Abstract
Listeria monocytogenes was screened from different seafood contact surfaces in five sampling sites of fishing harbour, fish landing centers, seafood processing plants, fish market, and fish curing yards of Tuticorin Coast of India. 115 swab samples were collected and tested for the occurrence of L. monocytogenes by conventional and molecular methods. Overall, 5.22% of swab samples collected were positive for L. monocytogenes. The fishing harbour had high incidence (10.3%) of L. monocytogenes followed by fish landing centers (5.9%), and seafood processing plants (4.1%). Boat deck, fish transport tricycle were the two seafood contact surfaces in fishing harbour, which had the occurrence of L. monocytogenes. The swab samples from fish market and fish curing yards were negative for L. monocytogenes. All the isolated colonies of L. monocytogenes were confirmed by PCR assay targeting virulent hlyA gene. The DNA of all the isolates yielded a product of 174 bp on PCR amplification in comparison with L. monocytogenes Type culture (MTCC 1143). The results clearly indicated the occurrence of L. monocytogenes in seafood contact surfaces along the Tuticorin Coast of India.
Keywords: Listeria monocytogenes, Seafood contact surfaces, PCR, hlyA
Introduction
Listeria monocytogenes is a gram-positive, catalase positive, motile and psychrotrophic pathogen. It is the causative agent of listeriosis, a severe disease with high hospitalization and fatality rates (Pilchova et al. 2014). Listeriosis causes serious symptoms, including septicemia, abortion, liver failure, and meningitis, especially among susceptible people like immuno-compromised individuals and pregnant women (Dhanashree et al. 2003). L. monocytogenes is a food-borne pathogen detected in various types of seafood, including shrimps, lobsters, crabs, mussels and fish (Elliot and Kvenberg 2000). In India, L. monocytogenes was first reported in 1996 from finfish and shellfish landed in the Mangalore along the West Coast of India (Jeyasekaran et al. 1996). L. monocytogenes has the ability to form biofilm in seafood contact surfaces and equipment in food-processing factories that can be transferred to food products (Harvey et al. 2007; Jeyasekaran et al. 2003). Biofilms not only present a considerable hygiene risk in the food industry, but also cause economic losses (Chorianopoulos et al. 2011).
PCR is one of the molecular methods that are reliable, rapid and less time consuming (Almeida and Almeida 2000). PCR has been used for the detection of L. monocytogenes from seafood and their contact surfaces (Park et al. 2012). One of the best ways to detect the pathogen is through targeting a virulence factor, listeriolysin O (LLO), produced by L. monocytogenes. The LLO-encoding gene (hlyA) is present only in virulent strains of the species and is a secreted protein toxin that can be easily detected (Churchill et al. 2006).
Tuticorin is located along the East Cost of India in latitude 8.8100°N and longitude 78.1400°E. The average water temperature is 28 °C, wind speed is 14 km/h, and relative humidity 69%. Seafood exports recorded a growth of 6.86% over the last year in quantity, 10.69% in rupee earnings and 10.05% growth in US dollar earnings. By way of logistic support, Tuticorin based V.O. Chidambaranar Port is contributing immensely in facilitating seafood exports. Shipments of seafood to various overseas destinations through VOC seaport from Tuticorin saw an increase of 6.72% over the previous fiscal in terms of quantity. It is therefore of paramount importance to understand the possible causes for bacterial contamination in the seafood landed and processed in this region. Different areas within seafood processing environment may have significant levels of L. monocytogenes contamination (Jami et al. 2014). The ability to form biofilm has been proposed as a crucial factor affecting the establishment of L. monocytogenes in seafood processing environment; and such biofilm formation poses a challenge for elimination of this pathogen (Gandhi and Chikindas 2007; Srey et al. 2013). In India, only few studies have been conducted to assess the presence of Listeria spp. and particularly L. monocytogenes in seafood (Kamat and Nair 1994; Jeyasekaran et al. 1996) and all the reports were from West Coast of India. Parihar et al. (2008), while studying the incidence of L. monocytogenes from tropical seafoods in Goa Coast of India, observed that the presence of L. monocytogenes also needs to be confirmed from other tropical areas of India. Since no work has been reported from the Tuticorin region on the East Coast of India, amidst high seafood export from this region, the present study was carried out to examine the occurrence of L. monocytogenes on the seafood contact surfaces, on which seafood are handled and processed prior to export.
Materials and methods
Samples from seafood contact surfaces (n = 115) were collected for this study from five sampling sites viz. fishing harbour, fish landing centers, fish processing plants, fish market and fish curing yards of Tuticorin Coast of India from October to March. Seafood contact surfaces examined from each sampling site under this study are given in Table 1 along with their sampling codes.
Table 1.
Swab samples collected from different sampling sites of Tuticorin coast of India
| Sl. no | Sampling sites | Sl. no | Seafood contact surfaces | No. of samples | Codes |
|---|---|---|---|---|---|
| I | Fishing harbour | 1 | Fish hold | 3 | FH 1.1,1.2,1.3 |
| 2 | Boat deck | 4 | FH 2.1,2.2,2.3,2.4 | ||
| 3 | Fish basket | 2 | FH 3.1,3.2 | ||
| 4 | Fish transport Tri cycle | 8 | FH 4.1,4.2,4.3,4.4,4.5,4.6,4.7,4.8 | ||
| 5 | Auction hall floor | 11 | FH 5.1,5.2,5.3,5.4,5.5,5.6,5.7,5.8, 5.9,5.10,5.11 |
||
| 6 | Fish cutting board | 1 | FH 6 | ||
| II | Fish landing centers | 1 | Fish hold | 3 | FLC 1.1,1.2,1.3 |
| 2 | Boat deck | 3 | FLC 2.1,2.2,2.3 | ||
| 3 | Fish basket | 3 | FLC 3.1,3.2,3.3 | ||
| 4 | Fish transport Tri cycle | 2 | FLC 4.1,4.2 | ||
| 5 | Auction hall floor | 4 | FLC 5.1,5.2,5.3,5.4 | ||
| 6 | Fisherman hand | 2 | FLC 6.1,6.2 | ||
| III | Seafood processing plants | 1 | Insulated vehicle | 1 | SFP 1 |
| 2 | Raw material receiving chute | 2 | SFP 2.1,2.2 | ||
| 3 | Fish basket | 5 | SFP 3.1,3.2,3.3,3.4,3.5 | ||
| 4 | Grading machine | 5 | SFP 4.1,4.2,4.3,4.4,4.5 | ||
| 5 | Grading table | 3 | SFP 5.1,5.2,5.3 | ||
| 6 | Soaking tank | 4 | SFP 6.1,6.2,6.3,6.4 | ||
| 7 | Weighing balance | 3 | SFP 7.1,7.2,7.3 | ||
| 8 | Weighing balance table | 2 | SFP 8.1,8.2 | ||
| 9 | IQF table | 2 | SFP 9.1,9.2 | ||
| 10 | Block freezing Conveyor belt | 5 | SFP 10.1,10.2,10.3,10.4,10.5 | ||
| 11 | Freezer pan | 5 | SFP 11.1,11.2,11.3,11.4,11.5 | ||
| 12 | Freezer | 1 | SFP 12 | ||
| 13 | Cooking boiler | 1 | SFP 13 | ||
| 14 | Depanning conveyor belt | 4 | SFP 14.1,14.2,14.3,14.4 | ||
| 15 | Wall tiles | 4 | SFP 15.1,15.2,15.3,15.4 | ||
| 16 | Cold store | 2 | SFP 16.1,16.2 | ||
| IV | Fish market | 1 | Fish hold | 2 | FM 1.1,1.2 |
| 2 | Fish basket | 1 | FM 2 | ||
| 3 | Fish display board | 2 | FM 3.1,3.2 | ||
| 4 | Fish cutting board | 3 | FM 4.1,4.2,4.3 | ||
| V | Fish curing yards | 1 | Fish basket | 4 | FCY 1.1,1.2,1.3,1.4 |
| 2 | Fish transport Tri cycle | 1 | FCY 2 | ||
| 3 | Brine tank | 4 | FCY 3.1,3.2,3.3,3.4 | ||
| 4 | Fish curing yard floor | 3 | FCY 4.1,4.2,4.3 | ||
| Total | 115 |
FH fishing harbour, FLC fish landing centers, SFP seafood processing plants, FM fish market, FCY fish curing yard
Swab samples were collected from the seafood contact surfaces to screen for the presence of L. monocytogenes based on FDA protocol following the conventional method (Liu et al. 2006). Contact area (50 cm2) was swabbed using sterile cotton swabs moistened with sterile saline and transferred into 10 ml of sterile physiological saline. All the samples were processed within 1 h of collection.
Genomic DNA was extracted using the Himedia Genomic DNA Extraction kit (Hi-Media Laboratories Pvt. Ltd., Mumbai, India) according to the manufacturer’s protocol and stored at − 20 °C until further use. Oligonucleotide primers for the PCR assay were selected based on the earlier published nucleotide sequence of hlyA gene (Ndahi et al. 2013; Park et al. 2012). Forward primer is 5′-GCATCTGCATTCAATAAAGA-3′ and reverse primer is 5′-TGTCACTGCATCTCCGTGGT- 3′ with a product size of 174 bp. The primers were synthesized by the Bioserve Biotechnologies (I) Pvt. Ltd., Hyderabad, India. The amplification was done using a Gradient Master Cycler (Eppendorf AG, Hamburg, Germany). The reaction mixture of 25 µl consisted of 2.5 µl reaction assay buffer (100 mM Tris (pH 9.0), 500 mM KCl, 15 mM MgCl2) 0.25 µl dNTP (100 mM), 19 µl of molecular grade water, 1.25 U of Taq DNA polymerase, 1 µl of each forward and reverse primers (10 pmol) and 1 µl of template DNA. The PCR protocol comprised of initial denaturation at 94 °C for 3 min followed by 30 cycles of denaturation at 94 °C for 60 s, annealing at 55 °C for 60 s, extension at 72 °C for 90 s and final extension at 72 °C for 10 min. The PCR products were analysed on 2% agarose gel using 0.5 X TAE buffer with ethidium bromide (0.5 µg/ml). A 100 bp DNA ladder was used as standard marker. Amplified DNA was visualized under UV transilluminator and photographed using gel documentation system (Alpha Innotech Co., San Leandro, USA) and analyzed using Alpha imager EC software (Version 3.4.0.0).
Results and discussion
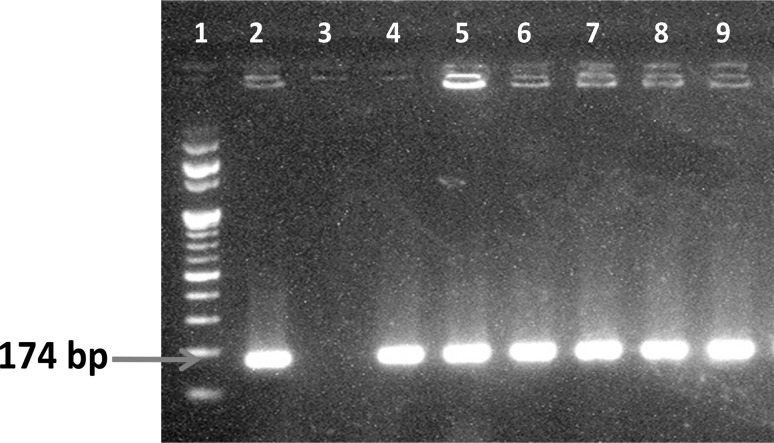
A total of 115 swab samples were collected from the seafood contact surfaces (Table 1). Of which, only six samples showed the presence of L. monocytogenes (Table 2). Twenty nine swab samples collected from fishing harbour were analysed for the occurrence of L. monocytogenes and three swab samples showed the presence of L. monocytogenes (Tables 1, 2). Two isolates were obtained from fish transport tricycle and one from boat deck. 17 swab samples collected from three different fish landing centers located in Tuticorin coast of India were tested for the incidence of L. monocytogenes (Table 1). Of which, one sample collected from boat deck was positive for L. monocytogenes (Table 2). 49 swab samples collected from four different seafood processing plants were examined for the incidence of L. monocytogenes (Table 1). Of which, two samples were positive for L. monocytogenes, and they were isolated from grading machine and freezer pan (Table 2). Eight swab samples collected from fish market of Tuticorin were analysed for the incidence of L. monocytogenes (Table 1). All the samples analysed were negative for L. monocytogenes (Table 2). Twelve swab samples collected from two different fish curing yards were tested for the incidence of L. monocytogenes and all the samples showed absence of L. monocytogenes (Tables 1, 2). All the positive isolates that showed typical colonies in PALCAM agar plates were confirmed by PCR. The amplified hlyA gene product of the isolates of L. monocytogenes viz., FH 2.1, FH 4.1, SFP 4.1, SFP 11.1, FLC 2.2, FH 4.7 taken from fishing harbour, fish landing centers and seafood processing plants is shown in Fig. 1. All the six isolates of L. monocytogenes were thus confirmed by PCR assay.
Table 2.
Occurrence of Listeria monocytogenes in different sampling sites of Tuticorin region
| Sl. no. | Sampling sites | No. of samples tested | No. of positive samples with percent of incidence |
|---|---|---|---|
| 1. | Fishing harbour | 29 | 3 (10.3%) |
| 2. | Fish landing centers | 17 | 1 (5.9%) |
| 3. | Seafood processing plants | 49 | 2 (4.1%) |
| 4. | Fish market | 8 | 0 |
| 5. | Fish curing yards | 12 | 0 |
| Total | 115 | 6 (5.22%) |
Fig. 1.
Detection of L. monocytogenes from seafood contact surfaces by PCR using hlyA gene. Lane 1—100 bp DNA marker; Lane 2—Positive control; Lane 3—Negative control; Lane 4—FH 2.1; Lane 5—FH 4.1; Lane 6—SFP 4.1; Lane 7—SFP 11.1; Lane 8—FLC 2.2; Lane 9—FH 4.7
L. monocytogenes is a bacterial pathogen capable of adhering to many surfaces (Galvao et al. 2012). Of 115 swab samples collected from different seafood contact surfaces of Tuticorin Coast of India, only 5.21% of swab samples were positive for L. monocytogenes (Tables 1, 2). The level of incidence of L. monocytogenes in seafood contact surfaces of Tuticorin region was slightly less when compared to earlier report on the incidence of L. monocytogenes along the West Coast of India (Jeyasekaran et al. 2003), as they have observed that 7.6% of environmental swab samples obtained from fishing harbour, fish landing centers, and seafood processing plants in Mangalore region along the West Coast of India were positive for L. monocytogenes.
The processing environment has proved to be particularly important source of L. monocytogenes contamination in food production. This pathogen has the ability to persist for years in food industry environments, including seafood premises leading to product contamination (Hoffman et al. 2003; Miettinen and Wirtanen 2006). However, Fallah et al. (2013) reported that 19.2% of environmental swab samples obtained from seafood markets and seafood processing plants of Iran were positive for L. monocytogenes, which is very high when compared to that of the present findings. Johansson et al. (1999) found higher incidence (15.4%) of L. monocytogenes from the environmental swab samples collected from fish processing plants in Finland. It has been reported that food production environmental areas can serve as source of L. monocytogenes contamination (Anonymous 1999).
Among the environmental swab samples collected from different seafood contact surfaces of Tuticorin region, the samples from fishing harbour had high incidence (10.3%) of L. monocytogenes followed by fish landing centers (5.9%) and seafood processing plants (4.1%) (Table 2). Boat deck and fish transport tricycle are the two important seafood contact surfaces in fishing harbour, where the incidence of L. monocytogenes was high. About 25 and 28.5% of swab samples from fish transport tricycle, and boat deck were positive for L. monocytogenes, respectively. However, Jeyasekaran et al. (2003) reported that 60% of swab samples collected from fishing harbour, 20% samples from fish landing center and 0.9% swab samples from seafood processing plants of Mangalore region were positive for L. monocytogenes. Cruz and Fletcher (2011) have clearly demonstrated that mussel processing plants in New Zealand harboured different population of L. monocytogenes.
The samples collected from fish market, and fish curing yards were positive for L. monocytogenes (Table 2). However, Akkaya et al. (2011) detected L. monocytogenes from swab samples of knives, cutting boards from fish market in Turkey. But, in a study conducted in Greece, none of the samples from fish markets were found to contain L. monocytogenes (Soultos et al. 2007).
The isolates of L. monocytogenes from different seafood contact surfaces of Tuticorin region, Tamil Nadu were confirmed by PCR targeting hlyA gene (Fig. 1). Detection of L. monocytogenes based on PCR amplification of the hlyA gene has earlier been reported by few workers (Furrer et al. 1991). Gawade et al. (2010) also reported that L. monocytogenes can be confirmed by PCR using hlyA gene as a marker to differentiate L. monocytogenes from the other Listeria spp. In the present study, the PCR amplification of hlyA gene for the identification of L. monocytogenes used a set of primers as described by Park et al. (2012) which yielded a product of 174 bp. The Type culture (MTCC 1143) and the isolates of L. monocytogenes from seafood contact surfaces swab yielded a similar product size of 174 bp on PCR amplification targeting hlyA gene (Fig. 1).
Conclusion
The isolates of L. monocytogenes from the seafood contact surfaces of Tuticorin region indicate the potential public health hazard. Diligent enforcement of sanitary conditions of food contact surfaces and handling areas, and personal hygiene practices shall reduce the potential contamination of seafood contact surfaces. An in-depth understanding of the physiology of the organism, establishment of HACCP in the food industry, regulated product sampling and testing, along with better detection and surveillance systems for reporting food borne disease outbreak is valuable in controlling the pathogen.
Acknowledgements
Authors wish to thank the financial support provided by the Indian Council of Agricultural Research, New Delhi through the Niche Area of Excellence programme on Fish Safety and Quality Assurance to the present study. First author also wishes to acknowledge the Tamil Nadu Fisheries University, as it is the part of the thesis of PG research works of the University.
References
- Akkaya L, Atabay HI, Gok V, Kara R. Detection of Listeria species in fresh fish and fish market environment by IMS technique in Turkey. Arch Lebensmittelhyg. 2011;62:16–19. [Google Scholar]
- Almeida PF, Almeida RC. A PCR protocol using inl gene as a target for specific detection of Listeria monocytogenes. Food Control. 2000;11(2):97–101. doi: 10.1016/S0956-7135(99)00067-5. [DOI] [Google Scholar]
- Anonymous . FAO expert consultation on the trade impact of Listeria in fish products. Amherst: Food and Agricultural Organization of the United Nations. Report; 1999. p. 34. [Google Scholar]
- Chorianopoulos NG, Tsoukleris DS, Panagou EZ, Falaras P, Nychas GJ. Use of titanium dioxide (TiO2) photocatalysts as alternative means for Listeria monocytogenes biofilm disinfection in food processing. Food Microbiol. 2011;28(1):164–170. doi: 10.1016/j.fm.2010.07.025. [DOI] [PubMed] [Google Scholar]
- Churchill RL, Lee H, Hall JC. Detection of Listeria monocytogenes and the toxin listeriolysin O in food. J Microbiol Methods. 2006;64(2):141–170. doi: 10.1016/j.mimet.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Cruz CD, Fletcher GC. Prevalence and biofilm-forming ability of Listeria monocytogenes in New Zealand mussel (Perna canaliculus) processing plants. Food Microbiol. 2011;28(7):1387–1393. doi: 10.1016/j.fm.2011.06.014. [DOI] [PubMed] [Google Scholar]
- Dhanashree B, Otta SK, Karunasagar I, Goebel W. Incidence of Listeria spp. in clinical and food samples in Mangalore, India. Food Microbiol. 2003;20(4):447–453. doi: 10.1016/S0740-0020(02)00140-5. [DOI] [Google Scholar]
- Elliot EL, Kvenberg JE. Risk assessment used to evaluate the US position on Listeria monocytogenes in seafood. Int J Food Microbiol. 2000;62(3):253–260. doi: 10.1016/S0168-1605(00)00344-5. [DOI] [PubMed] [Google Scholar]
- Fallah AA, Saei-Dehkordi SS, Mahzounieh M. Occurrence and antibiotic resistance profiles of Listeria monocytogenes isolated from seafood products and market and processing environments in Iran. Food Control. 2013;34(2):630–636. doi: 10.1016/j.foodcont.2013.06.015. [DOI] [Google Scholar]
- Furrer B, Candrian U, Hoefelein C, Luethy J. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. J Appl Microbiol. 1991;70(5):372–379. doi: 10.1111/j.1365-2672.1991.tb02951.x. [DOI] [PubMed] [Google Scholar]
- Galvao NN, Chiarini E, Destro MT, de Aguiar Ferreira M, Nero LA. PFGE characterisation and adhesion ability of Listeria monocytogenes isolates obtained from bovine carcasses and beef processing facilities. Meat Sci. 2012;92(4):635–643. doi: 10.1016/j.meatsci.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Gandhi M, Chikindas ML. Listeria: a foodborne pathogen that knows how to survive. Int J Food Microbiol. 2007;113(1):1–5. doi: 10.1016/j.ijfoodmicro.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Gawade L, Barbuddhe SB, Bhosle S. Isolation and confirmation of Listeria species from seafood off Goa region by polymerase chain reaction. Indian J Microbiol. 2010;50(4):385–389. doi: 10.1007/s12088-011-0064-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey J, Keenan KP, Gilmour A. Assessing biofilm formation by Listeria monocytogenes strains. Food Microbiol. 2007;24(4):380–392. doi: 10.1016/j.fm.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Hoffman AD, Gall KL, Norton DM, Wiedmann M. Listeria monocytogenes contamination patterns for the smoked fish processing environment and for raw fish. J Food Prot. 2003;66(1):52–60. doi: 10.4315/0362-028X-66.1.52. [DOI] [PubMed] [Google Scholar]
- Jami M, Ghanbari M, Zunabovic M, Domig KJ, Kneifel W. Listeria monocytogenes in aquatic food products-a review. Compr Rev Food Sci Food Saf. 2014;13(5):798–813. doi: 10.1111/1541-4337.12092. [DOI] [Google Scholar]
- Jeyasekaran G, Karunasagar I, Karunasagar I. Incidence of Listeria spp. in tropical fish. Int J Food Microbiol. 1996;31(1–3):333–340. doi: 10.1016/0168-1605(96)00980-4. [DOI] [PubMed] [Google Scholar]
- Jeyasekaran G, Karunasagar I, Karunasagar I. Occurrence of Listeria spp. in seafood handling environments. Ind J Fish. 2003;50(2):211–214. [Google Scholar]
- Johansson T, Rantala L, Palmu L, Honkanen-Buzalski T. Occurrence and typing of Listeria monocytogenes strains in retail vacuum-packed fish products and in a production plant. Int J Food Microbiol. 1999;47(1–2):111–119. doi: 10.1016/S0168-1605(99)00019-7. [DOI] [PubMed] [Google Scholar]
- Kamat AS, Nair PM. Incidence of Listeria species in Indian seafoods and meat. J Food Saf. 1994;14(2):117–130. doi: 10.1111/j.1745-4565.1994.tb00589.x. [DOI] [Google Scholar]
- Liu C, Duan J, Su YC. Effects of electrolyzed oxidizing water on reducing Listeria monocytogenes contamination on seafood processing surfaces. Int J Food Microbiol. 2006;106(3):248–253. doi: 10.1016/j.ijfoodmicro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- Miettinen H, Wirtanen G. Ecology of Listeria spp. in a fish farm and molecular typing of Listeria monocytogenes from fish farming and processing companies. Int J Food Microbiol. 2006;112(2):138–146. doi: 10.1016/j.ijfoodmicro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Ndahi MD, Kwaga JK, Bello M, Kabir J, Umoh VJ, Yakubu SE, Nok AJ. Prevalence and antimicrobial susceptibility of Listeria monocytogenes and methicillin-resistant Staphylococcus aureus strains from raw meat and meat products in Zaria, Nigeria. Lett Appl Microbiol. 2013;58(3):262–269. doi: 10.1111/lam.12183. [DOI] [PubMed] [Google Scholar]
- Parihar VS, Barbuddhe SB, Danielsson-Tham ML, Tham W. Isolation and characterization of Listeria species from tropical seafoods. Food Control. 2008;19(6):566–569. doi: 10.1016/j.foodcont.2007.06.009. [DOI] [Google Scholar]
- Park S, Jung J, Choi S, Oh Y, Lee J, Chae H, Ryu S, Jung H, Park G, Choi S, Kim B. Molecular characterization of Listeria monocytogenes based on the PFGE and RAPD in Korea. Adv Microbiol. 2012;2(04):605–616. doi: 10.4236/aim.2012.24079. [DOI] [Google Scholar]
- Pilchova T, Hernould M, Prévost H, Demnerová K, Pazlarová J, Tresse O. Influence of food processing environments on structure initiation of static biofilm of Listeria monocytogenes. Food Control. 2014;35(1):366–372. doi: 10.1016/j.foodcont.2013.07.021. [DOI] [Google Scholar]
- Soultos N, Abrahim A, Papageorgiou K, Steris V. Incidence of Listeria spp. in fish and environment of fish markets in Northern Greece. Food Control. 2007;18(5):554–557. doi: 10.1016/j.foodcont.2006.01.006. [DOI] [Google Scholar]
- Srey S, Jahid IK, Ha SD. Biofilm formation in food industries: a food safety concern. Food Control. 2013;31(2):572–585. doi: 10.1016/j.foodcont.2012.12.001. [DOI] [Google Scholar]