Abstract
During development, the endoderm initiates organ-restricted gene expression patterns in a spatiotemporally controlled manner. This process, termed induction, requires signals from adjacent mesodermal derivatives. Fibroblast Growth Factor (FGF) and Bone Morphogenetic Protein (BMP) emanating from the cardiac mesoderm and the septum transversum mesenchyme (STM), respectively, are believed to be simultaneously and uniformly required to directly induce hepatic gene expression from the murine endoderm. Using small molecule inhibitors of BMP signals during liver bud induction in the developing mouse embryo, we find that BMP signaling is not uniformly required to induce hepatic gene expression. Although BMP inhibition causes an overall reduction in the number of induced hepatoblasts, the STM-bounded posterior liver bud demonstrates the most severe loss of the essential hepatic transcription factor Hepatocyte nuclear factor 4-α (HNF4α) while the sinus venosus (SV)-lined anterior liver bud is less affected. We find that the posterior liver bud progenitors are anteriorly displaced and aberrantly activate pancreatobiliary markers including SOX9. Additionally, we find that ectopically expressed SOX9 inhibits HNF4α and that BMP is indirectly required for hepatoblast induction. Finally, because previous work demonstrated that FGF signals were essential for anterior but not posterior liver bud induction, we examined synchronous BMP and FGF inhibition and find that this leads to a massive loss of induced hepatoblasts.
Conclusions
BMP signaling is required to maintain the hepato-pancreatobiliary boundary, at least in part, by indirectly repressing SOX9 in the hepatic endoderm. BMP and FGF signals are each required for the induction of spatially complementary subsets of hepatoblasts. These results highlight the importance of studying early inductive processes in the whole embryo.
Keywords: anterior liver bud, posterior liver bud, HNF4α, induction, SOX9
Introduction
The liver bud emerges from the endoderm as a simple thickened epithelium wholly composed of the embryonic liver precursor, termed the hepatoblast (1). Hepatoblasts are induced at 7–8 somites (S) proximal to the developing heart. We previously identified two distinct populations of endoderm that give rise to the liver bud (2). One resides at the ventral-midline of the endoderm lip and is hence termed VMEL. The other population is present at two bilaterally symmetrical positions lateral to the endodermal lip. Although, lateral and VMEL progenitors converge at the endoderm lip to produce the nascent liver between 10–11S, fate mapping and ablation studies have shown that the lateral progenitors contribute to much of the middle and posterior liver bud while the VMEL progenitors contribute most pronouncedly to the anterior liver bud (2–4). By embryonic day (E) 9.0, a liver bud expressing liver-specific genes including α-fetoprotein (Afp) and Hepatocyte nuclear factor 4-α (Hnf4α) has formed. Explant experiments have suggested a role for secreted signals, including Fibroblast Growth Factors (FGFs) and Bone Morphogenetic Proteins (BMPs), in the induction of liver specific gene expression (5, 6).
In the canonical BMP pathway, BMP ligands bind the Type I and Type II BMP receptors to form a complex that results in receptor phosphorylation and activation. The activated receptor complex in turn phosphorylates receptor regulated SMAD-1, −5 or −8. Phosphorylated SMAD-1, −5 or −8 (pSMAD1/5/8) can bind to SMAD4, and the heteromeric complex translocates into the nucleus to regulate target gene expression. Because, SMAD1/5/8 activation is unique to the BMP pathway, pSMAD1/5/8 can be used as read-out for active BMP signaling (7, 8).
While a role for BMP signals for murine liver development has mainly been established in the context of pre-hepatic/mesoderm explants, a complete understanding is impeded by the redundancy of the pathway components and the requirement for this pathway in multiple earlier developmental processes (1). For example, a subset of BMP4 null embryos are delayed in liver budding (6), however, it is also clear that BMP4 is required to appropriately establish mesoderm derivatives that are involved in liver bud induction such as the cardiovascular system and STM (9, 10). Thus, the hepatic delay observed in BMP4 null embryos may be secondary to or exacerbated by its earlier requirement in mesoderm formation. Regardless of the tissue that utilizes BMP, a requirement for BMP in liver induction is evolutionarily conserved and has been established in zebrafish using BMP receptor I (Alk8) mutants and a dominant negative BMP receptor (11) and its role in liver induction supported by chick explants and ex vivo (12).
Herein, we use intact developing mouse embryos to gain a more complete understanding of BMP’s role in mammalian hepatic induction. Specifically, we use a whole embryo culture approach that allows for the isolation of embryos immediately prior to liver bud induction and for their normal development through liver budding. We utilize small molecule inhibitors that act as antagonists for BMP Type I receptors, preventing SMAD1/5/8 phosphorylation (13). This approach dampens BMP signals throughout the embryo in a temporally specific manner, bypassing earlier requirements for BMP signals. Using this technique, we find that the posterior liver bud is more affected by reduced BMP signals than the anterior liver bud and uncover a previously undescribed role for BMP in demarcating the hepato-pancreatobiliary boundary. Together, these experiments highlight the need to study the role of inductive signals in the context of the developing embryo.
Materials and Methods
Embryo culture and small molecule administration
All animal studies were approved by the Institutional Animal Care and Use Committee, University of Massachusetts, Amherst (protocol # 2015-0042). Embryo culture was performed as described (3). CD-1 animals (Charles River) were used to generate embryos. The small molecules used include 5 μM Dorsomorphin (Tocris), 12.5 μM DMH1 (Calbiochem), and 40 μM SU5402 (Tocris). DMH1 is capable of inhibiting signaling from ALK2 and ALK3 (13) whereas Dorsomorphin can inhibit signaling from ALK2, ALK3 and ALK6 (14). Each drug was dissolved in DMSO and control embryos of similar somite stages were provided with an equal volume of DMSO.
Histology and immunohistochemistry
Embryos were fixed overnight in 4% paraformaldehyde, dehydrated, cleared in xylene, embedded in paraffin (Paraplast Plus), serially sectioned (7.5 μm) and all subsequent analysis performed after dewaxing and rehydration. Immunofluorescence protocols and antibodies are described in the supplementary information. For analysis, sagittally sectioned liver buds were divided into anterior (liver bud bounded by the SV) and posterior (liver bud surrounded by the STM). Middle sections were all those that lack a “dorsal” component of the liver while lateral sections were all those remaining.
Section in situ hybridization
Section in situ hybridization was performed as described (15) (and supplementary information) using antisense probes for Afp (16) and Sox17 (IMAGE clone: 1529001).
Electroporation
7–9S embryos were used for electroporation as outlined (17), except as below. The electroporation chamber was produced with 0.007” thick platinum electrodes placed 0.5 cm apart and the chamber created using RTV615 silicone rubber (Momentive) cubes affixed to a glass slide with Epoxy (MG Chemicals). Embryos were soaked in 3–4 μg/μl plasmid for 5 min before transfer to the HBSS filled chamber. Embryos were oriented so that the hepatic endoderm is facing the anode. After electroporation, embryos were allowed to recover in equilibrated culture medium (75% rat serum, 25% DMEM) in a 37°C, 5% O2 and 5% CO2 incubator for 2 h and then transferred to rolling culture for ~30 h.
DiI-labeling
Embryos were dissected at 5–8S and individual embryos labeled with DiI (CM-DiI, Molecular Probes) as described (3). Bright field and fluorescent images were produced using an ANDOR Zyla camera and NIS Elements BR software to record the position of the labeled cells and cultured as documented above.
Data Analysis
All statistical analysis (p value generation) on samples was performed using the two-tailed student’s t-test unless otherwise noted.
Results
Inhibition of BMP signaling leads to loss of HNF4α in the posterior liver bud
Because of the stringent early developmental requirements for components of the BMP pathway and the genetic redundancy within the pathway, we used small molecule inhibitors in the context of the whole embryo to examine the role of BMP signaling in liver development. This method allows for strict temporal control, limits any secondary defects and promotes unbiased downregulation of the canonical signaling pathway. Embryos were cultured from pre-induction stages (4–7S) through liver bud stages (E9.25 or 19–21S; E9.75 or 26–27S) in either the BMP inhibitor DMH1 (13) or an equal volume of the vehicle (DMSO). The DMH1 concentration used (12.5 μM) was the maximum dose compatible with grossly normal embryonic development. To measure the efficacy of this dose, 7–8S embryos were cultured for 4, 8 or 12 h and pSMAD1/5 levels measured by Western blot. Compared with controls, DMH1 reduces pSMAD1/5 up to 80% (Figure S1A). Section immunofluorescence of treated embryos with an anti-pSMAD1/5/8 antibody demonstrates a uniform overall reduction in pSMAD1/5/8 compared with controls (Figure S1B).
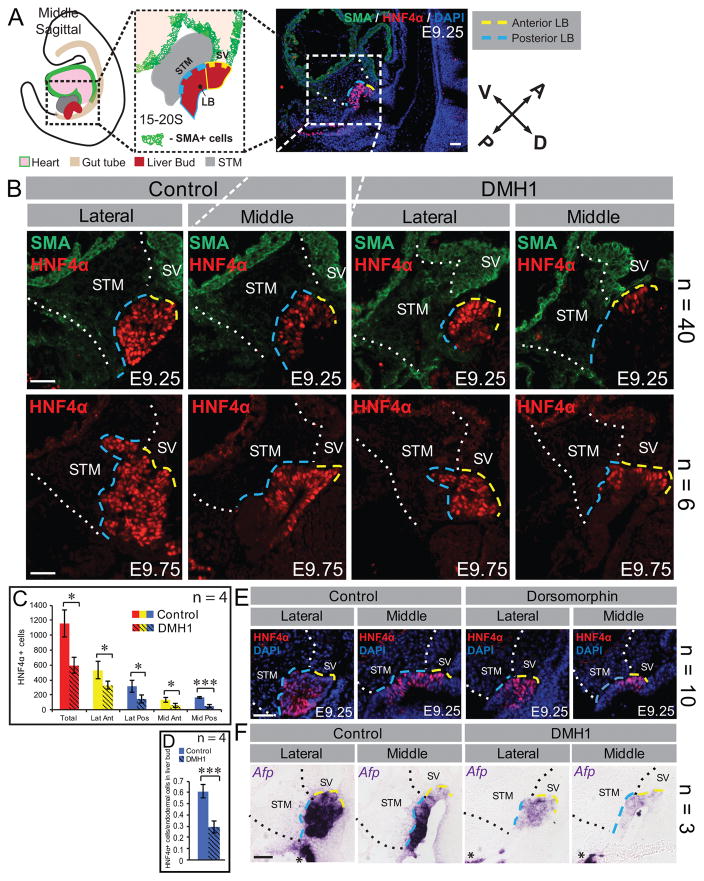
Despite looking grossly normal after culture (Figure S1C), careful histological analysis of sagittal sections revealed that DMH1 treatment alters liver bud and STM morphology (Figure S3A). The orientation of sections in relation to the embryonic axis and to the remainder of the embryo is described in more detail in Figure S2A, B and Figure 1A. Consistent with published results (6), BMP inhibition is accompanied by an overall reduction in the number of HNF4α positive (+) hepatoblasts compared with controls (Figure 1A–C). Intriguingly, HNF4α, an early and essential hepatoblast-specific transcription factor (18–20), is most dramatically reduced in the portion of the liver bud lined by the STM (Figure 1B, blue dashed line on the liver bud, LB), henceforth termed the posterior liver bud. The portion of the liver bud lined by the sinus venosus (SV) (Figure 1B, yellow dashed line on the liver bud, LB), henceforth termed the anterior liver bud, is less affected. These data are quantified in Figure 1C, D. Importantly, these results are recapitulated using the less specific BMP inhibitor, Dorsomorphin (13) (Figure 1E).
Figure 1. Loss of posterior liver bud upon BMP inhibition.
A) An illustrated sagittal view of an embryo depicting the orientation of the liver with respect to the whole embryo. Encased in the black dashed box is an illustration of sagittal section through the middle plane of the liver bud (LB) and highlights the anterior liver bud (yellow dashes), the posterior liver bud (blue dashes) and their surrounding mesoderm derivatives: the sinus venosus (SV) and the septum transversum mesenchyme (STM, gray). The SV walls and cardiac tissue (ventral to the STM) are smooth muscle actin (SMA) positive (+). A lower magnification of a sagittal section depicted in (B), but representative of most presented herein demonstrates that the anterior liver bud is bounded by the SMA+ sinus venosus (SV) while the posterior liver bud is bounded by the STM. The white dotted lines demarcate the anterior and posterior boundaries of the STM. The orientation of the anterior/posterior (A/P) and dorsal/ventral (D/V) axis of the embryo is represented by the compass. All sections are similarly oriented and illustrated unless otherwise specified. (B) Immunofluorescence with the indicated markers, performed on sagittal sections from control and DMH1 treated embryos. DMH1 treatment causes a reduction in liver bud size compared with control whether cultured to E9.25 (upper panels) or E9.75 (lower panels). In drug treated embryos, the most dramatic loss of HNF4α is in the posterior liver bud in the middle sections. (C, D) Quantification of HNF4α+ cells in the indicated liver bud regions of similarly staged control and DMH1-treated embryos reveals a significant reduction of HNF4α+ cells throughout the liver bud of the latter. The most dramatic reduction occurs in the middle posterior. Lat = lateral; Ant = anterior; Pos = posterior; Mid = Middle. (E) HNF4α immunofluorescence on sagittal sections from control and Dorsomorphin treated embryos. Like DMH1, Dorsomorphin treated embryos show a reduced liver bud with the most dramatic loss of HNF4α in the middle of the posterior liver bud. (F) In situ hybridization with an Afp antisense probe reveals a profound reduction of this marker throughout the posterior liver bud of DMH1 treated embryos. The black asterisks indicate Afp expression in the visceral endoderm of the yolk sac. The boundaries of the STM are marked with black dotted lines in this panel for clarity. The number of embryos (n) analyzed in each group is indicated on the right side of panel. * = p<0.05, *** = p<0.001. Error bars represent standard deviation. Each scale bar = 50 μM.
To further probe the abnormal distribution of hepatoblasts in DMH1 treated embryos, we examined the early non-essential liver marker Afp, which at this stage, is highly expressed in the posterior liver bud of controls (Figure 1F). BMP inhibition significantly hampers Afp in the presumptive posterior liver bud while the weaker expression in the anterior liver bud is not affected compared with controls (Figure 1F). Expression of the critical pan-endoderm pioneer factors FOXA1 and FOXA2 (Figure S3B, C) and the hepatopancreatobiliary domain marker PROX1 (21) (Figure S3D) were also examined. Other than putative liver bud size, no alterations between drug treated and control embryos are found. Together, these data suggest that BMP inhibition does not affect the pre-patterning of the hepatopancreatobiliary domain but is specifically required for appropriate hepatic emergence.
To tease apart the role for BMP signaling during hepatic induction from its role in hepatic growth, we treated progressively older embryos with DMH1 for at least 12 hours (Figure S4). We observe a consistent loss of posterior specification when the drug is administered to embryos 12S and younger (Figure S4A). When embryos are subjected to BMP inhibition at 13–15 or 16–18S a less severe posterior-restricted loss is observed (Figure S4B, C). Similar to that observed with younger embryos above, loss of BMP signals prior to 18S results in overall smaller liver buds. By 19–21S, BMP abrogation no longer restricts posterior induction nor overall growth (Figure S4D). Taken together, this data suggests that BMP signaling is vital between 4–12S for the induction of HNF4α+ hepatoblasts in the posterior liver bud and that BMP signals are simultaneously supporting hepatic maintenance or growth throughout the liver during these stages. Interestingly, these results are consistent with the role of BMP signaling on hepatic specification and growth in zebrafish (11).
BMP signaling is not required for survival or proliferation of posterior hepatoblasts
To assess whether the reduced number of HNF4α+ hepatoblasts in the liver bud was due to increased cell death or reduced cell proliferation, cleaved-CASPASE3 (CAS3) or phospho-histone H3 (pH3), respectively, were examined using immunofluorescence. DMH1 treatment does not affect apoptosis by E9.25 (Figure S5A). We surmised that perhaps the treated cells died earlier, and examined apoptosis levels after only 12 h of culture but did not observe any increased cell death in FOXA2+ cells in the nascent liver bud region (Figure S5B). Similarly, we do not observe statistically significant differences in the rate of cell proliferation of the HNF4α+ hepatoblasts in the drug treated versus the control embryos (Figure S5C, D). These results demonstrate that abrogating BMP signals affects neither hepatoblast proliferation nor cell death.
Inhibition of BMP signaling causes an anterior shift of the posterior liver bud
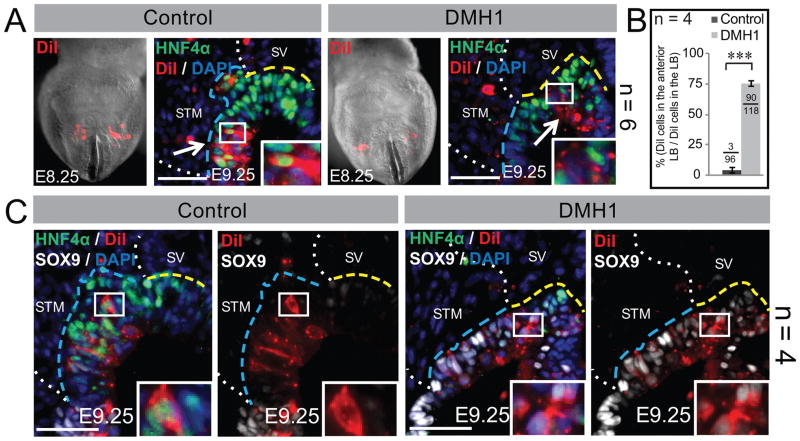
Because our results suggest that upon BMP inhibition endoderm normally fated to contribute to the hepatoblast population is present but not directed towards the hepatic lineage, we next sought to examine the fate of these cells. We previously demonstrated that the VMEL progenitors contribute mainly to the anterior liver bud while the lateral hepatic progenitors mainly contribute hepatoblasts that populate the middle and posterior liver bud (2–4). Hence, to assess the fate of the posterior liver bud, we labeled a subset of the lateral hepatic progenitors with the vital fluorescent dye, DiI, and cultured DMH1-treated and control embryos through the liver bud stage. As expected, all control embryos contain DiI-labeled HNF4α+ cells in the posterior liver bud (Figure 2A). All BMP-inhibited embryos contain DiI-labeled lateral progenitors that aberrantly localize to the anterior bud (Figure 2A). To quantify these differences, we counted the number of DiI-labeled cells in the anterior liver bud of control and BMP-inhibited embryos. After BMP inhibition, ~ 75% of the DiI-labeled lateral progenitors are localized to the anterior compared to only 3–4% in controls (Figure 2B). Furthermore, most DiI-labeled cells in the hepatic domain of drug treated embryos fail to express HNF4α but instead express the early pancreatobiliary marker, SOX9 (Figure 2C). These results support the observation that reduced BMP signaling does not result in cell death (Figure S5A, B) but rather causes a subset of lateral hepatic progenitors to be anteriorly displaced. To assess whether this anterior shift was specific to the lateral liver progenitors, we labeled the VMEL progenitors and found that DMH1 did not affect the localization of this liver progenitor population compared with controls (Figure S6). Together, these data highlight the progenitor-specific effects of BMP inhibition.
Figure 2. Anterior displacement of the putative posterior liver bud upon BMP inhibition.
A, C) Control or DMH1 treated embryos are DiI-labeled (red) and cultured. (A) Frontal views of whole embryos with DiI-labeled lateral hepatic progenitors at the onset of culture (E8.25). Section immunofluorescence after culture (E9.25) confirms that the lateral progenitors of control embryos mainly contribute to HNF4α+ hepatoblasts in the posterior liver bud. In contrast, the DiI-labeled lateral progenitors of DMH1 treated embryos are displaced towards the SV-bounded anterior liver bud and mainly do not express HNF4α. The arrows point to the bulk of the DiI-labeled cells and each inset is a higher magnification of the boxed area. (B) A quantification of the anteriorly displaced DiI-labeled cells as a percent of the total DiI-labeled cells in the whole liver bud. The fraction at the top of each bar depicts the number of the DiI-labeled cells counted in the anterior over the total number of DiI-labeled cells in the liver bud. (C) Section immunofluorescence of the DiI-labeled controls after culture, reveals that the DiI-labeled cells are not only restricted to the posterior liver bud but that these cells mainly express HNF4α and restrict the pancreatobiliary marker, SOX9. After DMH1 treatment, the anteriorly displaced DiI-labeled cells do not express HNF4α, but instead, aberrantly express SOX9. The insets reveal a higher magnification of the boxed area. The number of embryos (n) used for each experiment is listed to the right of each panel or within the graph. Each scale bar = 50 μM. Annotations are as in Figure 1.
BMP patterns the hepato-pancreatobiliary boundary
Numerous studies in zebrafish and in mouse support the hypothesis that the liver and pancreas arise from a common precursor pool that may be directed to one lineage or the other based on extrinsic cues (6, 22–24). Similarly, the data presented above (Figure 2) suggests the adoption of a pancreatobiliary fate by the presumptive posterior hepatoblasts upon BMP inhibition. To determine if reduced hepatogenic signals initiate a reciprocal increase in pancreatic fate, we examined markers of pancreas and gall bladder after BMP inhibition.
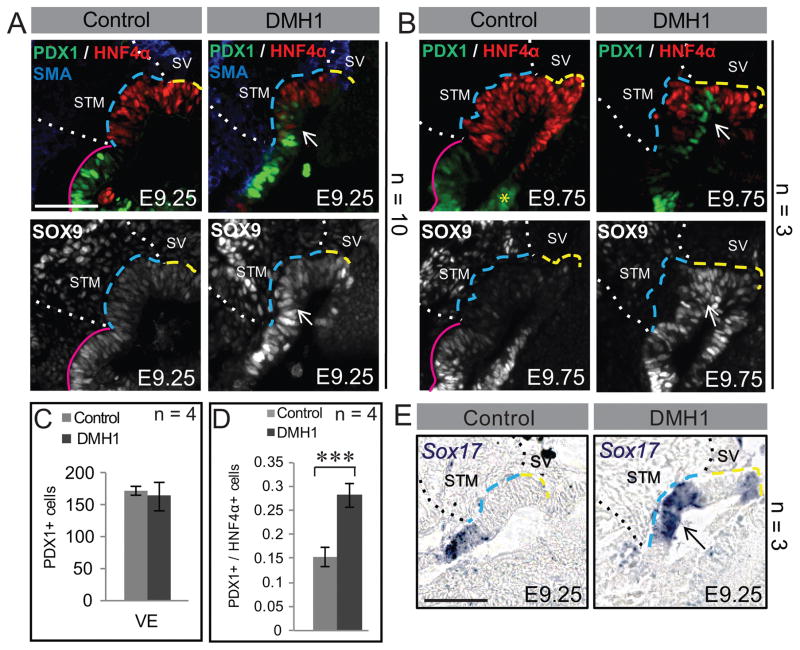
The early “ventral pancreas bud”, discernable as early as E9.0, contains both ventral pancreas and gall bladder progenitors (24). By E9.5 cells within the more aptly named pancreatobiliary bud begin sorting into an anteriorly situated PDX1-low/SOX17-high gall bladder primordium and the posteriorly located PDX1-high/SOX17-low ventral pancreas bud. By E10.5 sorting is complete and SOX17 now marks the gall bladder while PDX1 marks the ventral pancreas (25). Similar to SOX17, SOX9 is expressed throughout the early pancreatobiliary domain, but later becomes restricted to the ventral (and dorsal) pancreas bud (26). In E9.25 control embryos, PDX1 is expressed posterior to the hepatic domain, within the pancreatobiliary domain (Figure 3A, pink line). After drug treatment, PDX1+ cells are located more anteriorly than controls, and are found in the posterior liver bud domain (Figure 3A). Extending culture for another 12 h yields an increased number of PDX1+ cells in the posterior hepatic domain of treated embryos (Figure 3B). These results are complimented by the expansion of SOX9 throughout the liver bud of drug treated embryos over developmental time. In controls, SOX9 is most highly expressed in PDX1+ portion of the pancreatobiliary domain (Figure 3A, B).
Figure 3. Loss of hepato-pancreatobiliary boundary upon BMP inhibition.
A, B) Immunofluorescence of sagittally sectioned control and DMH1 treated embryos. In controls, the pancreatic markers SOX9 and PDX1 are restricted mostly to the ventral and dorsal pancreas buds at E9.25 (A) and E9.75 (B). DMH1 treated embryos fail to restrict these markers, displaying ectopic PDX1 and SOX9 in the hepatic domain (arrows). (C, D) Quantification of PDX1+ cells in the ventral endoderm (VE) at E9.25 indicates no changes in the absolute number of cells between control and drug treated embryos, resulting in a significantly increased ratio of PDX1+/HNF4α+ cells after DMH1 treatment (D). (E) Compared with controls, the gall bladder marker, Sox17, is ectopically expressed in the hepatic domain of DMH1 treated embryos. The yellow asterisk marks the visible dorsal pancreas bud. The number of embryos (n) used in each experiment is listed within or to the right of each panel. *** = p<0.001. Error bars represent standard deviation. Each scale bar = 50 μM. Annotations are as in Figure 1. VE = Ventral Endoderm.
Unlike the liver, the ultimate size of the pancreas is governed by the number of pancreatic progenitors (27). Thus, we opted to count the number of PDX1+ cells in the ventral foregut of drug treated and control embryos and we find that despite the spatial expansion, DMH1 treated embryos have no significant increase in the number of PDX1+ cells (Figure 3C). However, due to the decrease in hepatoblasts, the ratio of pancreatic/hepatic cells is significantly altered (Figure 3D). Finally, the early pancreatobiliary marker Sox17, displays aberrant expression in the posterior liver bud and in a portion of the anterior liver bud after DMH1 treatment (Figure 3E). Together, these data demonstrate that after BMP inhibition, the pancreatobiliary domain expands anteriorly into the region usually occupied by the posterior liver bud, suggesting that a role of BMP is to delineate the hepato-pancreatobiliary boundary.
SOX9 negatively regulates HNF4α
The anterior expansion of SOX9 after BMP inhibition suggests that a function of BMP is to exclude SOX9 from the hepatic domain. SOX9 negatively regulates the intestinal transcription factor CDX2 at the pancreato-duodenal boundary, restricting expansion of the duodenum into the pancreatic domain (28). Hence, we hypothesized that SOX9 may perform a similar function at the hepato-pancreatobiliary boundary by repressing HNF4α.
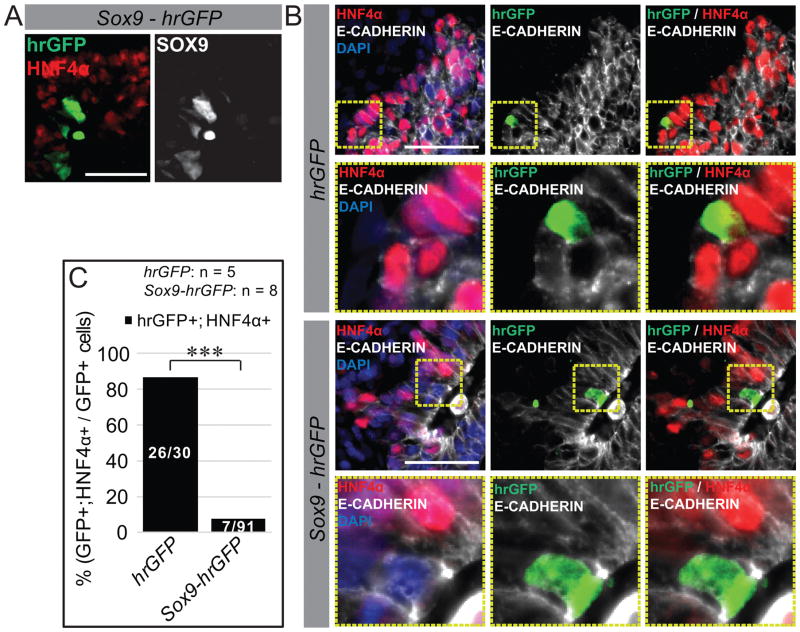
To test this hypothesis, we sought to overexpress SOX9 in the hepatic domain of wild type embryos. A SOX9 expression plasmid co-expressing GFP was constructed (Sox9-hrGFP; Figure S7A–C). Sox9-hrGFP or hrGFP was electroporated into the foregut of E8.5 embryos and cultured through E9.5. Detection of Sox9-hrGFP expressing cells was verified using the SOX9 and GFP antibodies (Figure 4A). After electroporation with hrGFP (control), approximately 85% of the GFP+ cells in the thickened hepatic domain also express HNF4α (Figure 4B, C). However, after electroporation with Sox9-hrGFP, only ~10% of the GFP+ cells in the thickened hepatic endoderm co-express HNF4α (Figure 4B, C). These data provide support for the hypothesis that SOX9 is sufficient to repress HNF4α in the hepatic domain.
Figure 4. SOX9 negatively regulates HNF4α in the liver bud.
A, B) Sox9-hrGFP or hrGFP plasmid was electroporated into pre-hepatic endoderm of 7–9S embryos and the embryos cultured to 24–25S. Immunofluorescence of sectioned Sox9-hrGFP (A, B) or hrGFP (B) embryos. (A) Anti-SOX9 robustly detects SOX9 from the Sox9-hrGFP plasmid, validating the expression of a properly folded SOX9. (B) While GFP and HNF4α often co-localize in the hepatic domain of controls, Sox9-hrGFP expressing cells mainly exclude HNF4α. The yellow boxes indicate the view magnified in the panel below. (C) Quantification (percentages) of hrGFP and Sox9-hrGFP electroporated hepatic domain cells that express both GFP and HNF4α (fractions within the bars of the graph depict actual cell counts) reveals that Sox9-hrGFP expression dramatically reduces the frequency with which cells express HNF4α compared to the control construct. The number of embryos (n) used in each experiment is listed. *** = p<0.001, p value was generated using Fisher’s exact test. Each scale bar = 50 μM.
To test whether SOX9 is sufficient to direct endoderm in the hepatic region to a pancreatic or biliary fate, we examined the expression of PDX1, an essential pancreatic gene, and SOX17, an essential early bile duct marker. Ectopic SOX9 failed to promote either PDX1 or SOX17 expression in the hepatic domain (data not shown), suggesting that SOX9 alone is not sufficient to promote pancreatic or biliary fate in these cells. Together, these results are similar to those of Shih et al (27) demonstrating that ectopic expression of SOX9 suppressed CDX2 in the duodenum but did not promote a pancreatic bud formation.
BMP signaling is not directly required for hepatoblast specification
To address which components of the BMP pathway are involved in hepatic induction, we examined the expression of BMP ligands in and around the prehepatic/hepatic domain. Based on their ability to induce hepatic gene expression in vitro and because they are expressed in a manner consistent with a role in hepatic induction, we examined the expression of Bmp2, Bmp4 and Bmp7 using section in situ hybridization (6, 29, 30). The BMP Type I receptors Alk3 (Bmpr1a), Alk2 (Acvr1) as well as the Type II receptor, Bmpr2, were also examined since null mutants of each display embryonic lethality and may play a role in hepatic induction (31–33). The embryonic stages examined included, 3–6S, representing pre-hepatic endoderm, 8–9S, coinciding with the onset of hepatic gene expression/induction, 11–12S, representing the onset of a morphologically thickened hepatic endoderm, and 14–15S, when the nascent liver bud can be readily identified. Bmp2 is expressed in the mesenchyme surrounding the anterior hepatic endoderm at the 8–9S and 11–12S, but absent from the hepatic endoderm at these stages (Figure S8A). It is surprising to note that Bmp4 is neither expressed in the mesoderm-derivatives surrounding the hepatic endoderm nor in the endoderm itself either before (3–6S) or at the liver induction stage (8–9S; Figure S8B). Bmp2 and Bmp4 are most strongly expressed in the anterior STM between 14–15S. Bmp7, on the other hand, is strongly expressed in the hepatic endoderm and proximal mesenchyme during induction stages (8–9S, Figure S8C). However, it is interesting to note that Bmp7 mutants do not have a liver phenotype (34, 35). Alk3 is expressed strongly in the liver bud at E8.75 (14–15S), however, weak expression is observed at the liver induction stages (8–9S, Figure S8D). Alk2 is strongly expressed in the hepatic endoderm at induction (8–9S) but is weakly expressed in the nascent liver bud (11–12S, 14–15S; Figure S8E). Bmpr2 is weakly expressed in the hepatic endoderm at 11–12S and 14–15S (Figure S8F).
Because the above expression analysis did not implicate specific pathway components in the direct transmission of BMP signals to the (pre-)hepatic endoderm immediately before or during hepatic induction, we next examined pSMAD1/5/8 levels. Interestingly, despite the widespread embryonic expression of Smad-1, 5 and 8 (36) we observe only weak levels of pSMAD1/5/8 in the hepatic endoderm at 4–6S and 8–9S stages using immunofluorescence (Figure 5A). However, a progressive increase in pSMAD1/5/8 intensity in the specified liver bud is observed between 11 and 14S (Figure 5A). Contrary to our expectations, this data suggests that BMP signals play an indirect role in the endoderm to liver bud transition (induction) while supporting a direct role for BMP in post-inductive liver bud growth.
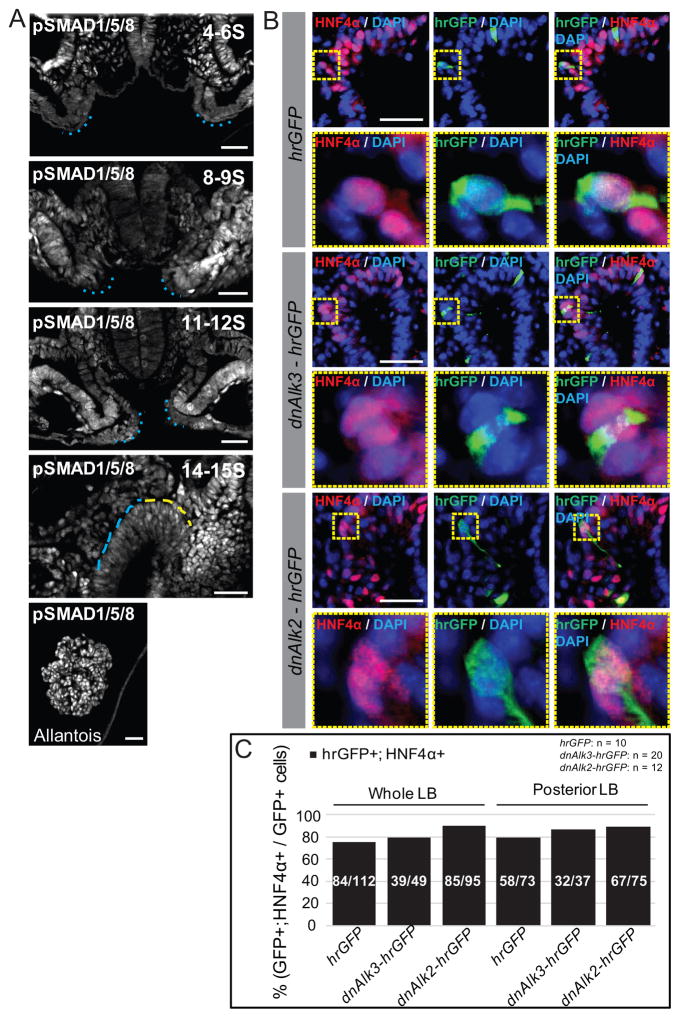
Figure 5. BMP signals are not required cell autonomously by the pre-hepatic endoderm for induction.
A) To assess which cells are actively responding to BMP signals immediately prior to and during liver bud induction and growth, pSMAD1/5/8 expression was examined in 4–6, 8–9, 11–12 and 14–15S embryos using immunofluorescence. Transverse sections through the lateral hepatic progenitors/posterior liver bud were examined at the pre-turning stages (4–6, 8–9, 11–12S), while the 14–15S embryos were analyzed in sagittal sections. No pSMAD1/5/8 activity is observed in the lateral hepatic progenitors (outlined with blue dotted lines) prior to or during specification (4–6 and 8–9S). Nearing the bud stage (11–12S and 14–15S), pSMAD1/5/8 expression gradually increases in hepatoblasts, however, the fluorescence intensity is considerably less compared to that of the surrounding mesoderm. At 14–15S, the blue dashed lines outline the posterior liver bud and the yellow dashed lines outline the anterior liver bud. The allantois is provided as a positive control for active BMP signaling. (B) To directly assess the cell-autonomous requirement for BMP signals during hepatic induction, dnAlk3-hrGFP, dnAlk2-hrGFP or hrGFP plasmid was electroporated into pre-hepatic endoderm of 8–10S embryos. Embryos were cultured through 24–25S and immunofluorescence for GFP and HNF4α examined. The lower panels correspond to a high magnification view of the dashed boxes in the top image highlighting single electroporated cells. (C) Quantification of the percentage of GFP+ cells in the hepatic domain that also express HNF4α (fractions within the bars of the graph depict actual cell counts). No significant differences are noted between the control and dnAlk3 or dnAlk2 expressing cells (p values were calculated using Fisher’s exact test). The number of embryos (n) used in each experiment is listed. Each scale bar = 50 μM. LB = Liver bud.
To further assess the requirement for BMP signals in hepatoblast induction, the pre-hepatic endoderm was targeted by electroporation with either a dominant-negative (dn) Alk3-hrGFP (37), a dnAlk2-hrGFP (38) or an hrGFP plasmid and the embryos cultured through E9.5 (dnAlk3-hrGFP and dnAlk2-hrGFP validations, Figure S9A–E). If BMP signals are directly required for hepatic specification, then electroporation of the DN constructs into pre-hepatic endoderm would preclude induction of HNF4α. Instead, similar to the hrGFP controls, approximately 80% of dnAlk3-hrGFP and approximately 90% of dnAlk2-hrGFP electroporated cells (GFP+) in the presumptive liver bud are also HNF4α+ (Figure 5B, C). These results further support the hypothesis that BMP signals are not directly required in the naïve murine endoderm for hepatoblast induction.
Inhibition of both BMP and FGF signaling leads to a dramatic loss of hepatic induction
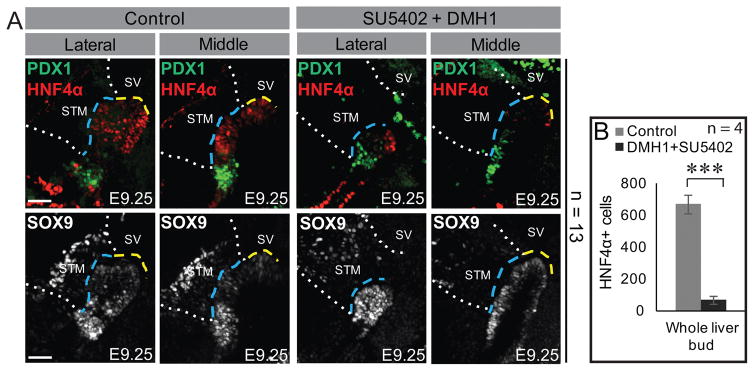
We previously demonstrated that small-molecule mediated inhibition of FGF signaling leads to a selective loss of HNF4α in the anterior but not posterior liver bud of cultured embryos (16). While E9.25 controls displayed pronouncedly thickened liver buds (39), loss of FGF signaling produced increased cell death throughout the bud and an abnormal thinning and loss of HNF4α in the anterior liver bud (16). Indeed, after simultaneous exposure to the BMP and FGF inhibitors, DMH1 and SU5402 respectively, HNF4α expression is almost completely lost, much of presumptive liver bud is abnormally thin and SOX9 is ectopically expressed throughout the hepatic domain (Figure 6A, B). These results support the hypothesis that BMP and FGF signaling act on complementary subsets of pre-hepatic endoderm. This novel view of hepatic induction is summarized in Figure 7. Finally, the small anterior population of HNF4α+ cells that remains after dual drug treatment (Figure 6A) is reproducible and may indicate the involvement of a third signaling pathway.
Figure 6. Inhibition of BMP and FGF signaling leads to a dramatic loss of liver bud.
A) Immunofluorescence of sagittally sectioned liver buds (lateral and middle sections) from control embryos and those treated with both the BMP inhibitor, DMH1, and the FGF inhibitor, SU5402. In E9.25 control embryos, HNF4α is expressed in much of the liver bud while SOX9 is most robustly expressed in the putative gall bladder and PDX1+ ventral pancreas bud. DMH1 and SU5402 treatment results in a drastic reduction of HNF4α+ cells and an increase of ectopic SOX9 throughout the hepatic domain. (B) Quantification of the number of HNF4α+ cells demonstrates a significant reduction of hepatoblasts after treatment with both inhibitors compared with controls. The number of embryos (n) used for each analysis is indicated. *** = p<0.001. Error bars represent standard deviation. Each scale bar = 50 μM. Annotations are as in Figure 1.
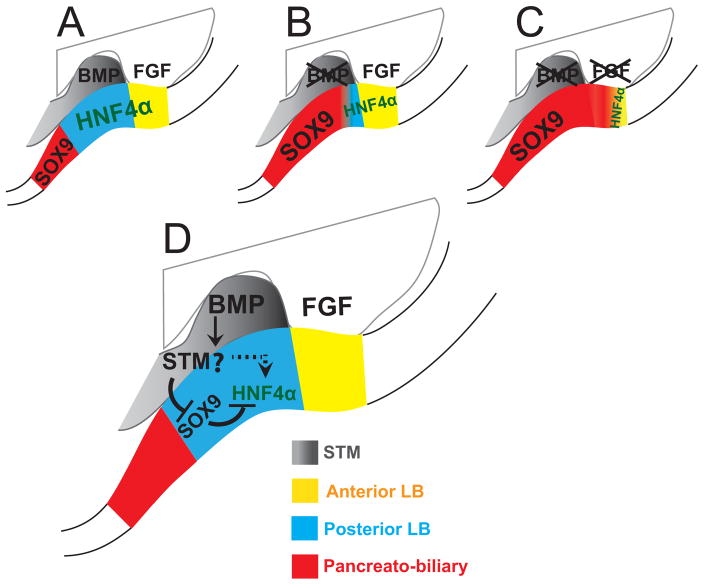
Figure 7. Working model for the role of BMP and FGF signals in hepatic induction.
A) Under the influence of normal BMP and FGF signaling the posterior foregut endoderm is divided into discrete domains: the anterior liver bud (yellow), in contact with the SV, the posterior liver bud (blue), in contact with the STM and the pancreatobiliary domain (red) which lies immediately posterior to the liver bud and mostly outside the STM. (B) Upon loss of BMP signals the hepato-pancreatobiliary boundary becomes diffuse; the posterior liver bud is lost and SOX9 is present in the posterior hepatic domain. (C) Upon loss of both BMP and FGF signaling almost all of the HNF4α+ liver bud is lost and ectopic SOX9 is observed throughout the hepatic domain. (D) Signaling events likely occurring in the hepatic domain and surrounding tissues. BMP promotes HNF4α in the posterior liver bud through indirect mechanism/s (possibly through the STM; the question mark indicates query). At least one indirect function of BMP is to repress SOX9 in the hepatic domain where SOX9 can repress HNF4α. Other unknown mechanisms may exist depicted by the dashed arrow. Annotations are as in Figure 1.
Discussion
The most persuasive studies implicating a role for BMP signals in murine hepatic induction include explant experiments that demonstrate a requirement for BMP in initiating the hepatic markers Albumin and Afp (6, 28). Herein, we expand on these studies and investigate the role BMP plays in hepatic induction in the context of the whole embryo. Given the radical tissue remodeling and distinct migrations that occur in each tissue layer during early post-gastrulation (40–42), including the pre-hepatic endoderm and adjacent mesoderm (JRA and KDT; unpublished data), static explant studies may yield results that differ from those in the dynamic environment of the developing embryo. Indeed, a global reduction of BMP signaling in the embryo during the period of hepatic induction results in an overall reduction in liver bud size that is most severe in the posterior liver bud. In agreement with the conclusions made by the explant experiments, our data supports a role for BMP signaling in liver specification, however, when performed in the context of an embryo an important role for BMP signals in defining the hepato-pancreatobiliary boundary is also apparent (Figure 7).
Inhibition of the BMP pathway profoundly disturbs the posterior liver bud. There are at least two possibilities to explain these novel results. The first is that the hepatic endoderm is differently induced along the anterior/posterior axis. Posterior hepatoblast induction requires BMP signals while anterior induction requires FGF signals (16). In agreement with this idea we demonstrate that the anterior and posterior hepatic endoderm is adjacent to distinct and potentially inductive, mesoderm-derivatives. Furthermore, simultaneous inhibition of the signals required for anterior and posterior induction result in a profound overall loss of hepatoblasts (Figure 6). An alternative possibility is that BMP is uniformly required for hepatic induction but DMH1 fails to inhibit anterior signals as efficiently as the posterior signals because BMP ligands are distributed in an anterior to posterior gradient. In support of this hypothesis, Bmp2 and 4 are expressed more highly in the anterior STM (Figure S8). To further examine this possibility, we cultured embryos in different concentrations of DMH1. If the gradient hypothesis were correct, it predicts that the boundary of HNF4α+ cells in the posterior liver bud would shift with the dose. However, at lower doses of DMH1 (6 μM), no effects on the liver bud are observed, and at a higher dose (24 μM), where there are more obvious changes in overall embryo morphology, a similar hepatic phenotype is found (data not shown). Thus, taken together, our data favors the idea that BMP signals above a particular threshold are required to induce the posterior liver bud. Whether this heterogeneity and the ability to respond to signaling pathways is intrinsic or guided by the availability of extrinsic factors is a topic for future investigations.
The discovery of distinct inductive requirements for subsets of hepatoblasts during normal development has implications for protocols designed to differentiate stem cells into hepatocytes. Generally, such protocols utilize Transforming Growth Factor-β members such as Activin-A and BMP4 to produce definitive endoderm from embryonic stem cells, followed by a cocktail of FGFs and BMPs to produce hepatoblast-like cells and then by Hepatocyte Growth Factor to produce hepatocyte-like cells (43–45). Thus far, only hepatocyte-like cells have been produced, indicating that the current regime requires refinement. Our data suggests that up to 3 populations of induced hepatoblasts are present in the liver bud – one that responds to FGF signaling, a second that requires factors downstream of BMP signaling and potentially a third, represented by those induced upon BMP and FGF inhibition, that responds to a yet unknown signaling pathway. A 2010 study used a cocktail of FGF10, retinoic acid and an inhibitor of Activin/Nodal receptor to obtain hepatic progenitors that were able to mature into hepatocytes (46), indicating retinoic acid might be the missing third signal that induces the small population of HNF4α+ cells that remain after DMH1 and SU5402 treatment.
BMP signaling is indirectly required for hepatoblast induction
Surprisingly, our data suggests that BMP signals indirectly support hepatoblast specification. Although, a requirement for active BMP signals by the pre-hepatic endoderm has been assumed, little direct evidence supports this hypothesis. In murine explants, the BMP inhibitor, Noggin, reduces expression of albumin and Afp (6). Because these explants contain both endoderm and mesoderm, the tissue directly responding to BMP is unclear. Furthermore, although pSMAD1/5/8 immunofluorescence is detected in the ventral midline of 3–9S embryos in whole mount (29), our section immunofluorescence indicates that pSMAD1/5/8 activity is most pronounced in the mesoderm adjacent to the VMEL and lateral liver progenitors (Figure 5A, Figure S1B, and data not shown). Furthermore, our in situ data suggests that the adjacent mesoderm rather than the hepatic endoderm is equipped to respond to BMP signals prior to and during hepatic induction (3–10S). Finally, we demonstrate that ectopic expression of dnAlk2 or dnAlk3 construct does not inhibit induction of HNF4α in the posterior liver bud (Figure 5). Instead, we propose that the STM or the endothelial cells associated with the hepatic endoderm are the tissues that respond to BMP signals. Indeed, endothelial cells not only support expansion of the liver bud (47), but also play a role in hepatic induction from mouse embryonic stem cells (48). These data suggest that an STM or endothelial derived secondary signal may be necessary for proper specification and maintenance of liver developmental program.
We propose that one way that BMP regulates liver specification is by indirectly suppressing SOX9 in the posterior liver bud. Furthermore, we suggest that SOX9 expression in the liver bud precludes activation of liver-specific genes such as HNF4α. In support of this hypothesis, BMP inhibition during hepatic induction stages produces ectopic SOX9 expression (Figure 3A, B) and we demonstrate that ectopic SOX9 alone is sufficient to prevent HNF4α expression throughout the liver bud, although its mechanism remains unclear (Figure 4). It is interesting to note that some SOX9+ cells in the anterior of the liver bud in both control and DMH1 treated embryos are also HNF4α+ (Figure 2B). In these cells, SOX9 expression is weaker compared to surrounding SOX9+ cells, suggesting that any co-expressing cell may be in transition and in the process of turning off SOX9 or that low levels of SOX9 is not necessarily sufficient to suppress HNF4α in the early liver bud.
In conclusion, we show that in the context of the developing embryo, BMP signals are required for appropriately inducing the liver bud and that these requirements are complimentary to those of FGF signals. Furthermore, we highlight a requirement for BMP signals in establishing the posterior hepatic boundary. Together, these experiments further underscore the importance of assessing complex embryonic processes in their native environment.
Supplementary Material
Acknowledgments
Financial Support: The work was supported by the R01DK087753 grant to KDT.
We thank Drs. Peter Chien and Kamal Joshi for help with the western blots and Kamal Joshi and members of the Tremblay and Mager labs for their insights and support. We would also like to thank the Alfandari, Peyton, Jerry and Farkas labs from UMASS, Amherst for supplying cells or cDNA.
List of Abbreviations
- FGF
Fibroblast Growth Factor
- BMP
Bone Morphogenetic Protein
- STM
septum transversum mesenchyme
- HNF4α
Hepatocyte Nuclear Factor 4-α
- SV
sinus venosus
- SOX17
Sex-Determining Region Y-Box 17
- SOX9
Sex-Determining Region Y-Box 9
- PDX1
Pancreatic And Duodenal Homeobox 1
- S
somite
- VMEL
Ventral-Medial of the Endoderm Lip
- E
embryonic day
- Afp
α-fetoprotein
- Alk8
Activin Receptor-like Kinase 8
- CD-1
Charles River
- DMSO
Dimethyl Sulfoxide
- DMH1
Dorsomorphin Homolog 1,4-[6-[4-(1-Methylethoxy) phenyl]pyrazolo[1,5-a]pyrimidin-3-yl]-quinoline
- H&E
Hematoxylin and Eosin
- PBT
Phosphate buffered saline with Tween 20
- DAPI
4′, 6-diaminido-2-phenylindole dihydrochloride
- dn/DN
dominant negative
- IRES-hrGFP2
Internal Ribosomal Entry Site-humanized recombinant Green Fluorescent protein 2
- HBSS
Hanks’ Buffered Salt Solution
- FOXA2
Forkhead Box A2
- FOXA1
Forkhead Box A1
- PROX1
Prospero Homeobox protein 1
- pH3
phospho histone H3
- CAS3
cleaved CASPASE 3
- CDX2
Caudal Type Homeobox 2
Footnotes
Supplementary data include supplemental figures and detailed experimental procedures.
References
Author names in bold designate shared co-first authorship.
- 1.Tremblay KD. Inducing the liver: understanding the signals that promote murine liver budding. J Cell Physiol. 2011;226:1727–1731. doi: 10.1002/jcp.22409. [DOI] [PubMed] [Google Scholar]
- 2.Tremblay KD, Zaret KS. Distinct populations of endoderm cells converge to generate the embryonic liver bud and ventral foregut tissues. Dev Biol. 2005;280:87–99. doi: 10.1016/j.ydbio.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Angelo JR, Guerrero-Zayas MI, Tremblay KD. A fate map of the murine pancreas buds reveals a multipotent ventral foregut organ progenitor. PLoS One. 2012;7:e40707. doi: 10.1371/journal.pone.0040707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angelo JR, Tremblay KD. Laser-mediated cell ablation during post-implantation mouse development. Dev Dyn. 2013;242:1202–1209. doi: 10.1002/dvdy.24017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung J, Zheng M, Goldfarb M, Zaret KS. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science. 1999;284:1998–2003. doi: 10.1126/science.284.5422.1998. [DOI] [PubMed] [Google Scholar]
- 6.Rossi JM, Dunn NR, Hogan BL, Zaret KS. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev. 2001;15:1998–2009. doi: 10.1101/gad.904601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang RN, Green J, Wang Z, Deng Y, Qiao M, Peabody M, Zhang Q, et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis. 2014;1:87–105. doi: 10.1016/j.gendis.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katagiri T, Watabe T. Bone Morphogenetic Proteins. Cold Spring Harb Perspect Biol. 2016:8. doi: 10.1101/cshperspect.a021899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 10.van Wijk B, Moorman AF, van den Hoff MJ. Role of bone morphogenetic proteins in cardiac differentiation. Cardiovasc Res. 2007;74:244–255. doi: 10.1016/j.cardiores.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 11.Shin D, Shin CH, Tucker J, Ober EA, Rentzsch F, Poss KD, Hammerschmidt M, et al. Bmp and Fgf signaling are essential for liver specification in zebrafish. Development. 2007;134:2041–2050. doi: 10.1242/dev.000281. [DOI] [PubMed] [Google Scholar]
- 12.Zhang W, Yatskievych TA, Baker RK, Antin PB. Regulation of Hex gene expression and initial stages of avian hepatogenesis by Bmp and Fgf signaling. Dev Biol. 2004;268:312–326. doi: 10.1016/j.ydbio.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 13.Hao J, Ho JN, Lewis JA, Karim KA, Daniels RN, Gentry PR, Hopkins CR, et al. In vivo structure-activity relationship study of dorsomorphin analogues identifies selective VEGF and BMP inhibitors. ACS Chem Biol. 2010;5:245–253. doi: 10.1021/cb9002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moorman AF, Houweling AC, de Boer PA, Christoffels VM. Sensitive nonradioactive detection of mRNA in tissue sections: novel application of the whole-mount in situ hybridization protocol. J Histochem Cytochem. 2001;49:1–8. doi: 10.1177/002215540104900101. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Rhee S, Palaria A, Tremblay KD. FGF signaling is required for anterior but not posterior specification of the murine liver bud. Dev Dyn. 2015;244:431–443. doi: 10.1002/dvdy.24215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierreux CE, Poll AV, Jacquemin P, Lemaigre FP, Rousseau GG. Gene transfer into mouse prepancreatic endoderm by whole embryo electroporation. JOP. 2005;6:128–135. [PubMed] [Google Scholar]
- 18.Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, et al. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- 19.Li JX, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4 alpha. Genes & Development. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- 20.DeLaForest A, Nagaoka M, Si-Tayeb K, Noto FK, Konopka G, Battle MA, Duncan SA. HNF4A is essential for specification of hepatic progenitors from human pluripotent stem cells. Development. 2011;138:4143–4153. doi: 10.1242/dev.062547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burke Z, Oliver G. Prox1 is an early specific marker for the developing liver and pancreas in the mammalian foregut endoderm. Mechanisms of Development. 2002;118:147–155. doi: 10.1016/s0925-4773(02)00240-x. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Seguel E, Mah N, Naumann H, Pongrac IM, Cerda-Esteban N, Fontaine JF, Wang Y, et al. Mutually exclusive signaling signatures define the hepatic and pancreatic progenitor cell lineage divergence. Genes Dev. 2013;27:1932–1946. doi: 10.1101/gad.220244.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nissim S, Sherwood RI, Wucherpfennig J, Saunders D, Harris JM, Esain V, Carroll KJ, et al. Prostaglandin E2 regulates liver versus pancreas cell-fate decisions and endodermal outgrowth. Dev Cell. 2014;28:423–437. doi: 10.1016/j.devcel.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung WS, Shin CH, Stainier DY. Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Dev Cell. 2008;15:738–748. doi: 10.1016/j.devcel.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spence JR, Lange AW, Lin SC, Kaestner KH, Lowy AM, Kim I, Whitsett JA, et al. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev Cell. 2009;17:62–74. doi: 10.1016/j.devcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seymour PA, Freude KK, Tran MN, Mayes EE, Jensen J, Kist R, Scherer G, et al. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci U S A. 2007;104:1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanger BZ, Tanaka AJ, Melton DA. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature. 2007;445:886–891. doi: 10.1038/nature05537. [DOI] [PubMed] [Google Scholar]
- 28.Shih HP, Seymour PA, Patel NA, Xie R, Wang A, Liu PP, Yeo GW, et al. A Gene Regulatory Network Cooperatively Controlled by Pdx1 and Sox9 Governs Lineage Allocation of Foregut Progenitor Cells. Cell Rep. 2015;13:326–336. doi: 10.1016/j.celrep.2015.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wandzioch E, Zaret KS. Dynamic signaling network for the specification of embryonic pancreas and liver progenitors. Science. 2009;324:1707–1710. doi: 10.1126/science.1174497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danesh SM, Villasenor A, Chong D, Soukup C, Cleaver O. BMP and BMP receptor expression during murine organogenesis. Gene Expr Patterns. 2009;9:255–265. doi: 10.1016/j.gep.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishina Y, Suzuki A, Ueno N, Behringer RR. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 1995;9:3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- 32.Beppu H, Kawabata M, Hamamoto T, Chytil A, Minowa O, Noda T, Miyazono K. BMP type II receptor is required for gastrulation and early development of mouse embryos. Dev Biol. 2000;221:249–258. doi: 10.1006/dbio.2000.9670. [DOI] [PubMed] [Google Scholar]
- 33.Gu Z, Reynolds EM, Song J, Lei H, Feijen A, Yu L, He W, et al. The type I serine/threonine kinase receptor ActRIA (ALK2) is required for gastrulation of the mouse embryo. Development. 1999;126:2551–2561. doi: 10.1242/dev.126.11.2551. [DOI] [PubMed] [Google Scholar]
- 34.Dudley AT, Lyons KM, Robertson EJ. A requirement for bone morphogenetic protein-7 during development of the mammalian kidney and eye. Genes Dev. 1995;9:2795–2807. doi: 10.1101/gad.9.22.2795. [DOI] [PubMed] [Google Scholar]
- 35.Jena N, Martin-Seisdedos C, McCue P, Croce CM. BMP7 null mutation in mice: developmental defects in skeleton, kidney, and eye. Exp Cell Res. 1997;230:28–37. doi: 10.1006/excr.1996.3411. [DOI] [PubMed] [Google Scholar]
- 36.Tremblay KD, Dunn NR, Robertson EJ. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development. 2001;128:3609–3621. doi: 10.1242/dev.128.18.3609. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki A, Thies RS, Yamaji N, Song JJ, Wozney JM, Murakami K, Ueno N. A truncated bone morphogenetic protein receptor affects dorsal-ventral patterning in the early Xenopus embryo. Proc Natl Acad Sci U S A. 1994;91:10255–10259. doi: 10.1073/pnas.91.22.10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo J, Tang M, Huang J, He BC, Gao JL, Chen L, Zuo GW, et al. TGFbeta/BMP type I receptors ALK1 and ALK2 are essential for BMP9-induced osteogenic signaling in mesenchymal stem cells. J Biol Chem. 2010;285:29588–29598. doi: 10.1074/jbc.M110.130518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bort R, Signore M, Tremblay K, Barbera JPM, Zaret KS. Hex homeobox gene controls the transition of the endoderm to a pseudostratified, cell emergent epithelium for liver bud development. Developmental Biology. 2006;290:44–56. doi: 10.1016/j.ydbio.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 40.Lawson KA, Meneses JJ, Pedersen RA. Cell Fate and Cell Lineage in the Endoderm of the Presomite Mouse Embryo, Studied with an Intracellular Tracer. Developmental Biology. 1986;115:325–339. doi: 10.1016/0012-1606(86)90253-8. [DOI] [PubMed] [Google Scholar]
- 41.Lawson KA, Pedersen RA. Cell Fate, Morphogenetic Movement and Population-Kinetics of Embryonic Endoderm at the Time of Germ Layer Formation in the Mouse. Development. 1987;101:627–652. doi: 10.1242/dev.101.3.627. [DOI] [PubMed] [Google Scholar]
- 42.Kinder SJ, Tsang TE, Quinlan GA, Hadjantonakis AK, Nagy A, Tam PPL. The orderly allocation of mesodermal cells to the extraembryonic structures and the anteroposterior axis during gastrulation of the mouse embryo. Development. 1999;126:4691–4701. doi: 10.1242/dev.126.21.4691. [DOI] [PubMed] [Google Scholar]
- 43.Mallanna SK, Duncan SA. Differentiation of hepatocytes from pluripotent stem cells. Curr Protoc Stem Cell Biol. 2013;26(Unit 1G):4. doi: 10.1002/9780470151808.sc01g04s26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauer V, Roy-Chowdhury N, Guha C, Roy-Chowdhury J. Induced pluripotent stem cells as a source of hepatocytes. Curr Pathobiol Rep. 2014;2:11–20. doi: 10.1007/s40139-013-0039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han S, Bourdon A, Hamou W, Dziedzic N, Goldman O, Gouon-Evans V. Generation of functional hepatic cells from pluripotent stem cells. J Stem Cell Res Ther. 2012;(Suppl 10):1–7. doi: 10.4172/2157-7633.S10-008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Touboul T, Hannan NR, Corbineau S, Martinez A, Martinet C, Branchereau S, Mainot S, et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–1765. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- 47.Matsumoto K, Yoshitomi H, Rossant J, Zaret KS. Liver organogenesis promoted by endothelial cells prior to vascular function. Science. 2001;294:559–563. doi: 10.1126/science.1063889. [DOI] [PubMed] [Google Scholar]
- 48.Han SY, Dziedzic N, Gadue P, Keller GM, Gouon-Evans V. An Endothelial Cell Niche Induces Hepatic Specification Through Dual Repression of Wnt and Notch Signaling. Stem Cells. 2011;29:217–228. doi: 10.1002/stem.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.