Abstract
Background
Ayurveda, an ancient Indian medicinal system, has categorized human body constitutions in three broad constitutional types (prakritis) i.e. Vata, Pitta and Kapha.
Objectives
Analysis of plasma metabolites and related pathways to classify Prakriti specific dominant marker metabolites and metabolic pathways.
Materials and methods
38 healthy male individuals were assessed for dominant Prakritis and their fasting blood samples were collected. The processed plasma samples were subjected to rapid resolution liquid chromatography–electrospray ionization–quadrupole time of flight mass spectrometry (RRLC–ESI–QTOFMS). Mass profiles were aligned and subjected to multivariate analysis.
Results
Partial least square discriminant analysis (PLS-DA) model showed 97.87% recognition capability. List of PLS-DA metabolites was subjected to permutative Benjamini–Hochberg false discovery rate (FDR) correction and final list of 76 metabolites with p < 0.05 and fold-change > 2.0 was identified. Pathway analysis using metascape and JEPETTO plugins in Cytoscape revealed that steroidal hormone biosynthesis, amino acid, and arachidonic acid metabolism are major pathways varying with different constitution. Biological Go processes analysis showed that aromatic amino acids, sphingolipids, and pyrimidine nucleotides metabolic processes were dominant in kapha type of body constitution. Fat soluble vitamins, cellular amino acid, and androgen biosynthesis process along with branched chain amino acid and glycerolipid catabolic processes were dominant in pitta type individuals. Vata Prakriti was found to have dominant catecholamine, arachidonic acid and hydrogen peroxide metabolomics processes.
Conclusion
The neurotransmission and oxidative stress in vata, BCAA catabolic, androgen, xenobiotics metabolic processes in pitta, and aromatic amino acids, sphingolipid, and pyrimidine metabolic process in kapha Prakriti were the dominant marker pathways.
Keywords: Ayurveda, Prakriti, Human metabolomics, RRLC–ESI–QTOFMS
1. Introduction
Ayurveda is an ancient Indian medicinal system and is in practice since 1500 BC. It is extensively used in India, besides the contemporary medicine introduced only 250 years back. In Ayurvedic philosophy, five elements combine in pairs to form three dynamic forces or interactions called ‘doshas’ which are considered as the governing principles of living beings [1]. The doshas are divided into three classes and are described, viz. vata, pitta and kapha. These doshas refer to functions of motion, digestion & metabolism, anabolic activity & accumulation. Ayurveda attributes these constitutional characteristics – Prakriti of an individual to the preponderance of certain “doshas”. All doshas are present in every human being; however, only one is dominant. On the basis of dominant dosha, Prakriti i.e. vata, pitta and kapha are determined. Prakriti can be further divided into seven sub-types or mixed types of Prakritis [1]. Ayurveda describes physical, physiological, psychological and behavioral traits, to determine Prakriti subtypes. As Prakritis are based on dosha, it describes predisposition of individual to disease and response to treatment. All these descriptions of Prakritis seem to be based on genetics, later on proved by several studies. Studies have correlated single nucleotide polymorphisms, inflammation and oxidative stress related genes with different Prakritis. Authors also claimed the correlation of genetic constitution and Prakritis [2], [3].
Ayurveda suggests that the origin of vata, pitta and kapha is motion, digestion and accumulation respectively. It shows that these functions are closely related to metabolic events in body and hence to phenotypic expressions. Moreover, instant energy reserves and neurotransmitters in the body are in the form of metabolites [4]. Therefore, as genome, metabolome of an individual can be directly correlated with its Prakriti [5], [6], in this study, metabolomic approach was used to develop discrimination models to assess the Prakritis of individuals and to identify the dominant metabolic pathways of different dominating Prakritis.
2. Materials and methods
2.1. Chemicals and reagents
Internal and external calibrants and ESI tuning solution were purchased from Agilent Technologies. Solvents used for sample processing and in LCMS assembly: acetonitrile, methanol, formic acid and water were of LC–MS grade (Sigma–Aldrich). Standards of metabolites were purchased from Sigma–Aldrich.
2.2. Ethics statement
Human Ethics Committee of the PDDYP Ayurveda College, Pune, India approved the study wide letter no. RRI/2011/HEC/2023 dated 18-02-2011. All volunteers signed an informed consent to be a part of this study. This clinical trial was registered with the Clinical Trials Registry-India (CTRI) vide registration no. CTRI/2016/08/007187.
2.3. Development of questionnaire for Prakriti assessment
A questionnaire for clinical phenotyping was designed on the basis of Ayurvedic literature on phenotypes and methods of Prakriti assessment (Table S1). The physiological as well as psychological assessment of volunteers was done. The phenotypic classification included criteria for defining anatomical features like body built, body frame, size and symmetry of body parts, physiology, physical endurance and aptitudes.
2.4. Study design and Prakriti assessment
On the basis of dominant Prakriti types out of 205, 38 healthy male individuals were selected for the study. Volunteers were free from diabetes, hyperlipidemia, hypertension, and other chronic systemic disorders, smoking and alcohol consumption habits. As per the assessment the volunteers were categorized in vata–pitta (VP), pitta–kapha (PK) and kapha–pitta (KP) dual Prakriti categories where the earlier term refers the dominant Prakriti. For the sake of simplicity the groups were labeled as vata (V), pitta (P) and kapha (K). Individuals were asked to take a decided vegetarian diet and restricted from any kind of medicine for one week before collection of blood samples. On seventh day morning, fasting blood samples were collected in K+-EDTA vacutainer tubes (BD). Plasma fractions were separated out by centrifugation at 3000g for 15 min at 15 °C. Plasma samples (100 μL) were mixed with equal amounts of acetone and stored overnight at −80 °C. In the morning samples were centrifuged at 10,000 rpm for 15 min and supernatants were collected. Solvent from supernatants was evaporated using lyophilizer under reduced pressure. Lyophilized samples were dissolved in 100 μL acetonitrile:water (80:20 v/v) and kept overnight at −80 °C, centrifuged at 10,000 rpm for 15 min at 15 °C and supernatants were collected for further analysis [7].
2.5. HPLC-Q-TOF-MS analysis
Samples were resolved on Agilent 1290 Infinity Series UPLC interfaced to an Agilent 6538 Accurate-Mass Q-ToF MS. A volume of 15 μL of each sample was injected into an assembly of C18 ZORBAX (4.6 × 12.5 mm) guard column followed by ZORBAX 300SB-C18 (4.6 × 150 mm) HPLC column of particle size 5.0 μm. The solvent system had (A) 0.1% formic acid–water and (B) 0.1% formic acid–acetonitrile. The gradient mode {concentration/time (%/min)} used for solvent B was {0%/0; 1%/5; 100%/25; 100%/32; 1%/37}, with 0.4 mL/min flow rate. The mass spectrometer was operated in positive ion polarity mode with parameters: ionization type electrospray ionization (ESI), gas temperature 350 °C, nebulizer 45 psi, gas flow 8 L/min, capillary voltage 3500 V, octopole voltage 750 V, skimmer voltage 65 V and fragmentor voltage 175 V, fixed collision energy of 25.0 V and acquisition rate 4 spectra per cycle. The quadrupole was operated in the extended dynamic range (1700 m/z, 2 GHz). To assure the mass accuracy of recorded data, continuous internal calibration was performed using standards of signals of standard compounds. Blank samples (solvent mixture of acetonitrile and water) were also injected between the sample runs. The chromatograms of blank samples were used to study the carry-over effects in LC–MS runs.
2.6. Multivariate analysis
Mass Hunter Qualitative Analysis (B.04.00, Agilent Technologies) software was used for preliminary quality check of raw MS/MS data. Profinder software was used for baseline correction, noise reduction, background contaminants removal and extraction of molecular features. Molecular features having abundance >5000 cycles per second (cps), minimum absolute counts 10, relative height (5%) of largest peak, peak space tolerance, i.e. 0.0025 m/z and 5.0 ppm were extracted from mass spectra to avoid false molecular features. Profinder reduces acquired data size and complexity through the removal of redundant and non-specific information by identifying the important variables (features). The extracted data was imported in Mass Profiler Professional software (MPP, B.12.02, Agilent Technologies) and all the molecular features were aligned using mass error < 5.0 ppm and retention time (RT) variation < 0.2 min. Compounds present in less than 75% of samples of any of three groups and having p > 0.05, fold change < 2.0 and co-efficient of variance < 15 were removed from data sets and subjected to generate Box–Whisker plot and principal component analysis (PCA). Partial least-squares-discriminant analysis (PLS-DA), a class prediction algorithms was performed to distinguish 3 Prakriti groups and explore unique features present across the groups. Finally, data were subjected to one way ANOVA with permutative Benjamini–Hochberg FDR correction and Tukey HSD post hoc to minimize the FDR, keeping p value cutoff < 0.05 and fold change > 2.0 {for p value cutoff < 0.05, false discovery rate (FDR) become <5%}. Based on changes in expression levels (upregulated or downregulated) metabolites were arranged with respect to K vs P, K vs V and P vs V classes of comparison. Differentially expressed metabolites were identified by comparison of fragmentation pattern of the metabolites with standard human metabolome in Metlin library, HMDB and NIST databases. The flowchart indicating important steps during the analysis has been shown in Supplementary Fig. 1 (Fig. S1).
2.7. Significant pathway and biological processes identification
Cytoscape 3.3 was used for further analysis of data and to find out significantly altered pathways in all the three Prakritis. Data were imported in Metascape, a plugin of Cytoscape 3.3 and subjected to pathway analysis. All the metabolic pathways generated were further subjected to enrichment and topological analysis using another plugin JEPETTO. Only the pathways having significant q value were selected and further subjected to identification of biological processes associated with them using another plugin ClueGo with standard parameters.
3. Results
3.1. Prakriti assessment
The questionnaire (Table S1) was validated by pre-testing and the results obtained were confirmed by the clinical assessment of Prakriti independently by two Ayurveda physicians. More than 90% concordance was observed in Prakriti assessment by the two clinicians and questionnaire. Based on the responses of volunteers to standard questionnaire and physical examination carried out by an Ayurvedic physician; 38 healthy volunteers were categorized in 3 Prakriti categories as pitta (n = 16), kapha (n = 10) and vata (n = 12). Individuals who had thin and narrow body frame, weakly developed body build, with irregular appetite, food and bowel habits, difficulty in gaining weight, quick at physical activities, dry skin and hair, and less tolerance for cold temperature were considered as vata Prakriti. Individuals with moderately developed build, high frequency of appetite and thirst, good digestive power, perspiration tendency higher than normal, tolerance for cold weather, moderately mobile with moderate physical strength were identified as pitta Prakriti. Individuals who had broad body frames with well developed body build, tendency to gain weight, low appetite and digestion, preferred to be less mobile, less forgetful and with good healing power and cool temperament, were selected as kapha individuals (Table S2). Less than 20 basal metabolic rate (BMR) was observed in Vata category while 24.0–25.0 and >25.0 were observed for Pitta and Kapha respectively. Kapha Prakriti individuals had highest level of HDL, TG and TC (Table 1a). The number of compounds obtained for each individual in each group has been represented (Table 1b).
Table 1a.
Basic characteristics of the data sets.
| Sr. no. | Parameters | Kapha | Pitta | Vata |
|---|---|---|---|---|
| 1 | Participants, n (%) | 10 (26.3) | 16 (42.1) | 12 (31.5) |
| 2 | Age (years), Mean ± SD | 36.77 ± 1.07 | 41.22 ± 1.38 | 39.66 ± 1.49 |
| 3 | Smoker, n (%) | 2 | 0 | 1 |
| 4 | BMI (kg m−2) | >25.0 | 24.0–25.0 | <20.0 |
| 5 | HDL (mg dl−1), Mean ± SD | 54.73 ± 1.98 | 20.92 ± 1.48 | 34.92 ± 1.42 |
| 6 | Triglyceride TG (mg dl−1), Mean ± SD | 190.58 ± 1.66 | 178.11 ± 1.51 | 176.49 ± 1.04 |
| 7 | Cholesterol (mg dl−1), Mean ± SD | 187.27 ± 1.36 | 171.53 ± 1.76 | 178.81 ± 1.57 |
Table 1b.
Total number of compounds extracted from each sample falling under 3 study categories.
| Sl. no. | Kapha (n = 10) | No. of compounds extracted | Pitta (n = 16) | No. of compounds extracted | Vata (n = 12) | No. of compounds extracted |
|---|---|---|---|---|---|---|
| 1 | Sample 1 | 2817 | Sample 1 | 2470 | Sample 1 | 3112 |
| 2 | Sample 2 | 2769 | Sample 2 | 2409 | Sample 2 | 2568 |
| 3 | Sample 3 | 2615 | Sample 3 | 2248 | Sample 3 | 2166 |
| 4 | Sample 4 | 2389 | Sample 4 | 2241 | Sample 4 | 2166 |
| 5 | Sample 5 | 2160 | Sample 5 | 2231 | Sample 5 | 3002 |
| 6 | Sample 6 | 2543 | Sample 6 | 2150 | Sample 6 | 2922 |
| 7 | Sample 7 | 2506 | Sample 7 | 2148 | Sample 7 | 2661 |
| 8 | Sample 8 | 2461 | Sample 8 | 2118 | Sample 8 | 2447 |
| 9 | Sample 9 | 2344 | Sample 9 | 2117 | Sample 9 | 2544 |
| 10 | Sample 10 | 2130 | Sample 10 | 2112 | Sample 10 | 2290 |
| 11 | Sample 11 | 2079 | Sample 11 | 2080 | ||
| 12 | Sample 12 | 2075 | Sample 12 | 2020 | ||
| 13 | Sample 13 | 2063 | ||||
| 14 | Sample 14 | 1956 | ||||
| 15 | Sample 15 | 2562 | ||||
| 16 | Sample 16 | 2501 |
3.2. Multivariate analysis of plasma metabolome
The uniform peak occurrence, spreading and lack of any outliers in chromatograms of individual groups indicated uniformity in metabolic composition in each sampling category and within group consistency of metabolites (Fig. S2, A–C). Further, only the compounds present universally in >75% of samples in each group were extracted for further processing to ensure the consistency of metabolomics profiles in each group. The chromatograms of blank samples contained only low intensity peaks, therefore, it was concluded that the carryover effect did not occur. The extracted base peak chromatograms (BPC) of kapha (K), pitta (P) and vata (V) samples showed distinct peaks (Fig. S3). Analysis of total ion chromatograms showed average 23 peaks containing around 2000 compounds. Variability in the peaks was normalized successfully using internal standards and Z-transforms. Reproducible and stable molecular features were subsequently used for statistical analysis. The well-established PLS-DA regression value 0.97 (R2 = 0.97) shows the accuracy of results. The result of the sample classification presented in terms of discrimination ability was found to be 97.87% accurate, representing the percentage of the samples correctly classified during model training and cross-validation. Data were further subjected to one way ANOVA with permutative (n = 100) and Benjamini–Hochberg multiple testing correction to validate the PLS-DA model and to further decrease false discovery rate (Table 2). Supervised PCA of P, V and K groups showed 46.0%, 7.3% and 5.96% variability along the X, Y and Z axes respectively. PCA is a most commonly used visualization to understand the metabolic differences in 3D space (Fig. 1A). The overall percent variance was represented as a box–whisker plot showing altered metabolites across all the 3 groups, i.e. P, V and K. Out of these, vata showed highest compound variability represented by elongated box shapes and whiskers towards either side to the grand mean (Fig. 1B). A Pearson heat map was also constructed to visualize the metabolomic variability among 3 study groups. All the groups showed considerable variability in metabolomic level and were clearly separated from each other (Fig. 2).
Table 2.
Partial least square discriminant analysis (PLS-DA) model of class prediction was applied to study categories.
| Kapha (predicted) | Pitta (predicted) | Vata (predicted) | Accuracy (%) | |
|---|---|---|---|---|
| Kapha (True) | 9 | 0 | 0 | 100.00 |
| Pitta (True) | 0 | 29 | 0 | 100.00 |
| Vata (True) | 1 | 0 | 11 | 91.66 |
| Overall accuracy (%) | 97.87 |
Fig. 1.
(A) PCA plot of variable metabolites obtained in positive mode showed significant differences among 3 sampling categories. X axis component value was 46% while for Y and Z it was 7.3%, 5.96% respectively. (B) The box-and-whisker plot is an exploratory graphic, used to show the distribution of metabolites in kapha (K), pitta (P) and vata (V) groups of individuals. The high variation in top whiskers of kapha and vata and low whisker of pitta, high variation in upper quartile of kapha and pitta and lower whisker of vata are representing metabolite variations in study groups, out of these vata has most diverse pool of metabolites.
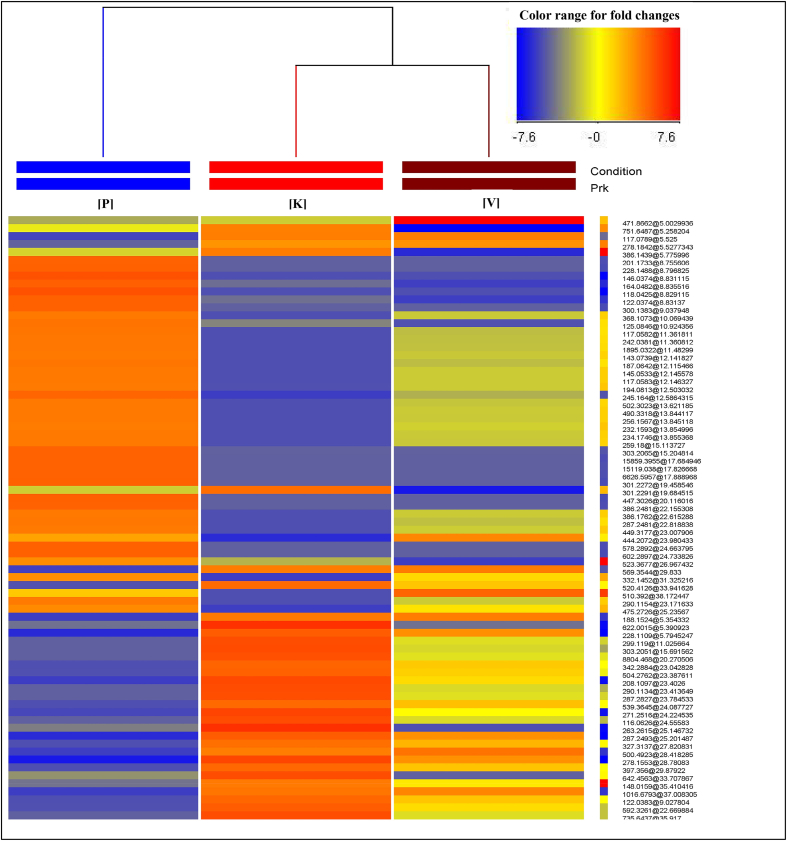
Fig. 2.
Pearson heat map alignment denoting fold changes of 76 listed metabolites across kapha (K), pitta (P) and vata (V) Prakritis. The columns correspond to different Prakriti categories and rows correspond to the altered metabolites. Correlation map is showing pitta (P) group as most discrete group and kapha (K) and vata (V) groups are more closely related.
3.3. Qualitative analysis and identification of variable metabolites
Metabolites with any missing values were filtered out from the statistically significant data obtained after analysis. Finally, 76 differentially expressed metabolites in all groups were obtained and identified using standard Metlin library and HMDB (Table 3). The variations in expression of these metabolites within K vs P, K vs V and P vs V classes were expressed in tabular format. These 76 metabolites were belonging to carnitines, nucleic acids, amino acids, vitamins and fatty acids/lipids. Out of these 76, 39 compounds had highest expression in pitta category, 6 were in vata while 31 were in kapha group. Four metabolites belonging to nucleotides class were 8-hydroxyguanine, an oxidative derivative of guanosine and biomarker of oxidative stress, 10-formyl tetrahydrofolyl l-glutamate is an useful intermediate in purine biosynthesis, 2′-deoxyuridine and 5-methylcytosine nucleotide analogues. 8-Hydroxyguanine and 2′-deoxyuridine were abundant in kapha while other two were in pitta Prakriti (Table 3). The lipid compounds were mostly belonging to kapha and pitta categories: PS(19:0/0:0), PA(14:0/14:0), DG(18:3(9Z,12Z,15Z)/20:2(11Z,14Z)/0:0), LysoPC(22:5(7Z,10Z,13Z,16Z,19Z)), 2,3-dinor-6-keto-prostaglandin F1α, a potent vasodilator and inhibitor of platelet aggregation whereas docosanamidoacetic acid and N-stearoylethanolamine, appear in various fatty acid oxidation disorders, were abundant in kapha Prakriti. 21-Hydroxypregnenolone useful in corticosterone and deoxycorticosterone synthesis, N-docosanoyl taurine, a novel taurine-conjugated fatty acids, LysoPC(18:0) useful in lipid signaling, lysoPE, (18:3(6Z,9Z,12Z)/0:0N-heptanoylglycine), appears in various fatty acid oxidation disorders, 2-hydroxy-13-O-d-glucuronoside-octadec-9Z-enoate is a linoleate intermediate were found upregulated in kapha Prakriti, whereas 16,17-epoxydeoxycorticosterone acetate; valerenic acid, a branched fatty acid; 2-methoxyestrone, a steroid intermediate; and glycochenodeoxycholate, a bile salt were abundant in pitta Prakriti. The 10,11-dihydro-20-trihydroxy-leukotriene B4, a metabolite of lipid omega-oxidation of leukotriene B4 and a diglyceride DG(14:0/14:1(9Z)/0:0) were abundant in vata category. Out of 11 differentially expressed amino acids and their derivatives, 6 were belonging to pitta category; l-valine is an essential branched chain amino acid (BCAA), critical to human life and is involved in stress, energy and muscle metabolism, L-2,4-diaminobutanoate an another BCAA metabolism component, 6-amino-2-oxohexanoate is a lysine degradation intermediate, capryloylglycine is used to diagnose disorders associated with mitochondrial inborn errors of metabolism. The dominant amino acid derivatives in kapha were argininosuccinic acid, a precursor for arginine and fumarate; kynurenine, a l-tryptophan breakdown product significant in numerous biological processes; and 2-oxopentanoic acid, an arginine catabolism intermediate (Table 3).
Table 3.
The list of plasma metabolites obtained in +ESI, processed through Profinder and analyzed in MPP are arranged in ascending order of their mass range. Metabolites from the PLS-DA model without missing values were ranked according to the significance and variability and identified using various databases.
| Sr. no. | Compounds | Mass (Da) | RT (min) | p Value (corrected) | FC (K vs P) | FC (K vs V) | FC (P vs V) | Abundance based order | ±PPM error |
|---|---|---|---|---|---|---|---|---|---|
| 1 | LysoPC(22:5(7Z,10Z,13Z,16Z,19Z)) | 569.3544 | 29.83 | 9.37E−04 | 2.75 | 1.00 | −2.75 | K = V > P | −3.81 |
| 2 | Homoarginine | 188.1524 | 5.35 | 9.37E−04 | 2.81 | 1.01 | −2.77 | K = V > P | −9.69 |
| 3 | Unknown | 1016.679 | 36.00 | 9.37E−04 | 2.85 | 1.05 | −2.70 | K = V > P | – |
| 4 | Unknown | 278.1842 | 5.52 | 0.009072 | 2.31 | −1.00 | −2.32 | K = V > P | – |
| 5 | Unknown | 622.0015 | 5.39 | 4.28E−04 | 3.12 | 3.12 | −1 | K > P = V | – |
| 6 | 1-O-Sinapoyl-beta-d-glucose | 386.1439 | 5.77 | 0.01693 | 1.80 | 2.95 | 1.64 | K > P > V | −7.64 |
| 7 | Sphinganine | 301.2291 | 19.68 | 0.00583 | 1.93 | 3.29 | 1.70 | K > P > V | 0.57 |
| 8 | DG(18:3(9Z,12Z,15Z)/20:2(11Z,14Z)/0:0) | 642.4563 | 33.70 | 0.002483 | 2.57 | 2.97 | 1.15 | K > P > V | 4.43 |
| 9 | Hydroxyvalerylcarnitine | 263.2615 | 25.14 | 3.74E−04 | 3.02 | 3.45 | 1.14 | K > P > V | 2.48 |
| 10 | Unknown | 751.6487 | 5.25 | 0.008485 | 1.66 | 3.28 | 1.97 | K > P > V | – |
| 11 | N-Stearoylethanolamine | 327.3137 | 27.82 | 0.002483 | 2.76 | 1.28 | −2.16 | K > V > P | −5.71 |
| 12 | d-Threitol | 122.0383 | 9.02 | 0.002113 | 2.85 | 1.40 | −2.03 | K > V > P | −0.34 |
| 13 | 21-Hydroxydydrogesterone glucuronide | 504.2762 | 23.38 | 0.001943 | 2.90 | 1.47 | −1.96 | K > V > P | 3.82 |
| 14 | 8-Hydroxyguanine | 299.119 | 11.02 | 0.001819 | 2.96 | 2.05 | −1.44 | K > V > P | −11.99 |
| 15 | 2-Deoxyuridine | 228.1109 | 5.79 | 1.90E−04 | 3.30 | 1.21 | −2.74 | K > V > P | 5.02 |
| 16 | PS(19:0/0:0) | 539.3645 | 24.08 | 0.002217 | 2.82 | 1.35 | −2.08 | K > V > P | −4.78 |
| 17 | 2,3-Dinor-6-keto-prostaglandin F1α | 342.2884 | 23.04 | 0.002003 | 2.87 | 1.42 | −2.01 | K > V > P | 2.56 |
| 18 | PA(14:0/14:0) | 592.3261 | 22.66 | 0.001766 | 2.95 | 1.56 | −1.88 | K > V > P | −0.54 |
| 19 | Docosanamidoacetic acid | 397.356 | 29.87 | 0.002113 | 2.84 | 1.38 | −2.05 | K > V > P | −2.41 |
| 20 | 16-Hydroxypalmitic acid | 271.2516 | 24.22 | 6.06E−04 | 3.31 | 1.91 | −1.74 | K > V > P | −3.77 |
| 21 | Dibutyl phthalate | 278.1553 | 28.78 | 1.45E−06 | 3.72 | 1.31 | −2.87 | K > V > P | −3.15 |
| 22 | 2-Oxopentanoic acid | 116.0626 | 24.55 | 0.001766 | 2.98 | 2.08 | −1.43 | K > V > P | −4.77 |
| 23 | Argininosuccinic acid | 290.1134 | 23.41 | 0.001757 | 2.98 | 2.09 | −1.42 | K > V > P | −3.34 |
| 24 | l-Kynurenine | 208.1097 | 23.40 | 4.40E−04 | 3.33 | 1.67 | −1.99 | K > V > P | −7.95 |
| 25 | 1α,25-dihydroxy-11-(4-hydroxymethylphenyl)-9,11-didehydrovitamin D3 | 520.4126 | 33.94 | 0.002678 | 2.79 | 1.37 | −2.02 | K > V > P | −4.17 |
| 26 | Unknown | 287.2827 | 23.78 | 0.001819 | 2.96 | 2.04 | −1.45 | K > V > P | – |
| 27 | 3-Hydroxyoctanoyl carnitine | 303.2051 | 15.69 | 0.001701 | 2.99 | 2.11 | −1.41 | K > V > P | 6.92 |
| 28 | 4-Methylthio-2-oxobutanoic acid | 148.0159 | 35.41 | 0.017991 | 2.40 | 1.47 | −1.63 | K > V > P | 0.56 |
| 29 | Paired-like homeodomain transcription factor 3 | 8804.468 | 20.27 | 0.001903 | 2.95 | 2.03 | −1.45 | K > V > P | 6.89 |
| 30 | l-Octanoylcarnitine | 287.2493 | 25.20 | 1.90E−04 | 3.30 | 1.20 | −2.74 | K > V > P | −7.77 |
| 31 | Unknown | 735.6437 | 35.91 | 0.001766 | 2.97 | 2.06 | −1.43 | K > V > P | – |
| 32 | 2-Methoxyestrone | 300.1383 | 9.037 | 9.37E−04 | −2.76 | 1 | 2.76 | P > K = V | −4.52 |
| 33 | N-Docosanoyl taurine | 447.3026 | 20.11 | 9.37E−04 | −2.76 | 1 | 2.76 | P > K = V | 4.68 |
| 34 | L-2,4-diaminobutanoate | 118.0425 | 8.82 | 1.90E−04 | −3.07 | 1 | 3.07 | P > K = V | −5.95 |
| 35 | Capryloylglycine | 201.1733 | 8.75 | 9.37E−04 | −2.76 | −1.00 | 2.76 | P > K = V | −4.84 |
| 36 | Malonuric acid | 146.0374 | 8.83 | 1.90E−04 | −3.07 | −1 | 3.07 | P > K = V | 5.92 |
| Transcription factor HFK1 | 15,119.04 | 17.82 | 9.37E−04 | −2.75 | −1 | 2.75 | P > K = V | 8.55 | |
| 38 | Unknown | 303.2065 | 15.20 | 9.37E−04 | −2.75 | −1 | 2.75 | P > K = V | – |
| 39 | Coumaric acid | 164.0482 | 8.83 | 9.37E−04 | −2.69 | 1.14 | 3.07 | P > K > V | −8.61 |
| 40 | 5-Methylcytosine | 125.0846 | 10.92 | 0.004456 | −2.33 | 1.16 | 2.71 | P > K > V | −8.74 |
| 41 | LysoPC(18:0) | 523.3677 | 26.96 | 0.017991 | −1.85 | 1.41 | 2.62 | P > K > V | −5.17 |
| 42 | Benzoic acid | 122.0374 | 8.83 | 9.37E−04 | −2.65 | 1.15 | 3.06 | P > K > V | −9.66 |
| 43 | Traumatic acid | 228.1488 | 8.79 | 9.37E−04 | −2.76 | −1.00 | 2.76 | P > V = K | 4.26 |
| 44 | 1,25-Dihydroxyvitamin D3 3-glycoside | 578.2892 | 24.66 | 9.37E−04 | −2.76 | −1 | 2.76 | P > V = K | 0.15 |
| 45 | (2E,6E,8E)N-Isobutyl-2,6,8-hexadecatriene-10-yneamide | 301.2272 | 19.45 | 9.37E−04 | −2.76 | −1 | 2.76 | P > V = K | 2.42 |
| 46 | Putative reverse transcriptase | 6626.596 | 17.88 | 9.37E−04 | −2.75 | −1 | 2.75 | P > V = K | 7.45 |
| 47 | Inositol cyclic phosphate | 242.0381 | 11.36 | 0.004514 | −2.70 | −1.39 | 1.94 | P > V > K | 0.09 |
| 48 | 12α-Hydroxyerosone | 368.1073 | 10.06 | 0.004862 | −2.67 | −1.42 | 1.87 | P > V > K | −2.51 |
| 49 | Guanidinoacetic acid | 117.0583 | 12.14 | 0.004862 | −2.66 | −1.43 | 1.86 | P > V > K | −8.61 |
| 50 | Glycochenodeoxycholate | 449.3177 | 23.00 | 0.005034 | −2.66 | −1.49 | 1.84 | P > V > K | 8.88 |
| 51 | Melatonin | 232.1593 | 13.85 | 0.005267 | −2.65 | −1.45 | 1.82 | P > V > K | −1.97 |
| 52 | 10-Formyl tetrahydrofolyl l-glutamate | 602.2897 | 24.73 | 9.37E−04 | −2.76 | −1 | 2.76 | P > V > K | 0.14 |
| 53 | 21-Hydroxypregnenolone | 332.1452 | 31.32 | 0.006784 | −2.61 | −2.01 | 1.29 | P > V > K | −8.83 |
| 54 | LysoPE(18:3(6Z,9Z,12Z)/0:0) | 475.2726 | 25.23 | 0.005976 | −2.64 | −1.93 | 1.36 | P > V > K | −3.17 |
| 55 | N-Heptanoylglycine | 187.0642 | 12.11 | 0.004414 | −2.71 | −1.37 | 1.97 | P > V > K | 9.5 |
| 56 | 2-Hydroxy-13-O-d-glucuronoside-octadec-9Z-enoate | 490.3318 | 13.84 | 0.004862 | −2.67 | −1.43 | 1.87 | P > V > K | −8.2 |
| 57 | Dehydrospermidine | 143.0739 | 12.14 | 0.004862 | −2.67 | −1.42 | 1.88 | P > V > K | −14.42 |
| 58 | 9-Hydroxyphenanthrene | 194.0813 | 12.50 | 0.005181 | −2.65 | −1.43 | 1.85 | P > V > K | 7.51 |
| 59 | Valine | 117.0582 | 11.36 | 0.004414 | −2.71 | −1.37 | 1.96 | P > V > K | −7.75 |
| 60 | 6-Amino-2-oxohexanoate | 145.0533 | 12.14 | 0.004862 | −2.66 | −1.43 | 1.86 | P > V > K | −4.15 |
| 61 | Valerenic acid | 234.1746 | 13.85 | 0.004862 | −2.67 | −1.42 | 1.87 | P > V > K | −8.3 |
| 62 | 2-(3-Carboxy-3-aminopropyl)-l-histidine | 256.1567 | 13.84 | 0.004862 | −2.67 | −1.42 | 1.87 | P > V > K | −7.36 |
| 63 | 1α-hydroxy-2β-(5-hydroxypentoxy) vitamin D3 | 502.3023 | 13.62 | 0.004862 | −2.68 | −1.41 | 1.90 | P > V > K | 1.98 |
| 64 | Unknown | 287.2481 | 22.81 | 0.004514 | −2.70 | −1.38 | 1.94 | P > V > K | – |
| 65 | l-Hexanoylcarnitine | 259.18 | 15.11 | 0.004862 | −2.67 | −1.42 | 1.88 | P > V > K | 3.78 |
| 66 | beta-Alanyl-l-arginine | 245.164 | 12.58 | 9.37E−04 | −3.02 | −1.39 | 2.16 | P > V > K | −6.56 |
| 67 | Splicing factor 1 isoform 4 | 15,859.4 | 17.68 | 9.37E−04 | −2.76 | −1 | 2.76 | P > V > K | 6.88 |
| 68 | Catechin | 290.1154 | 23.17 | 0.004862 | −2.66 | −1.43 | 1.86 | P > V > K | 3.65 |
| 69 | 16,17-Epoxydeoxycorticosterone acetate | 386.1762 | 22.61 | 0.004862 | −2.67 | −1.43 | 1.86 | P > V > K | 5.92 |
| 70 | Unknown | 1895.032 | 11.48 | 0.004537 | −2.69 | −1.39 | 1.93 | P > V > K | – |
| 71 | l-Aspartate 4-semialdehyde | 117.0789 | 5.52 | 0.001141 | 2.72 | −1.00 | −2.73 | V > K > P | −4.05 |
| 72 | 1α,25-dihydroxy-26,27-dipropylvitamin D3 | 500.4923 | 28.41 | 9.37E−04 | 2.76 | −1.03 | −2.86 | V > K > P | 6.72 |
| 73 | Unknown | 471.8662 | 5.00 | 0.005393 | 1.10 | −2.83 | −3.13 | V > K > P | – |
| 74 | Unknown | 444.2072 | 23.98 | 0.0033 | −2.56 | −2.84 | −1.11 | V > P > K | – |
| 75 | 10,11-Dihydro-20-trihydroxy-leukotriene B4 | 386.2481 | 22.15 | 9.37E−04 | −2.76 | −1 | 2.76 | V > P > K | −10.48 |
| 76 | DG(14:0/14:1(9Z)/0:0) | 510.392 | 36.17 | 0.013222 | −2.02 | −2.86 | −1.41 | V > P > K | 0.71 |
Abbreviations: RT – retention time, p – calculated probability and FC – fold change.
Out of 4 vitamin D derivatives observed, 1,25-Dihydroxyvitamin D3 3-glycoside and 1α-hydroxy-2β-(5-hydroxypentoxy) vitamin D3 had highest expression in pitta Prakriti. Most of the carnitines were predominantly expressed in kapha category; these include l-octanoylcarnitine, 3-hydroxyoctanoyl carnitine and hydroxyvalerylcarnitine while hexanoylcarnitine an unusual acylcarnitine indicator of metabolic disorders was dominant in pitta Prakriti.
m-Coumaric acid and benzoic acid are polyphenol metabolites formed by the gut microflora, guanidoacetic acid, a creatine precursor that causes cardiovascular and skeletal problems, melatonin, an antioxidant, nucleic acid protector and regulator of body clock and dehydrospermidine that reduces amount of aging in human immune cells were highest in pitta category. Sphinganine, a blocker of postlysosomal cholesterol transport was abundant in kapha category.
The carbohydrate compounds, d-threitol, a main end product of d-xylose metabolism and an indicator of carbohydrate metabolism derangements, 1-O-sinapoyl-beta-d-glucose and 21-hydroxydydrogesterone glucuronide were highest in kapha Prakriti. Overall 11 compounds remained unidentified of which majority belonged to kapha category (Table 3).
3.4. Pathway analysis
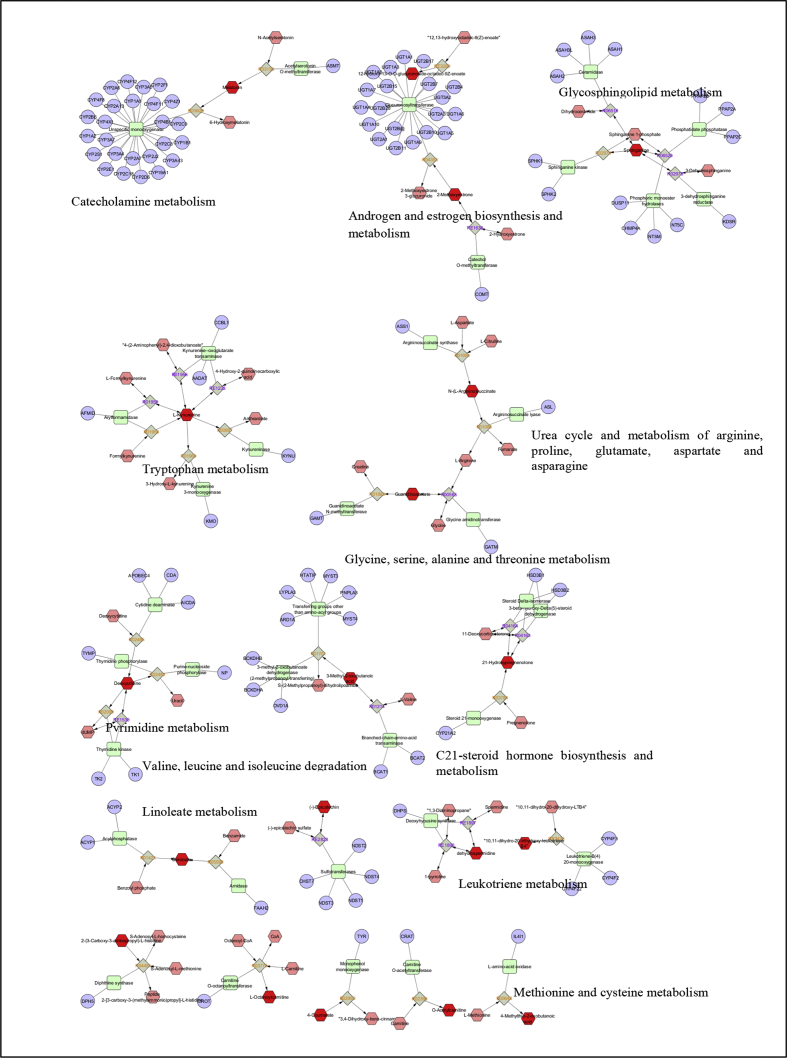
Category-wise the highest abundant upregulated and downregulated metabolites were segregated and used to look at the significant metabolic pathways associated with each Prakriti group. Metascape plugin in Cytoscape 3.3 was used to explore significantly affected metabolomics pathways in different Prakritis. In kapha Prakriti, pathways with q value < 0.01 were found to be sphingolipid, glycerophospholipid, ether lipid, alanine, aspartate, glutamate, arginine, proline, pyrimidine metabolism and Fc gamma R-mediated phagocytosis. Pitta Prakriti showed enhanced valine, leucine and isoleucine degradation. Other pathways were found to be equally affected in vata and kapha Prakritis. Vata Prakriti showed upregulated arachidonic acid, beta-alanine, butanoate, catecholamine and glycerolipid metabolism (Table 4, Fig. 3).
Table 4.
The list of significant metabolic pathways that are being regulated obtained from Cytoscape and its plugins (Metascape and JEPETTO) is represented. The metabolic pathways are arranged in ascending order of their respective q value, their impact in terms of XD score and regulation of pathways in different Prakritis have also been mentioned.
| Sr. No. | Pathway or process | XD-score | q-Value | Overlap/size | Regulation in different Prakritis |
|---|---|---|---|---|---|
| 1 | Steroid hormone biosynthesis | 1.29764 | 0.00001 | 5/15 | P = V |
| 2 | Metabolism of xenobiotics by cytochrome P450 | 0.96431 | 0.00001 | 5/20 | P = V |
| 3 | Sphingolipid metabolism | 0.8734 | 0.00002 | 5/22 | K |
| 4 | Tryptophan metabolism | 0.73354 | 0.00003 | 5/26 | P = V |
| 5 | Steroid hormone metabolism | 1.05522 | 0.00173 | 3/11 | P = V |
| 6 | Retinol metabolism | 0.96431 | 0.00196 | 3/12 | P = V |
| 7 | Glycerophospholipid metabolism | 0.42145 | 0.00243 | 4/35 | K |
| 8 | Ether lipid metabolism | 0.71431 | 0.00377 | 3/16 | K |
| 9 | Tyrosine metabolism | 0.67673 | 0.00377 | 3/17 | P = V |
| 10 | Drug metabolism – cytochrome P450 | 0.67019 | 0.00377 | 3/17 | K = P = V |
| 11 | Glycerolipid metabolism | 0.56431 | 0.0057 | 3/20 | V |
| 12 | Alanine, aspartate and glutamate metabolism | 0.53574 | 0.00611 | 3/21 | K |
| 13 | Valine, leucine and isoleucine degradation | 0.40876 | 0.01211 | 3/27 | P |
| 14 | Arginine and proline metabolism | 0.28864 | 0.02857 | 3/37 | K |
| 15 | Fc gamma R-mediated phagocytosis | 0.15943 | 0.03186 | 4/82 | K |
| 16 | Pyrimidine metabolism | 0.15255 | 0.03034 | 3/70 | K |
| 17 | Arachidonic acid metabolism | 1.04886 | 0.00000 | 7/26 | V |
| 18 | beta-Alanine metabolism | 0.77194 | 0.00148 | 3/15 | V |
| 19 | Butanoate metabolism | 0.54336 | 0.04580 | 2/14 | V |
| 20 | Catecholamine metabolic process | 0.53574 | 0.00377 | 3/21 | V |
Fig. 3.
Figure showing significant pathways found altered in different Prakritis. All the metabolites were imported to Cytoscape from MPP and analysis was done with help of Metascape plugin and further significance was calculated using JEPETTO plugin.
3.5. Go biological processes
Further, in order to explore the biological processes associated with differentially expressed metabolites, ClueGo plugin in Cytoscape 3.3 was used. In kapha Prakriti mitochondrial genome maintenance, pyrimidine, tryptophan, kynurenine, membrane lipid, aromatic amino acid family, NAD biosynthetic metabolism, and cellular biogenic amine catabolic processes were found to be dominant (Fig. 4A). In pitta Prakriti retinoid metabolic, aminoglycan biosynthetic, uronic acid metabolic, estrogen metabolic process, branched-chain amino acid metabolic, toxin metabolic, heparin biosynthetic and regulation of cellular ketone metabolic processes were found to be positively regulated. Whereas, cellular carbohydrate, lipid, and cellular glucuronidation metabolic processes regulation were found to be downregulated (Fig. 4B). In vata Prakriti, very long-chain fatty acid, sulfur amino acid, glutamate, catecholamine, leukotriene, estrogen metabolic, vitamin K, hydrogen peroxide metabolic processes and ketone, phylloquinone, dopamine, sodium ion homeostasis and cellular detoxification catabolic processes were found to be dominant (Fig. 4C) (Fig. S4, A–C).
Fig. 4.
Go Biological process in all the three Prakritis analyzed using Metscape and ClueGo modules of Cytoscape 3.3 (A) Kapha, (B) Pitta, and (C) Vata.
4. Discussion
Ayurveda is an ancient system of personalized medicine documented and practiced in India since 1500 B.C. In Ayurveda, it is believed that the human body constitution determines predisposition, prognosis of diseases, therapy and lifestyle regime. The entire description of human physiology in Ayurveda is based on the theory of Tridosha [8]. According to Ayurveda, vata, pitta, and kapha Prakriti are found to have unique metabolic activities, kapha is slow, pitta is fast, and vata is considered to have variable metabolism [9]. Vata is associated with bone, pitta with blood, while kapha is associated with other tissues related to structure and storage such as adipose tissue [10], [11].
Kapha governs the nervous and musculo-skeletal systems that maintain body mass, shape, and flexibility [12]. The association of kapha dosha with anabolic processes and co-ordination of genes delayed manifestation of aging and provides a longer life span [13], [14]. In the current study, mitochondrial genome maintenance, pyrimidine, membrane lipid, NAD biosynthetic processes were found to be dominant in kapha Prakriti (Fig. 4A). It leads to slow catabolism of lipids and therefore, storage, and weight gaining. Kapha Prakriti individuals are prone to disorders of the respiratory system and therefore, continuous mitochondrial genome maintenance is required. The results supported by Hankey who reported that the peptide coenzyme A is associated with lipid metabolism and dominant in kapha Prakriti [15]. As noticed in the present study (Table 1a, Table 1b), higher levels of markers of metabolic syndrome, such as triglyceride (TG), total cholesterol, LDL, VLDL and HDL were also reported in kapha Prakriti people [16]. Accumulations of lipids also lead to a variety of cardiovascular, neurological processes and diseases [17]. Increased levels of capryloylglycine, 8-hydroxyguanine, d-threitol, hexanoylcarnitine and Kynurenine; the well-established markers of cardiovascular diseases, metabolic irregularities and oxidative stress, confirm the biological processes established in the study (Table 3). Docosanamidoacetic acid, a known marker of fatty acid oxidation disorders was abundantly found in kapha people (Table 3). Amino acid derivatives 2-oxopentanoic acid, a metabolite of arginine catabolism; homoarginine, an inhibitor of liver alkaline phosphohydrolase; and argininosuccinic acid, an arginine precursor were observed in kapha. Presence of valine and arginine precursors may be linked with body's response to stress induced by kapha defects by enhancing BCAA and urea metabolism. Overall, lipid and pyrimidine metabolic and aromatic amino acid family catabolic processes are dominant in kapha Prakriti people (Fig. 4A).
Pitta is primarily responsible for metabolism, actions of enzymes, growth factors, hormones, thermo-regulation, energy homeostasis, digestion, and cellular or sub-cellular metabolism [18]. Ghodke et al., have shown that the extensive metabolizer (EM) genotype of the drug metabolizing enzyme CYP2C19 was found only in pitta Prakriti [19]. In the current study, enhanced metabolism of xenobiotics by cytochrome P450 (Table 4) and the toxin metabolic process was observed in pitta Prakriti (Fig. 4B). The pitta predominance Prakriti type individuals have a high basal metabolic rate (BMR) and energy consumption leading to tissue destruction and premature aging and average life span [9]. LysoPE(18:3(6Z,9Z,12Z)/0:0) and LysoPC(18:0) are useful in lipid signaling, 2-hydroxy-13-O-d-glucuronoside-octadec-9Z-enoate, a linoleate intermediate and N-heptanoylglycine, a marker of mitochondrial fatty acid beta-oxidation disorders were abundant in pitta Prakriti. Along with these, levels of glycochenodeoxycholate, a bile acid salt that solubilizes fats for absorption; were found to be high in pitta Prakriti people. All of these metabolites correspond to the enhanced lipid and energy metabolism and therefore, BMI.
The high metabolic turnover in pitta Prakriti was shown by valine, leucine, isoleucine biosynthesis as well as degradation and vitamin metabolism. The BCAA are critical to human life and are particularly involved in stress, energy and muscle metabolism, as valine goes solely to carbohydrates, and valine deficiency is marked by neurological defects in the brain [20]. At the same time, melatonin, a crucial hormone that decides the body's basal metabolism rate and sleep–wake cycle was also found to be highest in pitta Prakriti. Protein network analysis has shown that BCCA metabolic, heparin biosynthetic and cellular ketone metabolic processes were positively regulated in pitta Prakriti (Fig. 4B).
In pitta Prakriti, 2-methoxyestrone, a steroid intermediate of androgen and estrogen metabolic pathway that determines the physical constitution of humans was found to be upregulated. These steroid hormones are known to modify brain structures, implicated in behavioral conditions, and libido, hence can be first marker for Tridosha system. Moreover, in pitta Prakriti, retinoid metabolic, aminoglycan biosynthetic, uronic acid metabolic, cellular carbohydrate, lipid, and cellular glucuronidation metabolic processes were found to be affected (Fig. 4B).
Vata contributes to manifestation of shape, cell division, signaling, movement, excretion of wastes, cognition and also regulates the activities of kapha and pitta. Vata dosha is responsible for movement, cell growth and apoptosis. Vata body types can have propensity to develop neurological problems, dementia, movement and speech disorders, arrhythmias, and related chronic diseases [21]. In vata Prakriti very long-chain fatty acid, sulfur amino acid, glutamate, catecholamine, leukotriene, estrogen metabolic, vitamin K, hydrogen peroxide metabolic processes and ketone, phylloquinone, dopamine, sodium ion homeostasis and cellular detoxification catabolic processes were found to be dominant (Fig. 4C). In the present study, LysoPC(22:5(7Z,10Z,13Z,16Z,19Z)), lysophospholipid have role in lipid signaling, 10,11-dihydro-20-trihydroxy-leukotriene B4, a metabolite of lipid omega-oxidation of leukotriene B4 and a diglyceride DG(14:0/14:1(9Z)/0:0) were abundant in vata category. Amino acid metabolism and l-Aspartate 4-semialdehyde that is involved in lysine and homoserine synthesis also were most abundant in vata. Increased amounts of dopamine in vata Prakriti people may lead to neurological problems as well as insulin resistance. Insulin resistance, cytokines and inflammatory markers have got positive relation with V and K group both [15].
During the human metabolism studies, it is an established fact that average metabolite concentration varies by ±50% [22]. This variation is due to age, gender, genetics, health status, activity level and diurnal variations [23]. Ayurveda describes the predominance of kapha in childhood, pitta in young and vata in old age. It is apparent that children are more prone to certain type of diseases due to provocation of kapha. Similarly certain types of diseases are very common in younger and older age. This concept is called as vayoanupatinee Prakriti [24], [25]. Hence, the age may have a profounding effect on metabolic expression that can't be assessed in present setup and needs to be explored further. Ayurveda considers all these factors during Prakriti analysis. However, current methods for Prakriti analysis are inconsistent and some scientifically developed methods lack informational ground. Ayurmetabolomics may provide a platform to discriminate different Prakriti types with constitutional information. In our study, functional categories of metabolites showing differential expression among Prakriti types were significantly enriched in core biological processes like amino acids, vitamins, lipids and nucleic acid metabolism. The relative closeness between vata and kapha Prakritis has been confirmed in PCA plot and heat map (Figs. 1A and 2).
5. Conclusion
The present study, presented an idea that plasma level of some simple metabolites, i.e. carnitines, 4-aminohippuric acid, androgen, melatonin, prostaglandins, l-kynurenine, amino acids, vitamins and lipids; can classify the human phenotypes, i.e. Prakritis. Metabolites are the end players in body metabolism and signaling molecules to initiate metabolic and catabolic pathways in the body. Metabolomics deals with the levels of metabolites in the body fluids and tissues in different conditions. Phenotypes of individuals mainly depends on the body metabolism. Tridosha classification in Ayurveda mainly depends upon the phenotypic and psychological characteristics of individuals. Hence, integration of Ayurveda with metabolomics holds potential and promise for future predictive, individualized medicine. Present study indicates that dominating kapha Prakriti individual are predisposed to respiratory problems, hence continuous mitochondrial genome maintenance is the dominating biological process in response to stress slow lipid catabolism. It also leads to lipid storage and hence, weight gain. Moreover, upregulated NAD biosynthetic processes also confirmed the slow lipid metabolism. The increased lipid levels may lead to oxidative stress and cardiovascular problems. In the study, the levels of cardiovascular and neurological diseases markers i.e. capryloylglycine, 8-hydroxyguanine, d-threitol, hexanoylcarnitine and kynurenine were found to be increased. Hence, study showed the predisposition of kapha dominant people to cardiovascular and neurological diseases as described in Ayurveda, but continuous maintenance of respiratory system help to control these diseases. In pitta Prakriti metabolism of xenobiotics and toxins was observed through cytochrome P450. In this group, beta-oxidation disorders are more common, may be because of increased lipid absorption through the intestine as increased levels of bile acids were observed in the study. Melatonin and 2-methoxyestrone, hormones to maintain circadian biological clock, basal metabolism rate, and physical constitution, were found to be dominant. All these processes can be correlated with fast metabolism of pitta Prakriti. In vata Prakriti cellular detoxification process along with neurotransmitters i.e. dopamine and glutamate was found to be dominant. These processes are important to excrete waste and cognition. Hence, vata individuals may have predisposition to neurological problems and insulin resistance. The present study provides a framework to understand Prakriti in terms of metabolites and also to prognose associated disorders. Experimentally, this is limited data and needs further exploration with large data sets. However, data provides a useful benchmark and provides correlation in phenotype to metabolic signatures of different Prakritis.
Sources of funding
None.
Conflict of interest
None.
Acknowledgement
The authors are grateful to CCRAS, Ministry of AYUSH, Govt of India for their moral support.
Footnotes
Peer review under responsibility of Transdisciplinary University, Bangalore.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jaim.2017.05.002.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Rotti H., Raval R., Anchan S., Bellampalli R., Bhale S., Bharadwaj R. Determinants of prakriti, the human constitution types of Indian traditional medicine and its correlation with contemporary science. J Ayurveda Integr Med. 2014;5(3):167–175. doi: 10.4103/0975-9476.140478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patwardhan B., Joshi K., Chopra A. Classification of human population based on HLA gene polymorphism and the concept of Prakriti in Ayurveda. J Altern Complement Med. 2005;11(2):349–353. doi: 10.1089/acm.2005.11.349. [DOI] [PubMed] [Google Scholar]
- 3.Patwardhan B. The quest for evidence based Ayurveda: lesson learned. Curr Sci. 2012;102:1–12. [Google Scholar]
- 4.Gupta P.D. Pharmacogenetics, pharmacogenomics and ayurgenomics for personalized medicine: a paradigm shift. Indian J Pharm Sci. 2015;77(2):135–141. doi: 10.4103/0250-474x.156543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dettmer K., Aronov P.A., Hammock B.D. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26(1):51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joshi K., Ghodke Y., Shintre P. Traditional medicine and genomics. J Ayurveda Integr Med. 2010;1(1):26–32. doi: 10.4103/0975-9476.59824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikolai L. 2006. Blood extraction standard operating protocol. [SOP # 024.1 Versions # 1] [Google Scholar]
- 8.Tripathi N.S. Concept of formation of Prakriti in ayurveda. Ind J Res. 2011;5:1–5. [Google Scholar]
- 9.Dey S., Pahwa P. Prakriti and its associations with metabolism, chronic diseases, and genotypes: possibilities of new born screening and a lifetime of personalized prevention. J Ayurveda Integr Med. 2014;5(1):15–24. doi: 10.4103/0975-9476.128848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma H., Chandola H.M. Ayurvedic concept of obesity, metabolic syndrome, and diabetes mellitus. J Altern Complement Med. 2011;17(6):549–552. doi: 10.1089/acm.2010.0690. [DOI] [PubMed] [Google Scholar]
- 11.Sumantran V.N., Tillu G. Cancer, inflammation, and insights from ayurveda. Evid Based Complement Altern Med. 2012:11. doi: 10.1155/2012/306346. [Article ID 306346] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma P.V. Chaukhambha Orientalia; Varanasi: 2011. Caraka Samhita. [Reprint edition] [Google Scholar]
- 13.Sharma H.M. Fundamentals of complementary and alternative medicine. 4th ed. Saunders Elsevier; USA: 2011. Contemporary ayurveda; pp. 495–508. [Google Scholar]
- 14.Telles S., Pathak S., Kumar A., Mishra P., Balkrishna A. Ayurvedic doshas as predictors of sleep quality. Med Sci Monit. 2015;21:1421–1427. doi: 10.12659/MSM.893302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hankey A. A test of the systems analysis underlying the scientific theory of Ayurveda's Tridosha. J Altern Complement Med. 2005;11:385–390. doi: 10.1089/acm.2005.11.385. [DOI] [PubMed] [Google Scholar]
- 16.Prasher B., Negi S., Aggarwal S., Mandal A.K., Sethi T.P., Deshmukh S.R. Indian genome variation consortium, whole genome expression and biochemical correlates of extreme constitutional types defined in Ayurveda. J Transl Med. 2008;6(48):12. doi: 10.1186/1479-5876-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Upadhyay R.K. Emerging risk biomarkers in cardiovascular diseases and disorders. J Lipids. 2015:50. doi: 10.1155/2015/971453. [Article ID 971453] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahalle N.P., Kulkarni M.V., Pendse N.M., Naik S.S. Association of constitutional type of Ayurveda with cardiovascular risk factors, inflammatory markers and insulin resistance. J Ayurveda Integr Med. 2012;3(3):150–157. doi: 10.4103/0975-9476.100186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghodke Y., Joshi K., Patwardhan B. Traditional medicine to modern pharmacogenomics: Ayurveda Prakriti type and CYP2C19 gene polymorphism associated with the metabolic variability. Evid Based Complement Alternat Med. 2011:5. doi: 10.1093/ecam/nep206. [Article ID 249528] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Callaway E. Amino-acid deficiency underlies rare form of autism. Nat News. Sep 6, 2012:11375. [Google Scholar]
- 21.Tiwari S., Gehlot S., Tiwari S.K., Singh G. Effect of walking (aerobic isotonic exercise) on physiological variants with special reference to Prameha (diabetes mellitus) as per Prakriti. AYU. 2012;33:44–49. doi: 10.4103/0974-8520.100308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Psychogios N., Hau D., Peng J., Guo A., Mandal R., Bouatra S. The human serum metabolome. PLoS ONE. 2011;6(2):e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawton K.A., Berger A., Mitchell M., Milgram K.E., Evans A.M., Guo L. Analysis of the adult human plasma metabolome. Pharmacogenomics. 2008;9:383–397. doi: 10.2217/14622416.9.4.383. [DOI] [PubMed] [Google Scholar]
- 24.Sharma R., Malviya R., Saraswat V., Shenoy S., Rama Murthy A. Prakriti (human constitution) – an individual identity of a human. Ayurpharm Int J Ayur Alli Sci. 2012;1(7):151–158. [Google Scholar]
- 25.Purvya M.C., Meena M.S. A review on role of prakriti in aging. AYU. 2011;32(1):20–24. doi: 10.4103/0974-8520.85719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.