Abstract
The bacterium Streptomyces avermitilis is an industrial-scale producer of avermectins, which are important anthelmintic agents widely used in agriculture, veterinary medicine, and human medicine. During the avermectin fermentation process, S. avermitilis is exposed to organic peroxides generated by aerobic respiration. We investigated the role of MarR-family transcriptional regulator OhrR in oxidative stress response and avermectin production in S. avermitilis. The S. avermitilis genome encodes two organic hydroperoxide resistance proteins: OhrB1 and OhrB2. OhrB2 is the major resistance protein in organic peroxide stress responses. In the absence of organic peroxide, the reduced form of OhrR represses the expression of ohrB2 gene by binding to the OhrR box in the promoter region. In the presence of organic peroxide, the oxidized form of OhrR dissociates from the OhrR box and the expression of ohrB2 is increased by derepression. OhrR also acts as a repressor to regulate its own expression. An ohrR-deletion mutant (termed DohrR) displayed enhanced avermectin production. Our findings demonstrate that OhrR in S. avermitilis represses avermectin production by regulating the expression of pathway-specific regulatory gene aveR. OhrR also plays a regulatory role in glycolysis and the pentose phosphate (PP) pathway by negatively controlling the expression of pykA2 and ctaB/tkt2-tal2-zwf2-opcA2-pgl.
Keywords: Streptomyces avermitilis, avermectin biosynthesis, OhrR, organic hydroperoxide stress resistance, repression
Introduction
The organic peroxide-sensing repressor OhrR is a MarR family transcriptional regulator that regulates the expression of organic hydroperoxide resistance protein (Ohr) (Fuangthong et al., 2001; Sukchawalit et al., 2001). OhrR is widely distributed in Gram-positive and Gram-negative bacteria (Chuchue et al., 2006; Oh et al., 2007; Atichartpongkul et al., 2010; da Silva Neto et al., 2012; Saikolappan et al., 2015). Most ohrR genes are genetically linked to ohr genes, either expressed divergently from a bidirectional promoter region (Chuchue et al., 2006; Oh et al., 2007; Saikolappan et al., 2015) or co-transcribed in an operon (Sukchawalit et al., 2001; Atichartpongkul et al., 2010).
The DNA-binding sequence of OhrR (OhrR box) has been identified in some bacteria as a conserved AT-rich inverted repeat region. Examples of OhrR box consensus sequences are TACAATTNAATTGTA in Bacillus subtilis (Fuangthong and Helmann, 2002), TTnCAATT-(16/17)-AATTGnAA in Xanthomonas campestris (Mongkolsuk et al., 2002), TACAATTNAATTGTA in Agrobacterium tumefaciens (Chuchue et al., 2006), and GCAACTNAATTGC in Streptomyces coelicolor (Oh et al., 2007). In the absence of organic peroxide, the reduced form of OhrR binds to the OhrR box in the ohr promoter region and represses ohr expression (Panmanee et al., 2002; Chuchue et al., 2006; Oh et al., 2007; Saikolappan et al., 2015). Upon exposure to organic hydroperoxides, oxidation of the conserved cysteine (Cys) residue in the OhrR N-terminus leads to a major conformational change of OhrR, resulting in DNA dissociation and allowing RNA polymerase to bind to the promoter and initiate ohr gene transcription (Hong et al., 2005; Lee et al., 2007; Newberry et al., 2007; Soonsanga et al., 2007).
OhrRs are divided into two classes (single-Cys and multiple-Cys) according to presence vs. absence of additional C-terminal Cys residues (Panmanee et al., 2006). OhrRs from B. subtilis and S. coelicolor have a single N-terminal Cys residue (Hong et al., 2005; Oh et al., 2007). OhrRs from many other species, including X. campestris, A. tumefaciens, and Pseudomonas aeruginosa, have two or more Cys residues (Chuchue et al., 2006; Newberry et al., 2007; Atichartpongkul et al., 2010; Atichartpongkul et al., 2016). In B. subtilis, oxidation of the single Cys residue (Cys15) results in a sulfenic acid intermediate (Fuangthong and Helmann, 2002; Hong et al., 2005), which is further oxidized to generate mixed disulfides or a protein sulfenamide derivative, with consequent dissociation of OhrR from operator DNA (Lee et al., 2007). In X. campestris, upon organic hydroperoxide oxidation, the N-terminal Cys residue (Cys22) of OhrR forms a reversible intersubunit disulfide bond with C-terminal Cys127′ between the two subunits of OhrR dimer. This conformational change results in 28° rotation of each winged helix-turn-helix, and DNA dissociation (Panmanee et al., 2006; Newberry et al., 2007).
Soil-dwelling Streptomyces bacteria are characterized by complex morphological differentiation and secondary metabolism. To avoid harmful effects of various oxidants present in the environment or generated by aerobic metabolism, Streptomyces utilize specific antioxidant enzymes, notably alkyl hydroperoxide reductase (AhpCD), catalase (CatA), Ohr, and thioredoxin systems, which are mediated, respectively, by transcriptional regulators OxyR (Hahn et al., 2002; Liu et al., 2016), CatR (Hahn et al., 2000), OhrR (Oh et al., 2007), and σR (Kim et al., 2012). Upon exposure to organic hydroperoxides in S. coelicolor, OhrR acts as a repressor of ohrA and an activator of its own gene (Oh et al., 2007).
S. avermitilis is an industrial-scale producer of avermectins, a series of 16-membered macrocyclic lactone derivatives widely used as drugs or pesticides in agriculture, veterinary medicine, and human medicine (Ikeda and Omura, 1997). During the avermectin fermentation process, S. avermitilis is exposed to organic peroxides generated by aerobic respiration. We investigated the regulatory role of OhrR in oxidative stress response and avermectin production in S. avermitilis.
Materials and Methods
Strains, Plasmids, Culture Conditions, and Culture Media
Strains and plasmids used in this study are listed in Table 1. Culture conditions for S. avermitilis sporulation, protoplast preparation, and regeneration were as described previously (Macneil and Klapko, 1987). Seed medium and fermentation medium FM-I were used for avermectin production and for RNA isolation (Jiang et al., 2011). YEME were also used for RNA isolation with peroxide treatment. E. coli strains were grown at 37°C in LB medium.
Table 1.
Strains and plasmids used in this study.
| Strain or plasmid | Description | Source |
|---|---|---|
| S. avermitilis | ||
| ATCC31267 | Wild-type strain (WT) | Laboratory stock |
| DohrR | ohrR deletion mutant | This study |
| CohrR | ohrR complementation strain | This study |
| E. coli | ||
| JM109 | General cloning host | Laboratory stock |
| BL21 (DE3) | Host for protein overexpression | Laboratory stock |
| Plasmids | ||
| pKC1139 | Multiple-copy, temperature-sensitive E. coli–Streptomyces shuttle vector | Bierman et al., 1992 |
| pSET152 | Integrative E. coli–Streptomyces shuttle vector | Bierman et al., 1992 |
| pET-28a (+) | Vector for His6-tagged protein overexpression in E. coli | Novagen |
| pKCD-ohrR | ohrR deletion vector based on pKC1139 | This study |
| pSET-ohrR | ohrR complementation vector based on pSET152 | This study |
| pET-OhrR | ohrR overexpression vector based on pET-28a (+) | This study |
Gene Deletion and Complementation
An ohrR (SAV5090) gene deletion mutant was constructed using traditional homologous recombination strategy. Two DNA fragments flanking ohrR gene were amplified by PCR from S. avermitilis ATCC31267 genomic DNA. A 510-bp fragment upstream of ohrR (position -426 to +84 from start codon) was amplified by primers OR-up-Fw and OR-up-Rev, and a 431-bp fragment downstream of ohrR (position +445 to +875) was amplified by primers OR-dw-Fw and OR-dw-Rev (Supplementary Table S1 and Supplementary Figure S1). The two fragments were recovered, digested, respectively, by EcoRI/XbaI and BamHI/XbaI, and ligated together into pKC1139 (Bierman et al., 1992) to produce ohrR-deletion vector pKCD-ohrR. pKCD-ohrR was introduced into ATCC31267 protoplasts, and double-crossover mutants were selected as described previously (Zhao et al., 2007). The ohrR-deletion mutant (termed DohrR) was confirmed by PCR using OR-V-Fw/OR-V-Rev (external primers) and OR-V2-Fw/OR-V2-Rev (internal primers), followed by DNA sequencing (Supplementary Table S1 and Supplementary Figure S1).
For complementation analysis of DohrR, ohrR ORF and its promoter were amplified by PCR using primers OR-C-Fw and OR-C-Rev (Supplementary Table S1), and ligated into BamHI/EcoRI-digested pSET152 to produce complementation vector pSET-ohrR. The resulting pSET-ohrR was transformed into DohrR protoplasts to produce a complementation strain of DohrR (termed CohrR).
RNA Preparation and Quantitative Real-Time RT-PCR Analysis (qRT-PCR)
RNA was isolated from S. avermitilis mycelia grown in FM-I or YEME, using TRIzol reagent (Tiangen, China) as described previously (Guo et al., 2013). The chromosomal DNA contamination of RNA samples was removed by adding DNase I (TaKaRa, Japan). The concentrations of RNA were measured by NanoVue Plus spectrophotometer (GE Healthcare). Each RNA sample (2 μg) was reverse transcribed by random hexamers (25 μM), dNTP mixture (10 mM) and M-MLV (TaKaRa, Japan). qRT-PCR was performed to determine the transcription levels of various genes using primers listed in Supplementary Table S1. Each reaction system (20 μl) contains template cDNA, forward and reverse primers (each 300 nM) and 10 μl FastStart Universal SYBR Green Master (ROX). PCR protocol: 95°C for 10 min, 40 cycles of 95°C for 10 s/60°C for 30 s. The relative expression level was calculated using the comparative Ct method. hrdB gene was used as internal control.
Fermentation and HPLC Analysis of Avermectin Production
Fermentation of S. avermitilis strains and HPLC analysis of avermectin production were performed as described previously (Jiang et al., 2011).
Overexpression and Purification of His6-OhrR
For overexpression of the N-terminal His6-tagged OhrR in E. coli, ohrR ORF was amplified using primers His-ohrR-Fw and His-ohrR-Rev (Supplementary Table S1). The DNA fragment was purified, cut with EcoRI/HindIII, and cloned into pET28a (+) to generate the expression plasmid pET-OhrR, which was then introduced into E. coli BL21 (DE3) for overexpression. His6-OhrR was induced by treatment with 0.2 mM IPTG for 12 h at 16°C. Cells were collected and disrupted in lysis buffer (20 mM Tris base, 500 mM NaCl, 5 mM imidazole, 5% glycerol [pH 7.9]) by sonication on ice. His6-OhrR was purified from the lysate using Ni2+-NTA resin (Bio-works, Sweden) as per the manufacturer’s protocol.
Electrophoretic Mobility Gel Shift Assays (EMSAs)
Electrophoretic mobility gel shift assays (EMSAs) were performed using a DIG Gel Shift Kit (2nd Generation, Roche) as per the manufacturer’s protocol. DNA probes were amplified by PCR (Supplementary Table S1) and labeled with digoxigenin using recombinant terminal transferase. Binding reactions and detection conditions were as described previously (Liu et al., 2013). Various concentrations of tert-butyl hydroperoxide (tBHP) and dithiothreitol (DTT) were added to the mixture to evaluate the effects of oxidation on OhrR binding activity.
Determination of Transcriptional Start Sites (TSSs)
The TSSs of ohrR and ohrB2 were analyzed using a 5′/3′ RACE Kit (2nd Generation, Roche) as per the manufacturer’s protocol. Total RNA of wild-type strain ATCC31267 (WT) grown in FM-I for 6 days was used as the template. Primers used are listed in Supplementary Table S1.
DNase I Footprinting Assays
A fluorescence labeling method was used for these assays. DNA probes were obtained by PCR using FAM-labeled primers (Supplementary Table S1). Reaction mixtures (each 20 μL) containing labeled DNA fragments and various quantities of His6-OhrR were incubated for 30 min at 25°C. DNase I digestion was performed for 40 s at 37°C, and stopped by addition of 60 μM EDTA (pH 8.0). After phenol/chloroform extraction and ethanol precipitation, the samples were subjected to capillary electrophoresis. Data were analyzed using GeneMarker software v2.2.0.
Results
ohrB2 Expression Is Upregulated in DohrR
The annotated S. avermitilis genome revealed one ohrR gene (SAV5090) and two ohr genes, ohrB1 (SAV3610), and ohrB2 (SAV5091). ohrR and ohrB2 are transcribed from a bidirectional promoter region. OhrB1 and OhrB2 contain two highly conserved Cys residues, similar to Ohr proteins in other bacteria (Atichartpongkul et al., 2001). In contrast to S. coelicolor OhrR, which has a single Cys residue (Oh et al., 2007), S. avermitilis OhrR has a C-terminal Cys residue (Cys164) in addition to the N-terminal conserved Cys residue (Cys25).
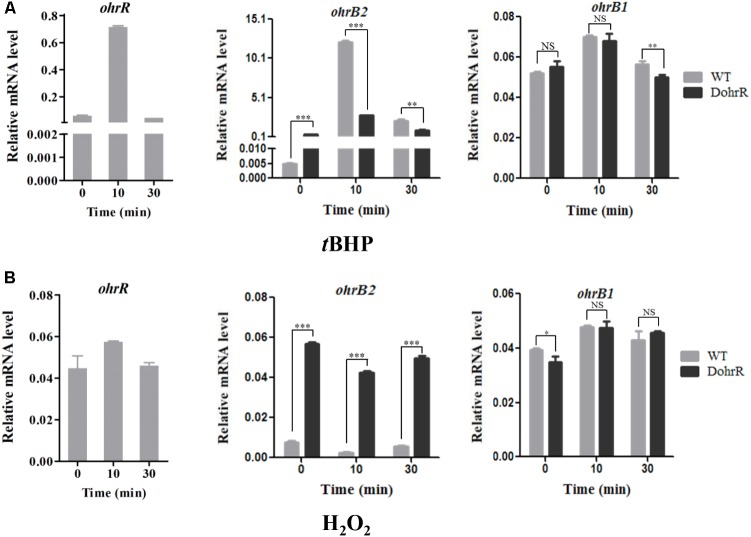
An ohrR-deletion mutant, DohrR, was constructed by homologous recombination as described in the section “Materials and Methods.” ohrR deletion had no effect on morphology, but slightly inhibited growth of S. avermitilis, and the decreased growth could be complemented by introduction of a single-copy ohrR gene to DohrR (Supplementary Figure S1). Sensitivity of DohrR to oxidants (0.2 mM tBHP or 1 mM H2O2) was the same as that of WT (Supplementary Figure S2). Transcription levels of ohrR, ohrB2, and ohrB1 in FM-I were analyzed by RT-qPCR for ATCC31267 and DohrR. ohrB2 expression in DohrR was much higher than in WT, increased by approximately 300-fold on day 2 (exponential phase) and 135-fold on day 6 (stationary phase), indicating a negative role of OhrR in regulation of ohrB2 expression (Figure 1A). ohrR transcription levels were increased by 18-fold on day 2 and 15-fold on day 6 in DohrR than in WT, indicating that OhrR also negatively regulates its own expression. ohrB1 transcription levels in DohrR were only slightly higher than those in WT, suggesting that ohrB1 is not tightly controlled by OhrR. The transcriptional levels of ohrB2 and ohrB1 were restored to WT level in CohrR strain, indicating that OhrR is responsible for the observed effects on transcription. However, the expression of ohrR was only partially complemented in CohrR strain, still higher than that of WT, probably because ohrR gene only functions properly in cis.
FIGURE 1.
Interaction of OhrR with ohrR–ohrB2 intergenic region and ohrB1 promoter. (A) qRT-PCR of ohr expression in DohrR, CohrR and WT. RNA was prepared from cells grown in FM-I for 2 or 6 days. Values shown are mean ± SD from three technical replicates. ∗∗P < 0.01; ∗∗∗P < 0.001; NS, not significant (Student’s t-test). (B) EMSAs of His6-OhrR with ohrR–ohrB2 intergenic region and ohrB1 promoter region. Competition assays were performed using 200-fold excess of specific (S) and non-specific (N) unlabeled DNAs. Top: concentrations of His6-OhrR and DTT. Bottom: probes used. Arrow: free probe.
Determination of OhrR Binding Site of ohrR–ohrB2 Intergenic Region
OhrR protein was overexpressed in E. coli with an N-terminal His6-tag, and purified for EMSAs. His6-OhrR bound specifically to the ohrR–ohrB2 intergenic region and the promoter region of ohrB1 (Figure 1B). The quantity of His6-OhrR required to generate intense shifted bands was greater for ohrB1 promoter than for ohrR–ohrB2 intergenic region, suggesting that OhrR interacts more weakly with ohrB1 promoter.
DNase I footprinting assay was performed to detect the OhrR binding site in the ohrR–ohrB2 intergenic region. A 28-nt protected region was detected in the presence of 0.4 μM and 0.8 μM His6-OhrR (Figure 2A). Two similar 13-nt sites (ACAATTCAGTTGT [site a], ACAACTTAATGGT [site b]) were found in the protected region (Figure 2B). The consensus sequence of OhrR box (ACAATTNAATTGT) in S. avermitilis (Figure 2C) was similar to that of OhrR box (GCAACTNAATTGC) in S. coelicolor (Oh et al., 2007). Identified OhrR boxes extend from positions -45 to -18 relative to TSS for ohrB2, and from positions -50 to -23 relative to TSS for ohrR. In both cases, the protected region is located in the promoter and contains the -35 region. OhrR Thus represses ohrB2 and ohrR transcription by blocking attachment of RNA polymerase to its promoter.
FIGURE 2.
DNase I footprinting assay of ohrR–ohrB2 intergenic region using His6-OhrR. (A) Fluorograms corresponding to control DNA and to protected reactions with 0.4 and 0.8 μM His6-OhrR. (B) Nucleotide sequences of ohrR–ohrB2 intergenic region. Non-shaded boxes: presumed –35 and –10 regions of ohrR and ohrB2. Shaded boxes: regions protected by His6-OhrR. Underlining: OhrR motif (site a and site b). Gray bent arrows: translational start codons. Black bent arrows: TSSs. (C) Consensus sequence of OhrR motif.
OhrR Mediates Organic Peroxide Induction of ohrB2 by Derepression
Effects of OhrR oxidation on binding affinity to target DNA were evaluated by adding various concentrations of tBHP and DTT to EMSA mixtures. tBHP inhibited DNA binding between His6-OhrR and the ohrR–ohrB2 intergenic region at very low concentration. When low concentration (40 nM) of His6-OhrR was used for EMSA, 1 μM tBHP was sufficient to eliminate binding, whereas when high concentration (120 nM) of His6-OhrR was used, 2 μM tBHP was required to eliminate binding (Figures 3A,B). tBHP had no effect on binding affinity of control DNA binding protein Rex to its target promoter cydA1, suggesting that OhrR specifically senses and responds to organic oxidant tBHP. In DTT experiments, binding increased slightly as DTT concentration increased, indicating that interaction with target DNA was stronger for reduced than for oxidized His6-OhrR (Figure 3C). The enhancing effect of DTT on binding affinity was more noticeable when the interaction was weak or when lower OhrR concentration was used for EMSA (Supplementary Figure S3).
FIGURE 3.
Effects of tBHP (A,B) and DTT (C) on in vitro binding of His6-OhrR to ohrR–ohrB2 intergenic region. Concentrations of His-OhrR, tBHP, and DTT are shown at top. Rex-His6 was used as DNA binding protein control.
ohr expression levels in peroxide-treated DohrR and WT were analyzed by qRT-PCR. S. avermitilis cells were cultured in YEME for 42 h and 0.4 mM tBHP or 1 mM H2O2 was added. Cells were treated for 10 or 30 min. Compared to untreated (0 min), 10 min tBHP treatment increased ohrR transcription to a maximal level in WT, and H2O2 treatment did not induce ohrR transcription in WT (Figure 4), indicating that OhrR responds specifically to organic peroxides. The transcription level of ohrB2 in WT increased by >1000-fold to its maximal value within 10 min of tBHP treatment, and decreased gradually with extended treatment. The transcription profile of ohrB2 in DohrR was similar to that in WT under tBHP treatment condition, only with a much lower increased fold at its maximal value. ohrB2 transcription was not induced by H2O2 treatments in both strains, but were much higher in untreated DohrR than in untreated WT. Thus, OhrR negatively regulated ohrB2 expression in the absence of peroxide, whereas in the presence of peroxide, OhrR interaction with ohrB2 promoter was eliminated and ohrB2 expression was improved by derepression. ohrB1 expression was only slightly induced by tBHP treatment in WT, and its expression was much lower than that of ohrB2 under tBHP treatment condition, indicating that OhrB2 is the major organic hydroperoxide resistance protein in S. avermitilis. ohrB1 expression was also slightly induced by H2O2 treatment, and ohrB1 levels under both treatments were similar for DohrR and WT, suggesting that the slight induction of ohrB1 by either treatment is not mediated by OhrR.
FIGURE 4.
qRT-PCR of oxidant-induced genes in WT and DohrR. RNA was prepared from cells grown in YEME and treated with 0.4 mM tBHP (A) or 1 mM H2O2 (B) for 10 or 30 min. No peroxide was added at 0 min. Values shown are mean ± SD from three replicates. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; NS, not significant (Student’s t-test).
OhrR Negatively Regulates Avermectin Production
ohrR deletion (DohrR mutant) caused a ˜2-fold increase in avermectin production (Figure 5A). Avermectin production was restored to WT level by re-introduction of ohrR gene to DohrR, indicating a negative regulatory role of OhrR in the process. To examine the causes of avermectin overproduction in DohrR, we measured the transcription levels of selected avermectin biosynthesis-related genes (pathway-specific regulatory gene aveR and biosynthetic gene aveA1) in DohrR and WT by qRT-PCR. RNA samples were prepared from cells grown in FM-I for 2 and 6 days. Transcription levels of aveR and aveA1 were higher in DohrR than in WT on both day 2 and day 6, and increased by >4-fold at day 6 (Figure 5B). The transcription levels of both genes were restored to WT levels in CohrR. These findings indicate that ave expression was repressed by OhrR, especially at the late stage of avermectin production.
FIGURE 5.
Effect of ohrR deletion on avermectin production. (A) Avermectin production in ohrR-related mutants. (B) qRT-PCR of aveR and aveA1 transcription levels in WT, DohrR and CohrR. RNA samples as in Figure 1. ∗P < 0.05; ∗∗∗P < 0.001; NS, not significant (Student’s t-test).
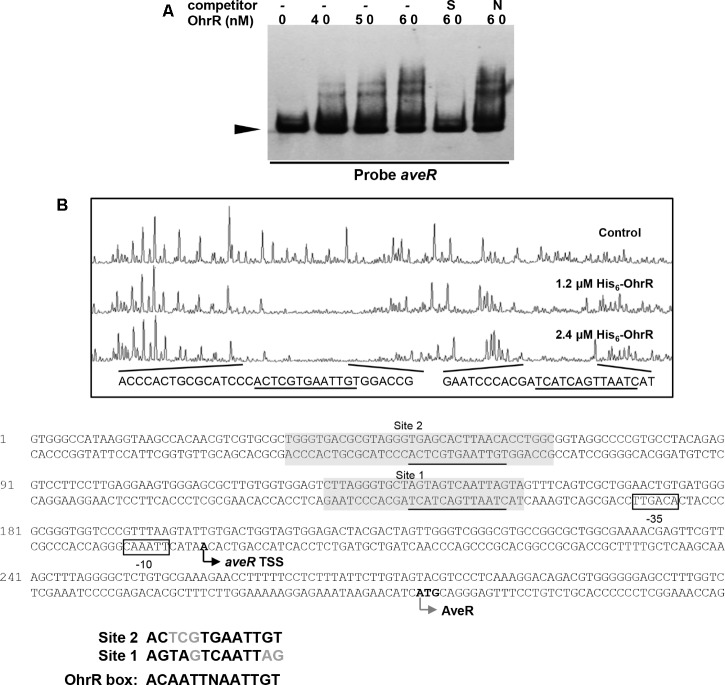
OhrR Binds to aveR Promoter Region
Electrophoretic mobility gel shift assays were performed using the promoter regions of ave genes and His6-OhrR to determine whether OhrR directly regulates transcription of the genes. With the aveR promoter region and His6-OhrR, two retarded bands were observed. The shifted bands were completed abolished by addition of unlabeled specific probe, but not by non-specific competitor probe (Figure 6A), indicating that the interaction between OhrR and the aveR promoter region was specific. With the aveA1 promoter region, no retarded band was observed (data not shown), indicating that OhrR represses avermectin production by regulating the expression of regulatory gene aveR.
FIGURE 6.
Binding of OhrR to aveR promoter region. (A) EMSA of His6-OhrR with aveR promoter region. (B) DNase I footprinting assay of aveR promoter region using His6-OhrR and nucleotide sequences of ohrR–ohrB2 intergenic region. EMSA and DNase I footprinting conditions as in Figures 1, 2.
To determine the OhrR binding site in the aveR promoter region, DNase I footprinting assays were performed using fluorescein-labeled aveR promoter region (position -426 to +51 from start codon) and His6-OhrR. In the presence of 1.2 or 2.4 μM His6-OhrR, two protected regions were observed, one extending 26 nt (site 1) and the other 35 nt (site 2) (Figure 6B), consistently with the two retarded bands observed in EMSA (Figure 6A). Two possible 11-nt OhrR binding sites (AGTAGTCAATTAG in site 1; ACTCGTGAATTGT in site 2) were found in the protected regions. The OhrR boxes are, respectively, located from positions -153 to -141 and -63 to -51 relative to the TSS of aveR (Zhuo et al., 2010). These findings, in combination with transcription results, suggest that OhrR possibly represses the transcription of aveR by impeding attachment of RNA polymerase to the promoters.
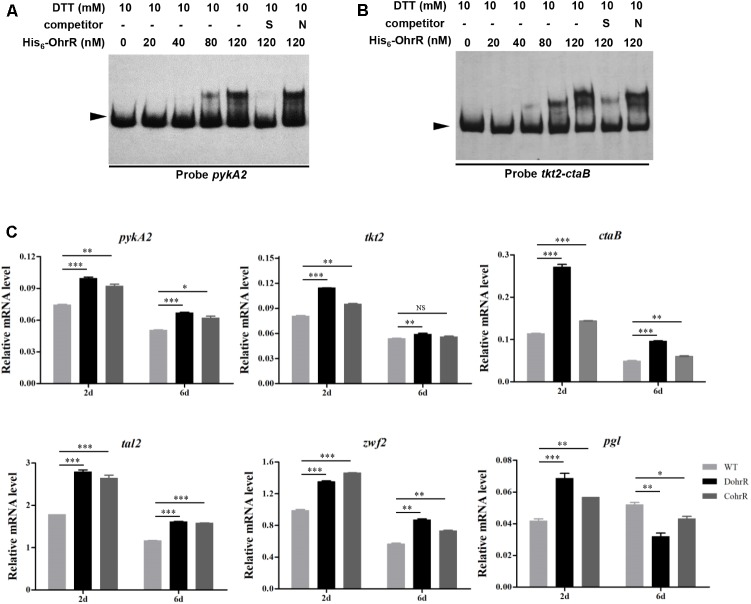
OhrR Binds to pykA2 and tkt2/ctaB Promoter Regions
In a search for additional putative OhrR targets in S. avermitilis, we scanned the S. avermitilis genome with the consensus sequence ACAATTNAATTGT by Virtual Footprint program (Munch et al., 2005). We found one annotated gene (pykA2) with two mismatches, and nine genes (sig7, aveR, ilvA, ohrB1, cspD3, paaA, tkt2/ctaB, fadE3, sig57) with three mismatches. In view of our finding that OhrR directly controls aveR and ohrB1, we performed EMSAs to determine whether OhrR binds to promoter regions of other putative targets. OhrR bound specifically only to the promoter region of pykA2 (encodes a putative pyruvate kinase) and the intergenic region of tkt2/ctaB (encodes a putative transketolase and protoheme IX farnesyltransferase) (Figure 7). tkt2 and its downstream genes tal2, zwf2, opcA2, and pgl have the same transcription direction with very short intergenic regions and may be located in the same transcription unit. The transcription levels of pykA2, ctaB, tkt2, tal2, zwf2, and pgl were higher in DohrR than in WT on both day 2 and day 6, indicating that the genes are under negative control of OhrR.
FIGURE 7.
Binding of His6-OhrR to pykA2 promoter region (A) and tkt2-ctaB intergenic region (B). EMSAs using His6-OhrR protein at the indicated concentrations. EMSA conditions as in Figure 1. (C) qRT-PCR of pykA2, tkt2, ctaB, tal2, zwf2, and pgl transcription levels in WT, DohrR, and CohrR. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; NS, not significant (Student’s t-test).
Discussion
Our findings demonstrate that OhrR in S. avermitilis acts as a repressor specifically in response to organic peroxide stress. OhrB2 is the major organic hydroperoxide resistance protein responsible for such stress. In the absence of organic peroxide stress, the reduced form of OhrR binds to the OhrR box of the ohrR–ohrB2 intergenic region and represses the expression of ohrB2 and its own. In the presence of such stress, oxidized OhrR dissociates from OhrR box, and ohrB2 expression is increased by derepression. Similar regulatory roles of OhrR have been reported for S. coelicolor and other bacterial taxa (Fuangthong et al., 2001; Panmanee et al., 2002; Oh et al., 2007), indicating a conserved role of OhrR in organic peroxide stress responses. Negatively regulation of ohrR itself provides a mechanism for rapidly reduced the expression of major organic hydroperoxide resistance protein in response to disappearance of organic peroxide stress.
Avermectin production in S. avermitilis was enhanced when ohrR gene was deleted. OhrR binds to two OhrR boxes located upstream of the aveR TSS, and represses aveR expression. OhrR thus represses avermectin production by controlling the expression of pathway-specific activator gene aveR. Several transcription factors have been shown to control avermectin production through direct regulation of aveR expression; these include phosphate metabolism regulator PhoP, pseudo γ-butyrolactone receptor homolog AvaR2, GBL receptor AvaR1, and Redox-sensing regulator Rex (Yang et al., 2015; Zhu et al., 2016, 2017; Liu et al., 2017). The present results demonstrate that avermectin production is directly regulated by OhrR. The site 1 of OhrR-binding sites in the aveR promoter region is located upstream -35 region (-63 to -51), and the site 2 is also the binding site of Rex which represses aveR expression. Therefore, OhrR regulates the expression of aveR possibly through blocking RNA polymerase to the promoter or interfering Rex or other regulator to bind to the aveR promoter region.
The genes pykA2 and ctaB/tkt2 are also directly regulated by OhrR. pykA2 encodes a putative pyruvate kinase that catalyzes the final step of glycolysis by transferring a phosphate group from phosphoenolpyruvate to adenosine diphosphate, to yield pyruvate and ATP (Ponce et al., 1995). tkt2 and its downstream genes tal2, zwf2, opcA2, and pgl may be located in the same transcription unit. These genes encode putative transketolase, transaldolase, glucose-6-phosphate dehydrogenase, oxppcycle protein, and 6-phosphogluconolactonase of the pentose phosphate (PP) pathway (Sprenger, 1995). ctaB encodes a putative protoheme IX farnesyltransferase (CtaB) involved in synthesis of heme containing terminal oxidases of the bacterial respiratory chain (Xu et al., 2016). In cells subjected to organic peroxide stress, oxidized OhrR activates expression of pykA2 and ctaB/tkt2-tal2-zwf2-opcA2-pgl by derepression, thereby enhancing glycolysis and PP pathway. Streptomyces have no glutathione/glutaredoxin system; instead, they use thioredoxin to reduce protein disulfide bonds formed during oxidative stress. The oxidized form of thioredoxin is reduced by thioredoxin reductase, with expenditure of NADPH (Stefankova et al., 2006; Koharyova et al., 2011). Enhancement of PP pathway generates new NADPH, which helps prevent oxidative stress. Intermediate metabolites generated during PP pathway, which include NADPH, ribose-5-phosphate, and erythrose-4-phosphate, are key precursors in the processes of cell growth, DNA biosynthesis, and DNA repair (Zhang et al., 2003). New ATP is produced by glycolysis, and protoheme IX farnesyltransferase overexpression promotes recovery from energy-depleted condition resulting from respiratory chain damage during oxidative stress. Enhancement of glycolysis and PP pathway may likewise stimulate avermectin production in S. avermitilis.
In summary, organic peroxide-sensing repressor OhrR functions as a repressor, mediating organic peroxide induction of ohr genes through derepression. OhrR represses avermectin production by directly regulating pathway-specific activator gene aveR. OhrR also has a negatively regulatory role in glycolysis and PP pathway by controlling the expression of pykA2 and ctaB/tkt2-tal2-zwf2-opcA2-pgl in S. avermitilis.
Author Contributions
ZC and MS designed the experiments. MS and ML performed the experiments. MS, ML, and ZC analyzed the data and wrote the manuscript. YW, YS, and JL contributed the study materials.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. S. Anderson for English editing of the manuscript.
Footnotes
Funding. This study was supported by the National Natural Science Foundation of China (Grant No. 31470190 to ZC).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01398/full#supplementary-material
References
- Atichartpongkul S., Fuangthong M., Vattanaviboon P., Mongkolsuk S. (2010). Analyses of the regulatory mechanism and physiological roles of Pseudomonas aeruginosa OhrR, a transcription regulator and a sensor of organic hydroperoxides. J. Bacteriol. 192 2093–2101. 10.1128/JB.01510-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atichartpongkul S., Loprasert S., Vattanaviboon P., Whangsuk W., Helmann J. D., Mongkolsuk S. (2001). Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. Microbiology 147 1775–1782. 10.1099/00221287-147-7-1775 [DOI] [PubMed] [Google Scholar]
- Atichartpongkul S., Vattanaviboon P., Wisitkamol R., Jaroensuk J., Mongkolsuk S., Fuangthong M. (2016). Regulation of organic hydroperoxide stress response by two OhrR homologs in Pseudomonas aeruginosa. PLoS One 11:e0161982. 10.1371/journal.pone.0161982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierman M., Logan R., Obrien K., Seno E. T., Rao R. N., Schoner B. E. (1992). Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116 43–49. 10.1016/0378-1119(92)90627-2 [DOI] [PubMed] [Google Scholar]
- Chuchue T., Tanboon W., Prapagdee B., Dubbs J. M., Vattanaviboon P., Mongkolsuk S. (2006). ohrR and ohr are the primary sensor/regulator and protective genes against organic hydroperoxide stress in Agrobacterium tumefaciens. J. Bacteriol. 188 842–851. 10.1128/JB.188.3.842-851.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Neto J. F., Negretto C. C., Netto L. E. (2012). Analysis of the organic hydroperoxide response of Chromobacterium violaceum reveals that OhrR is a cys-based redox sensor regulated by thioredoxin. PLoS One 7:e47090. 10.1371/journal.pone.0047090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuangthong M., Atichartpongkul S., Mongkolsuk S., Helmann J. D. (2001). OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183 4134–4141. 10.1128/JB.183.14.4134-4141.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuangthong M., Helmann J. D. (2002). The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl. Acad. Sci. U.S.A. 99 6690–6695. 10.1073/pnas.102483199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Zhang X., Luo S., He F., Chen Z., Wen Y., et al. (2013). A novel TetR family transcriptional regulator, SAV576, negatively controls avermectin biosynthesis in Streptomyces avermitilis. PLoS One 8:e71330. 10.1371/journal.pone.0071330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J. S., Oh S. Y., Chater K. F., Cho Y. H., Roe J. H. (2000). H2O2-sensitive Fur-like repressor CatR regulating the major catalase gene in Streptomyces coelicolor. J. Biol. Chem. 275 38254–38260. 10.1074/jbc.M006079200 [DOI] [PubMed] [Google Scholar]
- Hahn J. S., Oh S. Y., Roe J. H. (2002). Role of OxyR as a peroxide-sensing positive regulator in Streptomyces coelicolor A3(2). J. Bacteriol. 184 5214–5222. 10.1128/JB.184.19.5214-5222.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M., Fuangthong M., Helmann J. D., Brennan R. G. (2005). Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell 20 131–141. 10.1016/j.molcel.2005.09.013 [DOI] [PubMed] [Google Scholar]
- Ikeda H., Omura S. (1997). Avermectin biosynthesis. Chem. Rev. 97 2591–2610. 10.1021/cr960023p [DOI] [PubMed] [Google Scholar]
- Jiang L., Liu Y., Wang P., Wen Y., Song Y., Chen Z., et al. (2011). Inactivation of the extracytoplasmic function sigma factor Sig6 stimulates avermectin production in Streptomyces avermitilis. Biotechnol. Lett. 33 1955–1961. 10.1007/s10529-011-0673-x [DOI] [PubMed] [Google Scholar]
- Kim M. S., Dufour Y. S., Yoo J. S., Cho Y. B., Park J. H., Nam G. B., et al. (2012). Conservation of thiol-oxidative stress responses regulated by SigR orthologues in actinomycetes. Mol. Microbiol. 85 326–344. 10.1111/j.1365-2958.2012.08115.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koharyova M., Brynda J., Rezacova P., Kollarova M. (2011). Crystallization and diffraction analysis of thioredoxin reductase from Streptomyces coelicolor. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67 917–921. 10.1107/S1744309111021385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. W., Soonsanga S., Helmann J. D. (2007). A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. U.S.A. 104 8743–8748. 10.1073/pnas.0702081104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Cheng Y., Lyu M., Wen Y., Song Y., Chen Z., et al. (2017). Redox-sensing regulator Rex regulates aerobic metabolism, morphological differentiation, and avermectin production in Streptomyces avermitilis. Sci. Rep. 7:44567. 10.1038/srep44567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Sun M., Cheng Y., Yang R., Wen Y., Chen Z., et al. (2016). OxyR is a key regulator in response to oxidative stress in Streptomyces avermitilis. Microbiology 162 707–716. 10.1099/mic.0.000251 [DOI] [PubMed] [Google Scholar]
- Liu Y., Yan T., Jiang L., Wen Y., Song Y., Chen Z., et al. (2013). Characterization of SAV7471, a TetR-family transcriptional regulator involved in the regulation of coenzyme A metabolism in Streptomyces avermitilis. J. Bacteriol. 195 4365–4372. 10.1128/JB.00716-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macneil D. J., Klapko L. M. (1987). Transformation of Streptomyces-avermitilis by plasmid DNA. J. Ind. Microbiol. 2 209–218. 10.1007/Bf01569542 [DOI] [Google Scholar]
- Mongkolsuk S., Panmanee W., Atichartpongkul S., Vattanaviboon P., Whangsuk W., Fuangthong M., et al. (2002). The repressor for an organic peroxide-inducible operon is uniquely regulated at multiple levels. Mol. Microbiol. 44 793–802. 10.1046/j.1365-2958.2002.02919.x [DOI] [PubMed] [Google Scholar]
- Munch R., Hiller K., Grote A., Scheer M., Klein J., Schobert M., et al. (2005). Virtual footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21 4187–4189. 10.1093/bioinformatics/bti635 [DOI] [PubMed] [Google Scholar]
- Newberry K. J., Fuangthong M., Panmanee W., Mongkolsuk S., Brennan R. G. (2007). Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol. Cell 28 652–664. 10.1016/j.molcel.2007.09.016 [DOI] [PubMed] [Google Scholar]
- Oh S. Y., Shin J. H., Roe J. H. (2007). Dual role of OhrR as a repressor and an activator in response to organic hydroperoxides in Streptomyces coelicolor. J. Bacteriol. 189 6284–6292. 10.1128/Jb.00632-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panmanee W., Vattanaviboon P., Eiamphungporn W., Whangsuk W., Sallabhan R., Mongkolsuk S. (2002). OhrR, a transcription repressor that senses and responds to changes in organic peroxide levels in Xanthomonas campestris pv. phaseoli. Mol. Microbiol. 45 1647–1654. 10.1046/j.1365-2958.2002.03116.x [DOI] [PubMed] [Google Scholar]
- Panmanee W., Vattanaviboon P., Poole L. B., Mongkolsuk S. (2006). Novel organic hydroperoxide-sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J. Bacteriol. 188 1389–1395. 10.1128/Jb.188.4.1389-1395.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce E., Flores N., Martinez A., Valle F., Bolivar F. (1995). Cloning of the two pyruvate kinase isoenzyme structural genes from Escherichia coli: the relative roles of these enzymes in pyruvate biosynthesis. J. Bacteriol. 177 5719–5722. 10.1128/jb.177.19.5719-5722.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikolappan S., Das K., Dhandayuthapani S. (2015). Inactivation of the organic hydroperoxide stress resistance regulator OhrR enhances resistance to oxidative stress and isoniazid in Mycobacterium smegmatis. J. Bacteriol. 197 51–62. 10.1128/JB.02252-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soonsanga S., Fuangthong M., Helmann J. D. (2007). Mutational analysis of active site residues essential for sensing of organic hydroperoxides by Bacillus subtilis OhrR. J. Bacteriol. 189 7069–7076. 10.1128/JB.00879-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger G. A. (1995). Genetics of pentose-phosphate pathway enzymes of Escherichia coli K-12. Arch. Microbiol. 164 324–330. 10.1007/BF02529978 [DOI] [PubMed] [Google Scholar]
- Stefankova P., Perecko D., Barak I., Kollarova M. (2006). The thioredoxin system from Streptomyces coelicolor. J. Basic Microbiol. 46 47–55. 10.1002/jobm.200510036 [DOI] [PubMed] [Google Scholar]
- Sukchawalit R., Loprasert S., Atichartpongkul S., Mongkolsuk S. (2001). Complex regulation of the organic hydroperoxide resistance gene (ohr) from Xanthomonas involves OhrR, a novel organic peroxide-inducible negative regulator, and posttranscriptional modifications. J. Bacteriol. 183 4405–4412. 10.1128/JB.183.15.4405-4412.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Han J., Zhang J., Chen J., Wu N., Zhang W., et al. (2016). Absence of protoheme IX farnesyltransferase CtaB causes virulence attenuation but enhances pigment production and persister survival in MRSA. Front. Microbiol. 7:1625. 10.3389/fmicb.2016.01625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Liu X., Wen Y., Song Y., Chen Z., Li J. (2015). The PhoP transcription factor negatively regulates avermectin biosynthesis in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 99 10547–10557. 10.1007/s00253-015-6921-6 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu J., Wong T. (2003). The DNA excision repair system of the highly radioresistant bacterium Deinococcus radiodurans is facilitated by the pentose phosphate pathway. Mol. Microbiol. 48 1317–1323. 10.1046/j.1365-2958.2003.03486.x [DOI] [PubMed] [Google Scholar]
- Zhao J., Wen Y., Chen Z., Song Y., Li J. (2007). An adpA homologue in Streptomyces avermitilis is involved in regulation of morphogenesis and melanogenesis. Chin. Sci. Bull. 52 623–630. 10.1007/s11434-007-0105-4 [DOI] [Google Scholar]
- Zhu J., Chen Z., Li J., Wen Y. (2017). AvaR1, a butenolide-type autoregulator receptor in Streptomyces avermitilis, directly represses avenolide and avermectin biosynthesis and multiple physiological responses. Front. Microbiol. 8:2577. 10.3389/fmicb.2017.02577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Sun D., Liu W., Chen Z., Li J., Wen Y. (2016). AvaR2, a pseudo -butyrolactone receptor homologue from Streptomyces avermitilis, is a pleiotropic repressor of avermectin and avenolide biosynthesis and cell growth. Mol. Microbiol. 102 562–578. 10.1111/mmi.13479 [DOI] [PubMed] [Google Scholar]
- Zhuo Y., Zhang W., Chen D., Gao H., Tao J., Liu M., et al. (2010). Reverse biological engineering of hrdB to enhance the production of avermectins in an industrial strain of Streptomyces avermitilis. Proc. Natl. Acad. Sci. U.S.A. 107 11250–11254. 10.1073/pnas.1006085107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.