Abstract
Astrocytes play critical roles in central nervous system (CNS) homeostasis and are implicated in the pathogenesis of neurological and psychiatric conditions, including drug dependence. Little is known about the effects of chronic ethanol consumption on astrocyte gene expression. To address this gap in knowledge, we performed transcriptome-wide RNA sequencing of astrocytes isolated from the prefrontal cortex (PFC) of mice following chronic ethanol consumption. Differential expression analysis revealed ethanol-induced changes unique to astrocytes that were not identified in total homogenate preparations. Astrocyte-specific gene expression revealed calcium-related signaling and regulation of extracellular matrix genes as responses to chronic ethanol use. These findings emphasize the importance of investigating expression changes in specific cellular populations to define molecular consequences of chronic ethanol consumption in mammalian brain.
Introduction
Proper functioning of astrocytes is critical for normal brain function1. Originally considered supportive cells for neurons, astrocytes are now recognized as crucial regulators of synapse formation, elimination, and maintenance2. They contribute to the prevention of neuronal excitotoxicity and regulate synaptic plasticity by clearing and metabolizing extracellular neurotransmitters3,4. Astrocytes can sense and actively participate in neuronal activity via activation of G-protein-coupled-receptor (GPCR) signaling cascades and elevations in intracellular calcium5. In addition, astrocytes can secrete proteins to modify the extracellular matrix, shaping intercellular interactions and altering synaptic plasticity6.
Astrocyte function can affect ethanol self-administration. For example, stimulation of nucleus accumbens core astrocytes using Designer Receptors Exclusively Activated by Designer Drugs (DREADDs) decreases intermittent access self-administration, indicating that changes in astrocyte-specific activity, specifically Gαq-coupled GPCR signaling, are sufficient to modulate behavioral responses to ethanol7. Astrocytes utilize gap junction channels for communication via calcium signaling8. Astrocyte-specific gap junction inhibition increases motivation for ethanol self-administration7, and leads to a transient increase in ethanol preference9. These studies suggest that changes in astrocyte function play an important role in ethanol consumption and preference; however, astrocyte-specific molecular changes in response to chronic ethanol exposure are largely unknown.
Gene expression studies also suggest that astrocytes may be involved in functional adaptations in response to ethanol exposure. Microarray analysis of post-mortem alcoholic human brain identified ethanol-responsive gene categories associated with glial function10,11, and gene network analyses have identified astrocyte-related changes in ethanol-exposed rodent brain12–14. However, these and other studies have inferred cell-type enrichment based on gene expression data from total homogenate preparations, which likely fail to detect important cell-type specific responses to ethanol. In cultured astrocytes, acute ethanol exposure alters transcription of genes in functional categories such as calcium signaling, cytoskeleton remodeling, and extracellular matrix15,16. Although cultured astrocyte studies have provided insights to response to alcohol exposure, there are abundant gene expression and likely functional differences between cultured astrocytes and astrocytes isolated from mature brain17,18; thus, transcriptome analysis of acutely isolated cell-types should provide a deeper understanding of brain function and pathological states19–21.
We report the first transcriptome-wide analysis of astrocyte-specific gene expression changes in response to chronic ethanol consumption. Astrocytes were isolated from the prefrontal cortex (PFC) after every-other-day (EOD) drinking, a paradigm that promotes escalation of intake22. We selected the PFC because it is known to be involved in drug and alcohol addiction23. RNA-sequencing was used to profile EOD-induced gene expression changes in astrocytes and total homogenate preparations. EOD caused differential regulation for several molecular functions, including calmodulin binding and extracellular matrix binding in astrocytes. Little overlap was observed between astrocyte-specific and total homogenate differential expression in response to ethanol; emphasizing the importance of studying cell-type specific expression changes to more completely understand the molecular consequences of chronic ethanol exposure in the adult mammalian brain.
Materials and Methods
Mice
Adult (8 weeks) male C57BL/6J mice were purchased from Jackson Laboratory (Bar Harbor, ME) and allowed to habituate to individual housing for at least one week. Mice were housed in the Animal Resource Center at The University of Texas Austin with 12-hr light/dark cycles and kept on a standard laboratory diet and water ad libitum. All experiments were approved by The University of Texas at Austin Institute for Animal Care and Use Committee and conducted in accordance with NIH guidelines regarding use of animals in research.
Every-other-day (EOD) ethanol drinking paradigm
Mice (n=12/group) were subjected to an EOD 2-bottle choice paradigm for 72 days (at least 36 drinking days total). Sample sizes were determined based on previous alcohol-related gene expression studies performed by our laboratory and others12,24,25. The EOD model is a variation of the continuous 2-bottle choice paradigm that promotes higher ethanol intake22. The treatment group had access to 15% (v/v) ethanol every other day and water every day, and the non-ethanol drinking control group only had access to water. To control for position preferences, bottle positions were switched daily. To control for evaporation and spillage, two bottles in an empty cage were also measured daily; one bottle containing water, and one containing ethanol. Ethanol quantity consumed (g/kg body weight/24h) and preference (ethanol consumed divided by total fluid intake) for each mouse was calculated. Mice were sacrificed 24 hours following their last drinking day.
Astrocyte enrichment and RNA isolation
Mice were anaesthetized briefly with isoflurane and perfused with phosphate-buffered saline (PBS). The PFC was dissected by removing the olfactory bulbs, then cutting the foremost 2mm of the cortex from each side at an approximate 50-degree angle from the midline, as previously described12. Tissue was minced and separated for total homogenate (approximately 5%) and astrocyte specific preparations. Total homogenate tissue was flash frozen in liquid nitrogen and stored at -80°C until further use. For astrocyte preparation, tissue was dissociated into a single cell suspension using the Neural Tissue Dissociation Kit with Papain (Miltenyi Biotec, Bergisch Gladbach, Germany). Myelin was removed by centrifugation in a 25% Percoll gradient. Astrocyte enrichment was performed using astrocyte cell surface antigen-2 (ACSA2) magnetic MicroBeads (Miltenyi Biotec). This isolation technique was selected based on previous success in purifying astrocytes from adult rodent brain26–28. RNA was extracted using the RNeasy Micro Kit (Qiagen, Hilden, Germany) and examined on the Bioanalyzer (Agilent Technologies, Santa Clara, CA) with the Agilent Total RNA 6000 Pico Kit for quality and quantity. Three astrocyte samples were excluded due to low RNA concentrations. RNA concentrations of samples used for RNA-sequencing ranged from 128 - 2355 pg/μl, with RNA integrity number (RIN) scores ranging from 8.0–9.4.
In a separate experiment, we validated astrocyte specificity with whole brain isolation. RNA was isolated using the MagMAX-96 Total RNA Isolation Kit (Thermo Fisher Scientific, Rockford, IL), quantified with the NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific) and analyzed for quality using a TapeStation instrument (Agilent Technologies). Reverse transcription was performed using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). Quantitative real-time PCR (qRT-PCR) was performed using TaqMan Universal PCR Master Mix and primer pairs and probes (Thermo Fisher Scientific) for astrocyte markers including Gfap, Aldh1l1, Slc1a3, as well as Itgam and Rbfox3 to identify microglial and neuronal gene expression, respectively (Figure 2). 18S rRNA was used as an endogenous control gene.
Figure 2.
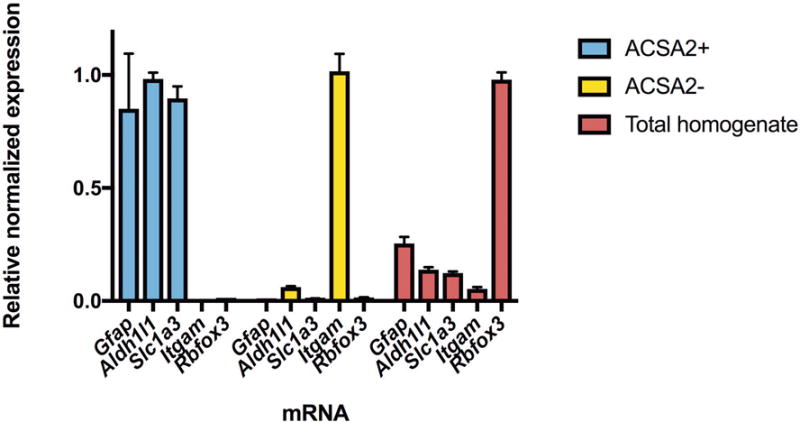
Analysis of cell fractions following astrocyte enrichment. The ACSA2+ fraction was isolated using the ACSA2 microbeads, while the ACSA2- fraction represents unbound cells. The total homogenate was not fractionated and contains all cells. qRT-PCR for astrocyte markers including Slc1a3, Aldh1l1 and Gfap confirm that the ACSA2+ fraction was enriched with astrocytes compared to the ACSA2- and total homogenate fractions. Markers for neurons (Rbfox3) and microglia (Itgam) show little neuronal or microglial gene expression in ACSA2+ cells. Expression levels are relative to 18S rRNA and normalized to the sample with highest expression for each marker. Values are mean ± SEM, for n=3.
RNA sequencing, read alignment, quality control, and quantification
A minimum of 1 ng RNA for each sample was submitted to the Genomic Sequencing and Analysis Facility at the University of Texas at Austin for mRNA selection with the MicroPoly(A) Purist Kit (Life Technologies, Carlsbad, CA) and library preparation with NEBNext Module Components (New England Biolabs, Ipswich, MA). Samples were sequenced on the Hi-Seq 4000 (Illumina) at a depth of ~20 million paired-end reads (150 base). Read quality was assessed using FASTQC (version 0.11.5)29. Adapter contamination was eliminated using Cutadapt (version 1.8), then reads were mapped to the mouse genome (UCSC mm10) with Bowtie2 (version 2.2.5)30. One sample was discarded due to poor mapping rate (< 50%). The final analyses included 20 ACSA2+ samples (n = 10/group) and 24 total homogenate samples (n = 12/group). Sorted BAM files were analyzed and duplicate reads filtered using Picard’s MarkDuplicates tool (version 1.141). Raw counts were quantified using HTSeq (version 0.5.3p9)31. FPKM were counted from sorted BAM files using Cufflinks (version 2.2.1)32. To ensure no perturbation of the transcriptome occurred due to the isolation procedure, we compared ratios of the FPKM values for generally accepted astrocyte markers Slc1a3 and Glul. Ratios of these markers were unchanged between ACSA2+ cells and total homogenate or between separate ACSA2+ samples, suggesting no marked transcriptome disruption occurs with astrocyte isolation. Raw and processed sequencing data from this study have been deposited to the Gene Expression Omnibus under accession number GSE92457.
Bioinformatics analysis
We utilized the phyper function of the R environment to determine whether there was significant overlap between our top 500 astrocyte-specific FPKM data and the top genes of a previously published astrocyte transcriptome20. Ethanol-mediated differential expression was determined using the Bioconductor package DESeq233. Differentially expressed genes (nominal p < 0.05) were entered into Enrichr34, an online tool that utilizes Fisher’s exact test to identify biological functions and pathways that are enriched in a list of genes. Genes differentially expressed below a nominal p-value threshold of 0.05 were included in Enrichr analysis to minimize type II error and include enough genes for functional enrichment. We also used gene set enrichment analysis (GSEA; version 2.2.2, Broad Institute)35, a ranked-based approach to identify enriched gene sets between control and ethanol-treated astrocytes. GSEA ranks all genes by expression level in both treatments and compares the pattern of expression in each treatment to established gene sets from the Molecular Signatures Database (version 5.1). Normalized counts obtained from DESeq2 were submitted to GSEA to calculate enrichment scores for gene sets. Specifically, Gene ontology (GO) molecular function gene sets (c5.mf.v5.1) were analyzed with GSEA to confirm Enrichr GO molecular function findings.
Immunohistochemistry
Mice were perfused with PBS and 4% paraformaldehyde (PFA), and brains were harvested. Brains were post-fixed in 4% PFA for 24 hours, then cryoprotected in 20% sucrose for 24 hours. Brains were frozen in optimal cutting temperature (OCT) and stored at -80C until sectioning. 20 μm sections were permeabilized in 0.05% Triton-X for 10 minutes and blocked in 10% goat serum for 1 hour at RT. Sections were incubated overnight at 4°C in primary antibody (mouse anti-Glutamine synthetase [GS] 1:1000 [Millipore, Billerica, MA, Catalog #MAB302] and rabbit anti-CAMK2G 1:100 [Life Technologies, Carlsbad, CA, Catalog #PA514035]). Following three washes in PBS, sections were incubated in secondary antibody (goat anti-mouse 594, goat anti-rabbit 488, Thermo Fisher Scientific, Rockford, IL) for 2 hours at RT. Sections were mounted in 0.2% gelatin, dehydrated, and cover slipped with mounting medium (Vector Labs, Burlingame, CA). Sections were visualized using a Zeiss Axiovert 200 M fluorescent light microscope (Zeiss, Thornwood, NY). Bilateral images of the PFC (Bregma +2.8 to +2.24) were captured using a 20× objective. Images were analyzed using ImageJ (version 1.50i). GS, CAMK2G, and dual-labeled cells were quantified in medial regions of the PFC in two regions of interest per section (box, 500 × 500 μm). Cells were considered co-localized when they expressed both red (GS) and green (CAMK2G). Percentages of co-localized cells were averaged between two sections per animal (n = 3).
Results
Ethanol consumption
Mice subjected to EOD drinking consumed an average of 174 g/kg ethanol total over the course of the experiment. Average daily ethanol intake and preference are displayed in Figure 1. Daily ethanol intake significantly differed over time, as indicated by a repeated-measures ANOVA (F(4.17, 55.90) = 11.34, p < 1.00e-04). These results are consistent with previous work establishing the EOD paradigm as a relevant model for human excessive alcohol consumption and escalation of drug use22.
Figure 1.
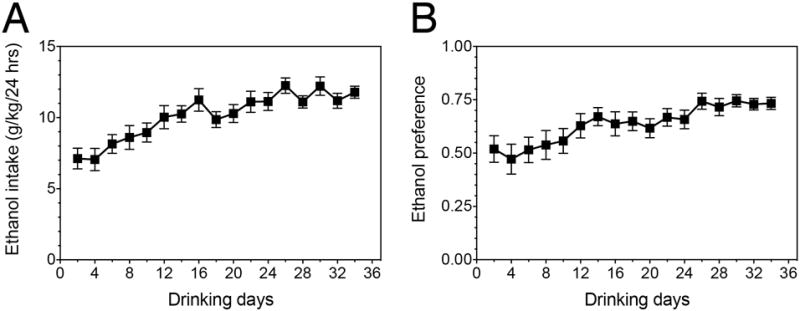
Ethanol consumption and preference of EOD drinking mice. (A) Average daily ethanol intake throughout the course of the experiment. (B) Average ethanol preference throughout the course of the experiment (amount of ethanol consumed divided by the total amount of fluids consumed per day). Values are mean ± SEM, for n=12.
Astrocyte isolation
We first verified that the astrocyte isolation technique selectively targeted astrocytes, as observed in prior studies26,36. We used qRT-PCR to confirm that astrocyte-specific genes were enriched when astrocytes were isolated using ACSA2+ magnetic beads (Figure 2). We observed robust enrichment of astrocyte markers Aldh1l1, Gfap, and Slc1a3 in ACSA2+ cells and negligible levels of the microglial marker Itgam and the neuronal marker Rbfox3, indicating an astrocyte-enriched population. In contrast, RNA from total homogenate tissue was enriched with Rbfox3 and contained both astrocyte and microglia markers. In addition, the 500 highest expressed genes (based on FPKM expression level) in ACSA2+ cells from the PFC were compared to the top 500 highest expressed genes in astrocytes, microglia, neurons, and three oligodendrocyte cell-types from a previously published cell-type specific transcriptome resource20. We observed the most overlap between ACSA2+ cells and astrocytes, with 225 genes shared between the top 500 genes of each transcriptome (p = 2.52e-214). Genes that did not overlap could be due to factors that were distinct for each experiment, such as regional heterogeneity of astrocyte gene expression, differences in isolation protocol, or age-specific differences in gene expression. The 225 overlapping genes were submitted to Enrichr34 for molecular function analysis and we identified categories including GTPase activity (p = 2.58e-08), guanyl nucleotide binding (p = 5.67e-05), and amino acid transmembrane transporter activity (p = 5.47e-05) as highly enriched in astrocytes (Supplementary Table 1).
Differential expression produced by chronic ethanol exposure
In response to EOD, a total of 800 genes were differentially expressed in PFC astrocytes (p < 0.05) (Figure 3A). In contrast, fewer genes (589) were differentially expressed in the total homogenate (p < 0.05) (Figure 3B). Differentially expressed genes are listed in Supplementary Tables 2 and 3. Only 24 genes were identified as differentially expressed in both preparations (Figure 3C); however, 15 of the overlapping genes (63%) changed in opposite directions, suggesting that in different cell-types, the same genes may have opposite ethanol-induced expression changes. In general, the magnitude of change in differentially expressed genes was greater in astrocytes compared to total homogenate (Figure 3D). The absolute value of log2-fold change in astrocytes was 0.27 (~20% change in expression, fold change=1.20), compared to 0.17 for total homogenate (~13% change, fold change=1.13). These results indicate that analyzing isolated astrocytes reveals unique genomic responses to ethanol that are not observed in total homogenate preparations.
Figure 3.
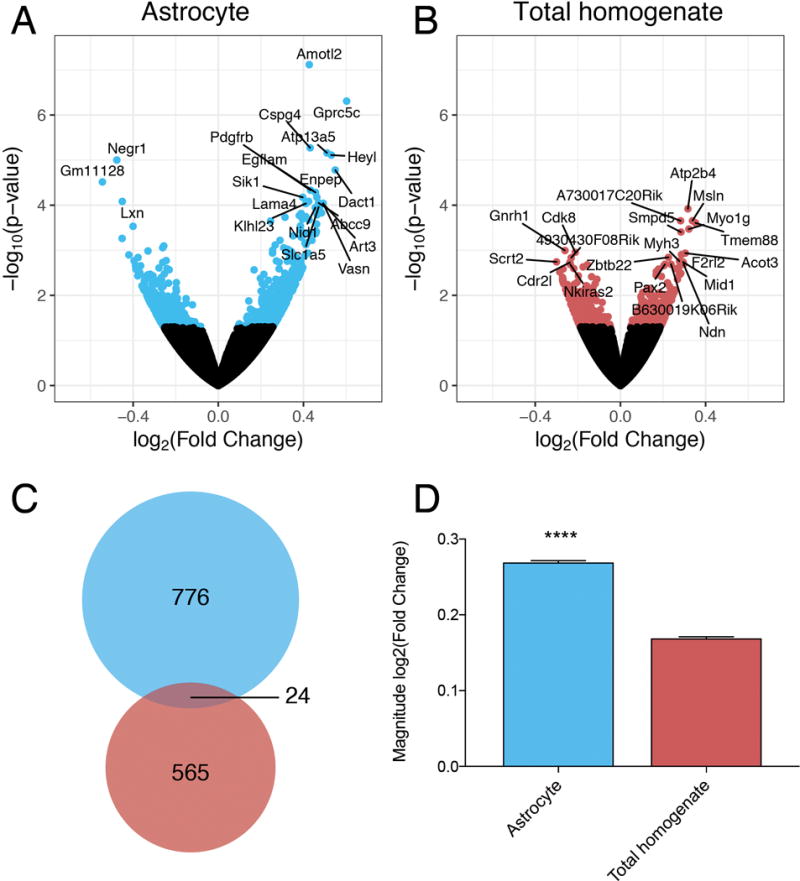
Differential gene expression in response to chronic ethanol exposure. Volcano plots show differentially expressed genes (p < 0.05) in (A) astrocytes (blue) and (B) total homogenate (red) after EOD. The top 20 differentially expressed genes for each preparation are labeled with gene symbols. (C) 24 genes (overlapping area) were differentially expressed in both the astrocytes (blue) and the total homogenate (red) after EOD. (D) Magnitude of fold change of differentially expressed genes in astrocyte and total homogenate preparations (*** indicates p < 0.0001, unpaired t-test)
Enrichr analysis of differentially expressed genes
Differentially expressed genes (p < 0.05) identified from astrocytes and total homogenate preparations were further characterized using the enrichment analysis tool Enrichr to identify biological functions and pathways regulated in response to ethanol exposure. Gene ontology (GO) molecular function terms associated with altered astrocyte expression after EOD are listed in Table 1 and Supplementary Table 4. The top-ranked GO term was calmodulin binding. Interestingly, calmodulin binding was also the top term identified in total homogenate preparations (p = 1.64e-04); however, the specific genes within the category were distinct from those in astrocytes. Results for total homogenate also included functionally distinct categories such as microfilament motor activity (GO:0000146, p = 3.09e-04) and coreceptor activity (GO:0015026, p = 8.43e-04) (Supplementary Table 5).
Table 1.
Molecular functions differentially regulated in astrocytes after EOD drinking.
| ID | Name | # DE/# in category | p-value |
|---|---|---|---|
| GO:0005516 | Calmodulin binding | 18/170 | 1.64e-04 |
| GO:0017075 | Syntaxin-1 binding | 5/17 | 4.19e-04 |
| GO:0005535 | Phosphatidylinositol phospholipase C activity | 6/28 | 7.12e-04 |
| GO:0005178 | Integrin binding | 12/102 | 7.69e-04 |
| GO:0050840 | Extracellular matrix binding | 8/51 | 8.77e-04 |
| GO:0004629 | Phospholipase C activity | 6/30 | 1.05e-03 |
| GO:0019838 | Growth factor binding | 13/123 | 1.30e-03 |
| GO:0045125 | Bioactive lipid receptor activity | 3/8 | 3.07e-03 |
| GO:0050839 | Cell adhesion molecule binding | 15/168 | 3.12e-03 |
| GO:0016298 | Lipase activity | 11/105 | 3.20e-03 |
| GO:0004879 | Ligand-activated sequence-specific DNA binding RNA polymerase II transcription factor activity | 7/50 | 3.56e-03 |
| GO:0070679 | Inositol 1,4,5 trisphosphate binding | 3/9 | 4.47e-03 |
| GO:0001618 | Virus receptor activity | 5/29 | 5.40e-03 |
| GO:0001540 | Beta-amyloid binding | 5/29 | 5.40e-03 |
| GO:0022890 | Inorganic cation transmembrane transporter activity | 32/497 | 5.78e-03 |
| GO:0008373 | Sialyltransferase activity | 4/20 | 7.38e-03 |
| GO:0003707 | Steroid hormone receptor activity | 7/57 | 7.41e-03 |
| GO:0008081 | Phosphoric diester hydrolase activity | 9/90 | 9.85e-03 |
| GO:0015077 | Monovalent inorganic cation transmembrane transporter activity | 23/343 | 1.12e-02 |
| GO:0005518 | Collagen binding | 7/62 | 1.16e-02 |
Enrichr was used to examine enrichment of biological pathways associated with astrocyte-specific differentially expressed genes, using multiple databases including Kegg, WikiPathways, and Reactome (Table 2). Consistent with the GO molecular function results described above for differential gene expression due to ethanol, pathway analysis identified the category “Calcium regulation in the cardiac cell”, further supporting a role for astrocyte-specific genes involved in calcium-related events occurring in response to ethanol. In addition, both molecular function and pathway analyses indicate changes in extracellular matrix genes in astrocytes (Table 1 and 2).
Table 2.
Pathways differentially regulated in astrocytes after EOD drinking.
| Database | Name | ID | #DE/# in category | p-value |
|---|---|---|---|---|
| KEGG | ECM-receptor interaction | Hsa04512 | 14/82 | 4.21e-06 |
| KEGG | cGMP-PKG signaling pathway | Hsa04022 | 19/167 | 4.11e-05 |
| KEGG | Focal adhesion | Hsa04510 | 20/202 | 1.83e-04 |
| KEGG | Vascular smooth muscle contraction | Hsa04270 | 13/120 | 1.03e-03 |
| KEGG | PI3-Akt signaling pathway | Hsa04151 | 26/341 | 1.32e-03 |
| WikiPathways | Calcium regulation in the cardiac cell | WP553 | 20/150 | 2.34e-06 |
| WikiPathways | Myometrial relaxation and contraction pathways | WP289 | 18/156 | 5.40e-05 |
| WikiPathways | Focal adhesion-Pi3K-Akt-mTOR signaling pathway | WP2841 | 26/303 | 2.26e-04 |
| WikiPathways | Focal adhesion | WP85 | 18/185 | 4.66e-04 |
| WikiPathways | Non-odorant GPCRs | WP1396 | 22/256 | 6.54e-04 |
| Reactome | Laminin organization | R-HSA-3000157 | 9/23 | 1.24e-07 |
| Reactome | Extracellular matrix organization | R-HSA-1474244 | 28/283 | 1.05e-05 |
| Reactome | Non-integrin membrane-ECM interactions | R-HSA-3000171 | 8/42 | 2.23e-04 |
| Reactome | ECM proteoglycans | R-HSA-3000178 | 9/55 | 3.05e-04 |
| Reactome | Integrin cell surface interactions | R-HSA-216083 | 10/62 | 3.12e-04 |
Gene Set Enrichment Analysis (GSEA)
Normalized counts for all identified genes were analyzed using GSEA35 to identify gene sets associated with chronic ethanol-exposed astrocytes. The highest ranked altered molecular function was identified as “calmodulin binding” (Figure 4A; p < 1.00e-03). Additional gene sets enriched in ethanol-exposed astrocytes include specific transcriptional repressor protein (p < 1.00e-03), protein complex binding (p = 0.02), and extracellular matrix structural constituent (Figure 4B; p = 0.03). In total homogenate preparations, gene sets such as phospholipid binding, nucleoside triphosphatase activity, and endonuclease activity were differentially regulated. Complete lists of molecular function gene sets enriched in astrocytes and total homogenate tissue isolated from chronic ethanol-drinking mice are included in Supplementary Tables 6 and 7. Overall, GSEA and Enrichr results demonstrate consistent evidence of an astrocyte-specific chronic ethanol response that includes altered calcium signaling and extracellular matrix remodeling.
Figure 4.
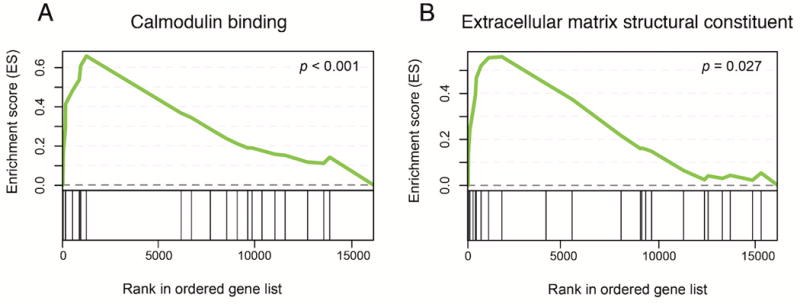
Enrichment plots from GSEA. GSEA ranks genes by correlating gene expression with a specific phenotype. In panels A and B, genes that are more highly correlated with ethanol treatment are ranked closer to 0, while those correlated with the control phenotype rank closer to 15 000 (x-axis). Enrichment score (ES; y-axis) for a given gene set increases as a gene within the set is present while walking down the rank ordered list. The greatest deviation from 0 on the plot represents the ES for a given gene set.
Astrocytic protein expression of a differentially expressed calmodulin signaling gene
We chose a differentially expressed candidate gene, Camk2g, to confirm astrocyte-specific protein expression. Camk2g encodes a Ca2+/calmodulin dependent protein kinase isoform and was down-regulated in astrocytes following every-other-day drinking. As published data regarding CAMK2G’s expression in astrocytes is lacking, we sought to verify that the protein was expressed in astrocytes in the mouse PFC. Co-localization analysis of CAMK2G and astrocyte-specific marker glutamine synthetase (GS) revealed that CAMK2G protein is expressed in about 30% of GS+ PFC astrocytes (Figure 5).
Figure 5.
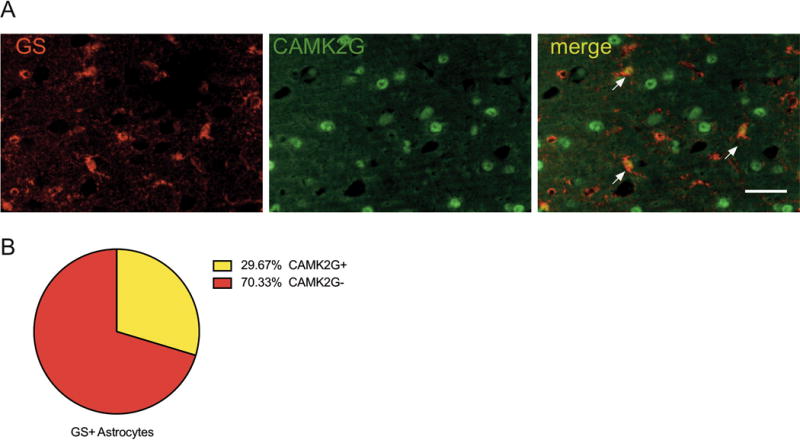
CAMK2G expression in astrocytes. (A) Representative image shows co-localization of CAMK2G (green) and GS astrocytes (red). Arrowheads point to examples of co-localization. Scale bar corresponds to 50μm. (B) 30% of GS+ astrocytes expressed CAMK2G. Cell counts were averaged between regions of interest from two sections per animal (n = 3).
Comparison of previous studies differentially expressed genes
We compared our astrocyte-specific differential expression data to a previously published ethanol-related astrocyte-specific gene expression dataset, as well as several total homogenate datasets to validate the current study’s ethanol-responsive gene expression changes (Table 3). The overlapping genes between our study and each compared previous study were submitted to Enrichr for GO molecular function analysis. The top two functional categories are listed for each in Table 3.
Table 3.
Comparison of differentially expressed genes in astrocytes with previous literature
| Tissue | Alcohol paradigm | Transcriptome profiling method | Number of overlapping differentially expressed genes | Top GO molecular function categories of overlapping genes | Study |
|---|---|---|---|---|---|
| Primary cultured mouse cortical astrocytes | Acute ethanol exposure | microarray | 62 | Peptidase regulator activity (GO:0061134), GTPase activity (GO:0003924) | Pignataro et al. 2013 |
| Human frontal cortex | Lifetime alcohol consumption | RNA-sequencing | 114 | calmodulin binding (GO:0005516), calmodulin-dependent protein kinase activity (GO:0004683) | Farris et al. 2015 |
| Human frontal cortex | Human alcoholic | microarray | 106 | Actin binding (GO:0003779), calcium ion binding (GO:0005509) | Ponomarev et al. 2012 |
| Mouse frontal cortex | Chronic intermittent drinking | microarray | 27 | Transcription corepressor activity (GO:0003714) | Osterndorff-Kahanek et al. 2013 |
Discussion
There is growing evidence that astrocyte function is linked to neurological disorders34,38; however, the potential roles of astrocytes in the neurobiological mechanisms of alcohol use disorder are largely unknown. Here, we found that chronic ethanol consumption leads to unique changes to the astrocyte transcriptome in the mouse prefrontal cortex (PFC). Our results suggest astrocyte-specific gene expression changes involving calcium signaling and extracellular matrix regulation may contribute to the brain adaptations driven by chronic ethanol consumption.
Previous studies11, 13 have attempted to parse out cell-type specific contributions to ethanol-induced gene expression changes observed in whole tissue, relying on the use of astrocyte-enriched gene lists that may not be accurate for astrocytes in each brain region or at every developmental stage. Our approach circumvents the need for predefined cell-type gene lists by isolating astrocytes from the adult mouse PFC and profiling ethanol-induced transcriptome alterations with RNA-sequencing. Our astrocyte transcriptome data shows significant overlap with that of a previously published astrocyte transcriptome study20, and functional enrichment analysis of the overlapping genes adds confidence to the cellular identity of the isolated cells. In addition, as most prior astrocyte-specific transcriptome profiling has been performed on astrocytes isolated from immature mice or from broadly defined brain regions17,20,39,40, our isolated astrocyte transcriptome data represent a new resource for researchers interested in biological functions of adult astrocytes in the mouse PFC.
Differential expression analysis revealed that most gene expression changes occurring in astrocytes isolated from mice with a history of chronic, voluntary ethanol consumption are not observable in total homogenate tissue. This is indicated by the small amount of overlap between differentially expressed genes in both astrocytes and total homogenate tissue. In addition, the differentially expressed genes common to both preparations do not consist of the most significantly differentially expressed genes in either. Therefore, not only are most ethanol-induced astrocyte gene expression changes not observed in total homogenate tissue, even the most significantly changing genes are not detected. A greater number of differentially expressed genes and slightly larger magnitude changes were observed in astrocytes compared to total homogenate tissue. This could be because gene expression changes observed in isolated astrocytes are not diluted by stably expressing transcripts in other cells that mask subtle differences in astrocytes. These results further emphasize the importance of examining isolated cellular populations to define changes in gene expression associated with alcohol abuse, and suggest that previous work examining total homogenate preparations likely underestimates astrocyte-specific effects.
We compared our astrocyte-specific expression data to published reports investigating ethanol-related gene expression changes, and found that 62 astrocyte-specific genes identified in the current study were also observed in response to acute ethanol in cultured astrocytes16. Importantly, these expression changes also overlapped significantly with genes identified in human alcoholic frontal cortex correlating to lifetime alcohol consumption (114 genes in common; p = 2.66e-06; Fisher’s exact test)41 and differentially expressed genes identified in a microarray study of human alcoholic frontal cortex11. This overlap could be explained by increased astrocyte representation in total homogenate human brain tissue, as human astrocytes have a much larger volume than mouse astrocytes42 and the human cerebral cortex may contain more astrocytes relative to neurons compared to mouse cortex43, although the ratio of astrocytes to neurons in both species has not been definitively established. There is also likely brain-region heterogeneity that has not been fully elucidated44. Recent evidence suggests the relative abundance of astrocytes in mouse cortex may be lower than previously thought45, possibly explaining the lack of agreement between astrocyte-specific and mouse total homogenate findings. Comparison with differential expression identified in a previously published microarray study of every-other-day drinking mice12 showed less overlap with astrocyte differentially expressed genes, consistent with the present study. Enrichr analysis identified calmodulin binding (p = 2.68e-05) and calmodulin-dependent protein kinase activity (GO:0004683; p = 9.67e-04) as common important functional categories shared between astrocyte and human alcohol differentially expressed genes, suggesting that additional astrocyte-specific studies will inform our understanding of molecular changes occurring in human alcoholics.
Enrichr analysis of differentially expressed genes as well as GSEA of normalized counts for all measured genes indicate changes in calmodulin binding in astrocytes after EOD exposure. Changes in calmodulin binding indicate altered Ca2+ activity, as calmodulin is activated with intracellular release of Ca2+ 46. Pathway analysis with Enrichr also suggested altered Ca2+ signaling in astrocytes after EOD. The ability for astrocytes to respond to neuronal activity through G-protein coupled receptor (GPCR) activation, intracellular Ca2+ fluctuations, and subsequent release of gliotransmitters is a mechanism underlying astrocyte modulation of synaptic transmission5. Previous studies have indicated an involvement for astrocyte Ca2+ signaling in a wide range of behaviors5, including motivation for alcohol consumption7. Our study provides the first evidence of chronic ethanol-mediated Ca2+ signaling modulation specifically in PFC astrocytes. GSEA analysis indicated an overall up-regulation of calmodulin binding related genes in astrocytes isolated from ethanol drinking mice. This suggests an increase in Ca2+-related signaling cascades in PFC astrocytes 24 hours after ethanol exposure, possibly mediated by GPCR activation. Indeed, genes coding for multiple members of the regulators of G-protein signaling (RGS) family had increased expression in astrocytes after ethanol consumption, including Rgs4, Rgs5, and Rgs16. Altered expression of RGS proteins, specifically Rgs4, has been implicated in neuropsychiatric disorders including drug addiction47. Other genes involved in calmodulin binding showed decreased expression in astrocytes after EOD, including the calcium/calmodulin dependent protein kinase Camk2g. Altered expression of Camk2g has been identified in other studies related to alcohol abuse16,48 including an astrocyte-specific transcriptome study16, however the role of Camk2g in astrocytes has not been characterized, nor has its protein expression pattern in mouse PFC. Double immunohistochemistry of CAMK2G and GS revealed astrocytic expression of the protein in a minority of astrocytes, indicating possible subpopulations of PFC astrocytes that may have functional heterogeneity, which warrants further investigation. In addition, while CAMK2G was expressed mainly in non-astrocyte cells, our study showed the RNA was differentially expressed in astrocytes but not total homogenate tissue. This illustrates the importance of how certain genes can be altered by alcohol only in specific cell populations; which may have been obscured in previous studies. Expression changes of a particular gene in one cell type may have different functional consequences than in another, and isolating cell populations is a way to isolate these cell-specific changes and interpret potential functional changes more accurately. In cultured cortical astrocytes, CaMKII inhibition can modulate glutamate uptake and lead to release of ATP, possibly altering synaptic activity through activation of adenosine receptors49, processes that can affect ethanol reward50,51. Therefore, changes in astrocyte calmodulin signaling specifically affecting calcium/calmodulin dependent protein kinases in astrocytes may have behavioral effects relevant to ethanol consumption.
Pathway analysis with Enrichr and GSEA identified a consistent overall up-regulation of extracellular matrix (ECM) genes in astrocytes following chronic ethanol exposure. The ECM and its associated proteins have been gaining attention as important factors in regulating plasticity-mediated addiction processes52,53. In addition, ECM genes were differentially expressed in cultured astrocytes in response to acute ethanol exposure16. One ECM gene with increased expression in astrocytes following chronic ethanol was Plat (tissue plasminogen activator [tPA]), a serine protease that regulates the structure of the ECM and can mediate many forms of synaptic plasticity54. tPA is elevated during ethanol withdrawal and its deletion in mice reduces chronic ethanol-induced withdrawal seizures55. The known roles of tPA in ethanol-related behaviors combined with our gene expression data hints at the intriguing possibility that PFC astrocytes may influence chronic ethanol consumption through the regulation of tPA. Many more ECM genes had increased expression in EOD-treated astrocytes, including laminin genes (Lama4, Lamb1, and Lamc1), collagen genes (Col4a2 and Col4a4), and integrin genes (Itga1, Itga7). Overall, these changes indicate complex alterations in the structure of the ECM and altered interactions between cells, which could influence synaptic plasticity and behavior56. While further research is needed to determine the possible consequences of these changes, our results point to an astrocyte-specific role in ECM remodeling after chronic ethanol exposure.
A major goal of transcriptome profiling is to elucidate gene pathways or networks that are perturbed by the disease state. We used several computational approaches to highlight potential new astrocytic targets that may be important for escalation of ethanol intake in an EOD paradigm, including Ca2+-related signaling and ECM components. It should be emphasized that changes in astrocytes from chronic ethanol consumption are only weakly and partially represented in the total homogenate, demonstrating the importance of transcriptome profiling in specific cell populations to better understand the cellular mechanisms of addiction. In addition, differentially expressed genes in astrocytes overlapped with changes observed in human alcoholic brain, indicating the relevance of these findings to human alcohol abuse. There is emerging interest in using gene expression profiles to predict therapeutics57, and the success of this approach for brain diseases will likely require expression profiles from astrocytes and other distinct brain cell populations. As our study is the first to profile the astrocyte-specific transcriptome in the adult mouse PFC, our data not only provides a framework for studying astrocyte responses in relation to chronic ethanol, it serves as a novel resource for future efforts in understanding the complexity of astrocyte function.
Supplementary Material
Acknowledgments
We thank Mendy Black and Adriana Dacosta for assisting with drinking experiments.
Funding
The work published in this paper represents original research, not previously submitted or published elsewhere. This work was supported by NIH/NIAAA INIA Consortium U01 AA13520, R01 AA06399, U01 AA020926, R01 AA012404, and a donation from June Waggoner.
Footnotes
Disclosure
The authors declare no conflict of interest.
References
- 1.Khakh BS, Sofroniew MV. Diversity of astrocyte functions and phenotypes in neural circuits. Nat Neurosci. 2015;18:942–952. doi: 10.1038/nn.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung WS, Allen NJ, Eroglu C. Astrocytes Control Synapse Formation, Function, and Elimination. Cold Spring Harb Perspect Biol. 2015;7:a020370. doi: 10.1101/cshperspect.a020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung W-S, Welsh CA, Barres BA, Stevens B. Do glia drive synaptic and cognitive impairment in disease? Nat Neurosci. 2015;18:1539–1545. doi: 10.1038/nn.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Šerý O, Sultana N, Kashem MA, Pow DV, Balcar VJ. GLAST But Not Least-Distribution, Function, Genetics and Epigenetics of L-Glutamate Transport in Brain-Focus on GLAST/EAAT1. Neurochem Res. 2015;40:2461–2472. doi: 10.1007/s11064-015-1605-2. [DOI] [PubMed] [Google Scholar]
- 5.Bazargani N, Attwell D. Astrocyte calcium signaling: the third wave. Nat Neurosci. 2016;19:182–189. doi: 10.1038/nn.4201. [DOI] [PubMed] [Google Scholar]
- 6.Jones EV, Bouvier DS. Astrocyte-secreted matricellular proteins in CNS remodelling during development and disease. Neural Plast. 2014;2014:321209–12. doi: 10.1155/2014/321209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bull C, Freitas KCC, Zou S, Poland RS, Syed WA, Urban DJ, et al. Rat nucleus accumbens core astrocytes modulate reward and the motivation to self-administer ethanol after abstinence. Neuropsychopharmacology. 2014;39:2835–2845. doi: 10.1038/npp.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett MV, Contreras JE, Bukauskas FF, Sáez JC. New roles for astrocytes: gap junction hemichannels have something to communicate. Trends Neurosci. 2003;26:610–617. doi: 10.1016/j.tins.2003.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miguel-Hidalgo J, Shoyama Y, Wanzo V. Infusion of gliotoxins or a gap junction blocker in the prelimbic cortex increases alcohol preference in Wistar rats. J Psychopharmacol. 2009;23:550–557. doi: 10.1177/0269881108091074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, et al. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31:1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- 11.Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osterndorff-Kahanek E, Ponomarev I, Blednov YA, Harris RA. Gene expression in brain and liver produced by three different regimens of alcohol consumption in mice: comparison with immune activation. PLoS ONE. 2013;8:e59870. doi: 10.1371/journal.pone.0059870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osterndorff-Kahanek EA, Becker HC, Lopez MF, Farris SP, Tiwari GR, Nunez YO, et al. Chronic ethanol exposure produces time- and brain region-dependent changes in gene coexpression networks. PLoS ONE. 2015;10:e0121522. doi: 10.1371/journal.pone.0121522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saba LM, Flink SC, Vanderlinden LA, Israel Y, Tampier L, Colombo G, et al. The sequenced rat brain transcriptome–its use in identifying networks predisposing alcohol consumption. FEBS J. 2015;282:3556–3578. doi: 10.1111/febs.13358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanco AM, Pascual M, Valles SL, Guerri C. Ethanol-induced iNOS and COX-2 expression in cultured astrocytes via NF-kappa B. Neuroreport. 2004;15:681–685. doi: 10.1097/00001756-200403220-00021. [DOI] [PubMed] [Google Scholar]
- 16.Pignataro L, Varodayan FP, Tannenholz LE, Protiva P, Harrison NL. Brief alcohol exposure alters transcription in astrocytes via the heat shock pathway. Brain Behav. 2013;3:114–133. doi: 10.1002/brb3.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG, et al. Genomic analysis of reactive astrogliosis. J Neurosci. 2012;32:6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srinivasan K, Friedman BA, Larson JL, Lauffer BE, Goldstein LD, Appling LL, et al. Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat Commun. 2016;7:11295. doi: 10.1038/ncomms11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melendez RI. Intermittent (every-other-day) drinking induces rapid escalation of ethanol intake and preference in adolescent and adult C57BL/6J mice. Alcohol Clin Exp Res. 2011;35:652–658. doi: 10.1111/j.1530-0277.2010.01383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldstein RZ, Volkow ND. Dysfunction of the prefrontal cortex in addiction: neuroimaging findings and clinical implications. Nat Rev Neurosci. 2011;12:652–669. doi: 10.1038/nrn3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Most D, Ferguson L, Blednov Y, Mayfield RD, Harris RA. The synaptoneurosome transcriptome: a model for profiling the emolecular effects of alcohol. Pharmacogenomics J. 2015;15:177–188. doi: 10.1038/tpj.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Repunte-Canonigo V, Shin W, Vendruscolo LF, Lefebvre C, van der Stap L, Kawamura T, et al. Identifying candidate drivers of alcohol dependence-induced excessive drinking by assembly and interrogation of brain-specific regulatory networks. Genome Biol. 2015;16:68. doi: 10.1186/s13059-015-0593-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt LM, Olsen ML. Novel Applications of Magnetic Cell Sorting to Analyze Cell-Type Specific Gene and Protein Expression in the Central Nervous System. PLoS ONE. 2016;11:e0150290. doi: 10.1371/journal.pone.0150290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kantzer GC, Boutin C, Herzig ID, Wittwer C, Reiß S, Tiveron MC, et al. Anti-ACSA-2 defines a novel monoclonal antibody for prospective isolation of living neonatal and adult astrocytes. Glia. 2017;101:8384–1004. doi: 10.1002/glia.23140. [DOI] [PubMed] [Google Scholar]
- 28.Batiuk MY, de Vin F, Duqué SI, Li C, Saito T, Saido T, et al. An immunoaffinity-based method for isolating ultrapure adult astrocytes based on ATP1B2 targeting by the ACSA-2 antibody. J Biol Chem. 2017;292:8874–8891. doi: 10.1074/jbc.M116.765313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010 bioinformatics.babraham.ac.uk. http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed 27 Sep2016)
- 30.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N, et al. Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci. 2015;18:1819–1831. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao X, Li L-P, Wang Q, Wu Q, Hu H-H, Zhang M, et al. Astrocyte-derived ATP modulates depressive-like behaviors. Nat Med. 2013;19:773–777. doi: 10.1038/nm.3162. [DOI] [PubMed] [Google Scholar]
- 38.Matos M, Shen HY, Augusto E, Wang Y, Wei CJ, Wang YT, et al. Deletion of adenosine A2A receptors from astrocytes disrupts glutamate homeostasis leading to psychomotor and cognitive impairment: relevance to schizophrenia. Biol Psychiatry. 2015;78:763–774. doi: 10.1016/j.biopsych.2015.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orre M, Kamphuis W, Osborn LM, Melief J, Kooijman L, Huitinga I, et al. Acute isolation and transcriptome characterization of cortical astrocytes and microglia from young and aged mice. Neurobiol Aging. 2014;35:1–14. doi: 10.1016/j.neurobiolaging.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 40.Simpson JE, Ince PG, Shaw PJ, Heath PR, Raman R, Garwood CJ, et al. Microarray analysis of the astrocyte transcriptome in the aging brain: relationship to Alzheimer’s pathology and APOE genotype. Neurobiol Aging. 2011;32:1795–1807. doi: 10.1016/j.neurobiolaging.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Farris SP, Arasappan D, Hunicke-Smith S, Harris RA, Mayfield RD. Transcriptome organization for chronic alcohol abuse in human brain. Mol Psychiatry. 2015;20:1438–1447. doi: 10.1038/mp.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oberheim NA, Takano T, Han X, He W, Lin JHC, Wang F, et al. Uniquely hominid features of adult human astrocytes. J Neurosci. 2009;29:3276–3287. doi: 10.1523/JNEUROSCI.4707-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herculano-Houzel S. The glia/neuron ratio: how it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia. 2014;62:1377–1391. doi: 10.1002/glia.22683. [DOI] [PubMed] [Google Scholar]
- 44.Chai H, Diaz-Castro B, Shigetomi E, Monte E, Octeau JC, Yu X, et al. Neural Circuit-Specialized Astrocytes: Transcriptomic, Proteomic, Morphological, and Functional Evidence. Neuron. 2017 doi: 10.1016/j.neuron.2017.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun W, Cornwell A, Li J, Peng S, Osorio MJ, Aalling N, et al. SOX9 Is an Astrocyte-Specific Nuclear Marker in the Adult Brain Outside the Neurogenic Regions. J Neurosci. 2017;37:4493–4507. doi: 10.1523/JNEUROSCI.3199-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braun A. The Multifunctional Calcium/Calmodulin-Dependent Protein Kinase: From Form to Function. Annual Review of Physiology. 1995;57:417–445. doi: 10.1146/annurev.ph.57.030195.002221. [DOI] [PubMed] [Google Scholar]
- 47.Roman DL, Traynor JR. Regulators of G protein signaling (RGS) proteins as drug targets: modulating G-protein-coupled receptor (GPCR) signal transduction. J Med Chem. 2011;54:7433–7440. doi: 10.1021/jm101572n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mulligan MK, Rhodes JS, Crabbe JC, Mayfield RD, Harris RA, Ponomarev I. Molecular profiles of drinking alcohol to intoxication in C57BL/6J mice. Alcohol Clin Exp Res. 2011;35:659–670. doi: 10.1111/j.1530-0277.2010.01384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashpole NM, Chawla AR, Martin MP, Brustovetsky T, Brustovetsky N, Hudmon A. Loss of calcium/calmodulin-dependent protein kinase II activity in cortical astrocytes decreases glutamate uptake and induces neurotoxic release of ATP. J Biol Chem. 2013;288:14599–14611. doi: 10.1074/jbc.M113.466235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee MR, Ruby CL, Hinton DJ, Choi S, Adams CA, Young Kang N, et al. Striatal adenosine signaling regulates EAAT2 and astrocytic AQP4 expression and alcohol drinking in mice. Neuropsychopharmacology. 2013;38:437–445. doi: 10.1038/npp.2012.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith KL, John CS, Sypek EI, Öngür D, Cohen BM, Barry SM, et al. Exploring the role of central astrocytic glutamate uptake in ethanol reward in mice. Alcohol Clin Exp Res. 2014;38:1307–1314. doi: 10.1111/acer.12361. [DOI] [PubMed] [Google Scholar]
- 52.Lasek AW. Effects of Ethanol on Brain Extracellular Matrix: Implications for Alcohol Use Disorder. Alcohol Clin Exp Res. 2016 doi: 10.1111/acer.13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith AC, Scofield MD, Kalivas PW. The tetrapartite synapse: Extracellular matrix remodeling contributes to corticoaccumbens plasticity underlying drug addiction. Brain Res. 2015;1628:29–39. doi: 10.1016/j.brainres.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melchor JP, Strickland S. Tissue plasminogen activator in central nervous system physiology and pathology. Thromb Haemost. 2005;93:655–660. doi: 10.1160/TH04-12-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pawlak R, Melchor JP, Matys T, Skrzypiec AE, Strickland S. Ethanol-withdrawal seizures are controlled by tissue plasminogen activator via modulation of NR2B-containing NMDA receptors. Proc Natl Acad Sci USA. 2005;102:443–448. doi: 10.1073/pnas.0406454102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frischknecht R, Gundelfinger ED. The brain’s extracellular matrix and its role in synaptic plasticity. Adv Exp Med Biol. 2012;970:153–171. doi: 10.1007/978-3-7091-0932-8_7. [DOI] [PubMed] [Google Scholar]
- 57.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


