Abstract
Marine Fungi are potent secondary metabolite producers. However, limited genetic information are available their biosynthetic gene clusters (BGCs) and their biotechnological applications. To overcome this lack of information, herein, we used next-generation sequencing methods for genome sequencing of two marine fungi, isolated from the German Wadden Sea, namely Calcarisporium sp. KF525 and Pestalotiopsis sp. KF079. The assembled genome size of the marine isolate Calcarisporium sp. KF525 is about 36.8 Mb with 60 BGCs, while Pestalotiopsis sp. KF079 has a genome size of 47.5 Mb harboring 67 BGCs. Of all BGCs, 98% and 97% are novel clusters of Calcarisporium sp. and Pestalotiopsis sp., respectively. Only few of the BGCs were found to be expressed under laboratory conditions by RNA-seq analysis. The vast majority of all BGCs were found to be novel and unique for these two marine fungi. Along with a description of the identified gene clusters, we furthermore present important genomic features and life-style properties of these two fungi. The two novel fungal genomes provide a plethora of new BGCs, which may have biotechnological applications in the future, for example as novel drugs. The genomic characterizations will provide assistance in future genetics and genomic analyses of marine fungi.
Introduction
Marine fungi have been much neglected for a long time, although first studies on marine fungi were published decades ago1. More recently, the phylogeny of some marine-derived fungi was elucidated by analysis of their small ribosomal RNA subunit sequences2. Thirty-six new marine lineages were isolated from six European near-shore sites. The isolates were dominated by chytrids, but also a few filamentous fungi and many ascomycetous and basidiomycetous yeasts were described2. Additionally, there is an effort to identify and catalog marine species in European marine environments3. Nevertheless, the number of cultivated marine fungi merely comprises some 470 species4 belonging to 244 genera. This would be less than one percent of all known fungi. Molecular analysis of rDNA sequences have contributed to a somewhat higher count of fungal species, but they still constitute a surprisingly low number of taxonomic units found in marine environments2.
Currently the estimated number of marine fungal species is over 10,0005, but may be much higher6. Factors that influence whether or not marine fungi are present in any particular location include the water temperature, water salinity, the water movement, the presence of suitable substrates for colonization, the presence of propagates in the water, interspecific competition, pollution and the oxygen content of the water7.
The biodiversity of marine fungal isolates is mirrored by the molecular diversity of their secondary metabolites8–12. Yet, these studies are mainly chemistry based, and marine fungi remain tremendously underexplored with regard to species, distribution and applications12,13. As such marine-derived fungi contain a treasure chest of secondary metabolites, of which a considerable number have promising biological or pharmaceutical properties14. However, so far, most studies did not use whole genome sequencing to discover the enormous potential of fungi for secondary metabolite production. A study on a marine fungus isolated from a sponge revealed the entire secondary metabolite cluster for scopularide A and B, which has anti-cancer activities15. Secondary metabolites are low-molecular-mass organic compounds that, unlike primary metabolites, are not directly involved in growth, development or reproduction of the producing organism. Up until 2014 about 170,000 natural products have been characterized from both marine and terrestrial organisms16. Fungal secondary metabolites can be divided into four main chemical classes: polyketides, terpenoids, shikimic acid-derived compounds, and non-ribosomal peptides. The majority of fungal secondary metabolites derives from either non-ribosomal peptide synthetases (NRPSs) or polyketide synthases (PKSs), A few compounds represent mixed polyketide–non-ribosomal peptide compounds called NRPS-PKS hybrids17. Furthermore, secondary metabolites can be found in microbes of diverse environments and even chemical biogeographic distribution maps for biomedically valuable families of natural products in the environment have been created18. A number of these compounds have important pharmacological applications and are used as antibiotics/antibacterial drugs19. Genome-mining efforts indicate that the capability of fungi to produce secondary metabolites has been substantially underestimated, because many of these gene clusters are silent under standard cultivation conditions19. This indicated a plethora of natural products remains to be discovered.
Here we report on the genomic sequences of two North Sea-derived fungal isolates, Calcarisporium sp. KF525 and Pestalotiopsis sp. KF079 by the use of two next-generation sequencing methods. The marine-derived Calcarisporium sp. KF525 has a 36.8 Mb genome with 60 biosynthetic gene clusters (BGCs) and Pestalotiopsis sp. KF079 has a genome size of ~47.5 Mb and harbors 67 BGCs. The majority of these clusters has not been previously identified and hence provide a great source of new potential natural products. Here we make a first attempt to characterize the genome content of these two marine fungi and their BGCs using high-throughput methods.
Results
Overview of genomic features and RNA-Seq statistics
We generated hybrid genome assemblies of two marine fungi using two different small read sequencing methods, Roche 454 and Illumina HiSeq. 2000. The estimated genome sizes for these marine strains of Calcarisporium KF525 and Pestalotiopsis KF079 are about 36.8 Mb and about 47.5 Mb, respectively (Table 1). Genomic coverages for Calcarisporium sp. KF0525 and Pestalotiopsis KF079 and are 11.5× and 9.3× using Roche 454 reads and 219× and 169× using Illumina HiSeq 2000 reads, respectively (Table 1). The estimated numbers of genes are 15,459 and 22,626, respectively (Table 1 and Figure S1). These data are within the range of genome sizes and corresponding gene contents of ascomycetes, which are rather variable in both land and marine fungi (Figure S1). The genome assembly is simplified by the fact that both Calcarisporium sp. KF525 and Pestalotiopsis sp. KF079 possess a limited number of repeated DNA sequences, which account for 1.28% and 0.97% of their respective genome sizes only (Table S1). The genomes of Calcarisporium KF525 and Pestalotiopsis KF079 contain tandem repeats with total genome size about 0.89% and 0.65% respectively. Low-complexity regions are comprised of biased composition with imperfect direct and inverted repeats (Table S1). Transposable elements make up 0.24% and 0.23%, in corresponding genomes of these two marine fungi. Among these transposable elements, retroelements are more frequent with 0.19% and 0.22% of genome size in the genomes of Calcarisporium KF525 and Pestalotiopsis KF079, respectively. Retrotransposons with long terminal repeats are the major component. Class II DNA transposons comprise only 0.05% of Calcarisporium genome with the Tc1-IS630-Pogo family as the major stakeholder. In contrast, Pestalotiopsis KF079 has a negligible number of class II DNA transposons. Most ascomycetic terrestrial fungi have a transposon content of 1–4% of fungal genome size20. By combing current data from terrestrial and two other marine fungi (S. brevicaulis LF58015 and Cadophora malorum Mo1221), it appears that marine fungi have a similar content of transposable elements.
Table 1.
Summary of hybrid genome assemblies of two marine fungi and statistical summary of reads from Roche 454 and Illumina.
| Calcarisporium sp. KF0525 | Pestalotiopsis sp. KF079 | ||
|---|---|---|---|
| Genome size | 36.8 Mb | 47.5 Mb | |
| Scaffolds | 2274 | 318 | |
| N25* (#scaffolds) | 162.8 kb (43) | 821.7 kb (11) | |
| N50* (#scaffolds) | 95.7 kb (115) | 429.3 kb (32) | |
| N75* (#scaffolds) | 51.3 kb (263) | 249.3 kb (67) | |
| GC content (%) | 50.6 | 52.1 | |
| No. of Genes | 15,459 | 22,626 | |
| Read Statistics | |||
| Roche 454 | No of reads | 773,371 | 824,828 |
| Total length | 441,946,919 | 424,413,772 | |
| Average read length | 571.5 | 514.5 | |
| Genomic coverage | 9.3× | 11.5× | |
| Illumina | No of reads | 77,557,838 | 79, 803, 010 |
| Total length | 7, 833,341,638 | 8, 060,104,010 | |
| Average read length | 101 | 101 | |
| Genomic coverage | 169× | 219× | |
*Length of the scaffold until which sum of lengths of scaffolds are reached to 25%, 50% and 75% of total assembled genome size are called N25, N50 and N75 respectively.
#Scaffolds – Number of scaffolds in the assembled genome that constitute particular N25 or N50 or N75.
Using homology-based genome annotation analyses, we were able to annotate 72% (Table S2) and 68.9% (Table S3) of encoded proteins from assembled genomes of Calcarisporium KF525 and Pestalotiopsis KF079, respectively.
We performed RNA-Seq with wild-type strains of Calcarisporium sp. KF525 and Pestalotiopsis sp. KF079 and we found a low number of only 4,401 and 6,658 genes, being expressed under laboratory conditions. This accounts for 28.5% and 29.43% of total genes in the corresponding genomes (Tables 2, S4–S5). Based on the reads per kilobase of transcript per million mapped reads (RPKM) values, we divided the expressed genes into nine tier (Tables 2, S4–S5).
Table 2.
Overview of RNA-Seq data of wild type of two marine fungi.
| Tiers | RPKM value | Calcarisporium sp. KF525 | Pestalotiopsis sp. KF079 | ||||
|---|---|---|---|---|---|---|---|
| No. of expressed genes | % age of expressed genes | % age of all genes | No. of expressed genes | % age of expressed genes | % age of all genes | ||
| Tier #1 | >3000 | 73 | 1.7 | 0.5 | 82 | 1.2 | 0.4 |
| Tier #2 | >1000 to <3000 | 90 | 2.1 | 0.6 | 112 | 1.7 | 0.5 |
| Tier #3 | >500 to <1000 | 83 | 1.9 | 0.5 | 97 | 1.5 | 0.4 |
| Tier #4 | >250 to <500 | 91 | 2.1 | 0.6 | 159 | 2.4 | 0.7 |
| Tier #5 | >100 to <250 | 172 | 3.9 | 1.1 | 304 | 4.6 | 1.2 |
| Tier #6 | >50 to <100 | 540 | 12.3 | 3.5 | 304 | 4.6 | 1.2 |
| Tier #7 | >10 to <50 | 654 | 14.9 | 4.2 | 1125 | 16.9 | 3.1 |
| Tier #8 | >1 to <10 | 2028 | 46.1 | 13.1 | 2063 | 31 | 9.1 |
| Tier #9 | >0 to <1 | 1000 | 22.7 | 6.5 | 2412 | 36.2 | 10.7 |
| Non-expressed genes | % age of all genes | Non-expressed genes | % age of all genes | ||||
| Tier #10 | 0 | 11058 | NA | 71.5 | 15968 | NA | 70.6 |
Phylogenomic relationships of the two marine fungi
Genome-wide annotations of Calcarisporium sp. and Pestalotiopsis sp. depicted that both marine fungi are members of the Sordariomycetes, and they are most closely related to Nectria and Fusarium species (Figure S2A). We also performed a phylogenomic analysis using the CVtree22, which revealed that Calcarisporium sp. is close to Metarhizium and Trichoderma. In contrast, Pestalotiopsis sp. has diverged early from other representative sordariomycetic fungi (Figure S2B) and is close to another marine fungus, S. brevicaulis15.
Overview of protein domains in the two marine fungi
Independently foldable structural protein units serve as protein domains, which share common functions that have been conserved during evolution. Generally, these protein domains are surveyed in newly sequenced genomes using two databases - Pfam (version 2723) and Interpro (version 4324). Calcarisporium sp. and Pestalotiopsis sp. genomes encode 2,599 and 3,613 different protein domains localized in 7,272 (Table S6) and 10,383 proteins (Table S7), respectively. The top 20 Pfam domains are summarized in Fig. 1. We identified 1,317 and 1,231 WD domain, G-beta repeat (WD40, Pfam ID - PF00400) in Calcarisporium sp. and Pestalotiopsis sp. genomes, respectively. WD40 domains are involved in the signal transduction and regulate fungal cell differentiation processes25. There were furthermore a total of 1,711 and 1,566 Ankyrin repeats (Ank, PF00023) in Calcarisporium sp. and Pestalotiopsis sp. genomes, respectively. Ankyrin repeats consists of 30–34 amino acid residues long protein motifs and assist protein-protein interactions26. Moreover, Calcarisporium sp. genome encodes two tetratricopeptide-repeat-carrying domains (TPR_1, PF00515 and TPR_2, PF07719), which are found in 418 TPR_1 and 569 TPR_2 proteins, while the Pestalotiopsis sp. genome contains 450 TPR_1 and 610 TPR_2 proteins. These TPR motifs function as protein interaction modules27. Additionally, Calcarisporium and Pestalotiopsis genomes encodes 316 and 443 zinc finger proteins of the C2H2 type (zf-C2H2, PF00096), 429 and 591 short chain dehydrogenases (adh_short, PF00106), 292 and 484 FAD dependent oxidoreductases (DAO, PF01266), 328 and 300 protein kinase domains (Pkinase, PF00069) and 287 and 242 methyltransferase domains (methyltransf_11, PF08241), respectively. The entire protein domain annotations are available in Tables S6 and S7 for Calcarisporium and Pestalotiopsis genomes, respectively.
Figure 1.
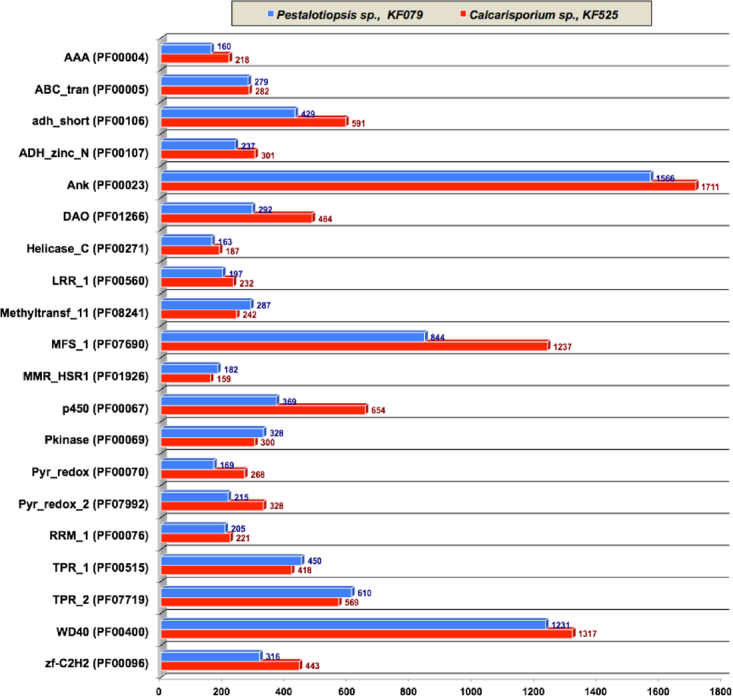
Overview of protein domains in the two marine fungal genomes. AAA (PF00004) - ATPase family associated with various cellular activities (AAA); ABC_tran (PF00005) - ABC transporter; adh_short (PF00106)-short chain dehydrogenases; ADH_zinc_N (PF00107) - Zinc-binding dehydrogenase; Ank (PF00023)- Ankyrin repeats;DAO (PF01266) - FAD dependent oxidoreductases; Helicase_C (PF00271) - Helicase conserved C-terminal domain; LRR_1 (PF00560) - Leucine Rich Repeat; Methyltransf_11 (PF08241) - methyltransferase domains; MFS_1 (PF07690) - Major Facilitator Superfamily; MMR_HSR1 (PF01926) - GTPase of unknown function; p450 (PF00067) - Cytochrome P450; Pkinase (PF00069) - Protein kinase domain; Pyr_redox (PF00070) - Pyridine nucleotide-disulphide oxidoreductase; Pyr_redox_2 (PF07992) - Pyridine nucleotide-disulphide oxidoreductase 2; RRM_1 (PF00076) - RNA recognition motif. (a.k.a. RRM, RBD, or RNP domain); TPR_1 (PF00515) - tetratricopeptide-repeat-carrying domain 1; TPR_2 (PF07719) - tetratricopeptide-repeat-carrying domain 2;WD40 (PF00400) - WD domain, G-beta repeats; zf-C2H2 (PF00096) - C2H2 type zinc finger proteins.
Biosynthetic gene clusters in Calcarisporium sp
Calcarisporium sp. has 60 BGCs named as mCaBGC1 to mCaBGC60 (Table S8), including 24 type I PKS (T1pks) clusters, 17 nonribosomal peptide synthetase (NRPS), seven terpene and two heterocyst-type glycolipid ketosynthase (Hglks) clusters. In addition, there are four hybrid clusters and seven putative clusters. Hybrid clusters include two NRPS-t1PKS and one each of Hglks-t1PKS and T1PKS-NRPS (Table S8). 59 BGCs are unique to Calcarisporium sp., as these BGCs have no homologs clusters in the MiBiG database (28 a database of known BGCs). Hence, more than 98% of detected BGCs of Calcarisporium sp. are by and large novel with some identities to uncharacterized BGCs in different species. Only one cluster, mCaBGC19, is known and has a homolog in the trichotecene gene cluster from Fusarium in the MiBiG database28, which is the database of known and characterized BGC.
PKS clusters in Calcarisporium sp
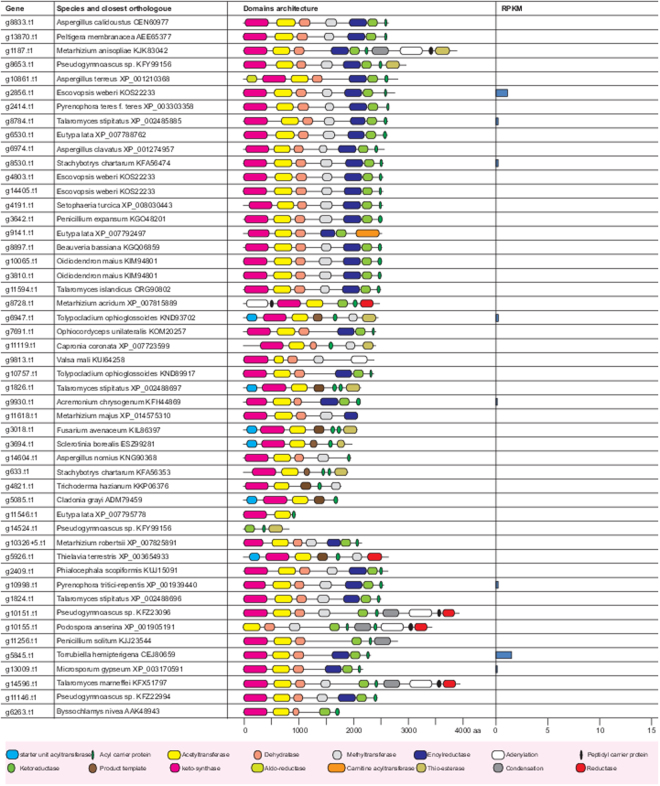
All the PKS genes and their expression based (on RNA-seq data) in standard growth media are shown in Fig. 2A comparison of 23 PKS gene clusters from Calcarisporium sp. with those from other fungi and bacteria is presented (Figure S3), while one cluster has no homologous clusters in any organisms. The PKS cluster mCaBGC21 (contig_169/39.7 kb) has 36% sequence identity with the previously uncharacterized BGC AM270194.1 c1 (localized on the contig An09c0050) from Aspergillus niger (Figure S3). The 43.8 kb long PKS cluster mCaBGC24 (contig178) shares 46% similarity with clusters NC015711.1 c13 from Myxococcus fulvus HW-1 and NC015953.1 c16 from Streptomyces sp. SirexAA-E (Figure S3). The PKS cluster mCaBGC25 (contig189/48.1 kb) has similarity to clusters in Streptomyces sp. and Kitasatospora setae KM-6054.
Figure 2.
Overview of PKS genes and their protein domains in genome of marine Calcariosporium genome. Only few genes are expressed in low quality with tiers 8–9.
The PKS cluster mCaBGC34 (contig_280/33.6 kb) has a counterpart in several fungal genomes such as Aspergillus, Metarhizium and Trichophyton (Figure S3). The cluster mCaBGC35 (contig282/48.7 kb) shares 45% of similarity with a bacterial cluster NC_012718.1_c1 (Burkholderia glumae BGR1) and it is also found in genomes of Cordyceps militaris and Glomerella graminicola (Figure S3). In a similar fashion, we found that mCaBGC40 (contig_358/28 kb) has homologous clusters in several strains of Streptomyces sp. (Figure S3) Likewise, mCaBGC41 (contig_367/32.7 kb) as similarities with clusters in Metarhizium strains, but at the same time, it has 30% similarity with a cluster from Nostoc punctiforme.
NRPS clusters in Calcarisporium
There are seventeen BGCs, representing NRPS clusters in the Calcarisporium genome (Fig. 3, Table S6 and Figure S3). Fifteen of these NRPS clusters have very low sequence identities (with any known clusters), while two NRPS clusters, mCaBGC44 (contig400/13.4 kb) and mCaBGC60 (contig1101/12.9 kb) have no homologs in the databases (Figure S3).
Figure 3.

Biosynthetic gene clusters encoding NRPS in Calcarisporium sp. KF525 genome.
Terpene clusters in Calcarisporium sp
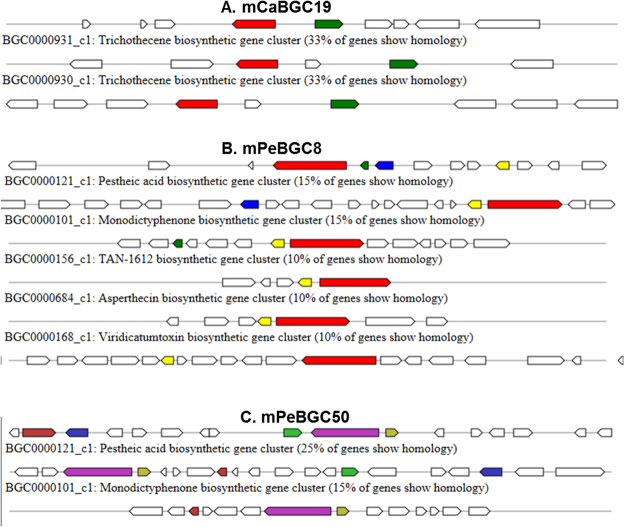
A trichotecene gene cluster (mCaBGC19, 21.2 kb) is localized on contig165 and it has homologs in several Fusarium strains (Fig. 4 and Figure S3). This cluster shares 33% of similarity with a trichothecene BGC from Fusarium graminearum (MIBiG BGC-ID - BGC0000931_c1, Table S8).
Figure 4.
Overview of known biosynthetic gene clusters in MiBiG database (A) mCaBGC19 from Calcarisporium sp. KF525 (B) mPeBGC8 from Pestalotiopsis sp. KF079 (C) mPeBGC50 from Pestalotiopsis sp. KF079.
Hybrid clusters in Calcarisporium sp
There are two heterocyst-type glycolipid ketosynthase (Hglks) clusters namely mCaBGC33 (contig274, size 27.4 kb) and mCaBGC46 (contig422, size 39.6 kb) in the Calcarisporium genome (Fig. 5). A cluster similar to mCaBGC33 is found in Saccharopolyspora spinosa (AY007564.1_c1) and Mycobacterium sp. MOTT36Y (NC_017904.1_c10), while the Hglks cluster mCaBGC46 has the top two hits in Streptomyces longisporoflavus (FJ462704.1_c1) and Amycolatopsis orientalis (DQ88475.1_c1, Figure S3).
Figure 5.
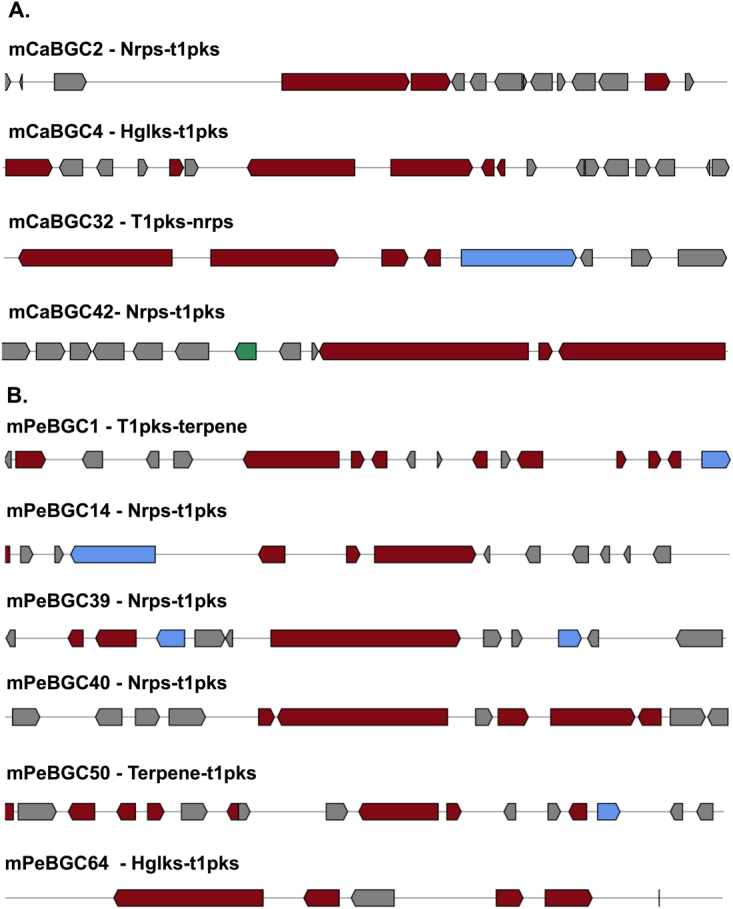
Genomic locations of hybrid biosynthetic gene clusters encoding genes in marine fungal genomes. (A) Calcarisporium sp. KF525 (B) Pestalotiopsis sp. KF079.
mCaBGC2 is a 52.2 kb long hybrid NRPS-t1PKS cluster (Fig. 5), which shares similarities with clusters in Metarhizium strains (GL698718.1_c1 and GL698473.1_c1), Mycobacterium thermoresistibils (NZ_AGVE01000050.1_c1) and Hydrogenophaga sp. PBC (NZ_AJWL01000058.1_c1). The hybrid Hglks-t1PKS cluster mCaBGC4 (size 54.8 kb) shares similarities with clusters in Stigmatella aurantiaca and Streptomyces viridochromogenes (Figure S3). Cluster mCaBGC32 is a 37.7 kb long hybrid NRPS-t1PKS cluster, which is similar to clusters in Vibrio mimicus and V. coralliilyticus. The 45.8 kb long cluster mCaBGC42 represents a hybrid NRPS-t1PKS with similarity to cluster CACQ02000519.1 c1 from Colletotrichum higginsianum (Figure S3).
Unknown types of BGCs in Calcarisporium sp
Unknown types of BGCs are putative BGCs, which were identified by using antiSMASH 3.029 under the category ‘others’29. We found seven unknown types of BGCs in the Calcarisporium sp. KF525 genome. One of them, mCaBGC1, is a 42.1 kb long cluster, sharing an overall sequence identity of 33% with AP007151.1 c1 from A. oryzae (RIB40) (Figure S3). The remaining unknown-type BGCs have no significant sequence identities with any known clusters in the databases (Figure S3).
Biosynthetic gene clusters in Pestalotiopsis sp
The 46 Mb assembled genome of Pestalotiopsis sp. KF079 contains 67 BGCs, which are named mPeBGC1 to mPeBGC67 (Table S9 and Figure S4). These clusters include 22 T1 PKS clusters, 12 nonribosomal peptide synthetase (NRPS) clusters, 9 terpene clusters and one each of type III PKS (T3pks) and lantipeptide clusters. Additionally, Pestalotiopsis KF079 has six clusters of hybrid nature such as three Nrps-t1pks and one each of Hglks-t1pks, T1pks-terpene and terpene-t1pks. On top of that sixteen clusters were detected with unknown types of biosynthetic genes (Table S9). Of all these BGCs only two are represented in the MiBiG database28 namely, mPeBGC8 and mPeBGC50. The remaining 65 BGCs are unique to Pestalotiopsis. sp., as there are no homologous clusters known or else similarity is too low. Hence, 65 out of 67 BGCs of Pestalotiopsis sp are previously uncharacterized, hence not found in MiBiG database28, a database of known BGCs.
PKS clusters in Pestalotiopsis sp
The single T3pks cluster found in the genome of Pestalotiopsis sp., mPeBGC2, is localized on the scaffold00001 within a fragment of 41.4 kb, and this cluster has similarities with clusters in Cordyceps militaris and Metarhizium anisopliae (Figure S4). There are further 22 BGCs encoding PKS gene products in the Pestalotiopsis sp. genome. Cluster mPeBGC4 is localized in the scaffold00002, spanning an about 45.8 kb fragment without significant homologies with any other known cluster (Figure S4). Spanning about 45.6 kb on the scaffold00007, cluster mPeBGC8 is known in several ascomycetes and also in different Streptomyces strains. The cluster mPeBGC8 has no homolog, however it has very limited similarities with four known clusters (Fig. 4), namely pestheic acid biosynthetic gene cluster (BGC0000121_c1, 15% identities), monodictyphenone biosynthetic gene cluster (BGC0000101_c1, 15% identities), asperthecin biosynthetic gene cluster (BGC0000684_c1, 10% identities), TAN-1612 biosynthetic gene cluster (BGC0000156_c1, 10% identities) and viridicatumtoxin biosynthetic gene cluster (BGC0000168_c1, 10% identities). However, due to the low similarities of 10–15%, these similarities may be random. Remaining PKS encoding BGC have no significant identities with BGCs in any other species (Figure S4).
NRPS clusters in Pestalotiopsis sp
There are total 11 BGCs (Fig. 6) capable of producing NRPS based compounds. The NRPS cluster mPeBGC18 (scaffold000016/46.3 kb) shares 53% homology with a bacterial cluster NC_010571.1_c1 from Opitutus terrae PB90–1 (Figure S4) and it is also related to lasalocid BGC (Streptomyces lasaliensis). Additionally, this cluster is also possessed by Metarhizium acridum. While mPeBGC49 (scaffold000071/46.5 kb) has top hits in Aspergillus and Trichophyton stains with 36% homology for the cluster AP007154.1_c2 from A. oryzae (Figure S4). All other NRPS clusters have no significant hits to known clusters in other organisms (Figure S4).
Figure 6.
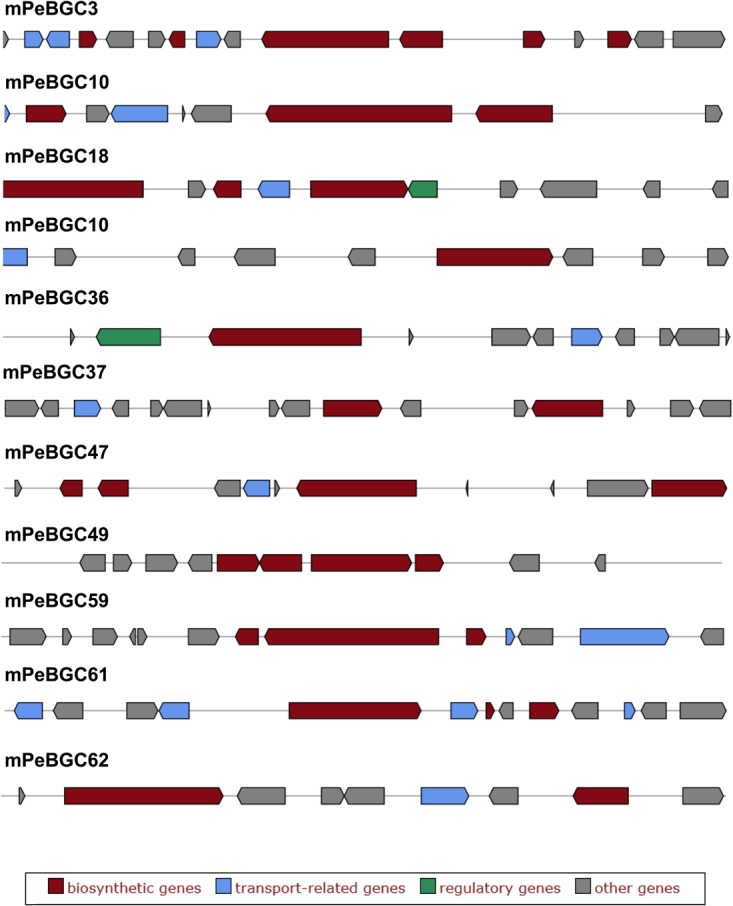
Biosynthetic gene clusters encoding NRPS genes in marine Pestalotiopsis genome.
Terpene clusters in Pestalotiopsis sp
There are nine terpene clusters in Pestalotiopsis genome and the first cluster mPeBGC9 is 22 kb long on the scaffold00007 (Figure S4). This terpene cluster shows roots with clusters from bacteria to fungi and is in agreement with another terpene cluster mPeBGC35 (scaffold000042/22.1 kb). The second terpene cluster mPeBGC17 is 22.1 kb long on the scaffold000016 and shares similarities with clusters in several ascomycetes, which is also the case for five other terpene clusters namely mPeBGC29 (scaffold000031/23 kb), mPeBGC31 (scaffold000036/22.6 kb), mPeBGC35 (scaffold000042/22.1 kb), mPeBGC43 (scaffold000057/23 kb) and mPeBGC63 (scaffold000156/18.3 kb). Interestingly, the mPeBGC63 cluster show similarities with trichothecene BGCs from several Fusarium strains. Two terpene clusters mPeBGC41 (scaffold000052/21.4 kb) and mPeBGC66 (scaffold000173/13.4 kb) have no significant hits in public databases (Figure S4).
Hybrid clusters in Pestalotiopsis sp
There are three Nrps-t1pks hybrid clusters in the Pestalotiopsis sp. genome (Fig. 5B) namely mPeBGC14 (scaffold000013/57.1 kb), mPeBGC39 (scaffold000049/54.2 kb), and mPeBGC40 (scaffold000051/52.5 kb). The remaining three hybrid clusters in the Pestalotiopsis genome (Fig. 5B) are of the types T1pks-terpene, Terpene-t1pks and Hglks-t1pks respectively, and are called as mPeBGC1 (scaffold0000/60.4 kb), mPeBGC50 (scaffold000072/50.4 kb) and mPeBGC64 (scaffold000158/30.7 kb), respectively. None of these hybrid clusters have significant hits, even with uncharacterized clusters in other organisms (Figure S4).
Unknown-type clusters in Pestalotiopsis sp
Unknown types of BGCs are putative BGCs identified by using antiSMASH 3.029 and put in the category of others29. We found sixteen unknown-type BGCs in Pestatiopsis genome (Figure S4), without significant cluster homology with other micro-organisms
Core PKS and NRPS proteins in the two marine fungi In the Calcarisporium KF525 genome, we identified 50 putative PKSs, including four PKS-NRPS hybrids, one PKS with an N terminal adenylation and PCP domain, six non-reducing PKSs and thirty-seven reducing PKSs (Fig. 2). The non-reducing PKS g3018 was found to be a possible orthologue of the proposed bikaverin synthase in Fusarium avenaceum (FAVG1_10226)30. Analyses of the remaining PKSs did not result in identification of orthologues with known products in other species.
Several bioactive compounds are identified from Calcarisporium sp
KF52531,32 and the most fascinating one is calcaride A, which has antibacterial properties. Calcaride A belongs to the group of macrocyclic polyesters, for which the biosynthetic routes are yet to be identified. However based on aromatic ring formation, calcaride A can be produced by two possible genes namely NR-PKS (g1826) and the HR-PKS (g1824). Nevertheless, these two genes are not expressed under laboratory conditions (Fig. 2).
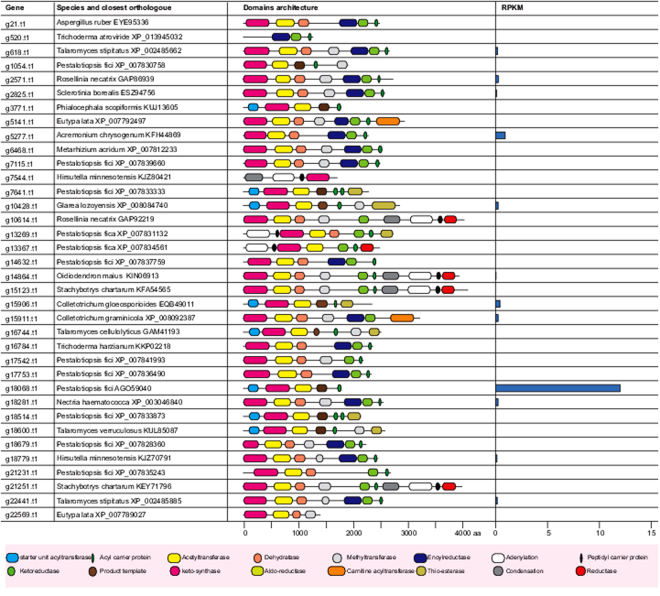
In Pestalotiopsis sp., putative 36 PKS were identified including four PKS-NRPS hybrides, two PKSs with N terminal adenylation and PCP domains, eight non-reducing PKSs and twenty-three reducing PKSs (Fig. 7). Twelve PKSs have an orthologue in the previously sequenced P. fici33 including an ortholog of the P. fici pestheic acid synthase (PtaA34). The non-reducing PKS g18514 shares 81% homology to the melanin polyketide synthase from Nodulisporium sp. ATCC74245 (AAD38786.1) and an orthologue in Pestalotiopsis microspore has previously been shown to be required for conidial pigmentation35. Another likely PKS for pigmentation was also identified in g7641, which shares 60–62% identity with fusarubin synthase (PKS3) from members of the Fusarium genus36. The non-reducing PKS g18600 shares 42% identity to AN6448 from Aspergillus nidulans, which produce 3-methylorsellinic acid37 and it seems therefore likely that Pestalotiopsis sp. can produce a similar compound.
Figure 7.
Summary of PKS genes and their protein domains in genome of marine Pestalotiopsis genome. Only few genes are expressed in low quality with tiers 8–9 with exception of g18068, which is in the tier 7.
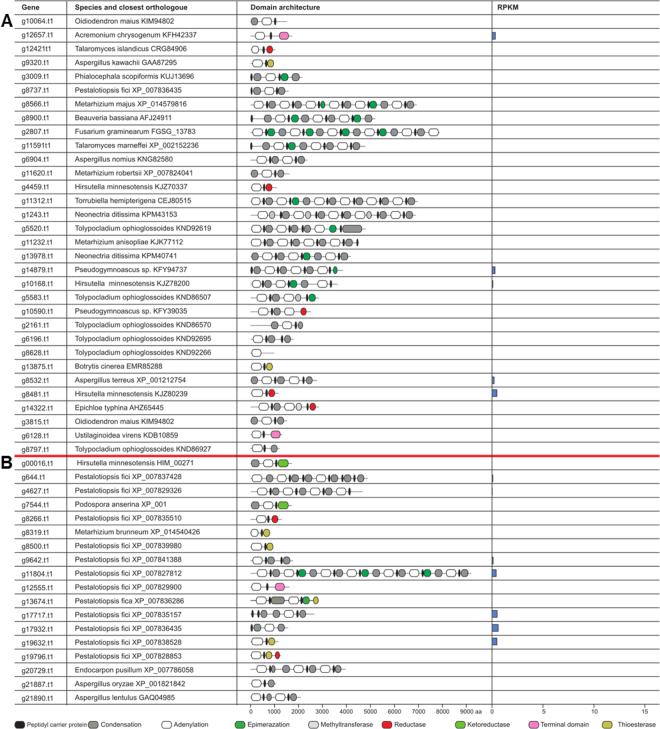
The genomes of both strains contain less NRPSs than PKSs, as we identified 32 and 18 putative NRPSs in Calcarisporium sp. and in Pestalotiopsis sp. (Fig. 8), respectively. We identified a putative iron-chelating siderophore synthetase of the ferricrocin type38 in both species - Calcarisporium sp. (g5520) and Pestalotiopsis sp. (g644). An additional siderophore synthetase is present in Pestalotiopsis sp. (g9642), as an orthologue of the SidE synthetase, which is responsible for fusarinine biosynthesis.
Figure 8.
Overview of NRPS genes and their protein domains present in two marine fungal strains. (A) Calcarisporium sp. KF525 (B) Pestalotiopsis sp. KF079.
An orthologue of the peramine synthase from Epichloe typhina was identified in Calcarisporium sp. (g14322). Peramine is a potent insect feeding deterrent synthesized by a two-module NRPS in Epichloë and has been suggested to be produced in a mutualistic interaction with its host plant perennial ryegrass in order to confer protection to against insect herbivory39.
Calcarisporium sp. has a MAT1–2 locus while Pestalotiopsis sp. has no detectable mating locus
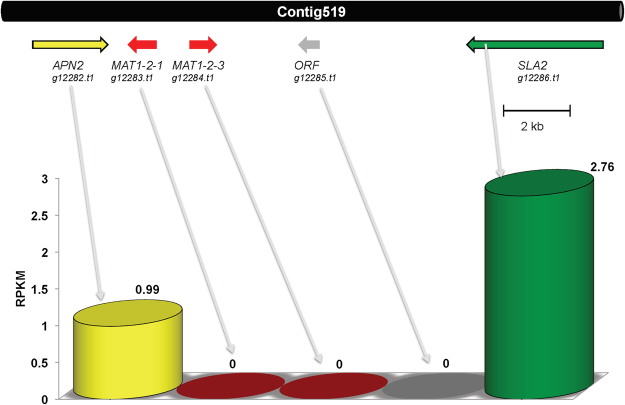
The genomic fragments that carry the mating type genes are usually designated as the mating-type (MAT) locus, which regulates sexual reproduction in different fungi40. In self-sterile (heterothallic) species, mating occurs between morphologically identical partners that are only distinguished by their MAT locus. In filamentous ascomycetes, the MAT locus consists of two different DNA segments in the mating partners termed the MAT1–1 and MAT1–2 idiomorphs41. In contrast to heterothallic species, the genome of self-fertile (homothallic) filamentous ascomycetes contains genes indicative of both mating types that can be either linked or unlinked42,43. During genome annotation process, detection of these loci and flanking gene identification is a standard process, which aids in determination of the mode of sexual reproduction in different fungi. So far little is known about the sexual life style of marine fungi. To unravel sexual lifes of these fungi, we have performed specialized homology detections of MAT genes and corresponding loci (explained in details in Suppl. section S1). A tBLASTN search with the MAT1–2–1 HMG domain mating type protein of S. macrospora and N. crassa revealed the presence of a MAT1–2–1 homolog (g12883.t1) in the genome sequence of Calcarisporium sp. The gene encodes a protein of 240 amino acids with a conserved HMG domain. Homology based extraction and explanation of these genes are provided in supplementary section 1 and in Figures S5 and S6. The APN2 gene encoding a putative DNA lyase and the SLA2 gene encoding a cytoskeleton assembly control factor have been reported neighboring MAT loci in many other ascomycetes44. Both genes, APN2 (g12882.t1) and SLA2 (g12886.t1) can be identified on the same contig_519 as the putative MAT1–2–1 gene (Fig. 9). Two further open reading frames are adjacent to MAT1–2–1 and flanked by APN2 and SLA2, g12884.t1 and g12885.t1 on contig519 (Fig. 9), which encode proteins of 242 and 133 amino acids, respectively. Transcriptomic data revealed that both mating types genes are not expressed in wild type condition, while flanking genes - APN2 (g12882.t1) and SLA2 (g12886.t1) were expressed (Fig. 9), in at low level within tiers 9 and 8 (Table 2), respectively.
Figure 9.
Overview of the mating-type locus MAT1–2 of Calcarisporium sp. KF525 on the contig519 with comparison with transcriptomic data. The positioning and transcriptional direction of the mating-type genes (red) is indicated by an arrow flanking genes APN2 and SLA2 are shown in yellow and green, respectively. A predicted ORF is indicated in grey.
Taken together, the genome data showed that the sequenced isolate Calcarisporium sp. has a MAT1–2 mating type. MAT1–1 sequences could not be identified suggesting that Calcarisporium sp. is heterothallic, providing that Calcarisporium spec is able to reproduce sexually. A novel MAT1–2 specific ORF could be identified which is related to the MAT-1–2–3 protein encoded in the MAT1–2 loci members of the Hypocreales45. We performed similar homology-based detection of mating type genes in Pestalotiopsis sp., which yielded no results, suggesting that the isolated Pestalotiopsis strain has no mating type genes. We have recently reported that another marine fungus S. brevicaulis LF580 was a MAT1–1 strain15. These finding corroborates that reproduction in marine environments can be varied nature like either vegetative and sexually or both.
Summary of carbohydrate active enzyme-encoding genes identified in the two novel marine fungal genomes
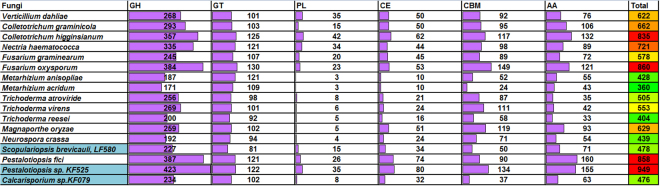
Little is known about the lifestyle and ecological importance of marine fungi. Many water environments are low in nutrients and therefore unlikely to favor organisms that feed primarily by attachment to larger physical substrates and osmotrophy2. Fixed carbon on land is largely invested in the construction of large and complex plant tissues rich in energy and nutrients. Digestion of these plant tissues requires a complex set of carbohydrate active enzymes. Calcarisporium and Pestalotiopsis strains used in this study were collected in the German Wadden Sea, which is an area of about 3475 square miles with one of the highest biological primary productions in the world. As such the analysis of carbohydrate active enzymes is highly relevant. Using annotation tools derived from the CAZy database (http://www.cazy.org/), we identified 949 and 476 CAZy genes in Pestalotiopsis sp. KF079 and Calcarisporium sp. KF525 genomes (Fig. 10 and Table S10), respectively, confirming that Pestalotiopsis species are very rich in CAZymes and that Calcarisporium sp. is rather close in number to the marine derived- fungus Scopulariopsis brevicaulis LF58046. For Pestalotiopsis sp., the proteins encoded by the corresponding genome are divided into six major classes, namely 423 glycoside hydrolases (GH), 122 glycosyltransferases (GT), 35 polysaccharide lyases (PL), 80 carbohydrate esterases (CE), 134 carbohydrate binding module (CBM) and 155 auxiliary activities (AA). In comparison, CAZymes candidates of Calcarisporium sp. are radically lower in number for each class except for GT. Comparing the various genomes from mainly plant pathogens, entomopathogens and other model fungi, we can observe that the number of GTs, enzymes involved in the biosynthesis of oligo- and polysaccharides, is rather stable across genomes while the classes involved in the degradation processes (GH, PL, CE, CBM and AA) are highly variable and depend on the lifestyle of the fungus. The total number of CAZymes is generally high for plant pathogens and saprophytes, with the exception of Trichoderma reesei, known to produce a very efficient enzyme cocktail but poorly diversified. Inversely, the entomopathogenic fungi, Metarhizium anisopliae and Metarhizium acridum, are rather poor in all these enzyme classes. The two Pestalotiopsis species (marine strain and P. fici) examined belong to the fungal group harboring the highest CAZyme number along with the plant pathogens, Colletotrichum higginsianum and Fusarium oxysporum. Calcarisporium sp., together with S. brevicaulis are in an intermediate position as they have slightly lower numbers of GH, CE, CBM or AA members compared to all pathogic fungi and a very low content of PLs as for fungi possessing a specific life style, like Trichoderma, or N. crassa. Further details of Cazyomes of these two fungi are provided in supplementary section S2.
Figure 10.
Summary of class-wise distribution of carbohydrate active enzymes in selected fungi. AA - auxiliary activities, GH - glycoside hydrolases, GT - glycosyltransferases, PL - polysaccharide lyases, CE - carbohydrate esterases and CBM - carbohydrate-binding modules.
Overview of MFS-type and sugar transporters encoded in the two novel genomes
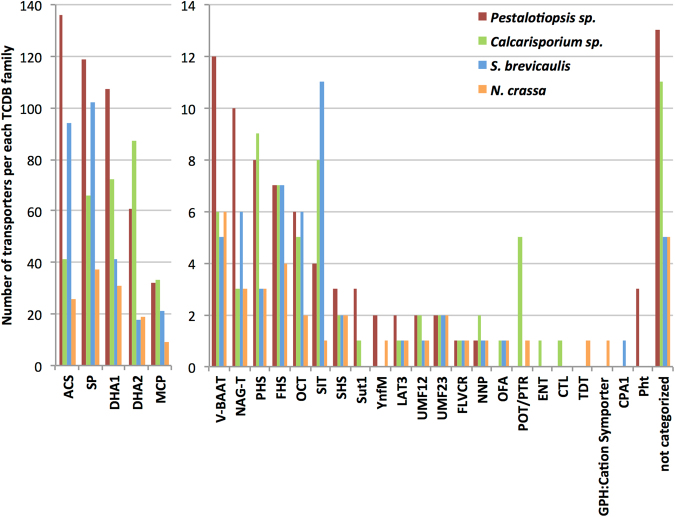
The genome sequence analysis of both Calcarisporium sp. and Pestalotiopsis sp. revealed that Pestalotiopsis sp. has the highest number of genes (534) encoding transport proteins predicted to have either a major facilitator superfamily (MFS) domain or a sugar transporter domain (Table S11). In Calcarisporium sp. on the other hand, the corresponding gene number (367) is rather close to the marine-derived S. brevicaulis strain LF580 (328 genes15). Comparing the distribution of the transporters into the categories defined by the Transporter Classification Database (TCDB; [2]), an overall similar distribution of the transporters of Pestalotiopsis sp., S. brevicaulis and N. crassa could be observed, while the distribution of Calcarisporium sp. transporters is slightly different (Fig. 11). We have provided complete details of transporter genes in the supplementary section S3.
Figure 11.
Comparative distribution of MFS-type and sugar transporter genes from Pestalotiopsis sp., Calcarisporium sp., S. brevicaulis and N. crassa into TCDB categories. The TCDB categories were ordered in descending fashion according to the number of Pestalotiopsis transporter genes present. The number of genes per category is presented as raw number for each organism. SP Family (TCDB category: 2.A.1.1); OFA Family (2.A.1.11); SHS Family (2.A.1.12); MCP Family (2.A.1.13); ACS Family (2.A.1.14); SIT Family (2.A.1.16); OCT Family (2.A.1.19); DHA1 Family (2.A.1.2); FLVCR Family (2.A.1.28); DHA2 Family (2.A.1.3); YnfM Family (2.A.1.36); LAT3 Family (2.A.1.44); V-BAAT Family (2.A.1.48); NAG-T Family (2.A.1.58); UMF12 Family (2.A.1.63); FHS Family (2.A.1.7); UMF23 Family (2.A.1.75); NNP Family (2.A.1.8); PHS Family (2.A.1.9); TDT Family (2.A.16); POT/PTR Family (2.A.17); GPH:Cation Symporter Family (2.A.2); CPA1 Family (2.A.36); Sut1 (2.A.2.6); ENT Family (2.A.57.5); CTL Family (2.A.92.1); Pht Family (2.A.1.53).
Summary of hydrophobins
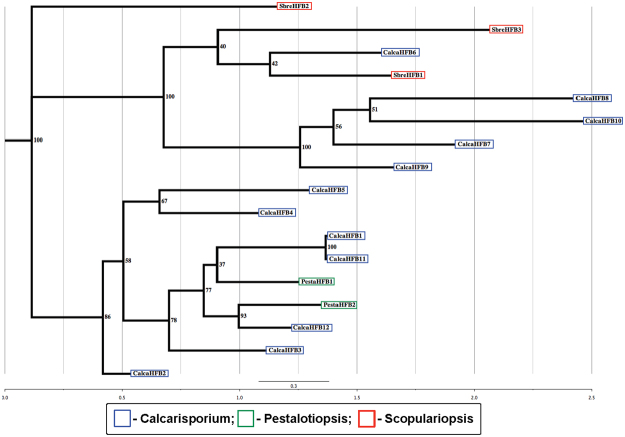
Hydrophobins are morphogenetic, small mass (20 kDa) secreted hydrophobic fungi-specific cell wall proteins and they assist in the construction of aerial structures (like spores or fruiting bodies) in fungi [43]. The recently sequenced marine fungus S. brevicaulis LF580 possesses three hydrophobin genes, named as SbreHPB1 (g5510.t1), SbreHPB2 (g7216.t1), and SbreHPB3 (g15602.t1)15. Calcarisporium sp. and Pestalotiopsis sp. genomes were found to possess 12 and 2 hydrophobins, respectively (Fig. 12 and Table S12). Variable number of hydrophobins were also found in previous analyses of marine fungi4, and may represent adaptions to salt stress.
Figure 12.
Phylogenetic analyses of hydrophins depict that there are different numbers of hydrophins are present in these marine fungi like 12, 2 and 3 in Calcarisporium sp. KF525, Pestalotiopsis sp. KF079 and S. brevicaulis KF580, respectively. Single copy of cryparin was found in two newly sequenced marine fungal genomes of Calcarisporium sp. KF525, and Pestalotiopsis sp. KF079.
Discussion
The oceans harbour enormous biochemical diversity in terms of natural products from various organisms. Marine fungi are potent producers of natural products however they have been under the least scientific focus. To bridge this gap, we used two marine fungal strains from the German Wadden Sea (Calcarisporium sp. KF525 and Pestalotiopsis sp. KF079) and performed genome wide scanning for the secondary metabolite genes and gene cluster (BGCs). We presented here genome sequences of two marine fungi namely Calcarisporium sp. KF525 and Pestalotiopsis sp. KF079 and the genome sizes of these two fungi are 35 Mb and ~46 Mb, respectively. Both genomes have a low content of total repeats (1%). Three other known marine fungal genomes, namely S. brevicaulis LF58015, C. malorum Mo1221 and Cryptococcus sp. Mo2921 have comparable repeat contents, which are close to 1%. This resides within the typical 1–4% of transposable elements in fungal genomes20. Exceptionally, only a few fungi have higher repeat contents like in a pezizomycete, Tuber melanosporum genome47 and several fungal genome of dothiodeomycetes48. However, these fungi typically have large expansions of the genome size like 125 Mb for T. melanosporum47.
Biosynthetic genes involved in the biosynthesis of a particular metabolite are organized in clusters and are often co-regulated and these clusters are known as biosynthetic gene clusters (BGCs)19,49. Exceptionally, more than one BGCs are involved in production of secondary metabolites like biosynthersis of meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans19. These clusters have one or two core genes and several accessary genes, which encode proteins required for biosynthesis of the metabolite. Often filamentous fungi have 25 BCGs but this number can be much higher (80–90)50. The two marine fungi analysed here possess a total of 127 BGCs. Most of them have no significant homologies to BGCs in other fungi as well as in the bacteria. Notably, only three of these gene clusters are well characterized BGCs (Fig. 4) and reported in MiBiG datasets28. Our genome analyses thus revealed a huge amount of novel BGCs, highlighting that marine fungi are an exceptional source for secondary metabolites. Future studies should be aimed at identifying conditions in which the BGCs are active or methods to activate them artificially so that their properties can be analysed in more detail. In the draft fungal genomes, genomic fragments are smaller and hence in a few cases are noted that possesses a single gene in the cluster like mCaBGC60 (Figure S3). The cluster mCaBGC60 is localized on the contig1101, which is only 11.9 kb in size. Hence, drawing a conclusion from this cluster will be difficult, yet kept as found, and further genome assembly improvements and/or resequencing can resolve issues of such clusters.
Pestalotiopsis and Calcarisporium species belong to the Xylariales and Hypocreales, respectively, and are distributed in tropical and temperate regions (for a review on Pestalotiopsis see51). Pestalotiopsis species typically are phytopathogens causing different diseases i.e. needle or tip blights, canker lesions or fruit rots. Pestalotiopsis sp. are also found as endophytes or saprophytes, as they were isolated from dead leaves, barks and twigs. In addition, some species were found in animal infections demonstrating that these fungi could have versatile nutritional habits, or are opportunistic. In general, Pestalotiopsis sp. appears to be host-specific and live in a wide range of substrates or associated with different hosts. Calcarisporium sp. have a widespread occurrence and they are generally isolated as endophytes or parasites from basidiomycetes52, or ascomycetes, or from wood53, but have occasionally been isolated from marine environments32. In a recent study of a mangrove fungus, Pestalotiopsis sp. was described along with its adaptation to sea salt54. By a proteomic analysis, it was demonstrated that the lignocellulolytic enzyme composition was considerably changed in salt conditions, with for instance, a great reduction of the oxidase abundance, while specific carbohydrate-active enzymes are secreted exclusively at high salt concentrations. Taking into account all these data related to the CAZyome and sugar transporter repertoires (Tables S10–S11), it can be suggested that Pestalotiopsis sp. KF079, similar to the marine-derived S. brevicaulis, possesses a metabolism focused on the breakdown of the terrestrial plant biomass rather than algal or animal biomass. On the other hand, Calcarisporium sp. has adopted a different (and reduced) CAZy repertoire specialized for algae and animal degradation that reflects a life style oriented either towards parasitism or endophytic growth or towards utilization of dead algae or animals.
Upon scanning MAT loci in these two marine fungi, we demonstrated that Calcarisporium sp. has a MAT1-2 mating type while no MAT locus was detected in Pestalotiopsis sp.
Altogether, we present two draft genomes of marine fungi with a high number of BGC most of them being new clusters not observed before. These computational predictions of the genomic data provide further insight into genetics of these two fungi. This study step platform for the designing experiments to confirm findings about BGCs and corresponding secondary metabolite biosynthesis. We provide a comprehensive annotation of these genomes.
Methods
Fungal strain collection, cultivation, and DNA isolation
These two marine fungal strains (Pestalotiopsis sp. KF079 and Calcarisporium sp. KF525) were isolated from the German Wadden Sea, which is the southeastern part of the North Sea. These strains were cultivated as previously described55. DNA isolation was per formed as described in the supplementary section S4.
Genome sequencing, assembly, genome and RNA-Seq analyses
Short-read DNA sequencings were performed using Roche 454 and Illumina HiSeq™ 2000 methods with starting samples of 20 µg genomic DNA for these two marine fungi at Macrogen (Korea). We constructed hybrid de novo genome assemblies of Roche 454 and Illumina HiSeq™ 2000 for Pestalotiopsis sp. KF079 and Calcarisporium sp. KF525 using the Newbler assembler56 and the CLCBio Genomic workbench57, respectively. Further details of genomics and RNA-Seq analyses are provided in the supplementary section S4.
Unraveling and characterization of biosynthetic gene clusters
Initially, putative genes that encoding for proteins which produce bioactive compounds are identified using BLAST58 with an E-value < 1e−3. Subsequently, this genome was analyzed using antiSMASH 3.029 for putative clusters and further examined by manually coupled with RNA-Seq data. The functional domains of PKSs and NRPSs were identified as previously described59, using a combinations of tools namely antiSMASH 3.029, NCBI Conserved Domain Database60, InterPro24 and the PKS/NRPS Analysis Web-site61.
Data access
Entire datasets used in the current work were publically available using BioSample accession IDs: SAMN06272793 and SAMN06272794 with corresponding BioProject accession IDs as PRJNA368776 and PRJNA368777 for Pestalotiopsis sp. KF079 and Calcarisporium sp. KF525, respectively.
Electronic supplementary material
Acknowledgements
We thank Hanna Schmidt for DNA and RNA isolation. This work was part of the EU consortium “Marine Fungi” funded in the European Union Seventh Framework Program (FP7/2007–2013 under grant agreement number 265926). We are grateful to Johannes Imhoff and Antje Labes (GEOMAR, Kiel) for providing strains of Calcarisporium and Pestalotiopsis. BH was funded by Agence Nationale de la Recherche (grant no. ANR-14-CE06-0020).
Author Contributions
Conceived and designed the experiments: A.K. and F.K. Performed the experiments: A.K., B.H., M.F.S., M.A., S.P. Analyzed the data: A.K., B.H., E.R., L.H., J.P.B., J.L.S., F.T.H., M.A., S.P., F.K. Contributed to the writing of the manuscript: A.K., B.H., E.R., J.P.B., J.L.S., S.P., F.K.
Competing Interests
The authors have declared that no competing interests exist. Concerning the role of VTT Technical Research Centre of Finland Ltd we emphasize that VTT as a whole is a not-for-profit organisation (http://www.vttresearch.com/about-us/co-operation-with-vtt/mission-and-mode-of-operations for further information). Biocomputing Platforms Ltd is the current employer of MFS and had no involvement in the study. During the time of the study MFS was employed by VTT Technical Research Centre of Finland Ltd.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-28473-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kohlmeyer, J. & Kohlmeyer, E. Marine Mycology: The higher fungi. 690 (Academic Press, New York, 1979).
- 2.Richards TA, Jones MD, Leonard G, Bass D. Marine fungi: their ecology and molecular diversity. Ann Rev Mar Sci. 2012;4:495–522. doi: 10.1146/annurev-marine-120710-100802. [DOI] [PubMed] [Google Scholar]
- 3.Landy ET, Jones GM. What is the fungal diversity of marine ecosystems in Europe? Mycologist. 2006;20:15–21. doi: 10.1016/j.mycol.2005.11.010. [DOI] [Google Scholar]
- 4.Kis-Papo T, et al. Genomic adaptations of the halophilic Dead Sea filamentous fungus Eurotium rubrum. Nature communications. 2014;5:3745. doi: 10.1038/ncomms4745. [DOI] [PubMed] [Google Scholar]
- 5.Jones EBG. Are there more marine fungi to be described? Botanica Marina. 2011;54:343–354. doi: 10.1515/bot.2011.043. [DOI] [Google Scholar]
- 6.Desjardins CA, et al. Comparative genomic analysis of human fungal pathogens causing paracoccidioidomycosis. PLoS genetics. 2011;7:e1002345–e1002345. doi: 10.1371/journal.pgen.1002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones, E. B. G. Marine fungi: some factors influencing biodiversity. Fungal Biodiversity (2000).
- 8.Konig GM, Kehraus S, Seibert SF, Abdel-Lateff A, Muller D. Natural products from marine organisms and their associated microbes. Chembiochem: a European journal of chemical biology. 2006;7:229–238. doi: 10.1002/cbic.200500087. [DOI] [PubMed] [Google Scholar]
- 9.Ebada, S. S. & Proksch, P. In Marine Pharmacognosy:Trends and Applications (ed S.K. Kim) 27-51 (CRC Press Taylor & Francis Group, LLC, Boca Raton, 2013).
- 10.Saleem M, et al. Marine natural products of fungal origin. Nat Prod Rep. 2007;24:1142–1152. doi: 10.1039/b607254m. [DOI] [PubMed] [Google Scholar]
- 11.Bugni TS, Ireland CM. Marine-derived fungi: a chemically and biologically diverse group of microorganisms. Nat Prod Rep. 2004;21:143–163. doi: 10.1039/b301926h. [DOI] [PubMed] [Google Scholar]
- 12.Rateb ME, Ebel R. Secondary metabolites of fungi from marine habitats. Nat Prod Rep. 2011;28:290–344. doi: 10.1039/c0np00061b. [DOI] [PubMed] [Google Scholar]
- 13.Debbab A, Aly AH, Lin WH, Proksch P. Bioactive compounds from marine bacteria and fungi. Microb Biotechnol. 2010;3:544–563. doi: 10.1111/j.1751-7915.2010.00179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montaser R, Luesch H. Marine natural products: a new wave of drugs? Future Med Chem. 2011;3:1475–1489. doi: 10.4155/fmc.11.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar A, et al. De Novo Assembly and Genome Analyses of the Marine-Derived Scopulariopsis brevicaulis Strain LF580 Unravels Life-Style Traits and Anticancerous Scopularide Biosynthetic Gene Cluster. PLoS One. 2015;10:e0140398. doi: 10.1371/journal.pone.0140398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seyedsayamdost MR, Clardy J. Natural Products and Synthetic Biology. ACS Synthetic Biology. 2014;3:745–747. doi: 10.1021/sb400025p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amoutzias, G. D., Chaliotis, A. & Mossialos, D. Discovery Strategies of Bioactive Compounds Synthesized by Nonribosomal Peptide Synthetases and Type-I Polyketide Synthases Derived from Marine Microbiomes. Marine drugs14, 10.3390/md14040080 (2016). [DOI] [PMC free article] [PubMed]
- 18.Charlop-Powers Z, et al. Global biogeographic sampling of bacterial secondary metabolism. Elife. 2015;4:e05048. doi: 10.7554/eLife.05048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brakhage AA. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 20.Linda, P. & Kempken, F. Fungal Transposable Elements. Vol. 2 (Springer International Publishing Switzerland, 2015).
- 21.Rédou V, et al. Draft Genome Sequence of the Deep-Sea Ascomycetous Filamentous Fungus Cadophora malorum Mo12 from the Mid-Atlantic Ridge. Genome Announc. 2016;4:1–2. doi: 10.1128/genomeA.00467-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Z, Hao B. CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic acids research. 2009;37:W174–W178. doi: 10.1093/nar/gkp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn RD, et al. Pfam: the protein families database. Nucleic acids research. 2014;42:D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic acids research. 2012;40:D306–312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poggeler S, Kuck U. A WD40 repeat protein regulates fungal cell differentiation and can be replaced functionally by the mammalian homologue striatin. Eukaryotic cell. 2004;3:232–240. doi: 10.1128/EC.3.1.232-240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Mahajan A, Tsai MD. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 27.Allan RK, Ratajczak T. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones. 2011;16:353–367. doi: 10.1007/s12192-010-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medema MH, et al. Minimum Information about a Biosynthetic Gene cluster. Nature chemical biology. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber T, et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic acids research. 2015;43:W237–243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown DW, Proctor RH. Insights into natural products biosynthesis from analysis of 490 polyketide synthases from Fusarium. Fungal genetics and biology: FG & B. 2016;89:37–51. doi: 10.1016/j.fgb.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 31.Silber J, Ohlendorf B, Labes A, Nather C, Imhoff JF. Calcaripeptides A-C, cyclodepsipeptides from a Calcarisporium strain. Journal of natural products. 2013;76:1461–1467. doi: 10.1021/np400262t. [DOI] [PubMed] [Google Scholar]
- 32.Silber J, Ohlendorf B, Labes A, Erhard A, Imhoff JF. Calcarides A-E, antibacterial macrocyclic and linear polyesters from a Calcarisporium strain. Marine drugs. 2013;11:3309–3323. doi: 10.3390/md11093309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, et al. Genomic and transcriptomic analysis of the endophytic fungus Pestalotiopsis fici reveals its lifestyle and high potential for synthesis of natural products. BMC Genomics. 2015;16:28. doi: 10.1186/s12864-014-1190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu X, et al. Identification of the first diphenyl ether gene cluster for pestheic acid biosynthesis in plant endophyte Pestalotiopsis fici. Chembiochem: a European journal of chemical biology. 2014;15:284–292. doi: 10.1002/cbic.201300626. [DOI] [PubMed] [Google Scholar]
- 35.Gu Y, et al. Greater taxol yield of fungus Pestalotiopsis hainanensis from dermatitic scurf of the giant panda (Ailuropoda melanoleuca) Appl Biochem Biotechnol. 2015;175:155–165. doi: 10.1007/s12010-014-1254-y. [DOI] [PubMed] [Google Scholar]
- 36.Studt L, Wiemann P, Kleigrewe K, Humpf HU, Tudzynski B. Biosynthesis of fusarubins accounts for pigmentation of Fusarium fujikuroi perithecia. Applied and environmental microbiology. 2012;78:4468–4480. doi: 10.1128/AEM.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahuja M, et al. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J Am Chem Soc. 2012;134:8212–8221. doi: 10.1021/ja3016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sørensen, J. L. et al. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites (eds Juan-Francisco Martín, Carlos García-Estrada, & Susanne Zeilinger) 317–339 (Springer New York, 2014).
- 39.Tanaka A, Tapper BA, Popay A, Parker EJ, Scott B. A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Mol Microbiol. 2005;57:1036–1050. doi: 10.1111/j.1365-2958.2005.04747.x. [DOI] [PubMed] [Google Scholar]
- 40.Fraser JA, Heitman J. Evolution of fungal sex chromosomes. Mol Microbiol. 2004;51:299–306. doi: 10.1046/j.1365-2958.2003.03874.x. [DOI] [PubMed] [Google Scholar]
- 41.Turgeon BG, Yoder OC. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal genetics and biology: FG & B. 2000;31:1–5. doi: 10.1006/fgbi.2000.1227. [DOI] [PubMed] [Google Scholar]
- 42.Poggeler S. Mating-type genes for classical strain improvements of ascomycetes. Appl Microbiol Biotechnol. 2001;56:589–601. doi: 10.1007/s002530100721. [DOI] [PubMed] [Google Scholar]
- 43.Debuchy, R. & Turgeon, B. G. In Growth, Differentiation and Sexuality (eds Ursula Kües & Reinhard Fischer) 293–323 (Springer Berlin Heidelberg, 2006).
- 44.Debuchy, R., Berteaux-Lecellier, V. & Silar, P. In Cellular and Molecular Biology of Filamentous Fungi. (eds K. Borkovich & D. Ebbole) 501–535 (ASM Press, 2010).
- 45.Martin SH, Wingfield BD, Wingfield MJ, Steenkamp ET. Structure and evolution of the Fusarium mating type locus: new insights from the Gibberellafujikuroi complex. Fungal genetics and biology: FG & B. 2011;48:731–740. doi: 10.1016/j.fgb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 46.Kumar, A. et al. de novo assembly and genome analyses of the marine sponge derived Scopulariopsis brevicaulis strain LF580 unravels life-style traits and anticancerous scopularide producing hybrid NRPS-PKS cluster. PLoS one (accepted) (2015). [DOI] [PMC free article] [PubMed]
- 47.Martin F, et al. Perigord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature. 2010;464:1033–1038. doi: 10.1038/nature08867. [DOI] [PubMed] [Google Scholar]
- 48.Ohm RA, et al. Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog. 2012;8:e1003037. doi: 10.1371/journal.ppat.1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keller NP, Turner G, Bennett JW. Fungal secondary metabolism - from biochemistry to genomics. Nature reviews. Microbiology. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 50.Clevenger KD, et al. A scalable platform to identify fungal secondary metabolites and their gene clusters. Nature chemical biology. 2017;13:895–901. doi: 10.1038/nchembio.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maharachchikumbura SS, Hyde KD, Groenewald JZ, Xu J, Crous PW. Pestalotiopsis revisited. Stud Mycol. 2014;79:121–186. doi: 10.1016/j.simyco.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watson P. Calcarisporium arbuscula living as an endophyte in apparently healthy sporophores of Russula and Lactarius. Transactions of the British Mycological Society. 1955;38:409–414. doi: 10.1016/S0007-1536(55)80044-7. [DOI] [Google Scholar]
- 53.Hoog GSd. The genera Blastobotrys, Sporothrix, Calcarisporium and Calcarisporiella gen. nov. Studies in Mycology. 1974;7:1–84. [Google Scholar]
- 54.Arfi Y, et al. Characterization of salt-adapted secreted lignocellulolytic enzymes from the mangrove fungus Pestalotiopsis sp. Nature communications. 2013;4:1810. doi: 10.1038/ncomms2850. [DOI] [PubMed] [Google Scholar]
- 55.Kramer A, Paun L, Imhoff JF, Kempken F, Labes A. Development and validation of a fast and optimized screening method for enhanced production of secondary metabolites using the marine Scopulariopsis brevicaulis strain LF580 producing anti-cancer active scopularide A and B. PLoS One. 2014;9:e103320. doi: 10.1371/journal.pone.0103320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knudsen, T. & Knudsen, B. CLC Genomics Benchwork 6. Available: http://www.clcbio.com. Accessed on 2013 Sept 20., 2013).
- 58.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hansen FT, et al. An update to polyketide synthase and non-ribosomal synthetase genes and nomenclature in Fusarium. Fungal genetics and biology: FG & B. 2015;75:20–29. doi: 10.1016/j.fgb.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 60.Marchler-Bauer A, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic acids research. 2011;39:D225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bachmann BO, Ravel J. Chapter 8. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 2009;458:181–217. doi: 10.1016/S0076-6879(09)04808-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.