Abstract
Rbpms (RNA-binding protein with multiple splicing) is a member of the RRM (RNA Recognition Motif) family of RNA-binding proteins, which is expressed as multiple alternatively spliced transcripts encoding different protein isoforms. We have shown earlier that Rbpms expression in the retina is restricted to retinal ganglion cells (RGCs), and have characterized this gene as a marker for RGCs. The aim of this study was to identify isoforms representing Rbpms in human retinas and to analyze its transcriptional regulation. We found that Rbpms is expressed as transcription variants 1 and 3 encoding isoforms A and C, respectively. These isoforms are encoded by the same first 6 exons but have different C-terminal ends encoded by exon 8 in variant 1 and exon 7 in variant 3. Computational analysis of the Rbpms 5′ untranslated and flanking regions reveals the presence of three CpG islands and four predicted promoter regions (PPRs). The effect of PPR 1 (− 1672/− 1420) and PPR2 (− 330/− 79) on transcriptional activation was minimal, whereas PPR 3 (− 73/+ 177) and PPR4 (+ 274/+ 524) induced the expression by ~ 7 and ninefold compared to control, respectively. The maximum activity, a 30-fold increase above the control level, was obtained from the construct containing both PPRs 3 and 4. Site-directed mutagenesis of several cis-elements within PPR3 and PPR4 including five for Sp1, one for AP1, and two for NF-kB showed that mutation of the first three and especially the first GC box resulted in a threefold downregulation of gene expression. AP1, NF-kB, and two downstream Sp1 sites had no significant effect on expression level. The possible involvement of the GC box 1 at position − 54 in transcriptional regulation of Rbpms was corroborated by EMSA, which showed formation of a DNA–protein complex in the presence of the oligonucleotide corresponding to this Sp1-binding site.
Keywords: Retina, Retinal ganglion cells, Transcription factors, CpG islands, Promoter
Introduction
Rbpms (RNA-binding protein with multiple splicing) is a member of the RRM (RNA Recognition Motif) family of RNA-binding proteins. The RRM, also known as RNA-binding domain (RBD) or ribonucleoprotein domain (RNP), is one of the most abundant protein domains in eukaryotes (Maris et al. 2005). In general, the RRM family members are involved in gene expression regulation, including pre-mRNA-processing (splicing, capping, and polyadenylation), RNA stability, transport, localization, and translational regulation. Two RNP consensus sequences, RNP1 and RNP2, that are involved in direct interaction with RNA have been identified in the RRM domain (Dreyfuss et al. 1988). A putative RRM domain of Rbpms is homologous to that of the Drosophila couch potato (cpo) and C. elegans mec-8 genes (Gerber et al. 1999). Mutations in cpo have been associated with several neurological phenotypes, including bang-sensitive paralysis, seizure susceptibility, and synaptic transmission defects, whereas mutations in Mec-8 affect mechanosensory and chemosensory neuronal function (Glasscock and Tanouye 2005; Lundquist et al. 1996). During the early development of frog, avian, and mammalian embryos, the highest level of Rbpms (hermes) expression was detected in the myocardial tissues of the heart (Gerber et al. 1999). In the Xenopus embryo, it was also detected in the vegetal cortex, kidney, epiphysis, and ganglion cell layer of the retina (Zearfoss et al. 2004). In the chicken embryo, in addition to the heart, hermes is expressed in the myotome, allantois, glomeruli of the pronephros and mesonephros, the gut, and notochord but not in the retina or eye (Wilmore et al. 2005). In human tissues, RBPMS was reported to be strongly expressed in the heart, prostate, intestine, and ovary (Shimamoto et al. 1996). In our earlier study, we have identified Rbpms and its paralog Rbpms2 as genes, expression of which in the retina is restricted to RGCs (Piri et al. 2006). This observation was further supported by comprehensive analysis of Rbpms expression in rodent retinas, which showed that Rbpms is specifically expressed in virtually 100% cells that are retrogradely labeled by the tracer, applied to the cut surface of the optic nerve, and thus could be used as a reliable marker for these cells (Kwong et al. 2010). Outside of the eye, we observed Rbpms and Rbpms2 expression mainly in the heart, liver, and kidney. The abundant expression of hermes in the rat liver and lung was not detected in the corresponding tissues of Xenopus or chicken. Furthermore, we did not observe strong expression of Rbpms in the prostate and intestine reported for human tissues. These data demonstrate significant interspecific and developmental variations in regulation of Rbpms expression. A fascinating, in our opinion, characteristic of the Rbpms expression profile is that although this gene is expressed in various tissues, in the adult CNS, it was restricted to RGCs. In a recent study to investigate the role of Rbpms in the retina, it has been shown that Rbpms loss of function in both Xenopus laevis and zebrafish embryos results in a significant reduction in retinal axon arbor complexity in the optic tectum (Hörnberg et al. 2013). At the same time, an increase in the density of presynaptic puncta was observed in these animals, suggesting that reduced arborization is accompanied by increased synaptogenesis to maintain synapse number.
The objective of the present study was to identify transcriptional variants expressed in the retina and gain insights into regulation of Rbpms expression. We found that Rbpms is represented by isoforms A and C in human retinas encoded by splicing variants 1 and 3, respectively. These isoforms differ by their C-terminal amino acid (aa) sequence. Four predicted promoter regions (PPRs) of this gene were identified and their functional relevance in transcriptional regulation was evaluated in human embryonic kidney 293T (HEK 293T) cells. Furthermore, the involvement of several cis-acting elements that were identified in the PPR3 and PPR4, including five GC boxes, an AP1 site, and two sites for NF-kB, in transcriptional regulation were analyzed by site-directed mutagenesis and gel shift assay.
Experimental procedures
Human retinal RNA and RT-PCR
The total retinal RNA from healthy human donor eyes that were obtained from the National Disease Research Interchange (Philadelphia, PA, USA) was generously provided by Dr. D. B. Farber (UCLA; Young et al. 2011). The RNA was DNase treated with Turbo DNA-free (Ambion, Austin, TX, USA) and purified with RNeasy MiniElute Cleanup kit (Qiagen, Valencia, CA, USA). The first-strand cDNA from 5 μgs of the total RNA was reverse transcribed with oligo-dT primer and M-MuLV (NEB, Beverly, MA, USA). Rbpms splicing variants were identified by PCR, with primers specific to Rbpms isoforms and first-strand retinal cDNA used as a template.
Expression constructs, cell lines, and transfection
Expression constructs containing human Rbpms 5′ flanking region (5′-FR) and 5′ untranslated region (5′-UTR) were generated by PCR with sequence-specific primers listed in Table 1 and human genomic DNA used as a template. PCR fragments were cloned into pGL3-Basic (pGL3-B) vector (Promega, Madison, WI, USA) containing the luciferase reporter gene for subsequent luciferase reporter gene assays. Site-directed mutagenesis was performed with the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA) with Rbpms-specific primers and the previously generated deletion construct in a pGL3-B vector used as a template. For transfection, the plasmid DNAs were isolated with Maxi-Prep Kit (Qiagen).
Table 1.
Primers used in PCR to obtain genomic DNA fragments for Rbpms expression constructs
| Constructs | Primers 5′–3′
|
|
|---|---|---|
| Forward | Reverse | |
| − 1691/+ 592 | GTATCTTGGCTCCATACCACTT | CTTCCCGGTCCTTCCTCGC |
| − 1691/− 75 | GTATCTTGGCTCCATACCACTT | GCTGCAGCCCGGAGGGA |
| − 344/+ 592 | AGGTCCCGGTCCAGGCC | CTTCCCGGTCCTTCCTCGC |
| − 72/+ 592 | GGCAGCCCACGTCCAGGGAGG | CTTCCCGGTCCTTCCTCGC |
| − 1691/− 1416 | GTATCTTGGCTCCATACCACTT | CACCCCCACCAGACATTCTT |
| − 344/− 75 | AGGTCCCGGTCCAGGCC | GCTGCAGCCCGGAGGGA |
| − 72/+ 180 | GGCAGCCCACGTCCAGGGAGG | CCGTCCCCTCCTTCCAGCGG |
| + 263/+ 525 | TTCTTCCTTCCTCCCCTGG | CACTGGAGCCTTCCCCG |
| − 72/+ 525 | GGCAGCCCACGTCCAGGGAGG | CACTGGAGCCTTCCCCG |
The HEK293T cell line was maintained in DMEM/F12 supplemented with 10% FBS and 100-U penicillin/0.1-mg/mL streptomycin. Transfection was performed in triplicates with Lipofectamine (Life Technologies, Grand Island, NY) according to the manufacturer’s protocol when cells were at 90–95% confluency. Cells were co-transfected with pGL3-Basic vectors containing the luciferase reporter gene and pSV-β-galactosidase control vector that was used as an internal normalization standard for transfection efficiency. Cells were lysed 24 h after transfection. Luciferase and β-galactosidase assays were subsequently performed, and reporter gene expression levels were normalized to the corresponding β-galactosidase activities.
Luciferase and β-galactosidase assays
Luciferase and β-galactosidase assays were carried out according to the manufacturer’s protocol (Promega). Briefly, for luciferase assays, cells were washed with 1X PBS buffer containing 137-mM NaCl, 2.7-mM KCl, 8.1-mM Na2HPO4, and 1.47-mM KH2PO4 and incubated with Reporter Lysis Buffer (Promega) for 15 min. Cells were collected and subjected to one rapid freeze/thaw cycle, and then immediately used for the firefly luciferase assay (Promega). The relative luciferase activities of the samples were counted with a luminometer at a 2-s measurement delay and 10-s measurement read in the presence of luciferin substrate.
For β-galactosidase assays, the cell extracts were incubated in a buffer containing 1.33 mg/mL of the substrate ONPG (O-nitrophenyl-D-galactopyranoside), 200-mM sodium phosphate buffer, 2-mM MgCl2, and 100-mM β-mercaptoethanol, at 37 °C for 30 min. The reaction was stopped with 1-M sodium carbonate and the absorbance was determined by spectroscopy at 420 nm.
Preparation of nuclear extract and electrophoretic mobility shift assay (EMSA)
HEK293T cells were allowed to grow to confluence. Cells were pelleted, resuspended in Cell Fractionation Buffer (Life Technologies) and incubated on ice for 10 min. Nuclei were collected from the crude lysate with centrifugation at 500g for 5 min at 4 °C. Nuclear fraction was then lysed in Cell Disruption Buffer (Life Technologies) on ice for 10 min. The nuclear extract was stored at − 80 °C prior to use in experiments. To prepare the double-stranded DNA fragments (20 bp) for EMSA, two single-stranded oligonucleotides, either 5′ end-labeled or unlabeled, were synthesized and annealed. The EMSA was used to assess DNA-binding properties of transcription factors (TFs) in nuclear extract, employing a LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, Waltham, MA). Briefly, 20 fmol of annealed biotin end-labeled DNA probe was incubated with 1 μL of crude nuclear extract and 1-μg poly(dI·dC) in a total volume of 20 μL for 20 min at room temperature. After the binding reaction, 20 μL of the binding mixture was electrophoresed on a 6% DNA retardation gel (Life Technologies) in 0.5× TBE buffer (45-mM Tris, 45-mM boric acid, 1-mM EDTA, pH 8.3) at 100 V for 60 min. The DNA or DNA–protein complex from the gel was transferred onto a positively charged nylon membrane using 0.5× TBE buffer. Following UV cross-linking of the DNA to the membrane and treatment of streptavidin–horseradish peroxidase, the biotinylated DNA probes on the membrane were detected by chemiluminescence.
Statistical analysis
Reporter gene activities are expressed as the mean ± SD of at least three separate transfection experiments performed in triplicates. Statistical differences were determined using the Student’s t test to define the significance of the differences between two groups. Statistical significance was determined at the p < 0.05 level.
Results
Rbpms isoforms A and C encoded by splicing variants 1 and 3 are expressed in human retina
Human Rbpms gene is known to be expressed as multiple, at least 11, alternatively spliced transcript variants encoding different protein isoforms (8). Most commonly expressed isoforms include A, B, and C. Transcript variant 1 (NM_001008710) encoding isoform A (196 aa) has been chosen as the ‘canonical’ sequence (http://www.uniprot.org/uniprot/Q93062). Isoform A is a product of variant 4 (NM_006867), which has an additional 3′ UTR exon compared to variant 1 (Fig. 1). Variant 2 (NM_001008711) encoding isoform B (204 aa) includes an alternate exon in the 3′ coding region, which results in a frameshift and an early stop codon, compared to variant 1. The isoform B has a longer and distinct C-terminus, compared to isoform A. Transcription variant 3 (NM_001008712) lacks several exons and its 3′-terminal exon extends past a splice site that is used in variant 1. The isoform C (219 aa) encoded by this transcript has a different C-terminus compared to isoform A. To identify Rbpms splicing variants expressed in human retinas, primers were designed to generate different size fragments representing variants 1, 2, 3, and 4 (Fig. 1a). Forward primer R1 (5′-CCTGCTCTACCTCCTCCTGCT) is located at 1070–1090 nt in all four transcription variants. An alternative forward primer R2 (5′-TACCTCAGTTCATTGCCAGAGAGC) is located at 963–986 nt in these four variants. The reverse primer R3 (5′-AACCAGTGTAAGATCGTCCATCC) is located at 1586–1564, 1672–1650 and 1792–1770 nt in transcripts 1, 2, and 4 respectively and is absent in the variant 3. PCR with R1 and R3 are expected to produce 517, 603, and 723 bp fragments, whereas a primer pair R2 and R3 will generate 624, 710, and 830 bp corresponding to variants 1, 2 and 4 respectively. No products representing a splice variant 3 are expected to be generated by these primers. The results of the PCR shown in Fig. 1b indicate a predominant expression of transcript 1 in human retinas. R1/R3 and R2/R3 pairs produced ~ 520 and 620 bp DNA fragments, respectively, which correlate with the expected size of variant 1 fragments. Sequence analysis of the PCR products confirmed the amplification of the transcript 1 DNA fragments. Among several minor bands that can be seen on the gel image, there are two products with approximate size of 750 and 620 bp amplified by R2/R3 and R1/R3 primers, respectively. These products appear to be larger than those calculated for variant 2 fragments. To determine whether Rbpms is also expressed as splicing variant 3, PCR with forward R2 and reverse R4 primers specific to transcript 3 (5′-TCTGCAGTAGGTTGGTATGTTACA; location 1227–1204 nt) was carried out. A product with the expected size of 264 nt was obtained (Fig. 1c). These results indicate that Rbpms in human retinas is represented by splicing variants 1 and 3 that encode isoforms A and C, respectively.
Fig. 1.
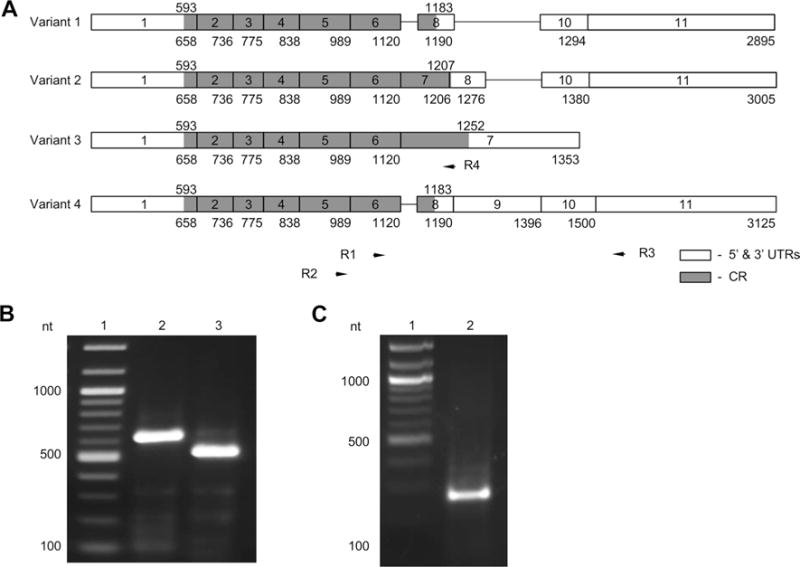
Rbpms splicing variants encoding isoforms A and C are expressed in human retina. a Splicing variants 1, 2, 3, and 4 are shown. Exons (boxes) are numbered from 1 to 11. Numbers under exons indicate the number of the last nucleotide of that exon. Positions of the coding regions (grey) are indicated above each splicing variant. Positions of primers (R1, R2, R3, and R4) used in PCR are indicated by arrows. b PCR products (lanes 2 and 3) corresponding to variant 1. PCR was performed with primer pairs R2/R3 (lane 2) and R1/R3 (lane 3). c PCR with primers R2/R4 produce a product corresponding to variant 3. Lane 1; DNA ladder
Computational analysis of Rbpms core promoter
To identify potential regulatory sequences that control RBPMS transcription, 5′-UTR and 5′-FR (~ 5000 nt upstream of translation initiation codon: includes 592 nt of 5′-UTR and 4370 nt of 5′-FR) were analyzed using several core promoter recognition software. First, the sequence was analyzed for GC content and the presence of CpG islands, genomic regions characterized by an exceptionally high CpG dinucleotide frequency (Bird 1986). CpG islands are among most important regulatory regions that have been shown to be associated with more than 75% of all known transcription start sites (TSS) and with 88% of active promoters (Kim et al. 2005; Bajic et al. 2006). Using GC-Profile software (http://tubic.tju.edu.cn/GC-Profile/), one segmentation point, which partitions a given genomic DNA sequence into composition-ally distinct domains, was identified at position − 610 bp (Fig. 2a). GC content downstream of the segmentation point was approximately 75%. Three CpG islands within this sequence were identified with CpG island prediction software (http://urogene.org/cgi-bin/methprimer/methprimer.cgi; Fig. 2b): island 1–141 bp (− 631/− 491; located in the 5′-FR), island 2–406 bp (− 412/− 7; located in the 5′-FR), and island 3–534 bp (+ 3/+ 536; located in the 5′-UTR). The following criteria were used to identify these regions: island size > 100, GC percent > 50.0, CpG observed-to-expected ratio > 0.60. We would like to note that the identified here island 1 would not qualify as a CpG island based on other criteria set for this type of sequence. For instance, according to Gardiner-Garden sequence criteria, a genomic region can be classified as a CpG island if its GC content is above 50%, an observed-to-expected CpG ratio is greater than 60% and length greater than 200 bp (Gardiner-Garden and Frommer 1987; Antequera 2003). Analysis of the Rbpms 5′-UTR and 5′-FR with Gardiner-Garden sequence criteria showed only one 1172 bp CpG island at − 685/+ 486. This sequence fully encompasses islands 1 and 2, as well as a large portion of island 3 identified above. In summary, the results of Rbpms epigenomic analysis reveals the high GC content and the presence of at least one CpG island within ~ 1300 nt upstream of translation initiation codon strongly suggesting the involvement of this region in transcriptional regulation. CpG sequence analysis of the Rbpms potential regulatory region was further complemented by core promoter prediction analysis, which revealed the presence of four potential promoter regions located at − 1672/− 1420, − 330/− 79, − 73/+ 177, and + 274/+ 524 (http://www-bimas.cit.nih.gov/molbio/proscan; Fig. 2c). Three of these PPR are located in the CpG islands.
Fig. 2.
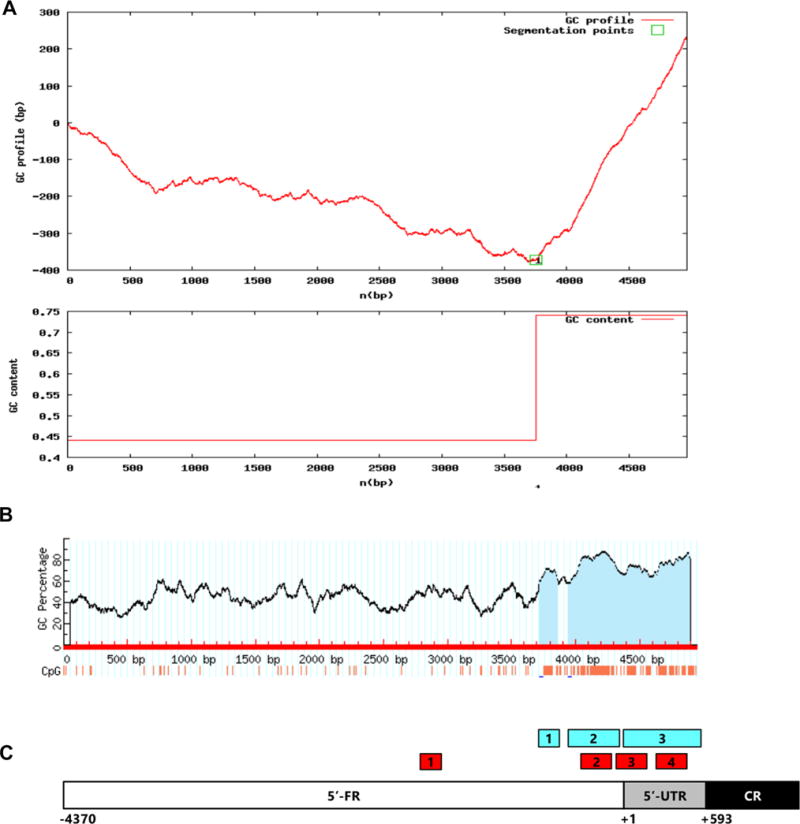
Promoter analysis of Rbpms 5′-UTR and 5′-FR. a GC-Profile software (http://tubic.tju.edu.cn/GC-Profile/) was used to analyze approximately 5 kb DNA sequence upstream of AUG translation initiation codon that includes Rbpms 5′-UTR and 5′-FR. One segmentation point was identified at position − 610 nt. GC content downstream of the segmentation point is approximately 75%. b Three CpG islands within downstream of − 610 nt sequence were identified with CpG island prediction software (http://urogene.org/cgi-bin/methprimer/methprimer.cgi): Island 1–141 bp (− 631/− 491; located in the 5′-FR), Island 2–406 bp (− 412/− 7; located in the 5′-FR) and Island 3–534 bp (+ 3/+ 536; located in the 5′-UTR). The following criteria were used to identify these regions: Island size > 100, GC Percent > 50.0, CpG observed-to-expected ratio > 0.60. c Schematic locations of CpG islands (blue boxes) and PPRs (red boxes) in Rbpms 5′-UTR and 5′-FR
The role of PPRs in regulation of gene expression
Based on the computational sequence analysis described above, four expression constructs − 1691/+ 592, − 1691/− 75, − 344/+ 592, and − 72/+ 592 were made in pGL3-B and tested in HEK 293 cells (Fig. 3). We would like to note here that the generation of these constructs was complicated by a limited choice in selection of sequence-specific primers for PCR due to the extremely high GC content of the region. The luciferase reporter gene expression from the − 1691/+ 592 construct containing all four PPRs was approximately 13-fold higher than that obtained from the promoter-less pGL3-B construct used in these experiments to set a baseline for luciferase activity. Interestingly, a − 1691/− 75 construct with 2 PPRs in the 5′-FR produced almost no reporter gene expression, whereas a − 72/+ 592 construct containing PPRs 3 and 4 had a 30-fold increase in luciferase activity compared to pGL3-B. The expression level from the − 344/+ 592 construct with PPRs 2, 3, and 4 was reduced from 30 fold produced by the − 72/+ 592 construct to about 22-fold. These data clearly indicate that DNA regions containing PPRs 3 and 4 are critical for activation of gene expression, whereas PPRs 1 and 2 are insufficient or even may have a negative impact on this process. Four additional constructs were designed to evaluate the role of individual PPRs on activation of reporter gene expression (Fig. 3). As expected, constructs − 1691/− 1416 and − 344/− 75 containing PPRs 1 and 2, respectively, produced a minimal luciferase expression: 1.5- and 3-fold higher than pGL3-B control. Expression from constructs with PPRs 3 and 4 (constructs − 72/+ 180 and + 263/+ 525) was induced ~ 7 and ninefold compared to pGL3-B control, respectively (Fig. 3). These results once again point to the positive role of the sequence within 5′-UTR and the adjacent 73 nt of the 5′-FR on activation of gene expression.
Fig. 3.
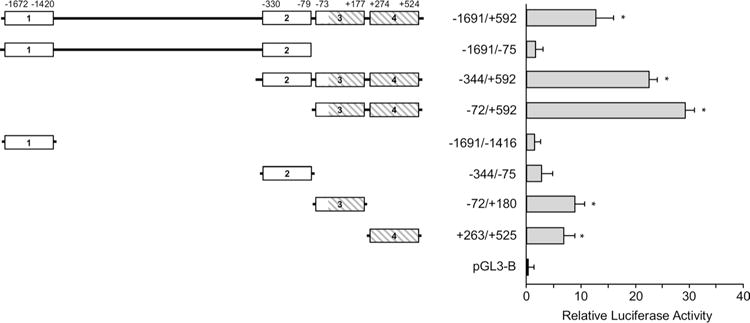
Effect of four PPRs identified in Rbpms gene on transcriptional activity. A schematic shows the four PPRs (white and hatched boxes) that were identified using promoter recognition software with their locations relative to TSS. Expression constructs containing these regulatory regions upstream of the luciferase reporter gene were evaluated in HEK293T cells and the produced luciferase activity is presented relative to that generated from promoter-less control construct. pGL3-B was used as the negative control and arbitrarily set to 1.0. The luciferase reporter gene expression from the − 1691/+ 592 construct containing all four PPRs was ~ 13-fold higher than that obtained from the pGL3-B. Constructs − 1691/− 75, − 1691/− 1416, and − 344/− 75 that do not contain PPRs 3 and 4 produced minimal expression levels, whereas constructs − 344/+ 592 and − 72/+ 592 containing both PPRs 3 and 4 had approximately 23- and 30-fold increase in luciferase activity, respectively, compared to pGL3-B. The data shown are the mean with SD from three independent transfection experiments. Each experiment ran in triplicate. *P < 0.05 compared with the pGL3-B control
Cis-elements in the PPRs 3 and 4 in regulation of Rbpms transcriptional activity
Sequence analysis of the PPRs 3 and 4 reveals a number of potential cis-elements (Table 2A, B). Since the consensus sequences for many of these elements are highly ambiguous, we focused in this study on several GC boxes, two NF-kB sites, and an AP1 site (Fig. 4a). To evaluate the involvement of these TF-binding sites (TFBS) in regulation of Rbpms expression, new constructs were generated by site-directed mutagenesis (Fig. 4b). Luciferase activity from a GC-1 construct in which the first GC box was mutated was approximately threefold lower than that produced by a wild-type − 72 + 525 plasmid. Expression from two other GC box mutant constructs, GC-2 and GC-3, was also significantly reduced (~ 1.7 to 1.8 fold) although not as much as from GC-1. Mutations in two other GC boxes, AP1 or NF-kB had a little effect on reporter gene expression (Fig. 4b).
Table 2.
Promoter regions 3 and 4 predicted on forward strand (A; 1 to 251 nt) and on reverse strand (B; 597 to 347 nt), respectively
| TFD | Strand | Location | Weight |
|---|---|---|---|
| A | |||
| JCV_rep_seq | + | 20 | 1.427000 |
| Sp1 | + | 20 | 3.013000 |
| Sp1 | + | 20 | 7.086000 |
| Sp1 | − | 25 | 3.061000 |
| AP-2 | − | 25 | 1.672000 |
| EARLY-SEQ1 | − | 27 | 5.795000 |
| Lam_B2_US | − | 58 | 5.378000 |
| AP-2 | − | 59 | 1.091000 |
| AP-2 | + | 160 | 1.108000 |
| GCF | + | 167 | 2.361000 |
| UCE.2 | − | 172 | 1.216000 |
| GCF | − | 175 | 2.284000 |
| EARLY-SEQ1 | + | 180 | 6.322000 |
| Sp1 | + | 182 | 3.292000 |
| JCV_rep_seq | − | 187 | 1.658000 |
| Sp1 | − | 187 | 3.361000 |
| Sp1 | − | 187 | 7.086000 |
| AP-1 | − | 224 | 1.052000 |
| B | |||
| AP-2 | − | 586 | 1.863000 |
| AP-2 | − | 586 | 1.108000 |
| NF-kB | − | 575 | 1.080000 |
| Sp1 | − | 564 | 3.013000 |
| Sp1 | − | 563 | 3.191000 |
| Sp1 | + | 559 | 3.061000 |
| Sp1 | + | 558 | 3.119000 |
| TTR_inv_rep | − | 557 | 2.151000 |
| T-Ag | − | 544 | 1.086000 |
| AP-2 | + | 498 | 1.091000 |
| NF-kB | − | 495 | 1.080000 |
| TTR_inv_rep | + | 481 | 3.442000 |
| Sp1 | − | 464 | 3.191000 |
| Sp1 | + | 459 | 3.119000 |
| AP-2 | + | 456 | 1.672000 |
| Myosin-spec | + | 455 | 1.115000 |
| T-Ag | − | 432 | 1.086000 |
| SIF | + | 430 | 1.161000 |
| GCF | − | 422 | 2.361000 |
| UCE.2 | − | 419 | 1.278000 |
| AP-2 | + | 376 | 1.672000 |
| Myosin-spec | + | 375 | 1.115000 |
| JCV_rep_seq | − | 363 | 1.427000 |
| Sp1 | − | 363 | 3.013000 |
| Sp1 | − | 362 | 3.191000 |
| Sp1 | + | 358 | 3.061000 |
| Sp1 | + | 357 | 3.119000 |
| EARLY-SEQ1 | + | 356 | 5.795000 |
| AP-2 | + | 348 | 1.091000 |
Fig. 4.
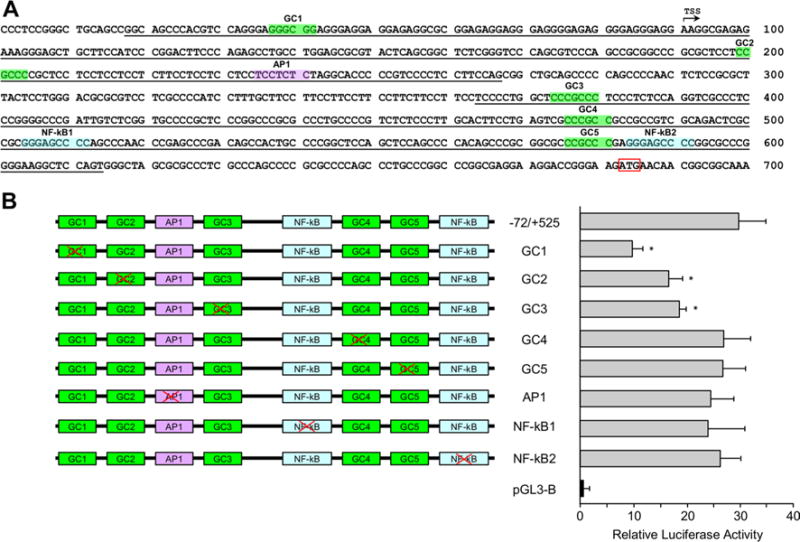
Sp1, AP1, and NF-kB TFBS in the PPRs 3 and 4 and their role in regulation of gene expression. a PPR3 in the 5-UTR and 5′-FR and PPR4 in the PPR4 are underlined. Predicted cis-elements for Sp1, AP1, and NF-kB are highlighted in green, purple, and blue, respectively. Transcription start site is indicated by an arrow and translation initiation codon is boxed. b Constructs generated by site-directed mutagenesis of individual predicted cis-element and reporter gene expression levels produced from these constructs in HEK293T cells relative to pGL3-B control. The most significant change in luciferase expression—approximately threefold decrease compared to the − 72/+ 525 construct—was produced by GC1 construct with first mutated GC box. The data shown are the mean with SD from three independent transfection experiments. Each experiment ran in triplicate. *P < 0.05 compared with the − 72/+ 525 construct
Since the PPRs 3 and 4 contain multiple GC boxes, it is possible that elimination of one consensus sequence in each construct does not fully reflect the contribution of Sp1 in regulation of Rbpms expression due to possible competition from the remaining intact GC motifs for TF binding. To address this issue, four additional constructs were made in which GC boxes were consequently mutated (Fig. 5). Expression levels from all these constructs (GC1/2, GC1/2/3, GC1/2/3/4, and GC1/2/3/4/5) were significantly lower compared to the − 72/+ 525 construct, but similar to GC1 with the first mutated GC box suggesting the critical role of this sequence in transcriptional activation.
Fig. 5.
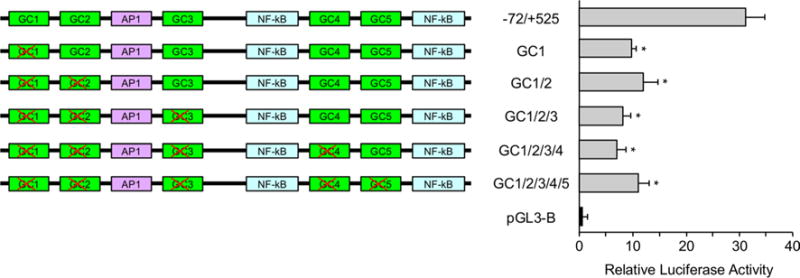
GC box 1 regulates transcriptional activity of Rbpms minimal promoter. Constructs were generated by subsequent mutation of all five GC boxes. Expression levels from all these constructs (GC1/2, GC1/2/3, GC1/2/3/4, and GC1/2/3/4/5) were significantly lower compared to the − 72/+ 525 construct, but similar to GC1 with the first mutated GC box suggesting the critical role of this sequence in transcriptional activation. The data shown are the mean with SD from three independent transfection experiments. Each experiment ran in triplicate. *P < 0.05 compared with the − 72/+ 525 construct
GC box interaction with nuclear proteins
To evaluate the ability of GC boxes described above to bind TFs, DNA–protein interactions were analyzed by EMSA with nuclear extract from HEK 293T and five labeled oligonucleotides spanning each of the GC motifs found in the RBPMS 5′-UTR. A “shifted” band indicating the formation of a DNA–protein complex was observed in a reaction containing labeled oligonucleotide with GC sequence 1 (Fig. 6, Lane 2). The specificity of the DNA–protein interaction within this complex was confirmed by competition experiments. Addition of a 100-molar excess of identical but unlabeled competitor oligonucleotide to the reaction completely abolished a signal corresponding to labeled DNA–protein complex. No DNA–protein complex was observed in reactions with four other labeled oligonucleotides corresponding to the predicted GC boxes 2, 3, 4, and 5. The results of the gel shift assay are in agreement with the reporter gene expression data obtained with wild-type − 72 + 525 and mutated GC1 constructs and further support the involvement of GC1 in transcriptional regulation of the RBPMS gene.
Fig. 6.
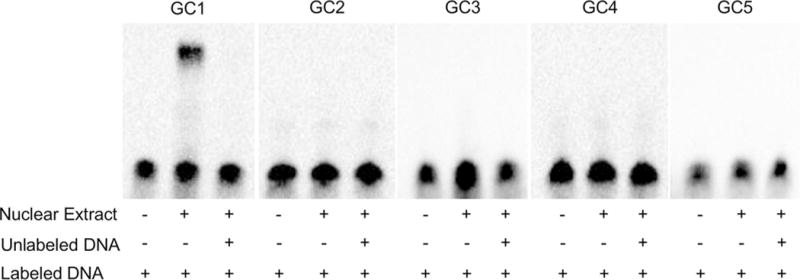
GC1 sequence forms a complex with nuclear protein extracted from HEK293 cells. EMSA was performed with labeled oligonucleotides corresponding to GC1 through GC5. Only one DNA–protein complex was detected in lane for GC1 labeled oligonucleotide. This interaction was competed out by an excess of unlabeled GC1 probe
Discussion
The present study delved into Rbpms isoforms expressed in human retinas as well as mechanisms of Rbpms transcriptional regulation. Rbpms, as evident from its name, can be represented by at least 11 mRNAs including 4 RefSeq transcription variants 1, 2, 3, and 4. According to the ECgene analysis (http://genome.ewha.ac.kr/ECgene/), this gene produces 77 transcript variants encoding 20 distinct proteins. Three most common isoforms include A, B, and C; isoform A is encoded by transcription variants 1 and 4, whereas isoforms B and C are encoded by splicing transcripts 2 and 3, respectively. In this study, we have determined that in human retinas, the Rbpms gene is expressed as isoforms A (196 aa) and C (219 aa). These proteins have identical N-terminal 176 aa encoded by the same six exons and different C-terminal aa sequences encoded by exon 8 in variants 1 or 4 and exon 7 in variant 3 (Fig. 2). The functional significance of this difference and consequently the reason for the Rbpms gene to be represented in the retina by two isoforms is yet to be determined. No conserved domains were found in the C-terminal region.
All 11 mRNAs reported for Rbpms including four RefSeq splicing variants share the same first exon, suggesting a common core promoter for transcriptional regulation. To identify Rbpms regulatory region, we first analyzed its 5′-UTR (562 nt) and the adjacent 5′-FR (~ 4400 nt) for GC content and the presence of CpG island. Despite the enormous variability in regulatory regions, all mammalian RNA Pol II promoters can be categorized into two classes based on the distribution of CpG dinucleotides: regions with average genome frequency of CpG, which is ~ 1%, and regions with CpG islands in which CpG content is about ten times higher than genome average (Bird 1986; Gardiner-Garden and Frommer 1987). In humans, approximately 60% of all promoters colocalize with CpG islands suggesting their involvement in transcriptional regulation (Gardiner-Garden and Frommer 1987; Bock et al. 2007; Bird 1987). Sequence analysis of Rbpms 5′-UTR and 5′-FR with GC-Profile and CpG island prediction software reveal the presence of three CpG islands: Island 1 (141 bp) and Island 2 (406 bp) in the 5′-FR and Island 3 (534 bp) in the 5′-UTR. The following criteria were used to identify these regions: island size > 100, GC Percent > 50.0, CpG observed-to-expected ratio > 0.60. It is noteworthy that criteria for CpG island prediction vary in different calculation methods. For instance, here, criteria for two widely accepted methods: the Gardiner-Garden filtered method uses 50% GC content, an 0.6 observed-to-expected CpG ratio and length 200 bp, whereas Takai Jones unmasked method uses GC content 55%, CpG observed-to-expected ratio 0.65, and CpG island length 500 bp (Gardiner-Garden and Frommer 1987; Takai and Jones 2002). With criteria set to satisfy Gardiner-Garden method, only one 1172 bp CpG island was identified for Rbpms gene. With stricter criteria for GC content (60%), CpG observed-to-expected ratio (0.7) and length (200 bp), we found three CpG islands: 200 bp and 437 bp islands in the 5′-FR and 405 bp island in the 5′-UTR. This analysis strongly suggests that the Rbpms regulatory region belongs to CpG island category and that the distribution of CpG islands can serve as a marker to predict and evaluate the identified promoter sequences for their transcriptional activity. Several promoter prediction algorithms were used to analyze Rbpms 5′-UTR and 5′-FR. In this study, we primarily focused on four promoter regions predicted by PROSCAN Version 1.7. Four PPRs were identified including three PPRs located within CpG islands. pGL3-B-based constructs with luciferase reporter gene placed under control of Rbpms PPRs were used to evaluate the role of these four regions in regulating the level of gene expression. The maximum luciferase activity (~ 30-fold higher than from the promoter-less construct) was obtained from constructs containing PPRs 3 and 4. Constructs with both PPRs 1 and 2, as well as with individual PPR1 and PPR 2, produced minimal or no expression. Furthermore, the activity produced by the construct with PPRs 3 and 4 was reduced when PPR 2 was included suggesting a negative effect of PPR2 on transcriptional activity drive by PPRs 3 and 4. Evaluation of gene expression from constructs with individual PPRs 3 and 4 showed an approximately 7- and 10-fold increase in luciferase activity compared to control, respectively. Results of this experiment demonstrate the importance of PPRs 3 and 4 located in the 5′-UTR and adjacent 73 nt of the 5′-FR on activation of gene expression. Although each of these PPRs had a significant positive effect on the level of gene expression, the maximum activation was observed when these two PPRs are located in the same construct. Several cis-elements within PPRs 3 and 4 were identified and tested for their involvement in transcriptional regulation. Among them were five Sp1-binding sites, two NF-kB, and one AP1-binding sites. Site-directed mutagenesis of AP1 and NF-kB elements had no effect on gene expression. The most significant reduction in expression level (threefold) was observed when the first GC box was mutated (located in the 5′-FR of PPR 3). A less dramatic effect was also observed when the second and third GC boxes were mutated. Elimination of two downstream GC boxes, GC4 and GC5, did not affect the level of reporter gene expression. We also address the possibility of remaining intact GC boxes substituting the role of the mutated GC box by generating constructs in which two, three, four, and all five GC boxes were mutated. However, we did not see a further significant decrease in activity when GC 2–5 were mutated in addition to GC1. These results indicate that among five GC boxes, an AP1 and two NF-kB binding sites identified in PPRs 3 and 4, GC1 and to a lesser degree GC2 and GC3 may be responsible for transcriptional activation. The involvement of GC1 was also confirmed by EMSA showing a formation of a DNA protein complex with oligonucleotide corresponding to GC1. No DNA–protein complex was observed when labeled oligonucleotides corresponding to four other Sp1-binding sites were used in reaction. It is important to note that since these experiments were performed in HEK 293 cells, the obtained results are valid for this cell line. It is quite possible, that in different environments, such as those mimicking heart tissue or retinal ganglion cells where Rbpms is most abundantly expressed, contribution of these or other TFBS on transcriptional activation may vary.
Unraveling mechanisms involved in regulation of Rbpms expression in RGCs is one of the important long-term objectives of this study. Although not tested experimentally yet, the analysis of the entire Rbpms gene (187,761 bp) using The Champion ChiP Transcription Factor Search Portal implicates Pax-6 in this process. The Portal is based on SABiosciences’ proprietary database DECODE, which combines SABiosciences’ Text Mining Application and data from the UCSC Genome Browser to assemble a list with over 200 predicted TFBS for every human gene. A single TFBS for Pax-6 (TATTATCTGAAACATGAAGCTG at position 30,249,274) was identified in intron 1 of Rbpms at 7330 nt from TSS (position 30,241,944). Pax-6 is a transcriptional master regulator involved in the eye morphogenesis (Gehring 2002). It is expressed in the optic primordium and later in cells of the prospective retina, pigmented epithelium, and lens epithelium (Walther and Gruss 1991; Grindley et al. 1995). Pax6-deficient mice completely lack eyes (Hill et al. 1991), whereas conditional knockout of this gene in the developing retina results in a failure in the specification of all cell types except amacrine cells (Marquardt et al. 2001). Pax-6 haploinsufficiency causes aniridia, which is frequently complemented by cataract, corneal opacification, and progressive glaucoma (Ton et al. 1991; Glaser et al. 1992). Mutations in the Pax6 have been associated with anophthalmia, nasal hypoplasia, and central nervous system defects (Glaser et al. 1994). In differentiated retinas Pax-6 is localized in the GCL (Jones et al. 1998; Steinmayr et al. 1998; Kaneko et al. 1999; Bhat et al. 2004), suggesting its role in the transcriptional regulation of genes expressed in the RGCs. Expression of Pax-6 has been shown to be regulated by POU factor Brn-3B, which in the retina is expressed exclusively in RGCs (Plaza et al. 1999). Deletion of Brn3b results in switching of differentiation of many cells from RGCs to amacrine or horizontal cells, up to 80% of RGC loss by adulthood, many surviving RGCs having axons with abnormal intraocular and extraocular trajectories as well as downregulation of multiple genes including Rbpms (Gan et al. 1996; Xiang 1998; Erkman et al. 2000; Pan et al. 2005). These observations suggest that Pax-6 could be involved in transcriptional activation of Rbpms in RGCs. Interestingly, similar to Rbpms, a promoter of another gene, Thy-1, expression of which in the retina is predominantly localized to RGCs, is located in a CpG island and its expression in neurons is directed by an element in the first intron (Vidal et al. 1990). Currently, for lack of a better one, Thy-1 promoter is commonly used to generate transgenic systems that target RGCs; however, both the cell specificity and efficiency of gene expression remain open issues. We believe that investigating Rbpms transcriptional machinery will help to better understand the regulation of this gene as well as contribute to developing a system that will ensure robust and specific expression of genes of interest in RGCs.
In summary, the Rbpms in human retinas is represented by two isoforms A and C encoded by transcription variants 1 and 3, respectively. Computational analysis of the Rbpms 5′-UTR and adjacent 5′-FR reveals the presence of 1 to 3 CpG islands depending on CpG island criteria. Four PPRs including three of them located in the CpG islands were identified. The effect of PPR 1 (− 1672/− 1420) and PPR2 (− 330/− 79) on reporter gene transcriptional activation was minimal, whereas PPR 3 (− 73/+ 177) and PPR4 (+ 274/+ 524) induced the expression by ~ 7 and ninefold compared to control, respectively. The maximum luciferase activity was obtained from the construct containing both PPRs 3 and 4. Site-directed mutagenesis of several TFBS located in PPR3 and PPR4 including five cis-elements for Sp1, one for AP1 and two sites for NF-kB showed irrelevance of AP1, NF-kB and two downstream Sp1 sites in regulation of gene expression in HEK 293T cells. Mutation of the first three and especially the first GC box resulted in up to a threefold downregulation of reporter gene expression. The possible involvement of the GC box 1 in transcriptional regulation was further corroborated by EMSA, which showed formation of a DNA–protein complex with the sequence corresponding to this Sp1-binding site.
Acknowledgments
Funding This work was supported by the National Institutes of Health (NIH) Grant EY018644 (NP).
References
- Antequera F. Structure, function and evolution of CpG island promoters. Cell Mol Life Sci. 2003;60:1647–1658. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic VB, Tan SL, Christoffels A, Schonbach C, Lipovich L, et al. Mice and men: their promoter properties. PLoS Genet. 2006;2:e54. doi: 10.1371/journal.pgen.0020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat SP, Rayner SA, Chau SC, Ariyasu RG. Pax-6 expression in posthatch chick retina during and recovery from form-deprivation myopia. Dev Neurosci. 2004;26:328–335. doi: 10.1159/000082274. [DOI] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird AP. CpG islands as gene markers in the vertebrate nucleus. Trends Genet. 1987;3:342–347. [Google Scholar]
- Bock C, Walter J, Paulsen M, Lengauer T. CpG island mapping by epigenome prediction. PLoS Comput Biol. 2007;3:e110. doi: 10.1371/journal.pcbi.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Swanson MS, Pinol-Roma S. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem Sci. 1988;13:86–91. doi: 10.1016/0968-0004(88)90046-1. [DOI] [PubMed] [Google Scholar]
- Erkman L, Yates PA, McLaughlin T, McEvilly RJ, Whisenhunt T, et al. POU domain transcription factor-dependent program regulates axon pathfinding in the vertebrate visual system. Neuron. 2000;28:779–792. doi: 10.1016/s0896-6273(00)00153-7. [DOI] [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, et al. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci USA. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. The genetic control of eye development and its implications for the evolution of the various eye-types. Int J Dev Biol. 2002;46:65–73. [PubMed] [Google Scholar]
- Gerber WV, Yatskievych TA, Antin PB, Correia KM, Conlon RA, et al. The RNA-binding protein gene, hermes, is expressed at high levels in the developing heart. Mech Dev. 1999;80:77–86. doi: 10.1016/s0925-4773(98)00195-6. [DOI] [PubMed] [Google Scholar]
- Glaser T, Walton DS, Maas RL. Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nature Genet. 1992;2:232–238. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, et al. Pax6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- Glasscock E, Tanouye MA. Drosophila couch potato mutants exhibit complex neurological abnormalities including epilepsy phenotypes. Genetics. 2005;169:2137–2149. doi: 10.1534/genetics.104.028357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley JC, Davidson DR, Hill RE. The role of Pax-6 in eye and nasal development. Development. 1995;121:1433–1442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, et al. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Hörnberg H, Wollerton-van Horck F, Maurus D, Zwart M, Svoboda H, et al. RNA-binding protein Hermes/RBPMS inversely affects synapse density and axon arbor formation in retinal ganglion cells in vivo. J Neurosci. 2013;33:10384–10395. doi: 10.1523/JNEUROSCI.5858-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Jomary C, Grist J, Thomas MR, Neal MJ. Expression of Pax-6 mRNA in the retinal degeneration (rd) mouse. Biochem Biophys Res Commun. 1998;252:236–240. doi: 10.1006/bbrc.1998.9631. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Matsumoto G, Hanyu Y. Pax-6 expression during retinal regeneration in the adult newt. Dev Growth Differ. 1999;41:723–729. doi: 10.1046/j.1440-169x.1999.00476.x. [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, et al. A highresolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong JM, Caprioli J, Piri N. RNA binding protein with multiple splicing: a new marker for retinal ganglion cells. Invest Ophthalmol Vis Sci. 2010;51:1052–1058. doi: 10.1167/iovs.09-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist EA, Herman RK, Rogalski TM, Mullen GP, Moerman DG, et al. The mec-8 gene of C. elegans encodes a protein with two RNA recognition motifs and regulates alternative splicing of unc-52 transcripts. Development. 1996;122:1601–1610. doi: 10.1242/dev.122.5.1601. [DOI] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, et al. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Pan L, Yang Z, Feng L, Gan L. Functional equivalence of Brn3 POU-domain transcription factors in mouse retinal neurogenesis. Development. 2005;132:703–712. doi: 10.1242/dev.01646. [DOI] [PubMed] [Google Scholar]
- Piri N, Kwong JM, Song M, Caprioli J. Expression of hermes gene is restricted to the ganglion cells in the retina. Neurosci Lett. 2006;405:40–45. doi: 10.1016/j.neulet.2006.06.049. [DOI] [PubMed] [Google Scholar]
- Plaza S, Hennemann H, Möröy T, Saule S, Dozier C. Evidence that POU factor Brn-3B regulates expression of Pax-6 in neuroretina cells. J Neurobiol. 1999;41:349–358. doi: 10.1002/(sici)1097-4695(19991115)41:3<349::aid-neu4>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Shimamoto A, Kitao S, Ichikawa K, Suzuki N, Yamabe Y, et al. A unique human gene that spans over 230 kb in the human chromosome 8p11-12 and codes multiple family proteins sharing RNA-binding motifs. Proc Natl Acad Sci USA. 1996;93:10913–10917. doi: 10.1073/pnas.93.20.10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmayr M, Andre E, Conquet F, Rondi-Reig L, Delhaye-Bouchaud N, et al. Staggerer phenotype in retinoid-related orphan receptor alpha-deficient mice. Proc Natl Acad Sci USA. 1998;95:3960–3965. doi: 10.1073/pnas.95.7.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton CC, Hirvonen H, Miwa H, Weil MM, Monaghan P, et al. Positional cloning and characterization of a paired box and homeobox containing gene from the aniridia region. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- Vidal M, Morris R, Grosveld F, Spanopoulou E. Tissue-specific control elements of the Thy-1 gene. EMBO J. 1990;9:833–840. doi: 10.1002/j.1460-2075.1990.tb08180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- Wilmore HP, McClive PJ, Smith CA, Sinclair AH. Expression profile of the RNA-binding protein gene hermes during chicken embryonic development. Dev Dyn. 2005;233:1045–1051. doi: 10.1002/dvdy.20392. [DOI] [PubMed] [Google Scholar]
- Xiang M. Requirement for Brn-3b in early differentiation of post-mitotic retinal ganglion cell precursors. Dev Biol. 1998;197:155–169. doi: 10.1006/dbio.1998.8868. [DOI] [PubMed] [Google Scholar]
- Young A, Jiang M, Wang Y, Ahmedli NB, Ramirez J, et al. Specific interaction of Gαi3 with the Oa1 G-protein coupled receptor controls the size and density of melanosomes in retinal pigment epithelium. PLoS One. 2011;6:e24376. doi: 10.1371/journal.pone.0024376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zearfoss NR, Chan AP, Wu CF, Kloc M, Etkin LD. Hermes is a localized factor regulating cleavage of vegetal blastomeres in Xenopus laevis. Dev Biol. 2004;267:60–71. doi: 10.1016/j.ydbio.2003.10.032. [DOI] [PubMed] [Google Scholar]


