Abstract
The hippocampus has been widely studied using neuroimaging, as it plays an important role in memory and learning. However, hippocampal subfield information is difficult to capture by standard magnetic resonance imaging (MRI) techniques. To facilitate morphometric study of hippocampal subfields, ADNI introduced a high resolution (0.4mm in plane) T2-weighted turbo spin-echo sequence that requires 8 minutes. With acceleration, the protocol can be acquired in 4 minutes. We performed a comparative study of hippocampal subfield volumes using standard and accelerated protocols on a Siemens Prisma 3T MRI in an independent sample of older adults that included 10 cognitively normal controls, 9 individuals with subjective cognitive decline, 10 with mild cognitive impairment, and 6 with a clinical diagnosis of Alzheimer’s disease (AD). The Automatic Segmentation of Hippocampal Subfields (ASHS) software was used to segment 9 primary labeled regions including hippocampal subfields and neighboring cortical regions. Intraclass correlation coefficients were computed for reliability tests between 4 and 8 minute scans within and across the four groups. Pairwise group analyses were performed, covaried for age, sex and total intracranial volume, to determine whether the patterns of group differences were similar using 4 versus 8 minute scans. The 4 and 8 minute protocols, analyzed by ASHS segmentation, yielded similar volumetric estimates for hippocampal subfields as well as comparable patterns of differences between study groups. The accelerated protocol can provide reliable imaging data for investigation of hippocampal subfields in AD-related MRI studies and the decreased scan time may result in less vulnerability to motion.
Keywords: Hippocampal subfields, magnetic resonance imaging, segmentation, volumetric analysis, Alzheimer’s disease
1. INTRODUCTION
Alzheimer’s disease (AD), as the most common type of age-related dementia, is widely studied using neuroimaging approaches with special emphasis on memory critical structures. The hippocampus plays a key role in learning and memory and is a particularly vulnerable region for AD-related neurodegeneration (Greicius et al., 2004). Hippocampal measures extracted from magnetic resonance imaging (MRI) scans have been established as key biomarkers related to AD and mild cognitive impairment (MCI, a prodromal stage of AD) (Petersen et al., 1999). Thus, hippocampal volumetry and morphometry have been employed to detect the presence and progression of cognitive disorders in quantitative neuroimaging. However, due to the limited resolution of conventional MRI scans, most studies cannot clearly capture the critical hippocampal subfields as well as their neighboring cortical subregions. Of note, the hippocampal subfields and the neighboring cortical structures are not uniformly affected by AD pathology or by the normal process of aging (Adler et al., 2014). Some regions (e.g., CA1 as reported in (Apostolova et al., 2006; Apostolova et al., 2010a; Apostolova et al., 2010b)) are selectively more vulnerable, and thus they have the potential to serve as sensitive biomarkers for early stage AD diagnosis. Due to the size, complexity, heterogeneity and folding anatomy of the hippocampus, acquiring volumetric and morphometric measures of hippocampal subfields usually presents not only technical challenges in quantitative neuroimaging but also analytical challenges.
Since T1-weighted sequences at conventional magnet strength (1.5 or 3T) often lack the contrast and necessary resolution for observing sufficient anatomical details of hippocampal subfields (see Fig. 1(a,d,g)), conventional MRI studies on subcortical structures typically examine the entire hippocampus as a single structure (e.g., (Patenaude et al., 2011)). In order to overcome the resolution limitation, existing subfield studies usually employed high magnetic field strength (4T and above) or high resolution 3T MRI techniques (Huang et al., 2013; Kirov et al., 2013; La Joie et al., 2013; Mueller et al., 2007; Mueller and Weiner, 2009; Olsen et al., 2013; Pluta et al., 2012b; Van Leemput et al., 2009; Winterburn et al., 2013b; Wisse et al., 2012; Wisse et al., 2016; Yassa and Stark, 2011), where, with the higher MRI resolution, hippocampal subfield layers could be better distinguished from one another. In these studies, manual (La Joie et al., 2013; Libby et al., 2012; Malykhin et al., 2010; Mueller and Weiner, 2009; Olsen et al., 2013; Pluta et al., 2012b; Winterburn et al., 2013b; Wisse et al., 2012) or semi-automated (Hunsaker and Amaral, 2014; Merkel et al., 2015; Yushkevich et al., 2010) methods were used to segment hippocampal subfields. However, these studies need long imaging acquisition times and tedious, labor insensitive work by anatomically-trained tracers and thus, are not practical for the analysis of large-scale datasets. At this point, the major challenges remaining are (1) insufficient anatomical details provided by T1-weighted MRI scans at conventional resolution, and (2) very few validated tools available for automated segmentation of hippocampal subfields.
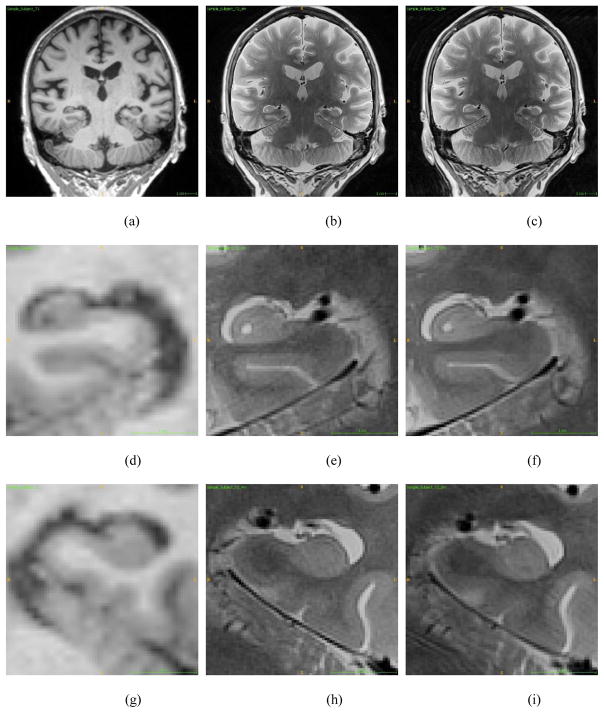
Fig. 1.
Coronal Views: (a–c) Conventional MRI, 4-min high resolution MRI, and 8-min high resolution MRI. (d–f) Left hippocampal area on conventional MRI, 4-min high resolution MRI, and 8-min high resolution MRI. (g–i) Right hippocampal area on conventional MRI, 4-min high resolution MRI, 8-min high resolution MRI.
To address the first challenge, rather than using only standard T1-weighted MRI scans (e.g., 1mm in plane resolution), this study employed multi-spectral analyses integrating conventional T1-weighted scans with high resolution T2-weighted at a widely available field strength (3T) (Bonnici et al., 2012; Mueller et al., 2010; Pluta et al., 2012a; Winterburn et al., 2013a). These scans have 0.4mm in plane resolution and enhanced contrast (see Fig. 1). As a result, the anatomical details of hippocampal subfields, which are not visible in conventional T1-weighted MRI scans, can be captured and utilized for extracting hippocampal subfields, as previously shown (Kirov et al., 2013; Yushkevich et al., 2015b). To address the second challenge, this study employs a previously established segmentation tool, Automatic Segmentation of Hippocampal Subfields (ASHS), (Yushkevich et al., 2015b) specifically designed for analyzing high resolution T2-weighted MRI scans. ASHS is a free, open-source software package, which employs latest label fusion approaches for multi-atlas segmentation. It reads in a T1-weighted scan and a high resolution T2-weighted scan, and automatically labels hippocampal subfields and a few subregions in medial temporal lobe.
The standard acquisition time of a T2-weighted high resolution MRI scan is 8 minutes. By activating generalized auto-calibrating partially parallel acquisitions, the acquisition time can be reduced to 4 minutes. Compared with the standard 8-min protocol, the accelerated 4-min protocol saves scanning time and reduces susceptibility to motion but at a cost to signal-to-noise ratio. In this study, we performed a comparison of the standard and accelerated protocols for hippocampal subfield volume measurement using the ASHS segmentation technique. Our goals were to: (1) evaluate automatic hippocampal subfield segmentation results using high resolution T2-weighted 3T-MRI scans; (2) compare the hippocampal subfield measures between a standard unaccelerated 8-min scanning protocol and an accelerated 4-min protocol; (3) investigate hippocampal subfield volume changes among normal control (CN), subjective cognitive decline (SCD), MCI and AD participants, and to determine whether the pattern of these differences were similar using hippocampal subfields segmented from the unaccelerated 8-min scan relative to those from the accelerated 4-min scan, using a cohort recruited at the Indiana Alzheimer Disease Center (IADC).
2. MATERIALS AND METHODS
2.1 Sample and Demographics
The sample (n=35) included research subjects from four categories: cognitively normal (CN, n=10), subjective cognitive decline (SCD, n=9), mild cognitive impairment (MCI, n=10), and Alzheimer’s disease (AD, n=6). All participants were recruited from the Clinical Core of the Indiana Alzheimer Disease Center (IADC). All procedures were approved by the Indiana University Institutional Review Board. All subjects signed a written informed consent form. Participant characteristics are shown in Table 1.
Table 1.
Participant characteristics regarding age, gender, and intracranial volume (ICV).
| Age | Gender | ICV | |
|---|---|---|---|
| (mean±std, in years) | (M, F) | (cm3) | |
| Cognitively normal control (CN, n=10) | 69.2±5.7 | 1, 9 | 1437±146 |
| Subjective cognitive decline (SCD, n=9) | 71.3±6.4 | 5, 4 | 1469±201 |
| Mild cognitive impairment (MCI, n=10) | 72.9±6.2 | 5, 5 | 1464±255 |
| Alzheimer’s disease (AD, n=6) | 64.5±12.9 | 2, 4 | 1407±174 |
| p-value | 0.197* | 0.157** | 0.993* |
One-way ANOVA.
Chi-square test.
2.2 Image Acquisition
MRI scans were acquired on a Siemens MAGNETOM Prisma 3T MRI scanner. The scanning protocols included a T1-weighted MPRAGE sequence with whole-brain coverage and a T2-weighted TSE sequence with partial-brain coverage and an oblique coronal slice orientation (positioned orthogonally to the main axis of the hippocampus). The following MRI sequence parameters were used: the MPRAGE had an acquisition matrix of 240 × 256 × 176 and voxel size 1.05 × 1.05 × 1.2 mm3; the T2 scan had an acquisition matrix of 448 × 448 × 30 and voxel size 0.4 × 0.4 × 2 mm3 with TR/TE 8020/50 ms, 30 interleaved slices with no gap. The acquisition time of the conventional protocol is 8 minutes and 11 seconds. The accelerated protocol uses Siemens parallel imaging implementation (integrated parallel imaging techniques - iPAT) with an acceleration factor of 2, and thus reduces the acquisition time to 4 minutes 18 seconds.
2.3 Segmentation of Hippocampal Subfields
Automatic Segmentation of Hippocampal Subfields (ASHS) is a software tool developed by Yushkevich et al. (2015b) for automatically segmenting hippocampal subfields and their adjoining structures in the medial temporal lobe (MTL). The software has been used in several prior studies (de Flores et al., 2015; Hindy et al., 2016). This technique uses T1-weighted and high resolution T2-weighted MRI scans as inputs, and performs multi-atlas segmentation by implementing Joint Label Fusion method (Wang et al., 2013) and Corrective Learning (Wang et al., 2011). ASHS has been shown to be able to produce accurate and reliable segmentation results in previous studies (de Flores et al., 2015; Yushkevich et al., 2015a; Yushkevich et al., 2015b). In this study, ASHS was used to segment the following hippocampal subfields and their adjoining regions from the unaccelerated and accelerated high resolution T2-weighted MRI scans coupled with the corresponding T1-weighted MRI scans (Fig. 2): cornu ammonis 1 (CA1), CA2, CA3, dentate gyrus (DG), subiculum (SUB), entorhinal cortex (ERC), Brodmann areas 35 and 36 (BA35 and BA36, which together form the perirhinal cortex), and collateral sulcus (CS).
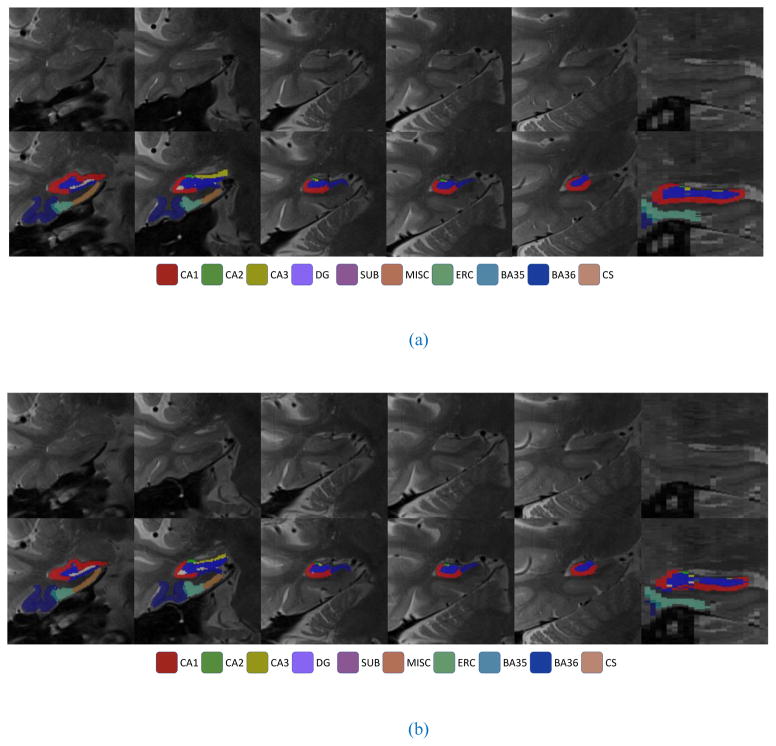
Fig. 2.
Examples of automatic segmentation results from (a) 4-min scans and (b) 8-min scans: Five coronal slices and one sagittal slice are shown.
2.4 Volumetric Analysis
Two types of comparative volumetric analyses were performed in this study including: (1) reliability tests to evaluate whether the measures extracted from the 8-min and 4-min scans are similar, and (2) statistical group analyses to see whether similar discriminative patterns can be discovered from the 8-min and 4-min scans. All the statistical analyses were performed using IBM SPSS 23 (SPSS Statistics 23, IBM Corporation, Somers, NY).
In our analyses, we examined primary hippocampal subfields and adjoining regions segmented directly from the ASHS software, as well as several composite regions of interest (ROIs). Specifically, we included the following nine primary regions: CA1, CA2, CA3, DG, SUB, ERC, BA35, BA36, and CS. In addition, we examined the following three composite regions: cornu ammonis (CA) containing CA1, CA2, and CA3, hippocampus (HIPP) containing CA, DG, and SUB, and perirhinal cortex (PRC) containing BA35 and BA36.
2.4.1 Reliability Analyses
Intraclass Correlation Coefficients (ICCs) were estimated within each diagnostic group and across all participants to measure reliability of the subfield volume estimates from the 8-min vs 4-min scans. ICCs are generally used to evaluate the consistency of quantitative measurements obtained by different acquisition protocols (Brebbia et al., 2011; Shrout and Fleiss, 1979).
In our study, ICCs were calculated to evaluate the reliability of the corresponding measures segmented from 8-min MRI scans versus those segmented from 4-min MRI scans. Given a variety of available ICC measures that may yield different values for the same data, we briefly describe below (1) the goal of this analytical study and (2) how to choose an appropriate ICC model for our reliability test to achieve the goal. The focus of this study is to examine the inter-rater reliability by comparing ASHS segmentation results from 8-min scans and 4-min scans. In our case, for each regional volume, we have estimates from two raters (i.e., volumes of regions segmented respectively from 8-min and 4-min MRI scans) and want to check whether they are consistent with each other. We employed a two-way mixed model, since the two acquisition protocols mentioned above were a fixed effect while the target ratings (e.g., all the regional volumes) were a random effect in our study. We tested the single measure reliability instead of the average measure reliability, because our goal was to evaluate the reliability of the ratings for a specific acquisition protocol (i.e., segmentation results from 4-min MRI scans) rather than the mean of all the ratings. We selected “consistency” as the model type instead of “absolute agreement”, since we were more interested in seeing the consistency of the relative standing of the measures over absolute agreement between two raters. In summary, the SPSS configurations of ICC analysis can be described as “two-way mixed model of single measure intraclass correlation with consistency type”. This type of ICC analysis belongs to “Case 3”, and can be denoted as “ICC(3,1)” based on (McGraw and Wong, 1996; Shrout and Fleiss, 1979).
ICC values range from 0 to 1 (from worse agreement to better agreement). In our study, an ICC value higher than 0.9 is considered as good agreement; an ICC value between 0.75 and 0.9 is considered as borderline or acceptable. For the convenience of discussion, in the rest of the paper, a result with an ICC value under 0.75 is characterized as “less reliable”.
2.4.2 Statistical Group Analyses
In addition to comparing the reliability of segmentation results from 8-min MRI scans versus 4-min MRI scans, we performed analyses to evaluated differences between diagnostic groups using SPSS General linear model (GLM). Specifically, our goal was to investigate whether there were significant regional volume differences between normal control (CN), subjective cognitive decline (SCD), MCI, and AD participants. Further, we evaluated whether the pattern of differences between groups using subfield volumetric estimates from ASHS were similar using data from 8-min and 4-min scans. In our experiments, we employed a multivariate regression model with diagnosis (DX) as fixed factor; age, sex, and total intracranial volume (ICV) as covariates; and primary and composite regional volumes as dependent variables.
To further examine the volume based morphometric differences between diagnostic groups, pairwise comparisons of effect sizes were performed for CN, SCD, MCI, and AD groups. Effect sizes were calculated using Cohen’s d (Cohen, 1988). The effect size of each group difference was computed after covarying for age, sex, and ICV.
3. RESULTS
3.1 Reliability Analyses
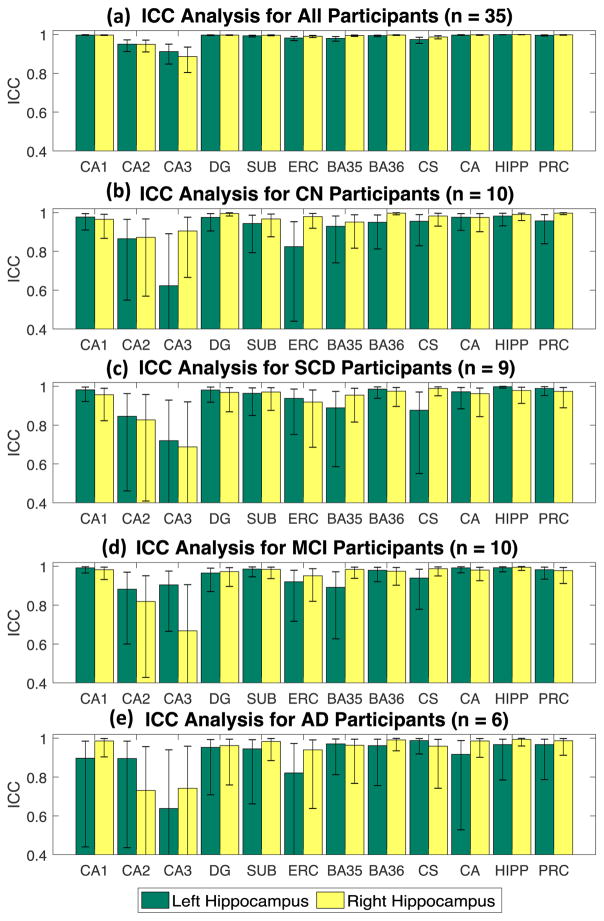
Table 2 shows inter-rater reliability results for the single measure ICC for the primary and composite regions both within each diagnostic group and across all groups. Fig. 3 shows the ICC results for all the primary and composite regions, within diagnostic groups, where error bars indicate the 95% confidence interval.
Table 2.
Intraclass Correlation Coefficients analysis (ICCs) results for all participants (n=35), cognitively normal control (CN, n=10), subjective cognitive decline (SCD, n=9), mild cognitive impairment (MCI, n=10), and Alzheimer’s disease (AD, n=6).
| Subfield | All (n=35) | CN (n=10) | SCD (n=9) | MCI (n=10) | AD (n=6) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| ICC | p | ICC | p | ICC | p | ICC | p | ICC | p | |
| Primary Regions | ||||||||||
| L_CA1 | 0.997 | <0.001 | 0.977 | <0.001 | 0.982 | <0.001 | 0.992 | <0.001 | 0.897 | 0.003 |
| R_CA1 | 0.997 | <0.001 | 0.965 | <0.001 | 0.957 | <0.001 | 0.983 | <0.001 | 0.986 | <0.001 |
| L_CA2 | 0.95 | <0.001 | 0.865 | <0.001 | 0.846 | <0.001 | 0.883 | <0.001 | 0.896 | 0.003 |
| R_CA2 | 0.949 | <0.001 | 0.872 | <0.001 | 0.827 | 0.002 | 0.819 | <0.001 | 0.731 | 0.031 |
| L_CA3 | 0.912 | <0.001 | 0.623 | 0.020 | 0.72 | 0.009 | 0.905 | <0.001 | 0.638 | 0.062 |
| R_CA3 | 0.886 | <0.001 | 0.905 | <0.001 | 0.688 | 0.014 | 0.668 | .012 | 0.742 | 0.028 |
| L_DG | 0.997 | <0.001 | 0.975 | <0.001 | 0.981 | <0.001 | 0.966 | <0.001 | 0.953 | <0.001 |
| R_DG | 0.997 | <0.001 | 0.995 | <0.001 | 0.969 | <0.001 | 0.973 | <0.001 | 0.962 | <0.001 |
| L_SUB | 0.993 | <0.001 | 0.944 | <0.001 | 0.964 | <0.001 | 0.986 | <0.001 | 0.945 | <0.001 |
| R_SUB | 0.996 | <0.001 | 0.967 | <0.001 | 0.971 | <0.001 | 0.984 | <0.001 | 0.983 | <0.001 |
| L_ERC | 0.982 | <0.001 | 0.824 | <0.001 | 0.938 | <0.001 | 0.921 | <0.001 | 0.821 | 0.012 |
| R_ERC | 0.991 | <0.001 | 0.979 | <0.001 | 0.919 | <0.001 | 0.952 | <0.001 | 0.94 | <0.001 |
| L_BA35 | 0.98 | <0.001 | 0.929 | <0.001 | 0.889 | <0.001 | 0.892 | <0.001 | 0.971 | <0.001 |
| R_BA35 | 0.994 | <0.001 | 0.951 | <0.001 | 0.955 | <0.001 | 0.984 | <0.001 | 0.964 | <0.001 |
| L_BA36 | 0.994 | <0.001 | 0.95 | <0.001 | 0.986 | <0.001 | 0.98 | <0.001 | 0.962 | <0.001 |
| R_BA36 | 0.997 | <0.001 | 0.997 | <0.001 | 0.975 | <0.001 | 0.975 | <0.001 | 0.991 | <0.001 |
| L_CS | 0.974 | <0.001 | 0.955 | <0.001 | 0.877 | <0.001 | 0.94 | <0.001 | 0.988 | <0.001 |
| R_CS | 0.987 | <0.001 | 0.982 | <0.001 | 0.989 | <0.001 | 0.988 | <0.001 | 0.959 | <0.001 |
| Composite Regions | ||||||||||
| L_CA | 0.997 | <0.001 | 0.976 | <0.001 | 0.972 | <0.001 | 0.992 | <0.001 | 0.917 | 0.002 |
| R_CA | 0.998 | <0.001 | 0.975 | <0.001 | 0.963 | <0.001 | 0.981 | <0.001 | 0.986 | <0.001 |
| L_HIPP | 0.999 | <0.001 | 0.982 | <0.001 | 0.998 | <0.001 | 0.993 | <0.001 | 0.967 | <0.001 |
| R_HIPP | 0.999 | <0.001 | 0.99 | <0.001 | 0.979 | <0.001 | 0.995 | <0.001 | 0.994 | <0.001 |
| L_PRC | 0.996 | <0.001 | 0.957 | <0.001 | 0.989 | <0.001 | 0.983 | <0.001 | 0.967 | <0.001 |
| R_PRC | 0.998 | <0.001 | 0.997 | <0.001 | 0.974 | <0.001 | 0.978 | <0.001 | 0.987 | <0.001 |
Abbreviations: L (left hemisphere), R (right hemisphere), cornu ammonis 1/2/3 (CA1/2/3), dentate gyrus (DG), subiculum (SUB), entorhinal cortex (ERC), Brodmann area 35/36 (BA35/36), collateral sulcus (CS), cornu ammonis (CA, including CA1, CA2 and CA3), hippocampus (HIPP, including CA, DG, and SUB), and perirhinal cortex (PRC, including BA35 and BA36).
Fig. 3.
Results of Intraclass Correlation Coefficients (ICC) analysis for the following groups: (a) CN participants, (b) SCD participants, (c) MCI participants, and (d) AD participants. Error bar indicates the 95% confidence interval.
Across all participants (n=35), all the ICCs were significant at levels ranging from 0.886 (right CA3) to 0.999 (left and right HIPP) (Fig. 3), indicating good or acceptable agreement and reliability. For the CN group (n=10), the ICCs range from 0.623 (left CA3) to 0.997 (right BA36, right PRC). The only less reliable one in this group is the left CA3 region; all of the other regions have ICCs ranging from 0.865 (left CA2) to 0.997 (right BA36, right PRC). For the SCD group (n=9), the ICCs range from 0.688 (right CA3) to 0.998 (left HIPP). The only less reliable ones are the left and right CA3 regions; all of the other regions have ICCs ranging from 0.827 (right CA2) to 0.998 (left HIPP). For the MCI group (n=10), the ICCs range from 0.668 (right CA3) to 0.995 (right HIPP). The only less reliable one is the right CA3; all of the other areas have ICCs ranging from 0.819 (right CA2) to 0.995 (right HIPP). For the AD group (n=6), the ICCs range from 0.638 (left CA3) to 0.994 (right HIPP). The only less reliable ones are the right CA2, left CA3, and right CA3; all of the other regions have ICCs ranging from 0.821 (left ERC) to 0.994 (right HIPP).
Overall, the most frequent less-reliable measures are located at the left and right CA3 regions (occurring 3 out of 5 times) and the right CA2 (occurring once). Specifically, these outlying cases are listed below: ICCL_CA3 = 0.623 in the CN set, ICCL_CA3 = 0.72 in the SCD set, ICCR_CA3 = 0.688 in the SCD set, ICCR_CA3 = 0.668 in the MCI set, ICCR_CA2 = 0.731 in the AD set, ICCL_CA3 = 0.638 in the AD set, and ICCR_CA3 = 0.742 in the AD set.
3.2 Statistical Group Analyses
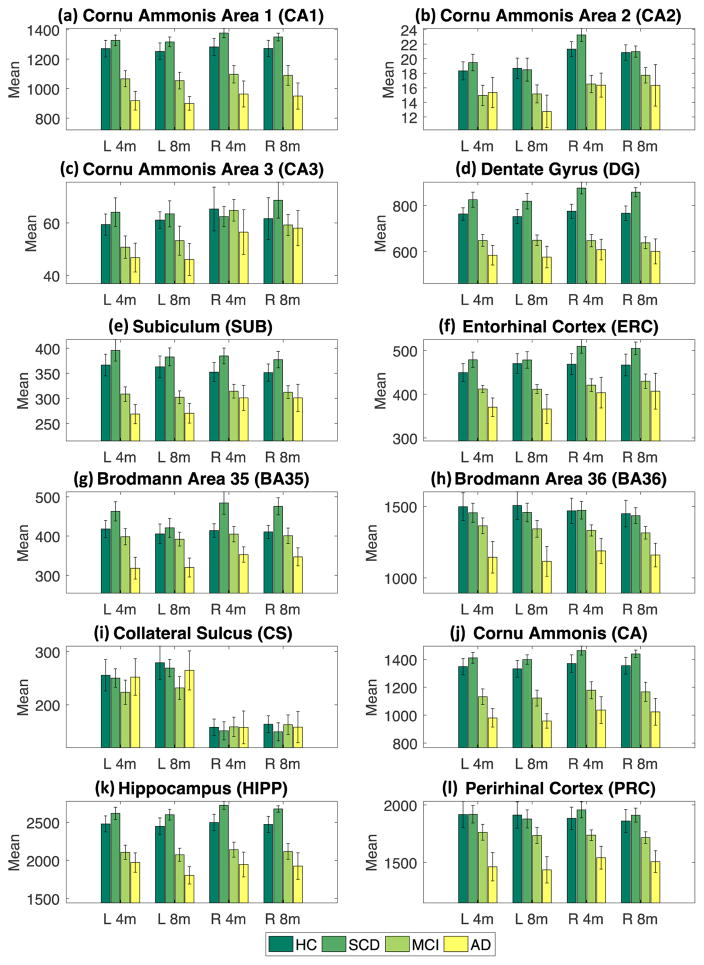
Fig. 4 shows mean volume for each sub-region adjusted by age, sex and ICV by DX group, where error bars indicate standard error. By visual inspection, only CA2 and CA3 show a minor inconsistency between 4-min scan and 8-min scan, while the mean and standard error in other comparison groups show great consistency. Table 3 shows the p-values of pairwise group comparison covaried for age, sex, and ICV for both the 4-min and 8-min scan data. Given our modest sample size, we used the nominal p<0.05 as the threshold to identify significant regions (see red cells in Table 3). Based on Table 3, we observe similar patterns (e.g., significant regions detected) between results generated from 4-min and 8-min scan data. In terms of the identified significant regions, among six pairwise comparisons, two comparisons (CN vs. SCD, CN vs. MCI) showed 100% agreement between 4-min scans and 8-min scans, two comparisons (SCD vs. MCI, MCI vs. AD) had 2 out of 24 mismatches, the comparison of CN vs. AD had 4 out of 24 mismatches, and the comparison of SCD vs. AD had 5 out of 24 mismatches. These mismatches are attributed to the following regions: two from left CA2, two from right CA2, one from left CA3, one from right SUB, one from left ERC, two from left BA35, two from right BA35, one from right BA36, and one from left PRC.
Fig. 4.
Group comparison for each of primary labels and compound labels. Mean and standard error (as error bar) are shown for each group (HC, SCD, MCI, or AD).
Table 3.
P value of diagnosis effect on subfield volume covaried for age, gender and ICV.
| Subfield | CN vs SCD | CN vs MCI | CN vs AD | SCD vs MCI | SCD vs AD | MCI vs AD | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 min | 8 min | 4 min | 8 min | 4 min | 8 min | 4 min | 8 min | 4 min | 8 min | 4 min | 8 min | |
| L_CA1 | .162 | .115 | .137 | .147 | .009 | .006 | .003 | .003 | .002 | .000 | .197 | .140 |
| R_CA1 | .087 | .106 | .233 | .260 | .026 | .024 | .002 | .006 | .012 | .010 | .251 | .259 |
| L_CA2 | .098 | .420 | .267 | .413 | .203 | .045 | .038 | .163 | .371 | .443 | .546 | .669 |
| R_CA2 | .401 | .641 | .124 | .473 | .030 | .098 | .001 | .033 | .020 | .918 | .963 | .731 |
| L_CA3 | .459 | .793 | .467 | .589 | .104 | .025 | .110 | .255 | .245 | .340 | .592 | .358 |
| R_CA3 | .941 | .311 | .782 | .949 | .478 | .613 | .710 | .255 | .841 | .978 | .377 | .897 |
| L_DG | .141 | .153 | .091 | .236 | .009 | .021 | .001 | .001 | .025 | .036 | .223 | .161 |
| R_DG | .031 | .030 | .099 | .105 | .026 | .040 | .000 | .000 | .008 | .015 | .424 | .521 |
| L_SUB | .128 | .194 | .132 | .118 | .033 | .049 | .008 | .004 | .010 | .018 | .209 | .324 |
| R_SUB | .084 | .107 | .254 | .232 | .231 | .229 | .009 | .014 | .047 | .115 | .694 | .791 |
| L_ERC | .165 | .397 | .385 | .248 | .030 | .026 | .005 | .012 | .032 | .154 | .070 | .213 |
| R_ERC | .128 | .112 | .452 | .788 | .135 | .237 | .001 | .004 | .183 | .332 | .622 | .602 |
| L_BA35 | .213 | .716 | .737 | .642 | .043 | .115 | .082 | .476 | .038 | .178 | .016 | .020 |
| R_BA35 | .048 | .035 | .632 | .654 | .156 | .152 | .054 | .034 | .033 | .016 | .048 | .062 |
| L_BA36 | .895 | .836 | .399 | .320 | .066 | .051 | .385 | .278 | .037 | .019 | .107 | .087 |
| R_BA36 | .394 | .457 | .277 | .302 | .076 | .083 | .063 | .089 | .055 | .030 | .179 | .160 |
| L_CS | .965 | .848 | .279 | .165 | .691 | .683 | .338 | .169 | .558 | .617 | .538 | .402 |
| R_CS | .751 | .785 | .859 | .809 | .667 | .511 | .935 | .766 | .672 | .835 | .973 | .987 |
| L_CA | .152 | .123 | .130 | .142 | .009 | .006 | .002 | .002 | .002 | .000 | .201 | .123 |
| R_CA | .107 | .094 | .261 | .280 | .030 | .029 | .002 | .006 | .016 | .014 | .251 | .273 |
| L_HIPP | .104 | .117 | .107 | .143 | .009 | .011 | .001 | .001 | .003 | .002 | .187 | .130 |
| R_HIPP | .047 | .046 | .192 | .202 | .037 | .040 | .000 | .000 | .010 | .014 | .327 | .372 |
| L_PRC | .872 | .929 | .517 | .457 | .047 | .047 | .219 | .275 | .021 | .020 | .051 | .047 |
| R_PRC | .191 | .236 | .382 | .393 | .069 | .069 | .023 | .033 | .019 | .009 | .085 | .080 |
Abbreviations: L (left hemisphere), R (right hemisphere), cornu ammonis 1/2/3 (CA1/2/3), dentate gyrus (DG), subiculum (SUB), entorhinal cortex (ERC), Brodmann area 35/36 (BA35/36), collateral sulcus (CS), cornu ammonis (CA, including CA1, CA2 and CA3), hippocampus (HIPP, including CA, DG, and SUB), and perirhinal cortex (PRC, including BA35 and BA36).
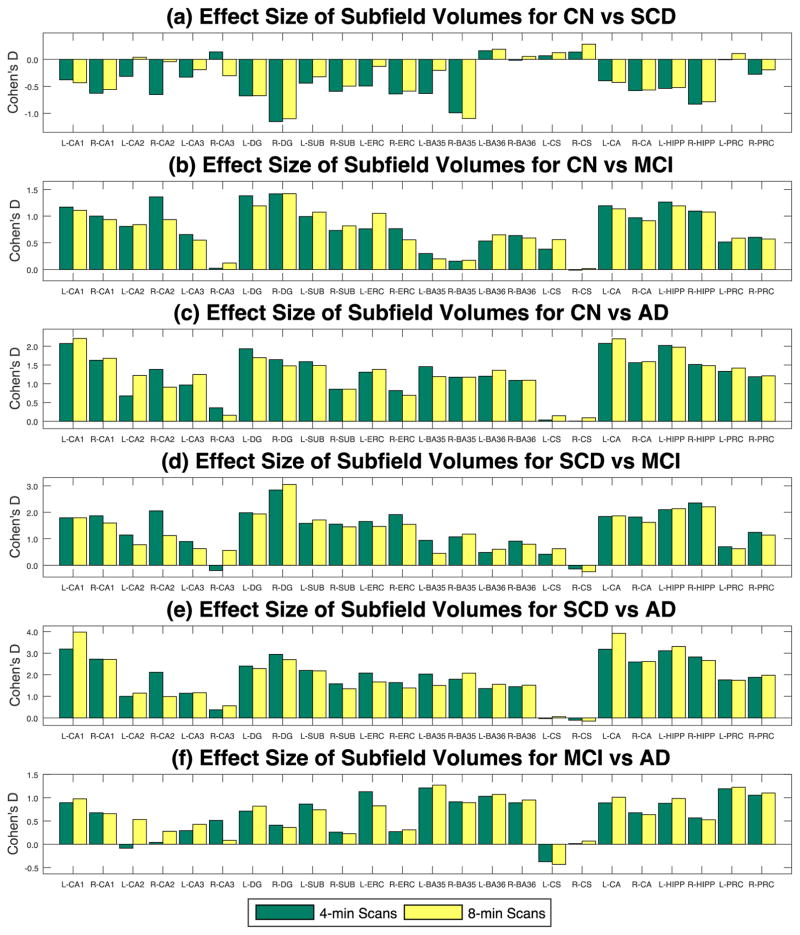
Table 4 show the effect size results (Cohen’s d) for each pairwise comparison among four diagnosis groups (CN, SCD, MCI, and AD). According to (Sawilowsky, 2009), an effect size with d≥1.2 is considered to be “very large”, and thus is colored in red in Table 4. The effect size pattern shown in Table 4 is very similar to the significance pattern described in Table 3. Graphical effect size results are shown in Fig. 5, indicating strong agreement between 4-min scans (green) and 8-min scans (yellow).
Table 4.
Effect size (Cohen’s d) of diagnosis effect on subfield volume covaried for age, gender and ICV, where |d|≥1.2 is shown as red, indicating “very large” effect size.
| Subfield | CN vs SCD | CN vs MCI | CN vs AD | SCD vs MCI | SCD vs AD | MCI vs AD | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 min | 8 min | 4 min | 8 min | 4 min | 8 min | 4 min | 8 min | 4 min | 8 min | 4 min | 8 min | |
| L_CA1 | −0.38 | −0.43 | 1.17 | 1.11 | 2.07 | 2.21 | 1.80 | 1.79 | 3.19 | 3.98 | 0.89 | 0.98 |
| R_CA1 | −0.63 | −0.56 | 1.00 | 0.93 | 1.63 | 1.68 | 1.87 | 1.59 | 2.73 | 2.71 | 0.68 | 0.66 |
| L_CA2 | −0.31 | 0.04 | 0.81 | 0.84 | 0.68 | 1.22 | 1.14 | 0.78 | 1.00 | 1.14 | −0.09 | 0.53 |
| R_CA2 | −0.65 | −0.04 | 1.36 | 0.94 | 1.38 | 0.91 | 2.06 | 1.12 | 2.11 | 0.99 | 0.04 | 0.28 |
| L_CA3 | −0.33 | −0.19 | 0.65 | 0.55 | 0.97 | 1.25 | 0.90 | 0.63 | 1.14 | 1.17 | 0.29 | 0.43 |
| R_CA3 | 0.14 | −0.30 | 0.02 | 0.12 | 0.36 | 0.16 | −0.20 | 0.56 | 0.37 | 0.56 | 0.51 | 0.09 |
| L_DG | −0.68 | −0.67 | 1.38 | 1.19 | 1.93 | 1.70 | 1.98 | 1.94 | 2.40 | 2.28 | 0.71 | 0.82 |
| R_DG | −1.15 | −1.10 | 1.42 | 1.42 | 1.64 | 1.47 | 2.84 | 3.05 | 2.94 | 2.70 | 0.41 | 0.36 |
| L_SUB | −0.44 | −0.32 | 0.99 | 1.08 | 1.59 | 1.49 | 1.58 | 1.71 | 2.20 | 2.18 | 0.86 | 0.74 |
| R_SUB | −0.59 | −0.50 | 0.73 | 0.82 | 0.85 | 0.86 | 1.56 | 1.45 | 1.58 | 1.35 | 0.26 | 0.23 |
| L_ERC | −0.49 | −0.13 | 0.76 | 1.05 | 1.31 | 1.38 | 1.65 | 1.46 | 2.08 | 1.67 | 1.13 | 0.83 |
| R_ERC | −0.64 | −0.59 | 0.76 | 0.56 | 0.82 | 0.69 | 1.92 | 1.55 | 1.63 | 1.38 | 0.27 | 0.31 |
| L_BA35 | −0.63 | −0.20 | 0.30 | 0.20 | 1.46 | 1.19 | 0.94 | 0.45 | 2.03 | 1.50 | 1.21 | 1.27 |
| R_BA35 | −0.99 | −1.10 | 0.16 | 0.17 | 1.18 | 1.17 | 1.07 | 1.18 | 1.79 | 2.07 | 0.91 | 0.90 |
| L_BA36 | 0.16 | 0.19 | 0.53 | 0.65 | 1.20 | 1.36 | 0.49 | 0.60 | 1.36 | 1.55 | 1.03 | 1.07 |
| R_BA36 | −0.02 | 0.06 | 0.64 | 0.59 | 1.09 | 1.09 | 0.91 | 0.79 | 1.45 | 1.51 | 0.89 | 0.95 |
| L_CS | 0.07 | 0.13 | 0.38 | 0.56 | 0.04 | 0.15 | 0.42 | 0.62 | −0.03 | 0.06 | −0.37 | −0.43 |
| R_CS | 0.14 | 0.28 | −0.01 | 0.02 | 0.00 | 0.09 | −0.14 | −0.24 | −0.11 | −0.15 | 0.01 | 0.07 |
| L_CA | −0.40 | −0.43 | 1.19 | 1.14 | 2.08 | 2.20 | 1.84 | 1.86 | 3.18 | 3.92 | 0.89 | 1.01 |
| R_CA | −0.58 | −0.57 | 0.97 | 0.91 | 1.56 | 1.59 | 1.82 | 1.62 | 2.59 | 2.61 | 0.68 | 0.64 |
| L_HIPP | −0.54 | −0.52 | 1.26 | 1.19 | 2.02 | 1.97 | 2.10 | 2.14 | 3.11 | 3.31 | 0.88 | 0.98 |
| R_HIPP | −0.83 | −0.78 | 1.09 | 1.08 | 1.51 | 1.48 | 2.36 | 2.21 | 2.83 | 2.66 | 0.57 | 0.53 |
| L_PRC | −0.01 | 0.11 | 0.51 | 0.59 | 1.33 | 1.42 | 0.70 | 0.62 | 1.76 | 1.74 | 1.19 | 1.23 |
| R_PRC | −0.28 | −0.19 | 0.60 | 0.57 | 1.19 | 1.21 | 1.24 | 1.14 | 1.88 | 1.98 | 1.05 | 1.10 |
Abbreviations: L (left hemisphere), R (right hemisphere), cornu ammonis 1/2/3 (CA1/2/3), dentate gyrus (DG), subiculum (SUB), entorhinal cortex (ERC), Brodmann area 35/36 (BA35/36), collateral sulcus (CS), cornu ammonis (CA, including CA1, CA2 and CA3), hippocampus (HIPP, including CA, DG, and SUB), and perirhinal cortex (PRC, including BA35 and BA36).
Fig. 5.
Effect sizes (Cohen’s d) of the pair-wised comparisons between four diagnosis groups (CN, SCD, MCI, and AD) evaluated for hippocampal subfield volumes. Subfield volumes are adjusted by age, gender and ICV by DX group.
4. DISCUSSION
We performed reliability tests within and across diagnostic groups, via computing ICC between measures obtained from the 8-min and 4-min scans. Our results demonstrated that the volumetric measures of hippocampal subfields and adjoining regions extracted from the 4-min and 8-min scans showed strong consistency. The ICCs of all primary regions except CA2 and CA3 ranged from 0.821 to 0.997, suggesting majority of ASHS hippocampal subfield segmentations from both imaging modalities have either acceptable (ICC>0.75) or good (ICC>0.9) consistency. This finding indicates that the accelerated 4-min scanning protocol provides commensurate hippocampal subfield segmentation results to the standard 8-min protocol with the added benefit of shorter scan time and likely reduced motion artefact. Of note, the investigation in (Yushkevich et al., 2015b) was performed on 8-min scans of a CN/MCI cohort, and was focused on comparing ASHS and manual segmentation results. In this work, our focus was to compare the ASHS segmentation results generated from accelerated 4-min scans and those from standard 8-min scans, using a cohort containing CN, SCD, MCI and AD participants.
The CA2 and CA3 subfields yielded suboptimal ICCs ranging from 0.731 to 0.95 and 0.623 to 0.912 respectively. Some of these ICC levels are relatively low, which could be caused by a combination of several factors. CA2 and CA3 were the smallest hippocampal subfields as shown in Fig. 4(b, c). Given the fixed imaging resolution, small regions with complex anatomical structures tend to present more challenges in automated segmentation process and statistical analyses. As reported in (Yushkevich et al., 2015b), comparing ASHS with manual segmentation results, CA2 and CA3 also had lowest ICCs (0.4 and 0.484 respectively) and dice coefficient values (0.552 and 0.525 in a cohort with N=29).
One strategy to overcome this limitation is to examine a composite region by combining several primary regions together. As shown in Table 2, after we combined all CA areas, improved ICCs were obtained. After combining all the CA regions, ICCCA increased to 0.976 and 0.975 for left and right CAs in the CN group (n=10), 0.992 and 0.981 for left and right CAs in the MCI group (n=10), and 0.997 and 0.998 for left and right CAs across all participants (n=35). These findings are similar to those reported in (Yushkevich et al., 2015b), as they observed similar improvements were observed using this strategy.
Given the nature of this study, we have only a limited number of subjects who have been scanned twice using 4-min and 8-min protocols respectively. Despite the modest sample size, the results are quite promising. When we included all participants (n=35), there were no obvious relatively low ICCs. However, when the group sizes were reduced to n=10 (CN and MCI) and n=9 (SCD), CA3 were observed as less reliable. For the smallest set (AD, n=6), even more less reliable regions were observed (right CA2, left and right CA3s). However, the results of all the other segmented regions showed satisfactory agreement between 8-min scans and 4-min scans. The above impact on the consistency results of the CA2 and CA3 regions could be attributed to not only the small sample size, but also the attenuated variance within each diagnostic group.
To test whether 4-min and 8-min scans could have similar sensitivity to detect significant volumetric differences between diagnostic groups, we applied general linear model (GLM) to examine the regional volume changes related to diagnosis while covarying for age, sex, and ICV. This pair-wise comparison study was performed among four diagnostic groups: CN, SCD, MCI, and AD, for both the 8-min scans and 4-min scans. Tables 3–4 and Figs. 4–5 demonstrate that similar patterns (red areas) were observed in both scan sets (8-min and 4-min), and a majority of significant regions detected from 4-min scans were in accordance with the significant ones from 8-min scans. Overall, the results demonstrated strong agreement between 8-min scans and 4-min scans in detecting regional volume changes related to AD or at-risk stages of AD.
Given the small sample size in this study, we adopted a nominal significance level of p<0.05 to evaluate the group differences. While comparing CN and SCD, a majority of regions demonstrate a trend of increased volume in the SCD group (Fig. 4), some of which were statistically significant (i.e., right DG, right BA35, and right HIPP; see the CN vs SCD column in Table 3). These findings suggest that this group might demonstrate increased regional volumes, potentially as a compensatory mechanism to delay the process of conversion into MCI for this group of subjects with relatively high risk. Similar statistical patterns were observed in (Risacher et al., 2015).
The major affected regions in AD include CA1, dentate gyrus, left subiculum, and left entorhinal cortex, as well as the composite CA, hippocampus and left perirhinal cortex. No regional significance between CN and SCD was identified. Also, we did not detect any regional significance between CN and MCI, unlike in (Yushkevich et al., 2015b), which demonstrated significant volumetric reductions in the dentate gyrus. Our lack of replication may be due to the small sample size of our study, making it under-powered to detect subtle changes.
Compared with previous results (Braak and Braak, 1995; Mueller et al., 2010; Yushkevich et al., 2015b), our analyses confirmed the importance of CA1, BA35, subiculum, and dentate gyrus as AD biomarkers. For example, in our study, BA35 turned out to be the only marker showing large difference between MCI and AD. This region was reported in (Braak and Braak, 1995) as the first cortical site affected by neurofibrillary tangle pathology. Also, we found that CA1 and the left subiculum could distinguish AD from CN and SCD, which is in accordance with the findings in (Mueller et al., 2010). In addition, we identified the dentate gyrus as a marker that distinguishes AD from CN and SCD. The dentate gyrus was identified in (Yushkevich et al., 2015b) as a marker showing differences between CN and MCI and the present results indicate sensitivity of this region to early through later stages. One potential application of a time efficient and robust scan acquisition enabling hippocampal subfield investigation is the area of MRI markers related to adult neurogenesis, a process that interacts with AD pathophysiology (Horgusluoglu et al., 2017).
A recent study compared the accelerated and non-accelerated T1-weighted MRI scans for brain volume and boundary shift integral measures of volume changes using the data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort (Manning et al., 2017). This study found that the accelerated scan protocol yielded fewer scan pairs with serious motion artefacts while not significantly changing the main results which included rate of volume changes. Similarly, our findings comparing the hippocampal subfield volumetric differences using the accelerated and non-accelerated high resolution T2-weighted MRI protocols also demonstrated positive findings supporting the use of the accelerated sequence.
In summary, our reliability analysis demonstrated that two scanning protocols (i.e., standard 8-min one and accelerated 4-min one), evaluated with the ASHS segmentation method, yield similar volume estimates for most hippocampal subfields and adjoining regions. In addition, our group analysis showed that these scan protocols produce similar statistical patterns in detecting significant regions between cognitively normal older adults and those with MCI and/or AD. CA2 and CA3 showed some minor inconsistency in our studies, which is not unexpected based on previous studies, as these are very challenging regions to segment given the limited scan resolution in relation to their small sizes. This limitation can be addressed to some extent by grouping the three CA regions together into a composite measure. In summary, our results suggest that the accelerated 4-min high-resolution hippocampal scan protocol can provide reliable imaging data for investigation of hippocampal subfields in AD-related MRI studies and may be less vulnerable to motion.
Acknowledgments
Data analysis was supported in part by the following grants from the National Institutes of Health: R01 EB022574 and R01 LM011360 to LS, P30 AG10133, R01 AG19771 and U01 AG024904 (IU Subcontract) to AJS, R01 AG040770 to LA, and K01 AG049050 to SLR.
References
- Adler DH, Pluta J, Kadivar S, Craige C, Gee JC, Avants BB, Yushkevich PA. Histology-derived volumetric annotation of the human hippocampal subfields in postmortem MRI. Neuroimage. 2014;84:505–523. doi: 10.1016/j.neuroimage.2013.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Dutton RA, Dinov ID, Hayashi KM, Toga AW, Cummings JL, Thompson PM. Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch Neurol. 2006;63:693–699. doi: 10.1001/archneur.63.5.693. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Mosconi L, Thompson PM, Green AE, Hwang KS, Ramirez A, Mistur R, Tsui WH, de Leon MJ. Subregional hippocampal atrophy predicts Alzheimer’s dementia in the cognitively normal. Neurobiol Aging. 2010a;31:1077–1088. doi: 10.1016/j.neurobiolaging.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Thompson PM, Green AE, Hwang KS, Zoumalan C, Jack CR, Jr, Harvey DJ, Petersen RC, Thal LJ, Aisen PS, Toga AW, Cummings JL, Decarli CS. 3D comparison of low, intermediate, and advanced hippocampal atrophy in MCI. Hum Brain Mapp. 2010b;31:786–797. doi: 10.1002/hbm.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnici HM, Chadwick MJ, Kumaran D, Hassabis D, Weiskopf N, Maguire EA. Multi-voxel pattern analysis in human hippocampal subfields. Front Hum Neurosci. 2012;6:290. doi: 10.3389/fnhum.2012.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiology of aging. 1995;16:271–278. doi: 10.1016/0197-4580(95)00021-6. [DOI] [PubMed] [Google Scholar]
- Brebbia CA, EGLITE M, LATVIA IK, Latvia RM, POPOV V. Environmental Health & Biomedicine 2011 [Google Scholar]
- Cohen J. Statistical power analysis for the behavioral sciences. Lawrence Earlbaum Associates; Hillsdale, NJ: 1988. pp. 20–26. [Google Scholar]
- de Flores R, La Joie R, Chetelat G. Structural imaging of hippocampal subfields in healthy aging and Alzheimer’s disease. Neuroscience. 2015;309:29–50. doi: 10.1016/j.neuroscience.2015.08.033. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindy NC, Ng FY, Turk-Browne NB. Linking pattern completion in the hippocampus to predictive coding in visual cortex. Nat Neurosci. 2016;19:665–667. doi: 10.1038/nn.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgusluoglu E, Nudelman K, Nho K, Saykin AJ. Adult neurogenesis and neurodegenerative diseases: A systems biology perspective. Am J Med Genet B Neuropsychiatr Genet. 2017;174:93–112. doi: 10.1002/ajmg.b.32429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Coupland NJ, Lebel RM, Carter R, Seres P, Wilman AH, Malykhin NV. Structural changes in hippocampal subfields in major depressive disorder: a high-field magnetic resonance imaging study. Biological psychiatry. 2013;74:62–68. doi: 10.1016/j.biopsych.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Hunsaker MR, Amaral DG. A semi-automated pipeline for the segmentation of rhesus macaque hippocampus: validation across a wide age range. PLoS One. 2014;9:e89456. doi: 10.1371/journal.pone.0089456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov II, Hardy CJ, Matsuda K, Messinger J, Cankurtaran CZ, Warren M, Wiggins GC, Perry NN, Babb JS, Goetz RR. In vivo 7Tesla imaging of the dentate granule cell layer in schizophrenia. Schizophrenia research. 2013;147:362–367. doi: 10.1016/j.schres.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Joie R, Perrotin A, de La Sayette V, Egret S, Doeuvre L, Belliard S, Eustache F, Desgranges B, Chetelat G. Hippocampal subfield volumetry in mild cognitive impairment, Alzheimer’s disease and semantic dementia. Neuroimage Clin. 2013;3:155–162. doi: 10.1016/j.nicl.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby LA, Ekstrom AD, Ragland JD, Ranganath C. Differential connectivity of perirhinal and parahippocampal cortices within human hippocampal subregions revealed by high-resolution functional imaging. The Journal of Neuroscience. 2012;32:6550–6560. doi: 10.1523/JNEUROSCI.3711-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malykhin N, Lebel RM, Coupland N, Wilman AH, Carter R. In vivo quantification of hippocampal subfields using 4.7 T fast spin echo imaging. Neuroimage. 2010;49:1224–1230. doi: 10.1016/j.neuroimage.2009.09.042. [DOI] [PubMed] [Google Scholar]
- Manning EN, Leung KK, Nicholas JM, Malone IB, Cardoso MJ, Schott JM, Fox NC, Barnes J Alzheimer’s Disease Neuroimaging, I. A Comparison of Accelerated and Non-accelerated MRI Scans for Brain Volume and Boundary Shift Integral Measures of Volume Change: Evidence from the ADNI Dataset. Neuroinformatics. 2017 doi: 10.1007/s12021-017-9326-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw KO, Wong SP. Forming inferences about some intraclass correlation coefficients. Psychological methods. 1996;1:30. [Google Scholar]
- Merkel B, Steward C, Vivash L, Malpas CB, Phal P, Moffat BA, Cox KL, Ellis KA, Ames DJ, Cyarto EV. Semi-automated hippocampal segmentation in people with cognitive impairment using an age appropriate template for registration. Journal of Magnetic Resonance Imaging. 2015;42:1631–1638. doi: 10.1002/jmri.24966. [DOI] [PubMed] [Google Scholar]
- Mueller S, Stables L, Du A, Schuff N, Truran D, Cashdollar N, Weiner M. Measurement of hippocampal subfields and age-related changes with high resolution MRI at 4T. Neurobiol Aging. 2007;28:719–726. doi: 10.1016/j.neurobiolaging.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Schuff N, Yaffe K, Madison C, Miller B, Weiner MW. Hippocampal atrophy patterns in mild cognitive impairment and Alzheimer’s disease. Hum Brain Mapp. 2010;31:1339–1347. doi: 10.1002/hbm.20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW. Selective effect of age, Apo e4, and Alzheimer’s disease on hippocampal subfields. Hippocampus. 2009;19:558–564. doi: 10.1002/hipo.20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen RK, Palombo DJ, Rabin JS, Levine B, Ryan JD, Rosenbaum RS. Volumetric analysis of medial temporal lobe subregions in developmental amnesia using high-resolution magnetic resonance imaging. Hippocampus. 2013;23:855–860. doi: 10.1002/hipo.22153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56:907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- Pluta J, Yushkevich P, Das S, Wolk D. In vivo analysis of hippocampal subfield atrophy in mild cognitive impairment via semi-automatic segmentation of T2-weighted MRI. J Alzheimers Dis. 2012a;31:85–99. doi: 10.3233/JAD-2012-111931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta J, Yushkevich P, Das S, Wolk D. In vivo analysis of hippocampal subfield atrophy in mild cognitive impairment via semi-automatic segmentation of T2-weighted MRI. Journal of Alzheimer’s Disease. 2012b;31:85–99. doi: 10.3233/JAD-2012-111931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risacher SL, Kim S, Nho K, Foroud T, Shen L, Petersen RC, Jack CR, Beckett LA, Aisen PS, Koeppe RA. APOE effect on Alzheimer’s disease biomarkers in older adults with significant memory concern. Alzheimer’s & Dementia. 2015;11:1417–1429. doi: 10.1016/j.jalz.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawilowsky SS. New effect size rules of thumb 2009 [Google Scholar]
- Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychological bulletin. 1979;86:420. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- Van Leemput K, Bakkour A, Benner T, Wiggins G, Wald LL, Augustinack J, Dickerson BC, Golland P, Fischl B. Automated segmentation of hippocampal subfields from ultra-high resolution in vivo MRI. Hippocampus. 2009;19:549–557. doi: 10.1002/hipo.20615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Das SR, Suh JW, Altinay M, Pluta J, Craige C, Avants B, Yushkevich PA Initiative A.s.D.N. A learning-based wrapper method to correct systematic errors in automatic image segmentation: consistently improved performance in hippocampus, cortex and brain segmentation. Neuroimage. 2011;55:968–985. doi: 10.1016/j.neuroimage.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Suh JW, Das SR, Pluta JB, Craige C, Yushkevich PA. Multi-atlas segmentation with joint label fusion. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 2013;35:611–623. doi: 10.1109/TPAMI.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterburn JL, Pruessner JC, Chavez S, Schira MM, Lobaugh NJ, Voineskos AN, Chakravarty MM. A novel in vivo atlas of human hippocampal subfields using high-resolution 3 T magnetic resonance imaging. Neuroimage. 2013a;74:254–265. doi: 10.1016/j.neuroimage.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Winterburn JL, Pruessner JC, Chavez S, Schira MM, Lobaugh NJ, Voineskos AN, Chakravarty MM. A novel in vivo atlas of human hippocampal subfields using high-resolution 3T magnetic resonance imaging. Neuroimage. 2013b;74:254–265. doi: 10.1016/j.neuroimage.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Wisse L, Gerritsen L, Zwanenburg JJ, Kuijf HJ, Luijten PR, Biessels GJ, Geerlings MI. Subfields of the hippocampal formation at 7T MRI: In vivo volumetric assessment. Neuroimage. 2012;61:1043–1049. doi: 10.1016/j.neuroimage.2012.03.023. [DOI] [PubMed] [Google Scholar]
- Wisse LE, Kuijf HJ, Honingh AM, Wang H, Pluta JB, Das SR, Wolk DA, Zwanenburg JJ, Yushkevich PA, Geerlings MI. Automated hippocampal subfield segmentation at 7T MRI. American Journal of Neuroradiology. 2016;37:1050–1057. doi: 10.3174/ajnr.A4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassa MA, Stark CE. Pattern separation in the hippocampus. Trends Neurosci. 2011;34:515–525. doi: 10.1016/j.tins.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Amaral RSC, Augustinack JC, Bender AR, Bernstein JD, Boccardi M, Bocchetta M, Burggren AC, Carr VA, Chakravarty MM, Chételat G, Daugherty AM, Davachi L, Ding SL, Ekstrom A, Geerlings MI, Hassan A, Huang Y, Iglesias JE, La Joie R, Kerchner GA, LaRocque KF, Libby LA, Malykhin N, Mueller SG, Olsen RK, Palombo DJ, Parekh MB, Pluta JB, Preston AR, Pruessner JC, Ranganath C, Raz N, Schlichting ML, Schoemaker D, Singh S, Stark CEL, Suthana N, Tompary A, Turowski MM, Van Leemput K, Wagner AD, Wang L, Winterburn JL, Wisse LEM, Yassa MA, Zeineh MM. Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: Towards a harmonized segmentation protocol. Neuroimage. 2015a;111:526–541. doi: 10.1016/j.neuroimage.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Pluta JB, Wang H, Xie L, Ding SL, Gertje EC, Mancuso L, Kliot D, Das SR, Wolk DA. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Human brain mapping. 2015b;36:258–287. doi: 10.1002/hbm.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Wang H, Pluta J, Das SR, Craige C, Avants BB, Weiner MW, Mueller S. Nearly automatic segmentation of hippocampal subfields in in vivo focal T2-weighted MRI. Neuroimage. 2010;53:1208–1224. doi: 10.1016/j.neuroimage.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]