Abstract
Background
South Africa has one of the highest rates of HIV-1 (HIV) infection world-wide, with the highest rates among young women. We analyzed the molecular epidemiology and evolutionary history of HIV in young women attending high school in rural South Africa.
Methods
Samples were obtained from the HPTN 068 randomized controlled trial, which evaluated the effect of cash transfers for school attendance on HIV incidence in women aged 13–20 years (Mpumalanga province, 2011–2015). Plasma samples from HIV-infected participants were analyzed using the ViroSeq HIV-1 Genotyping assay. Phylogenetic analysis was performed using 200 pol gene study sequences and 2,294 subtype C reference sequences from South Africa. Transmission clusters were identified using Cluster Picker and HIV-TRACE, and were characterized using demographic and other epidemiological data. Phylodynamic analyses were performed using the BEAST software.
Results
The study enrolled 2,533 young women who were followed through their expected high school graduation date (main study); some participants had a post-study assessment (follow-up study). Two-hundred-twelve of 2,533 enrolled young women had HIV infection. HIV pol sequences were obtained for 94% (n = 201/212) of the HIV-infected participants. All but one of the sequences were HIV-1 subtype C; the non-C subtype sequence was excluded from further analysis. Median pairwise genetic distance between the subtype C sequences was 6.4% (IQR: 5.6–7.2). Overall, 26% of study sequences fell into 21 phylogenetic clusters with 2–6 women per cluster. Thirteen (62%) clusters included women who were HIV-infected at enrollment. Clustering was not associated with study arm, demographic or other epidemiological factors. The estimated date of origin of HIV subtype C in the study population was 1958 (95% highest posterior density [HPD]: 1931–1980), and the median estimated substitution rate among study pol sequences was 1.98x10-3 (95% HPD: 1.15x10-3–2.81x10-3) per site per year.
Conclusions
Phylogenetic analysis suggests that multiple HIV subtype C sublineages circulate among school age girls in South Africa. There were no substantive differences in the molecular epidemiology of HIV between control and intervention arms in the HPTN 068 trial.
Introduction
South Africa has one of the highest rates of human immunodeficiency virus type 1 (HIV) infection in the world [1]. The highest HIV prevalence rates have been reported in the KwaZulu-Natal and Mpumalanga provinces (18% and 15.2%, respectively; ages 15–49; 2016) [2]. Adolescent girls and young women are at increased risk of HIV infection. In 2012, an estimated four million women in South Africa aged 15 and over were living with HIV/AIDS, with HIV prevalence rates of 5.6% among those aged 15–19 and 17.4% among those aged 20–24 [1]. In this region, young women acquire HIV infection earlier and have higher HIV incidence rates compared to young men [3, 4]. Several studies have evaluated HIV infection among high school students in South Africa. Studies of young women attending high-school in rural KwaZulu-Natal found higher HIV prevalence and incidence among those women than their male peers [3, 5].
Phylogenetic analysis of HIV sequences provides insights into viral transmission dynamics independent of self-reported risk behaviors and HIV prevalence data [6]. The number of available HIV sequences in public databases from sub-Saharan Africa is limited considering the size of its epidemic. Available sequence data have been used to elucidate the origin of HIV and its spread from central Africa, to identify transmission clusters, and for surveillance of HIV drug resistance [7, 8]. Phylogenetics has also been used to estimate the efficacy of interventions for HIV prevention in randomized controlled trials [9, 10], including trials evaluating antiretroviral therapy for HIV prevention [10–12].
More intensive sampling of local African epidemics is underway to gain insight into community-level transmission dynamics and to identify strategic targets for HIV prevention interventions [13]. For example, a recent phylogenetic study of young women in Kwa-Zulu Natal, South Africa showed high levels of viral diversity among and few large clusters. This study also showed that sequences from older men and young women tended to cluster, suggesting a possible role for age-disparate partnerships in the African epidemic [14]. Phylogenetic data from Uganda also show high levels of viral diversity in village communities with limited spatial clustering of incident HIV cases and local background sequences, implying geographically-dispersed transmission networks and frequent community-level viral introductions [15].
In this report, we examine molecular epidemiology and evolutionary history of HIV among adolescent girls and young women in Mpumalanga province, South Africa. Individuals 13–20 years of age were enrolled in the HIV Prevention Trials Network (HPTN) 068 study. Specifically, we sought to identify phylogenetic clusters, which are groups of genetically similar viruses, presumably close together along a transmission chain [16]. In this study, phylogenetic clusters are representations of partially sampled, indirectly linked HIV transmission chains because only women were included in the analysis [17]. HPTN 068 was conducted within the Agincourt Health and socio-Demographic Surveillance System (Agincourt HDSS) site in the rural Bushbuckridge sub-District in Mpumalanga province of South Africa [18]. In 2010–2012, HIV prevalence in the study area was over 45% among men and women who were 35–39 years old [19]. Migration for work purposes is common in this area; as many as 60% of adult men and 30% of women migrate from rural to urban areas to find work in any given year [20].
Materials and methods
Study cohort
HPTN 068 was a phase 3, randomized controlled trial (NCT01233531) in rural Mpumalanga province (Bushbuckridge sub-district), South Africa (enrollment period: March 2011 to December 2012) [17]. The study evaluated the effect of cash transfer for school attendance on HIV incidence among young women attending high school (enrollment age: 13–20 years). Participants were excluded if they were married or pregnant, had no parent or legal guardian living in the household, or for other reasons that might have impacted the participant’s health, well-being or study conduct. Participants were randomized 1:1 to one of two study arms: (1) 1,225 received a monthly cash transfer based on school attendance (≥80% of school days per month, intervention arm), and (2) 1,223 did not receive a cash transfer (control arm). Participants were tested for HIV infection at enrollment and annually after enrollment until the end of the trial or their expected high-school graduation date, whichever came first. The study found no significant difference in HIV incidence between study arms [17]. At the end of the main study, all eligible participants who were HIV-uninfected and agreed to participate in the follow-up study were tested for HIV infection at a post-study visit 1–2 years later. The study enrolled 81 HIV-infected and 2,448 HIV-uninfected young women in school grades 8 to 11, and followed them through their expected high school graduation date; some participants had a post-study follow-up visit 1–2 years later. Annual HIV incidence in the main study was 1.8% [17].
HIV testing
HIV testing was performed at the study site at all visits through graduation. All samples were retrospectively tested at the HPTN Laboratory Center (Baltimore, MD, USA) to confirm HIV status. Methods used for HIV testing in the main study are described in a previous report [17]. The same methods were used in the follow-up study, with one exception: the Geenius HIV ½ Supplemental Assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used for retrospective confirmation of HIV seroconversion, rather than Western blot testing.
HIV genotyping
Plasma samples from HIV-infected participants with viral loads >400 copies/mL were analyzed using the ViroSeq HIV-1 Genotyping assay, version 2.8 (Abbott Molecular, Des Plaines, IL, USA). This system generates sequences encoding HIV protease and amino acids 1–335 of HIV reverse transcriptase (1,302 base pairs, pol gene, corresponding to nucleotides 2252–3554 in the HXB2 K03455 reference strain). HIV drug resistance was assessed using software provided with the ViroSeq system. HIV drug resistance mutations were considered as major according to the Stanford HIV drug resistance database [21].
Other laboratory testing
CD4 cell count testing was performed at the study sites for participants with HIV infection. HIV viral load testing was performed retrospectively at the HPTN Laboratory Center using the RealTime HIV-1 Viral Load assay (Abbott Molecular, Des Plaines, IL). Pregnancy history was collected as described [22]. Herpes simplex virus type 2 (HSV-2) testing was performed as described [17].
Phylogenetic analysis
HIV subtyping was performed using the REGA HIV Subtyping tool v3.0 [23]. Subtyping results were confirmed with COMET HIV-1 and by approximately maximum-likelihood phylogenetic methods using FastTree v2.1.9 [24] with HIV subtype reference sequences from the Los Alamos National Laboratory’s (LANL) HIV Sequence Database [25]. Phylogenetic analysis was performed using HIV pol sequences from HPTN 068 participants and HIV pol reference sequences. HIV reference sequences from South Africa were obtained from the LANL HIV Sequence Database [25]. Reference sequences were selected using the following search criteria: subtype C; genomic region: positions 2252 to 3554; geographic region: South Africa (country code ZA). One reference sequence per individual in LANL was included in the analysis. Sequences were aligned using MAFFT software [26]; minimal manual editing was performed. Recombination analyses were conducted for reference and study sequences using RDP [27], Maxchi [28], Chimaera [29], Bootscan [30], and Siscan [31] integrated into Recombination Detection Program v4 (RDP4) [32], and suspected inter-subtype recombinant viruses were excluded from subsequent analysis. RDP4 software was run using default settings, with the following exception: the window size was set to 60 for RDP, 120 for Maxchi and Chimera, and 500 for Bootscan and Siscan tests. Approximately maximum-likelihood phylogenetic trees were reconstructed using FastTree v2.1.9 [24] with the GTR+CAT model of nucleotide substitution and Shimodaira-Hasegawa (SH) test for clade support. We also reconstructed phylogenetic trees using RAxML v8.2.10 [33] with the GTR+CAT model of nucleotide substitutions and 1,000 bootstrap iterations for confirmation of clusters. All trees were visualized with FigTree v1.4.2 [34]. Molecular genetic networks were also constructed using the HIV-TRACE software as previously described [35]. Transmission clusters were identified from phylogenetic trees using Cluster Picker v1.2.3 [36] and from phylogenetic networks using HIV-TRACE [37]. Clusters were defined using a 4.5% maximum genetic distance threshold between all sequences in the cluster and as well as a 90% minimum clade support threshold with the Cluster Picker software. Sensitivity analyses were conducted at 2.5% and 1% genetic distance thresholds. For HIV-TRACE, a genetic distance threshold of 2.5% based on genetic pairwise distances was used. These threshold values have been previously applied in other studies of HIV transmission clusters [14, 16, 36]. Phylogenetic trees were displayed using Interactive Tree of Life (iTOL) v4.1 [38].
Phylodynamics of HIV subtype C
To explore the origination date and genetic diversity of 200 HIV subtype C pol gene study sequences, we conducted a Bayesian Markov Chain Monte Carlo (MCMC) phylogenetic analysis using the BEAST v1.8.4 software [39, 40]. Analyses were performed under the HKY+Γ4 and GTR+Г4 substitution models and log-normal uncorrelated relaxed molecular clock model. A Gaussian Markov random field (GMRF) Bayesian Skyride coalescent model was used to reconstruct viral population dynamics. Two independent runs were performed for 5x107 steps with sampling every 1,000 generations. Convergence was assessed using Tracer v1.6 [41]. The cut-off for effective sample size (ESS) >200 was used for all parameters.
Statistical analysis
Logistic regression analysis was used to estimate associations between demographic and epidemiological characteristics. Variable assessed included age and school grade at enrollment, study arm, timing of HIV infection (enrollment, main study, and follow-up study), viral load and CD4 cell count at the first HIV-positive visit, the presence of the major drug resistance mutations, HSV-2 infection, pregnancy history, and the probability of study sequence clustering. We also assessed excess co-clustering by study arm and other categorical epidemiological characteristics by analyzing the probability of two sequences sharing a genetic cluster (<4.5 genetic distance threshold) also shared the same characteristic relative to the probability that a random pair of sequences from two girls which were not clustered also shared that same characteristic [15]. In the case of no excess co-clustering, we expect this relative probability to be one. Analyses were performed using R v3.3.2 [42] “base” and “ape” packages.
Nucleotide sequence accession numbers
Study sequences were submitted to GenBank under accession numbers KY883695-KY883762, KY888784-KY888875, KY921717-KY921757.
Ethics statement
Written consent for participation in the HPTN 068 study was provided by all study participants and their parents/guardians. The study was approved by ethical review board at the University of the Witwatersrand and the University of North Carolina.
Results
Summary of study sequences
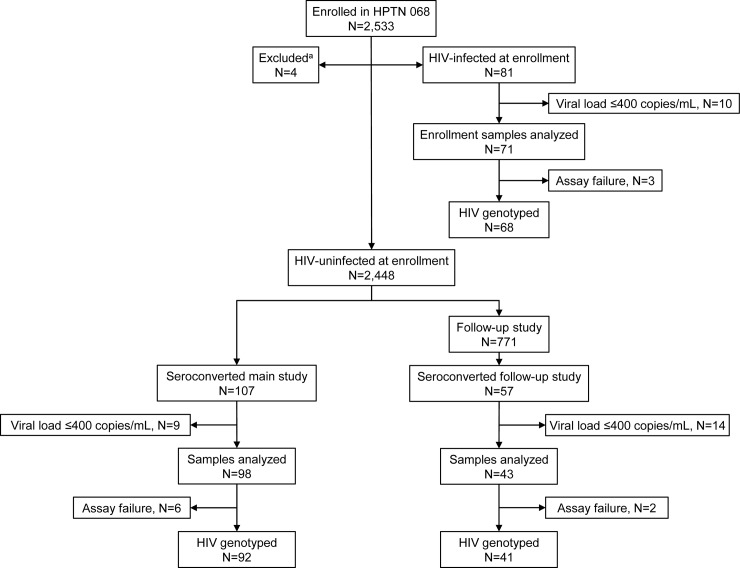
In HPTN 068, 245 HIV infections were documented: 81 participants were HIV infected at enrollment; 107 acquired HIV infection in the main study (between the enrollment visit and final main study visit), and 57 acquired HIV infection in the follow-up study (between the final main study visit and the post-study visit) (Fig 1). Plasma samples with HIV viral loads >400 copies/mL were available from 212 (86.5%) of the 245 HIV-infected participants (one sample per participant, collected at enrollment or the first HIV-positive visit; 33 samples had viral loads ≤400 copies/mL). HIV genotyping was successful for 201 (94.8%) of the 212 samples (68 who had HIV infection at enrollment; 92 who acquired HIV infection during the main study, 41 who acquired HIV infection during the follow-up study). Two hundred (99.5%) of the 201 sequences were HIV subtype C. One sequence was HIV subtype A; this sequence was excluded from further phylogenetic analysis. Drug resistance mutations identified using the ViroSeq HIV-1 Genotyping system were detected in 20 (10%) of the 200 sequences. At least one major non-nucleoside reverse transcriptase inhibitor (NNRTI)-resistance mutation (K103N, V106M, Y181C, G190A/S) was detected in 19 sequences; five of those sequences also had the M184V nucleoside reverse transcriptase inhibitor (NRTI)-resistance mutation. One sample had the L210W NRTI resistance mutation.
Fig 1. HPTN 068 study cohort flowchart.
The figure provides an overview of the study cohort, including the number of samples tested and the number of HIV genotyping results obtained in each participant group: infected at enrollment, infected during the main study (between enrollment and their expected graduation date), or infected during the follow-up study (after their expected graduation date). Abbreviations: mL: milliliter. Footnote for Fig 1: aFour participants were excluded due to unknown HIV status.
Identification of phylogenetic clusters
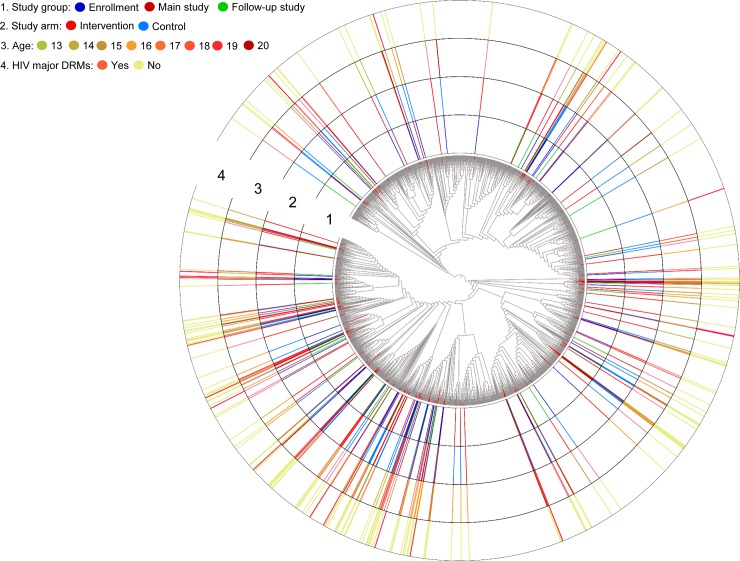
Phylogenetic analysis was performed using 200 pol sequences from the study participants and 2,294 HIV subtype C reference sequences from South Africa; four reference sequences were excluded from the analysis (three were identified as inter-subtype recombinant; one had a high level of sequence ambiguity, >5%) (Fig 2). Overall, median pairwise genetic distance between all pol gene sequences including references was 6.4% (interquartile range [IQR]: 5.7–7.2%). Using Cluster Picker software with genetic distance and clade support thresholds of 4.5% and 90% respectively, 52 sequences (26% of total) from study participants clustered with one or more sequences from other study participants (Table 1). No large clusters (over 10 individuals per cluster) were identified. Most of the clusters were pairs (n = 17); clusters of four participants (N = 3) and six participants (N = 1) were also identified. Thirteen (62%) of these 21 clusters included women who were HIV infected at enrollment (Fig 3). Median genetic pairwise distance among sequences sharing a cluster was 1.1% (IQR: 0.7–2.6%) (S1 Fig). There were no reference sequences in these clusters; however, seven women (3.5%) clustered with one or more reference sequences in other clusters. Thirty-five (67.3%) of the 52 clustered sequences were in the intervention arm and of these sequences, 18 (51.4%) clustered with control arm sequences. As expected, the number of clusters decreased when more stringent genetic distance thresholds were applied (Table 1).
Fig 2. Approximately maximum-likelihood phylogenetic tree of 2,494 HIV subtype C pol sequences.
Study sequences are indicated in the phylogeny with red branches. Abbreviation: DRM: drug resistance mutation.
Table 1. Phylogenetic clusters containing two or more study sequences detected using different tree reconstruction and cluster detection algorithms.
| Phylogenetic tree reconstruction method | Cluster detection software | Clade support threshold | Maximum genetic distance threshold | Total number of clusters detected | Cluster size distribution (N)a |
|---|---|---|---|---|---|
| FastTree | Cluster Picker | 90% | 4.5% | 21 | 2 (17), 4 (3), 6 (1) |
| RAxML | Cluster Picker | 90% | 4.5% | 21 | 2 (18), 4 (2), 6 (1) |
| FastTree | Cluster Picker | 90% | 2.5% | 16 | 2 (15), 6 (1) |
| RAxML | Cluster Picker | 90% | 2.5% | 16 | 2 (15), 6 (1) |
| FastTree | Cluster Picker | 90% | 1.0% | 6 | 2 (6) |
| RAxML | Cluster Picker | 90% | 1.0% | 6 | 2 (6) |
| - | HIV-TRACE | - | 2.5% | 16 | 2 (15), 6 (1) |
aCluster size distribution (number) is shown for clusters containing two or more study sequences.
Abbreviation: N: number.
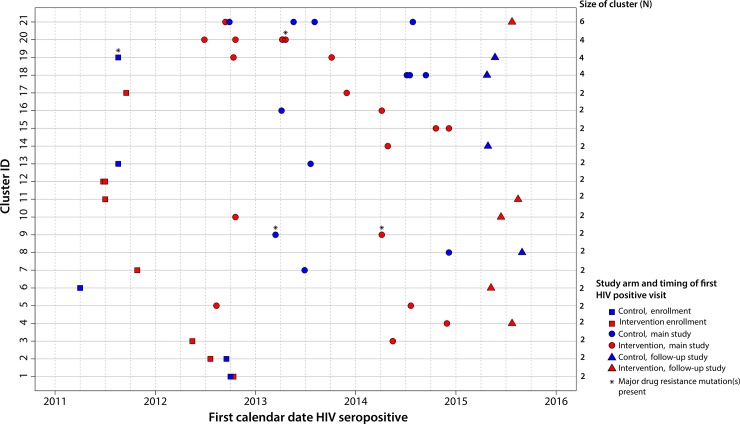
Fig 3. Transmission clusters from approximately maximum-likelihood phylogenetic tree.
Phylogenetic clusters were detected in the approximately maximum-likelihood phylogenetic tree (FastTree) at a 4.5% genetic distance threshold. Each row represents one of the twenty-one phylogenetic clusters. Symbols representing participants in clusters are colored in red (intervention) or blue (control). Symbols represent the timing of participants’ first positive HIV test (enrollment, main study, follow-up study). Data from participants are shown on the x-axis (calendar time) according to the date of their first HIV-positive visit in the study. Drug resistant viruses are denoted with an asterisk.
The largest cluster of six included participants who seroconverted during the main (n = 5) and follow-up (n = 1) studies. The mean genetic distance in this cluster was 0.8% with an estimated time to the most recent common ancestor (tMRCA) 2006 (95% highest posterior density [HPD]: 1988–2010). Four of the participants were in the control arm and two were in the intervention arm (Fig 3). Drug resistance mutations were not detected in any of the participants within this cluster. One cluster of four participants contained women from the control arm only. Three participants within this cluster were HIV-infected during the main study; one woman was infected during the follow-up study (Fig 3). A second cluster of four women included participants from intervention arm only (Fig 3); all four were infected during the main study and were in the same school grade (10th) at study enrollment. A single drug resistance mutation was detected in one woman. The third cluster of four women included participants from both study arms who were HIV-infected at different stages in the study (enrollment, main study, follow-up study) of the trial (Fig 3). One drug resistance mutation was detected in one woman in this cluster. Study sequences were otherwise intermingled with reference sequences from South Africa throughout the tree.
In sensitivity analyses, the vast majority of phylogenetic clusters identified with FastTree were also identified with RAxML at a genetic distance threshold of 4.5%. The same clusters of viral sequences were identified at genetic distance thresholds of 2.5% and 1% (Table 1). Identical clusters were also found using Cluster Picker and HIV-TRACE at a genetic distance threshold of 2.5% (Table 1).
Associations of demographic and clinical characteristics with HIV clustering are shown in Table 2. There was also no statistically significant excess co-clustering by study arm, drug resistance mutations detected, or any other study characteristic.
Table 2. Associations between demographic and epidemiological characteristics of 200 HIV-infected study participants and phylogenetic clustering (4.5% genetic distance threshold).
| Characteristic | Total number; N = 200 N (%) |
Clustered; N = 59 N (%) |
Odds ratio (95% CI) |
P-value |
|---|---|---|---|---|
| Age (years) | ||||
| 15 and younger | 69 (34.5%) | 22 (37.3%) | 1.0 (ref) | … |
| 16 | 46 (23%) | 13 (22%) | 0.84 (0.36–1.89) | 0.68 |
| 17 | 44 (22%) | 11 (18.7%) | 0.71 (0.3–1.64) | 0.43 |
| 18 and older | 41 (20.5%) | 13 (22%) | 0.99 (0.43–2.26) | 0.99 |
| School grade | ||||
| 8 | 39 (19.5%) | 9 (15.3%) | 1.0 (ref) | … |
| 9 | 41 (20.5%) | 15 (25.4%) | 1.92 (0.43–5.28) | 0.19 |
| 10 | 71 (35.5%) | 19 (32.2%) | 1.22 (0.5–3.14) | 0.67 |
| 11 | 49 (24.5%) | 16 (27.1%) | 1.62 (0.36–4.33) | 0.32 |
| Study arm | ||||
| Control | 95 (47.5%) | 24 (40.7%) | 1.0 (ref) | … |
| Intervention | 105 (52.5%) | 35 (59.3%) | 1.48 (0.8–2.76) | 0.21 |
| First HIV positive test | ||||
| Enrollment | 68 (34%) | 16 (27.1%) | 1.0 (ref) | … |
| Main study | 92 (46%) | 32 (54.2%) | 1.73 (0.87–3.27) | 0.65 |
| Follow-up study | 40 (20%) | 11 (18.6%) | 1.23 (0.5–3) | 0.13 |
| Viral load, copies/mL | ||||
| 400–9,999 | 61 (30.5%) | 22 (37.3%) | 1.0 (ref) | … |
| 10,000–99,000 | 112 (56%) | 27 (45.8%) | 0.56 (0.29–1.11) | 0.1 |
| >100,000 | 27 (12.5%) | 10 (16.9%) | 1.04 (0.39–2.65) | 0.93 |
| CD4 cell count, cells/mm3,a | ||||
| >500 | 92 (46%) | 30 (50.9%) | 1.0 (ref) | … |
| < = 500 | 75 (37.5%) | 19 (32.2%) | 0.70 (0.35–1.37) | 0.31 |
| < = 250 | 15 (7.5%) | 4 (6.8%) | 0.75 (0.2–2.4) | 0.65 |
| Major drug resistance mutation | ||||
| No | 180 (90%) | 55 (93.2) | 1.0 (ref) | … |
| Yes | 20 (10%) | 4 (6.8%) | 0.57 (0.16–1.63) | 0.33 |
| HSV-2 infectionb | ||||
| No | 90 (45%) | 25 (42.4%) | 1.0 (ref) | … |
| Yes | 69 (34.5%) | 23 (39%) | 1.3 (0.65–2.57) | 0.45 |
| Ever pregnantc | ||||
| No | 122 (61%) | 37 (62.7%) | 1.0 (ref) | … |
| Yes | 49 (24.5%) | 15 (25.4%) | 1.01 (0.48–2.06) | 0.971 |
aCD4 cell count data was not available for 18 women
bHSV-2 infection status was not available for 41 women
cPregnancy history was not available for 29 women.
Abbreviations: N: number; CI: confidence intervals; ref: reference.
Phylodynamics of HIV subtype C in the trial population
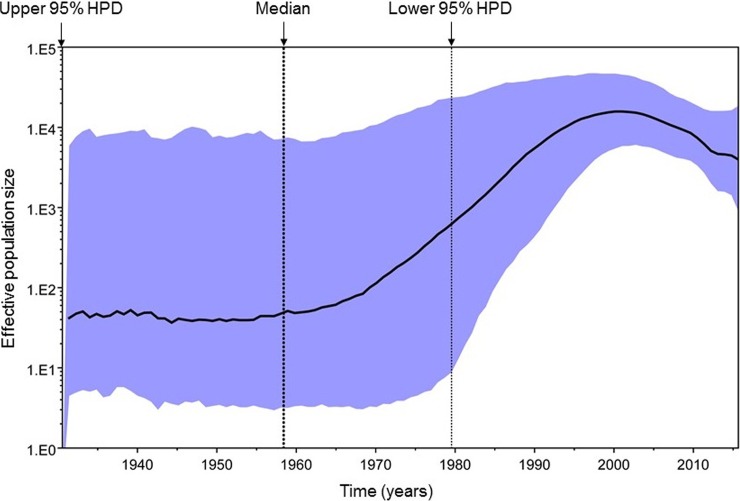
Phylodynamic reconstruction of the epidemic with Bayesian MCMC analysis revealed that the date of origin of HIV subtype C circulating in the study population was 1958 (95% HPD: 1931–1980) (Fig 4). There appeared to be a rapid growth in the effective size of the viral population between the 1970s and 1990s with growth peaking in the early 2000s and declining somewhat thereafter; however, changes in effective population size over time were not statistically significant. The median estimated substitution rate (i.e., the rate at genetic differences were accrued over time) among study pol sequences was 1.98x10-3 per site per year (95% HPD: 1.15x10-3–2.81x10-3). The estimated dates of origin and substitution rates using HKY+G4 and GTR+G4 substitution models were highly consistent. There were no substantive differences in population size trajectories between intervention and control arms of the trial population (data not shown).
Fig 4. GMRF Bayesian Skyride plot of HIV subtype C.
The GMRF Bayesian Skyride plot was reconstructed from the 200 pol gene sequences. Bold black line indicates the median effective population size through time; blue shaded area represents the 95% highest posterior density (HPD) interval. The vertical dotted lines represent the estimated date of origin (1958) of HIV subtype C, and lower and upper 95% HPD intervals (1931–1980).
Discussion
In this study, we analyzed indirect female-to-female HIV transmission chains among young women with HIV subtype C infection who were attending high school in rural South Africa. All but one of the 201 HPTN 068 participants with genotyping results had HIV subtype C infection; one participant had HIV subtype A infection. Phylogenetic analysis revealed small distinct HIV transmission clusters among study sequences scattered across the subset of subtype C reference sequences from South Africa. Clustering was not statistically significantly associated with demographic and select epidemiological characteristics of study participants. Results in this report and results of another study from the same region (Bushbuckridge sub-district, Mpumalanga province) [43] indicate that there are multiple HIV subtype C sublineages circulating in the population of this area.
Phylogenetics is widely used to describe evolution and origins of viruses [44], to provide information about circulating genetic variants of pathogens [44], and to identify transmission clusters [6, 8]. Each cluster is most often a representation of a partially sampled transmission chain identified based on similarity of the viral sequences [6, 45]. In the HPTN 068 trial, we found that a low proportion (26%) of participants clustered in small groups no larger than six women. Low levels of clustering between HIV infections and small cluster sizes are commonly observed in heterosexual African HIV epidemics, probably because the sampling fraction of transmission networks is low [15, 45, 46]. In this report, we sampled 2,533 individuals, which was less than 1% of the total population residing in the study area (Bushbuckridge sub-district, South Africa).
The lack of structured HIV subtype C phylogeny [47] and high genetic diversity of Southern African HIV strains has been demonstrated in previous studies [45, 48]. Phylogenetic reconstruction of subtype C infections in South Africa also suggest that there have been multiple introductions of the virus to the region [49, 50] and extensive intrasubtype recombination [51]. In our study, the median pairwise genetic distance among analyzed HIV pol sequences was 6.4%. Similar levels of genetic diversity were found in previous studies of HIV subtype C infection in the general population in African countries [15, 45, 48, 52].
The estimated year of HIV origin into the region was 1958, based on the subset of 200 pol HIV subtype C sequences. The HIV subtype C effective population size significantly increased between the 1970s and 1990s, with the peak of infections in the early 2000s. A decline of the effective population size was observed thereafter. These results are compatible with previous studies of origin and evolution of HIV subtype C viruses in South Africa [49, 50] and other countries [52, 53]. The rate of molecular evolution among HPTN 068 HIV subtype C pol sequences was estimated as 1.98x10-3 per site per year. Similar mutation rates in the pol gene were reported previously for HIV subtypes C [50, 53] and B [54]. High migration rates likely play a substantial role in the dynamics of the epidemic [50] and probably contribute to the high level of HIV diversity in South Africa. This report provides insight into the HIV epidemic among young women in rural South Africa who were enrolled in a randomized controlled trial. We observed high HIV prevalence and multiple HIV subtype C sublineages circulating among young women in the study. There was no evidence of distinct sub-epidemics among young women by study arm, age, school grade, or timing of infection (i.e., if they were infected at enrollment, in the main study, or in the follow-up study). There was also limited evidence for super-spreading events or large, highly connected networks similar to those observed in outbreaks of HIV among persons who inject drugs in the United States [55] or HIV transmission networks among men who have sex with men [56].
Supporting information
Boxplots represent the median and interquartile ranges of pol pairwise genetic distances obtained for participants who did or did not share a phylogenetic cluster in the approximately maximum-likelihood phylogenetic tree at a genetic distance threshold of 4.5%. The violin plots represent the distribution and density of pairwise genetic distances in each group.
(TIF)
Acknowledgments
The authors thank the HPTN 068 study team and study participants for their contributions to the HPTN 068 study. The authors also thank the laboratory investigators and study staff for their assistance with sample management and laboratory testing.
Data Availability
Study sequences are available from GenBank under accession numbers KY883695-KY883762, KY888784-KY888875, KY921717-KY921757.
Funding Statement
This work was supported by Award Numbers K01AI125086, UM1AI068613, (HPTN Laboratory Center), UM1AI068619 (HPTN Leadership and Operations Center), and UM1AI068617 (HPTN Statistical and Data Management Center), from the National Institute of Allergy and Infectious Diseases, the National Institute of Mental Health and the National Institute on Drug Abuse of the National Institutes of Health. This work was also supported by NIMH R01 (R01MH087118) and the Carolina Population Center and its NIH Center grant (P2C HD050924). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funder did not have any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the "author contributions" section.
References
- 1.UNAIDS. 2015. Accessed: January 12, 2017. Available from: http://www.unaids.org/en/regionscountries/countries/southafrica.
- 2.SANAC. The South African National AIDS Council. HIV Statistics. 2016. Accessed: January 14, 2017. Available from: http://ivizard.org/sanac/.
- 3.Kharsany AB, Mlotshwa M, Frohlich JA, Yende Zuma N, Samsunder N, Abdool Karim SS, et al. HIV prevalence among high school learners—opportunities for schools-based HIV testing programmes and sexual reproductive health services. BMC Public Health. 2012;12:231 doi: 10.1186/1471-2458-12-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramjee G, Daniels B. Women and HIV in sub-Saharan Africa. AIDS Res Ther. 2013;10:30 doi: 10.1186/1742-6405-10-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kharsany AB, Buthelezi TJ, Frohlich JA, Yende-Zuma N, Samsunder N, Mahlase G, et al. HIV infection in high school students in rural South Africa: role of transmissions among students. AIDS Res Hum Retroviruses. 2014;30:956–65. doi: 10.1089/AID.2014.0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grabowski MK, Redd AD. Molecular tools for studying HIV transmission in sexual networks. Curr Opin HIV AIDS. 2014;9:126–33. doi: 10.1097/COH.0000000000000040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ratmann O, Hodcroft EB, Pickles M, Cori A, Hall M, Lycett S, et al. Phylogenetic tools for generalized HIV-1 epidemics: findings from the PANGEA-HIV methods comparison. Mol Biol Evol. 2017;34:185–203. doi: 10.1093/molbev/msw217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dennis AM, Herbeck JT, Brown AL, Kellam P, de Oliveira T, Pillay D, et al. Phylogenetic studies of transmission dynamics in generalized HIV epidemics: an essential tool where the burden is greatest? J Acquir Immune Defic Syndr. 2014;67:181–95. doi: 10.1097/QAI.0000000000000271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell MS, Mullins JI, Hughes JP, Celum C, Wong KG, Raugi DN, et al. Viral linkage in HIV-1 seroconverters and their partners in an HIV-1 prevention clinical trial. PLoS One. 2011;6:e16986 doi: 10.1371/journal.pone.0016986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eshleman SH, Hudelson SE, Redd AD, Wang L, Debes R, Chen YQ, et al. Analysis of genetic linkage of HIV from couples enrolled in the HIV Prevention Trials Network 052 trial. J Infect Dis. 2011;204:1918–26. doi: 10.1093/infdis/jir651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, et al. Antiretroviral Therapy for the Prevention of HIV-1 Transmission. N Engl J Med. 2016;375:830–9. doi: 10.1056/NEJMoa1600693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodger AJ, Cambiano V, Bruun T, Vernazza P, Collins S, van Lunzen J, et al. Sexual Activity Without Condoms and Risk of HIV Transmission in Serodifferent Couples When the HIV-Positive Partner Is Using Suppressive Antiretroviral Therapy. Jama. 2016;316:171–81. doi: 10.1001/jama.2016.5148 [DOI] [PubMed] [Google Scholar]
- 13.Pillay D, Herbeck J, Cohen MS, de Oliveira T, Fraser C, Ratmann O, et al. PANGEA-HIV: phylogenetics for generalised epidemics in Africa. Lancet Infect Dis. 2015;15:259–61. doi: 10.1016/S1473-3099(15)70036-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Oliveira T, Kharsany AB, Graf T, Cawood C, Khanyile D, Grobler A, et al. Transmission networks and risk of HIV infection in KwaZulu-Natal, South Africa: a community-wide phylogenetic study. Lancet HIV. 2017;4:e41–e50. doi: 10.1016/S2352-3018(16)30186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grabowski MK, Lessler J, Redd AD, Kagaayi J, Laeyendecker O, Ndyanabo A, et al. The role of viral introductions in sustaining community-based HIV epidemics in rural Uganda: evidence from spatial clustering, phylogenetics, and egocentric transmission models. PLoS Med. 2014;11:e1001610 doi: 10.1371/journal.pmed.1001610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Vu S, Ratmann O, Delpech V, Brown AE, Gill ON, Tostevin A, et al. Comparison of cluster-based and source-attribution methods for estimating transmission risk using large HIV sequence databases. Epidemics. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pettifor A, MacPhail C, Hughes JP, Selin A, Wang J, Gomez-Olive FX, et al. The effect of a conditional cash transfer on HIV incidence in young women in rural South Africa (HPTN 068): a phase 3, randomised controlled trial. Lancet Glob Health. 2016;4:e978–e88. doi: 10.1016/S2214-109X(16)30253-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kahn K, Collinson MA, Gomez-Olive FX, Mokoena O, Twine R, Mee P, et al. Profile: Agincourt health and socio-demographic surveillance system. Int J Epidemiol. 2012;41:988–1001. doi: 10.1093/ije/dys115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez-Olive FX, Angotti N, Houle B, Klipstein-Grobusch K, Kabudula C, Menken J, et al. Prevalence of HIV among those 15 and older in rural South Africa. AIDS Care. 2013;25:1122–8. doi: 10.1080/09540121.2012.750710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collinson MA, White MJ, Bocquier P, McGarvey ST, Afolabi SA, Clark SJ, et al. Migration and the epidemiological transition: insights from the Agincourt sub-district of northeast South Africa. Glob Health Action. 2014;7:23514 doi: 10.3402/gha.v7.23514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang MW, Liu TF, Shafer RW. The HIVdb system for HIV-1 genotypic resistance interpretation. Intervirology. 2012;55:98–101. doi: 10.1159/000331998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pettifor A, MacPhail C, Selin A, Gomez-Olive FX, Rosenberg M, Wagner RG, et al. HPTN 068: A randomized control trial of a conditional cash transfer to reduce HIV infection in young women in South Africa-study design and baseline results. AIDS Behav. 2016;20:1863–82. doi: 10.1007/s10461-015-1270-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Oliveira T, Deforche K, Cassol S, Rambaut A, Vandamme A-M. REGA HIV-1 subtyping tool 2014. Accessed: December 7, 2016. Available from: http://dbpartners.stanford.edu:8080/RegaSubtyping/stanford-hiv/typingtool/.
- 24.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490 doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.HIV Sequence Database: Los Alamos National Laboratory; 2016. Accessed: December 7, 2016. Available from: https://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html.
- 26.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–8. doi: 10.1093/nar/gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin D, Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–3. [DOI] [PubMed] [Google Scholar]
- 28.Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–9. [DOI] [PubMed] [Google Scholar]
- 29.Posada D, Crandall KA. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci U S A. 2001;98:13757–62. doi: 10.1073/pnas.241370698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin DP, Posada D, Crandall KA, Williamson C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses. 2005;21:98–102. doi: 10.1089/aid.2005.21.98 [DOI] [PubMed] [Google Scholar]
- 31.Gibbs MJ, Armstrong JS, Gibbs AJ. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics. 2000;16:573–82. [DOI] [PubMed] [Google Scholar]
- 32.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003 doi: 10.1093/ve/vev003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rambaut A. FigTree v1.4.3: Molecular evolution, phylogenetics and epidemiology; 2007. updated October 4, 2016. Accessed: December 7, 2016. Available from: http://tree.bio.ed.ac.uk/software/figtree/.
- 35.Wertheim JO, Kosakovsky Pond SL, Forgione LA, Mehta SR, Murrell B, Shah S, et al. Social and Genetic Networks of HIV-1 Transmission in New York City. PLoS Pathog. 2017;13:e1006000 doi: 10.1371/journal.ppat.1006000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ragonnet-Cronin M, Hodcroft E, Hue S, Fearnhill E, Delpech V, Brown AJ, et al. Automated analysis of phylogenetic clusters. BMC Bioinformatics. 2013;14:317 doi: 10.1186/1471-2105-14-317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wertheim JO, Leigh Brown AJ, Hepler NL, Mehta SR, Richman DD, Smith DM, et al. The global transmission network of HIV-1. J Infect Dis. 2014;209:304–13. doi: 10.1093/infdis/jit524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–8. doi: 10.1093/bioinformatics/btl529 [DOI] [PubMed] [Google Scholar]
- 39.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. doi: 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214 doi: 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rambaut A, Suchard MA, Xie D, Drummond AJ. Tracer v1.6 2014. Available from: http://tree.bio.ed.ac.uk/software/tracer/.
- 42.R Core Team. R: A language and environment for statistical computing: R Foundation for Statistical Computing, Vienna, Austria; 2016. Available from: https://www.R-project.org/. [Google Scholar]
- 43.Msimanga PW, Vardas E, Engelbrecht S. HIV-1 diversity in an antiretroviral treatment naive cohort from Bushbuckridge, Mpumalanga Province, South Africa. Virol J. 2015;12:24 doi: 10.1186/s12985-015-0244-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lam TT, Hon CC, Tang JW. Use of phylogenetics in the molecular epidemiology and evolutionary studies of viral infections. Crit Rev Clin Lab Sci. 2010;47:5–49. doi: 10.3109/10408361003633318 [DOI] [PubMed] [Google Scholar]
- 45.Novitsky V, Bussmann H, Logan A, Moyo S, van Widenfelt E, Okui L, et al. Phylogenetic relatedness of circulating HIV-1C variants in Mochudi, Botswana. PLoS One. 2013;8:e80589 doi: 10.1371/journal.pone.0080589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Oliveira T, Kharsany AB, Graf T, Cawood C, Khanyile D, Grobler A, et al. Transmission networks and risk of HIV infection in KwaZulu-Natal, South Africa: a community-wide phylogenetic study. Lancet HIV. 2017;4:e41–e50. doi: 10.1016/S2352-3018(16)30186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomson MM, Fernandez-Garcia A. Phylogenetic structure in African HIV-1 subtype C revealed by selective sequential pruning. Virology. 2011;415:30–8. doi: 10.1016/j.virol.2011.03.021 [DOI] [PubMed] [Google Scholar]
- 48.Gordon M, De Oliveira T, Bishop K, Coovadia HM, Madurai L, Engelbrecht S, et al. Molecular characteristics of human immunodeficiency virus type 1 subtype C viruses from KwaZulu-Natal, South Africa: implications for vaccine and antiretroviral control strategies. J Virol. 2003;77:2587–99. doi: 10.1128/JVI.77.4.2587-2599.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilkinson E, Rasmussen D, Ratmann O, Stadler T, Engelbrecht S, de Oliveira T. Origin, imports and exports of HIV-1 subtype C in South Africa: A historical perspective. Infection, Genetics and Evolution. 2016;46:200–8. doi: 10.1016/j.meegid.2016.07.008 [DOI] [PubMed] [Google Scholar]
- 50.Wilkinson E, Engelbrecht S, de Oliveira T. History and origin of the HIV-1 subtype C epidemic in South Africa and the greater southern African region. Sci Rep. 2015;5:16897 doi: 10.1038/srep16897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rousseau CM, Learn GH, Bhattacharya T, Nickle DC, Heckerman D, Chetty S, et al. Extensive intrasubtype recombination in South African human immunodeficiency virus type 1 subtype C infections. J Virol. 2007;81:4492–500. doi: 10.1128/JVI.02050-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Novitsky V, Wang R, Lagakos S, Essex M. HIV-1 subtype C phylodynamics in the global epidemic. Viruses. 2010;2:33–54. doi: 10.3390/v2010033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delatorre EO, Bello G. Phylodynamics of HIV-1 subtype C epidemic in East Africa. PLoS One. 2012;7:e41904 doi: 10.1371/journal.pone.0041904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bello G, Aulicino PC, Ruchansky D, Guimaraes ML, Lopez-Galindez C, Casado C, et al. Phylodynamics of HIV-1 circulating recombinant forms 12_BF and 38_BF in Argentina and Uruguay. Retrovirology. 2010;7:22 doi: 10.1186/1742-4690-7-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peters PJ, Pontones P, Hoover KW, Patel MR, Galang RR, Shields J, et al. HIV Infection Linked to Injection Use of Oxymorphone in Indiana, 2014–2015. N Engl J Med. 2016;375:229–39. doi: 10.1056/NEJMoa1515195 [DOI] [PubMed] [Google Scholar]
- 56.Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008;5:e50 doi: 10.1371/journal.pmed.0050050 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Boxplots represent the median and interquartile ranges of pol pairwise genetic distances obtained for participants who did or did not share a phylogenetic cluster in the approximately maximum-likelihood phylogenetic tree at a genetic distance threshold of 4.5%. The violin plots represent the distribution and density of pairwise genetic distances in each group.
(TIF)
Data Availability Statement
Study sequences are available from GenBank under accession numbers KY883695-KY883762, KY888784-KY888875, KY921717-KY921757.