI of the lacrimal gland and has the potential to lead to more precise therapy as well as new insights into pathogenesis.
Tweet
Gene expression shows nonspecific lacrimal gland inflammation may be a limited form of a known disease.
Acknowledgments
Funding/Support: This study was supported in part by grants EY020249, EY010572, and RR024140 from the National Health Institute and by Research to Prevent Blindness, the William and Mary Bauman Foundation, the Mas Family Foundation, and the Stan and Madelle Rosenfeld Family Trust.
Role of the Funder/Sponsor: The funding organizations had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author Contributions: Drs. Rosenbaum and Planck had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Rosenbaum, Choi, Harrington, Planck.
Acquisition, analysis, or interpretation of data: Rosenbaum, Choi, Harrington, Wilson, Grossniklaus, Sibley, Ng, Dailey, Steele, Hayek, Craven, Edward, Maktabi, Hussain, White, Dolman, Czyz, Foster, Harris, Bee, Tse, Alabiad, Dubovy, Kazim, Selva, Yeatts, Korn, Kikkawa, Silkiss, Sivak, Stauffer, Planck, Salek
Drafting of the manuscript: Rosenbaum, Choi, Harrington, Sibley, Planck.
Critical revision of the manuscript for important intellectual content: Rosenbaum, Choi, Harrington, Wilson, Grossniklaus, Sibley, Ng, Dailey, Steele, Hayek, Craven, Edward, Maktabi, Hussain, White, Dolman, Czyz, Foster, Harris, Bee, Tse, Alabiad, Dubovy, Kazim, Selva, Yeatts, Korn, Kikkawa, Silkiss, Sivak, Stauffer, Planck, Salek
Statistical analysis: Choi, Sibley
Obtained funding: Rosenbaum, Choi, Planck.
Administrative, technical, or material support: Rosenbaum, Harrington, Wilson, Grossniklaus, Ng, Dailey, Steele, Hayek, Craven, Edward, Maktabi, Hussain, White, Dolman, Czyz, Foster, Harris, Bee, Tse, Alabiad, Dubovy, Kazim, Selva, Yeatts, Korn, Kikkawa, Silkiss, Sivak, Stauffer, Planck
Study supervision: Rosenbaum, Harrington, Planck.
Conflict of Interest Disclosures: Dr. Rosenbaum has consulted for Genentech and was a coinvestigator on a study funded by Genentech to evaluate the use of rituximab for orbital inflammatory diseases. No other disclosures were reported.
Previous Presentations: Some of these data will be presented at the annual meeting of the Association for Research in Vision and Ophthalmology; May 2017; Baltimore, Maryland. The raw and normalized gene expression microarray data will be uploaded to the GEO database.
Additional Contributions: RNA extraction and microarray assays were performed in the Oregon Health & Science University Gene Profiling Shared Resource. Kristina Vartanian, BS, Integrated Genomics Laboratory, Oregon Health & Science University, provided excellent technical support for the microarray work. There was no financial compensation.
References
- 1.Mombaerts I. The many facets of dacryoadenitis. Curr Opin Ophthalmol. 2015;26(5):399–407. doi: 10.1097/ICU.0000000000000183. [DOI] [PubMed] [Google Scholar]
- 2.Andrew NH, McNab AA, Selva D. Review of 268 lacrimal gland biopsies in an Australian cohort. Clin Exp Ophthalmol. 2015;43(1):5–11. doi: 10.1111/ceo.12371. [DOI] [PubMed] [Google Scholar]
- 3.Andrew NH, Kearney D, Sladden N, et al. Idiopathic Dacryoadenitis: Clinical Features, Histopathology, and Treatment Outcomes. Am J Ophthalmol. 2016;163:148–153 e141. doi: 10.1016/j.ajo.2015.11.032. [DOI] [PubMed] [Google Scholar]
- 4.Wallace ZS, Deshpande V, Stone JH. Ophthalmic manifestations of IgG4-related disease: single-center experience and literature review. Semin Arthritis Rheum. 2014;43(6):806–817. doi: 10.1016/j.semarthrit.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 5.Koturovic Z, Knezevic M, Rasic DM. Clinical significance of routine lacrimal sac biopsy during dacryocystorhinostomy: A comprehensive review of literature. Bosn J Basic Med Sci. 2016 doi: 10.17305/bjbms.2016.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong AJ, Planck SR, Choi D, et al. IgG4 immunostaining and its implications in orbital inflammatory disease. PLoS One. 2014;9(10):e109847. doi: 10.1371/journal.pone.0109847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeremenko N, Noordenbos T, Cantaert T, et al. Disease-specific and inflammation-independent stromal alterations in spondylarthritis synovitis. Arthritis Rheum. 2013;65(1):174–185. doi: 10.1002/art.37704. [DOI] [PubMed] [Google Scholar]
- 8.Wen T, Stucke EM, Grotjan TM, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology. 2013;145(6):1289–1299. doi: 10.1053/j.gastro.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lassner D, Kuhl U, Siegismund CS, et al. Improved diagnosis of idiopathic giant cell myocarditis and cardiac sarcoidosis by myocardial gene expression profiling. European heart journal. 2014;35(32):2186–2195. doi: 10.1093/eurheartj/ehu101. [DOI] [PubMed] [Google Scholar]
- 10.Sharma SM, Choi D, Planck SR, et al. Insights in to the pathogenesis of axial spondyloarthropathy based on gene expression profiles. Arthritis Res Ther. 2009;11(6):R168. doi: 10.1186/ar2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenbaum JT, Pasadhika S, Crouser ED, et al. Hypothesis: sarcoidosis is a STAT1-mediated disease. Clinical immunology. 2009;132(2):174–183. doi: 10.1016/j.clim.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenbaum JT, Choi D, Wilson DJ, et al. Orbital pseudotumor can be a localized form of granulomatosis with polyangiitis as revealed by gene expression profiling. Exp Mol Pathol. 2015;99(2):271–278. doi: 10.1016/j.yexmp.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenbaum JT, Choi D, Wong A, et al. The Role of the Immune Response in the Pathogenesis of Thyroid Eye Disease: A Reassessment. PLoS One. 2015;10(9):e0137654. doi: 10.1371/journal.pone.0137654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenbaum JT, Sibley CH, Choi D, Harrington CA, Planck SR. Molecular diagnosis: Implications for ophthalmology. Prog Retin Eye Res. 2016;50:25–33. doi: 10.1016/j.preteyeres.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenbaum JT, Choi D, Wilson DJ, et al. Molecular diagnosis of orbital inflammatory disease. Exp Mol Pathol. 2015;98(2):225–229. doi: 10.1016/j.yexmp.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenbaum JT, Choi D, Wilson DJ, et al. Parallel Gene Expression Changes in Sarcoidosis Involving the Lacrimal Gland, Orbital Tissue, or Blood. JAMA Ophthalmol. 2015;133(7):770–777. doi: 10.1001/jamaophthalmol.2015.0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraley C, Raftery AE. Model-based clustering, discriminant analysis, and density estimation. J Am Stat Assoc. 2002;97:611–631. [Google Scholar]
- 18.Vartanian K, Slottke R, Johnstone T, et al. Gene expression profiling of whole blood: comparison of target preparation methods for accurate and reproducible microarray analysis. BMC Genomics. 2009;10(1):2. doi: 10.1186/1471-2164-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Team RC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2016. [Google Scholar]
- 20.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 21.Kaufman L, Rousseeuw PJ. Finding groups in data: An introduction to cluster analysis. 2005 [Google Scholar]
- 22.Hartigan JA, Wong MA. A k-means clustering algorithm. Applied Statistics. 1979;28:100–108. [Google Scholar]
- 23.Schwarz GE. Estimating the dimension of a model. Annals of Statistics. 1978;6(2):461–464. [Google Scholar]
- 24.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stan MN, Garrity JA, Carranza Leon BG, Prabin T, Bradley EA, Bahn RS. Randomized controlled trial of rituximab in patients with Graves’ orbitopathy. J Clin Endocrinol Metab. 2015;100(2):432–441. doi: 10.1210/jc.2014-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segerberg-Konttinen M. A postmortem study of focal adenitis in salivary and lacrimal glands. J Autoimmun. 1989;2(4):553–558. doi: 10.1016/0896-8411(89)90188-1. [DOI] [PubMed] [Google Scholar]
- 27.Aakalu VK, Parameswaran S, Maienschein-Cline M, et al. Human Lacrimal Gland Gene Expression. PLoS One. 2017;12(1):e0169346. doi: 10.1371/journal.pone.0169346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ubels JL, Gipson IK, Spurr-Michaud SJ, Tisdale AS, Van Dyken RE, Hatton MP. Gene expression in human accessory lacrimal glands of Wolfring. Invest Ophthalmol Vis Sci. 2012;53(11):6738–6747. doi: 10.1167/iovs.12-10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video 1: A rotating view of the 3-dimensional MDS plot shown in Figure 2.


