Abstract
Background
Developmental programming of the embryo is controlled by genetic information but also dictated by epigenetic information contained in spermatozoa. Lifestyle and environmental factors not only influence health in one individual but can also affect the phenotype of the following generations. This is mediated via epigenetic inheritance i.e., gametic transmission of environmentally-driven epigenetic information to the offspring. Evidence is accumulating that preconceptional exposure to certain lifestyle and environmental factors, such as diet, physical activity, and smoking, affects the phenotype of the next generation through remodeling of the epigenetic blueprint of spermatozoa.
Scope of Review
This review will summarize current knowledge about the different epigenetic signals in sperm that are responsive to environmental and lifestyle factors and are capable of affecting embryonic development and the phenotype of the offspring later in life.
Major conclusions
Like somatic cells, the epigenome of spermatozoa has proven to be dynamically reactive to a wide variety of environmental and lifestyle stressors. The functional consequence on embryogenesis and phenotype of the next generation remains largely unknown. However, strong evidence of environmentally-driven sperm-borne epigenetic factors, which are capable of altering the phenotype of the next generation, is emerging on a large scale.
Keywords: Sperm, Spermatozoa, Epigenetic, Epigenetic inheritance, Small RNA, DNA methylation, Histone
1. Introduction
Lifestyle factors such as diet, physical activity, smoking, and alcohol consumption, are well known to influence the predisposition to obesity, type 2 diabetes, cardiovascular disease, and cancer, which represent an extraordinary disease burden worldwide. While one's lifestyle clearly affects health and lifespan at the individual level, recent epidemiological studies have provided evidence that the lifestyle of one generation can modify the risk of developing chronic diseases in subsequent generations through so-called parental effects. In fact, the plausible influence of preconceptional environmental factors on the next generations' phenotype is not a new idea. The evolutionary theories of both Jean-Baptiste Lamarck and Charles Darwin have long suggested that, at the population level, environmental factors select for particular phenotypes. However, what represents a paradigm shift is the discovery that parental effects can affect the successive generation's offspring, through mechanisms that seem independent from genetic factors. The separate investigation of paternal effects (where the male only is exposed to a specific environment before conception), has provided further evidence indicating that sperm-borne factors responsive to changes in lifestyle can modulate the developmental programming of the offspring by so-called epigenetic inheritance – a term referring to the direct modification of the gametic epigenome by the environment and subsequent transmission to the next generation [1].
Environmentally-driven epigenetic modifications of gametes provide a potential molecular basis to explain the transmission of developmental plasticity across generations, as well as a mechanism to understand “missing” heritability factors observed with certain diseases. Indeed, in the context of metabolic diseases, all or part of the unsolved heritability of obesity and type 2 diabetes may be ascribed to epigenetic inheritance. This is supported by the epidemiological observation that food availability in childhood and adolescence influences the risk of developing cardiovascular diseases in the offspring [2]. It should be emphasized that the second- and not the first-generation offspring is affected. Moreover, transmission occurs through the paternal line, thereby circumventing possible maternal or in utero effects, which is at the origin of the hypothesis that a non-genetic message is transmitted to the following generations through gametes [2]. Animal models of paternal inheritance have provided definitive evidence that dietary factors introduced before conception can affect the metabolism of the offspring through epigenetic inheritance [3], [4], [5]. For example, paternal overnutrition increases body weight, and adiposity and impairs glucose tolerance and insulin sensitivity in adult female offspring [4]. In a follow-up study, using the same animal model of diet-induced obesity in the fathers, high-fat diet feeding reprograms the epigenome of spermatozoa, thereby providing further evidence to support the hypothesis that nutritional factors modify the metabolic phenotype of the offspring through epigenetic inheritance [6]. In humans, nutritional status and physical activity levels were associated with dynamic epigenetic changes in spermatozoa [7], [8], [9], providing evidence to hypothesize that lifestyle factors prior to conception can modulate the health of the offspring through epigenetic inheritance in humans as well. In addition to nutritional factors, numerous prominent laboratories find that other environmental factors, such as exercise, endocrine-disruptors, as well as traumatic stress, influence the developmental plasticity of phenotypes through epigenetic inheritance (Figure 1) [10], [11], [12].
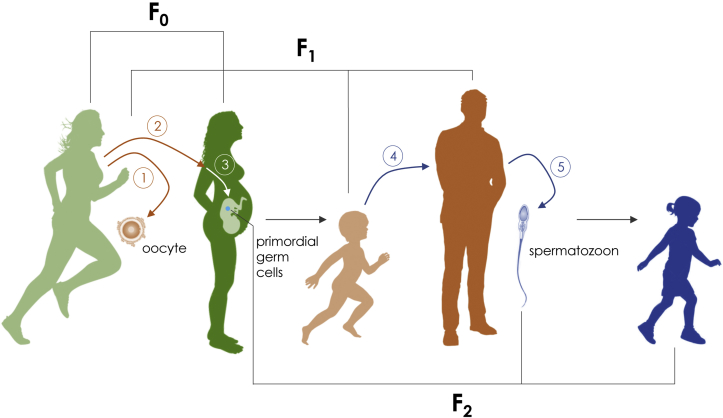
Figure 1.
Lifestyle and environmental influences across generations. Exercise in the F0 generation may induce epigenetic reprogramming of the oocyte (1), and/or change whole body physiology (2) which, if still persistent when a pregnancy occurs, may have consequences on the extracellular milieu in utero (3). The developing embryo could be exposed to the exercise effects, thereby affecting not only the F1 (the embryo itself) but also the primordial germ cells developing in the embryo. Primordial germ cells represent, in part, the second-generation offspring, or F2. Exercise in the F0 may also alter behavior and metabolism in the F1 to influence aerobic capacity or inclination to exercise in the F1, which in turn induces programming of the spermatozoa through serial programming. Alternatively, exercise in the F0 may stably reprogram gametes throughout generations (F0, F1, … ), leading to true transgenerational epigenetic inheritance. Likely, the F2 generation is an integration of all epigenetic reprogramming that occurs throughout ancestors.
For obvious technical limitations, few studies have investigated the effect of environmental factors on the oocyte epigenome [13], [14]. Therefore, this review focuses on the sperm epigenome, about which greater knowledge exists. When addressing epigenetic inheritance experimentally, paternal models are primarily used, as they require less experimental resources and confounding factors are easier to exclude. In models of maternal exposure, environmental factors, even if only present before conception, may later influence the developmental milieu of the embryo (e.g. by altering placental function). This constitutes an important source of bias, as the resulting phenotype of the offspring might be affected by gametic influences, and observed effects may simply be of pure intergenerational origin as compared to transgenerational. In addition, both F1 and F2 generation are under maternal influence during in utero development, as the germ cells of F1 are developing at the embryonic state. Consequently, to determine the effect of in utero exposure on epigenetic inheritance in a transgenerational fashion, investigations need to be extended to the F3 generation (Figure 1) [5]. However, it is sufficient to study the F2 generation in paternal models, as the aforementioned in utero influences are not at play.
Paternal models are not void of possible confounding factors, however, and are not self-sufficient to prove gametic inheritance (Figure 1). For example, it is speculated that contamination of maternal microbiota by the male at time of mating may impact the in utero environment [15]. In addition, the seminal fluid may send signals to the maternal tract and ultimately affect embryo development (reviewed in [16]). Approaches using in vitro fertilization (IVF) may represent a gold standard, with several groups successfully replicating respective parental effects by IVF/ICSI or microinjection [10], [17], [18], [19], [20]. However, caution should be applied when interpreting results from studies using prior handling of gametes, as the procedures themselves may induce significant epigenetic alterations with potential to affect offspring phenotype (reviewed in [21]). Nevertheless, in this review, we discuss current evidence supporting a role of the spermatozoal epigenome, in particular DNA methylation, chromatin, and small RNA expression, as a potential carrier of epigenetic inheritance under lifestyle influences.
2. DNA methylation in spermatozoa
DNA methylation controls numerous cell processes including cell differentiation and embryonic development. During embryonic development, DNA methylation participates in the regulation of gene expression, silencing of transposons, and endogenous retroviral sequences, X chromosome inactivation and genomic imprinting [22], [23]. Methylation of DNA is under the control of DNA methyltransferases (DNMTs) and enzymes of the demethylation pathway such as Ten-Eleven Translocation (TET), as well as the thymine–DNA–glycosylase (TDG) and the DNA base excision repair (BER) [24], [25]. The vast majority of DNA methylation occurs on cytosines in the genomes within a CpG context [26]. However, methylation in a non-CpG context (particularly CpA) has been described to account for up to 25% in some cell types [27], [28], [29].
Sperm cells harbor both CpG and non-CpG methylation, with non-CpG methylation accumulating within and around B1 SINE transposon sequences in male germ cells during mouse fetal development [30]. Non-CpG methylation is also detected at paternally methylated regions and some CpG islands, where maximum methylation is achieved at birth [30]. Such patterns of dynamic methylation change (de novo methylation followed by methylation loss towards the mature spermatozoa stage) is in striking contrast with methylation dynamics on CpGs. Indeed, during mammalian development, early germ line cells are subjected to nearly global DNA methylation erasure [31], a clearing process that is thought to erase cellular memory and to allow developmental totipotency. Primordial germ cells (PGCs) undergo global demethylation in a biphasic way. A first wave of DNA methylation loss of about 70% occurs after PGCs have colonized the developing gonadal region at embryonic day (E) 11.5 in the mouse [32] and presumably E37 in humans [33]). Global DNA demethylation is further increased and peaks at E13.5 in the mouse, with global CpG methylation levels down to 3–4% [34], and 8% in humans, where the peak is observed at week 7 of development [35]. DNA methylation loss is then followed by de novo DNA methylation throughout all stages of sperm cell maturation, with global CpG methylation levels of 90% in mouse- and 70% in fully mature human spermatozoa, yielding approximately 4% of total cytosines methylated [36], [37]. For comparison purpose, methylation levels in sperm are in the lower range compared to somatic cells, for example thymus and brain, which exhibit 15–20% more methylation [38]. Mature spermatozoa are therefore “hypomethylated cells,” with global methylation levels comparable to those in cancer cells [39].
The incomplete erasure of DNA methylation marks during epigenetic reprogramming opens a biological time window, allowing transfer of environmental influences from one generation to the next. Several genomic features escape epigenetic reprogramming, for example L1HS transposons, which are highly methylated at all stages of germline development [35]. However, retrotransposon sequences appear relatively depleted of genomic regions escaping reprogramming (also coined as escapees) [40]. While the role of methylation at transposons or repeated sequences is unknown and difficult to predict by nature, the functional implication of methylation at escapees located at the proximal end of protein-coding regions is obviously easier to appreciate. In human primordial germ cells, functional analysis of genes near escapees revealed an enrichment for genes expressed in the brain and controlling neural development [40]. Several studies, including those from our group, observed that lifestyle factors induce differential DNA methylation in sperm, in close vicinity to genes related to the control of neurogenesis and development of the central nervous system (Figure 2) [7], [8]. A three-month endurance training intervention in humans altered methylation of genes related to the development of the central nervous system, neurogenesis, and neuron differentiation [7]. A similar enrichment for nervous system development genes was found in sperm from obese men before and after gastric-bypass induced weight loss [8]. In an earlier study investigating the variation of DNA methylation in somatic cells obtained from inbred mice, similar genes referenced in ontologies related to embryonic development and neurogenesis were identified [41]. Taken together, these studies indicate that genes involved in the development of the central nervous system are susceptible to stochastic epigenetic variation. It is therefore legitimate to hypothesize that different lifestyle or environmental factors induce DNA methylation changes on neurological gene loci. Consistently, results from our group support specific effects of lifestyle on epigenetic changes on genes controlling the development of the central nervous system [9]. Spermatozoa collected before and after a six-week endurance exercise intervention, as well as after three months without exercise training, showed that DNA methylation changes occur close to genes related to neurogenesis and, noticeably, with a higher enrichment at the trained, compared to the untrained state [9]. This reveals that time is not the main factor driving epigenetic remodeling and suggests that epigenetic changes are induced by external influences. This is consistent with the previously formulated theory that cells respond to environmental stressors via increased epigenetic variation [41]. Yet, it is legitimate to question whether a technical bias is not at the origin of these results, since studies reporting epigenetic changes on genes related to the development of the central nervous system exclusively used DNA methylation arrays or Reduced Representation Bisulfite Sequencing [7], [8], [41], [42], two techniques that overestimates CpG-rich regions. Results could then be explained by the fact that genes referenced in gene ontology terms related to the development of the central nervous system have high CpG density regions (referred to as CpG islands) compared to the rest of the genome. However, gene ontology analysis corrected by the overestimation of genes with high CpG density did not abolish enrichment for the ontology term nervous system development in obese men [8], which rules out technical biases. Other causes of misinterpretation, such as the accuracy of genes referenced in ontology terms, should be explored and systematic use of whole genome bisulfite sequencing, which does not enrich for CG rich regions, should permit a conclusion on the actual existence of environmentally-induced epigenetic variation at genes related to brain development in sperm. However, it is disputed in the field whether epigenetic variation causes genetic variation or vice versa. In a pioneering study examining the link between genetic variation and DNA methylation, evidence for allele-specific gene expression outside of imprinted regions was found [43]. Others have suggested that genotypes influence the majority of the heritable regions of differential methylation. Investigations of a three-generation family as well as unrelated individuals discovered that heterozygous single nucleotide polymorphisms (SNPs) associate with different methylation patterns, correlating to gene expression [44]. Surprisingly, when examining the effects of genotype on DNA methylation, genetic variants appeared to have greater impact than imprinting [44]. Moreover, a mathematical model supports the hypothesis that epigenetic variation may be genetically selected [41]. Of course, the role of genetic mutations in epigenetic modifiers should not be discounted for either, as it has the potential to cause profound changes in the overall epigenetic machinery of cells (reviewed in [45]). Altogether, it is quite difficult at this stage to disentangle causality in the relationship between genetics and epigenetics. Indubitably, genetics and epigenetics do interfere with one another, but further investigations in this area are warranted.
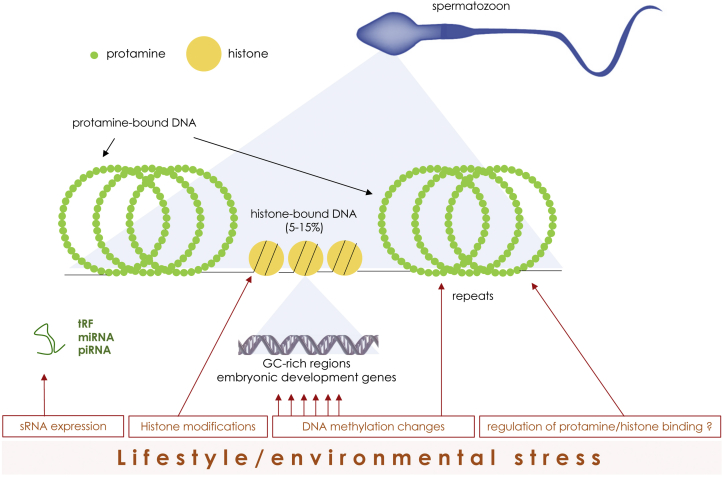
Figure 2.
Overview of epigenetic marks susceptible to be remodeled with environmental insult. A simplified secondary structure of the sperm genome is represented, with the histone-bound DNA fraction accounting for less than 15% of the genome. DNA methylation remodeling is enhanced at CG rich, histone-bound fractions in sperm and is also found at repetitive elements. The positioning of histone relative to protamines may also be regulated by environmental factors. Histone modifications at specific loci are also changed after nutritional stress. Expression of small RNA (sRNA) such as tRNA fragments (tRF), microRNA (miRNA) and PIWI-interacting RNA (piRNA) is affected by lifestyle or environmental stress.
Environmentally-induced methylation changes can also be quite gene-specific. For instance, in a model of type 2 diabetes, spermatozoa harbored DNA methylation changes at genes related to glucose metabolism and type 2 diabetes [46]. In another example, odor fear conditioning with acetophenone induced hypomethylation of the acetophenone receptor gene Olfr151 [10]. These studies suggest that environmental factors (or at least certain types) can trigger a very specific epigenetic response serving a specific physiological adaption. In addition, these findings challenge the theory that epigenetic response to an environmental insult is exclusively non-specific, to allow for, at the individual organism or cell level, a distinct physiological response compared to the rest of the population. Conversely, other studies have reported that environmental insults are associated with DNA methylation changes with unclear function-specific changes. Notably, exposure of plastic-derived compounds bisphenol-A (BPA), bis(2-ethylhexyl)phthalate (DEHP) and dibutyl phthalate (DBP) to several generations of males was associated with differential methylation at 197 promoters, but without enrichment of specific gene pathways [11]. Similarly, mice on a low protein diet exhibited DNA methylation changes in sperm that were not associated with any specific gene pathway [3]. Sperm from animals undernourished in utero showed a marked DNA hypomethylation profile, with enrichment in intergenic regions and CpG islands and underrepresentation of DNA methylation changes in long interspersed nuclear elements (LINE) and short interspersed nuclear elements (SINE) [5]. Hypomethylation was present at the germline level, supporting that in utero undernutrition disrupted DNA remethylation during germline reprogramming, but no clear coordinated changes in any specific gene pathway were identified [5]. High fat diet challenge in rats also induces remodeling of the sperm DNA methylome on 92 genomic regions located at proximity of genes enriched for unspecific gene function such as cellular localization, transport, and metabolic processes [6]. Eighteen regions were differentially methylated in both sperm of founders on HFD and their offspring, notably at the proximity of the Slc3a2 (solute carrier family 3 (amino acid transporter heavy chain), member 2), Tbrg4 (transforming growth factor beta regulator 4) and Mfsd7 (major facilitator superfamily domain containing 7), but, in this case, the paucity of genes in common between the two generations did not allow for gene enrichment analysis [6].
The diversity of epigenetic responses under environmental influences could be caused by many factors including timing and type of environmental stimulus, the methodology used to detect DNA methylation changes, the bioinformatic parameters used, the species, the site of sperm collection (epididymis vs. ejaculates), purification or not of motile spermatozoa, etc. Alternatively, the diversity in response to dietary influences could indicate that the signal detected corresponds to noise levels as suggested in a genome-wide investigation of the effect of high-fat diet on sperm DNA methylation in the mouse at extreme sequencing depth [47]. This conclusion is based on the assumption that methylation differences below 10% cannot account for the penetrance of phenotype, i.e. the phenotypic consistency in the next generation offspring [47]. However, another model by which modest epigenetic changes occurring at distinct loci are integrated into a unified phenotypic response remains possible, particularly when focusing on fundamental cell processes like metabolic processes. Indeed, the interconnection of energy metabolites combined with the fact that all cellular processes are dictated by energy metabolism implies that a perturbation of only few genes is likely to have metabolic implications. Of interest, the observation that psychological stress in fathers perturbs metabolism in the offspring supports that various environmental stimuli are integrated into a non-specific, yet consistent, metabolic response in the offspring [18]. Conversely, nutritional challenge induces epigenetic variation at chromatin-modifying genes in sperm that, in turn, globally affects the adipose tissue transcriptome of the offspring and leads to a homogenous body weight phenotype [48]. It is noteworthy that many intergenerational studies have focused on metabolic readouts on the offspring such as glucose tolerance and gene expression changes in metabolic organs [3], [4], [5], [6], [18], [46]. While the main purpose of these studies was to test the hypothesis that paternal influences affect the offspring phenotype through epigenetic inheritance, we believe that the nature of the phenotypic readout (metabolic) could not allow for exact determination of the specificity of the response. It is possible that other phenotypic responses, for example behavioral characteristics, are differently affected by the various environmental exposures used. Expanding the panel of phenotypic characterization in offspring in intergenerational studies, with a paralleled profiling of the epigenetic blueprint in spermatozoa, will allow the determination of the relationship between specific epigenetic changes in sperm and the implication in the offspring.
3. Sperm chromatin
In comparison to somatic cells, spermatozoa harbor a very distinct chromatin structure and organization. DNA in sperm is bound to protamines, which replace the vast majority of histones and allow a DNA packaging structure six times more dense, protecting spermatozoal DNA to extracellular stressors (Figure 2) [49]. Protamination during spermatogenesis thus results in removal of most epigenetic information carried by histones. Yet, remaining histone proteins are essential and may poise genes for early embryonic development.
3.1. Histone retention and modification
Replacement of histone by protamines results from a stepwise process during spermatogenesis, which gradually leads to removal of 85–95% of histones, depending on the species [50]. Replacement of histone by protamines is facilitated by transition proteins and H2A.L.2, which enables loading of protamines onto nucleosomes. Protamines, in turn, are responsible for histone eviction [50]. Histone hyperacetylation participates in histone removal and bioavailability of acyl-CoA has been proposed to impact sperm genome compaction [51]. Butyrylation, another post-translational modification of histones, can occur concomitantly to histone hyperacetylation during spermatogenesis, preventing acetylation-dependent histone removal and ultimately delaying replacement by protamines, leading to the modulation of chromatin compaction [50], [52], [53]. Thus, environmental factors could control genomic structure in mature spermatozoa through the regulation of acyl-CoA availability, acetylation and butyrylation.
Analysis of endonuclease-resistant regions of the sperm genome has revealed information about the packaging level of chromatin. When used in association with the detection of the H4K12ac mark, this analysis has provided evidence that nucleosomes are not randomly retained but instead located at specific genomic locations in the sperm genome [54]. Seminal studies have reported that histones are retained at genic regions, preferably at genes controlling embryonic development [55], [56]. However, comparison of nuclease accessibility between mouse embryonic stem cells and spermatozoa showed that most retained histones in sperm are primarily located in gene-poor regions [57]. This was confirmed by an independent group who showed that histone retention occurs mainly at distal intergenic regions, repeats, and retrotransposons [58]. Technical reasons, potentially related to the concentration of endonuclease or duration of digestion, could be at the origin of the discrepancies in the estimation of the specific genomic distribution of histones retention in sperm, as suggested [59]. Regardless of these technical considerations, a proportion of histones remain at promoter sequences, notably sequences recognized by the CCCTC-binding factor (CTCF) and near genes involved in embryonic development [54], [57]. Using antibody staining for histone 3 replication variants, it was found that protamines are replaced by maternal nucleosomes after fusion of the gametes, whereas the retained paternal histones remained associated to the paternal genome [60]. Interestingly, histone retention occurs predominantly at regions of high-CpG density and low DNA methylation, for instance at promoters of housekeeping and development-regulating genes [8], [55]. Of interest, histones are also retained at genes involved in sensory perception [54], which may provide a molecular mechanism by which environmental insults in the father alter sensory perception in the next generation [10]. Consistently, genomic regions with retained histones were enriched for differentially methylated regions in spermatozoa from obese as compared to lean men [8]. Together, these findings support a specific role of histone retention in embryonic development and also a potential effect of lifestyle and environmental factors, which could affect developmental programming through modulation of histone positioning in sperm.
Diet-induced variation of specific histone marks in mature sperm has also been reported [3]. Intensity of the H3K27me3 at the mitochondrial gene Monoamine oxidase a (Maoa) and elongation factor Tu GTP binding domain containing 1 (Eftud1a) is higher in sperm from mice who were fed a three-month low-protein diet [3]. Repetitive regions in the sperm genome are described to be enriched in the H3K27me3 mark, suggesting that silencing of repetitive elements is, at least in part, under the control of histone retention and modification [57]. This notion was later demonstrated in the context of early gene expression in the embryo [61]. The finding that histones are randomly retained in infertile patients, and that both H3K4me and H3K27me marks are decreased, further supports that histone positioning and modification is important for normal sperm function and definitely disqualifies the idea by which remaining histones are non-functional remnants of spermatogenesis [56].
Gradual protamination of sperm DNA during spermatogenesis passively erases epigenetic signals that were carried by the removed histones. Thereby, protamination participates in epigenetic reprogramming in much the same way as DNA demethylation during spermatogenesis. The environmental influences modifying protamination and protamine positioning, therefore, may constitute an epigenetic signal in itself that is equally important to histone modification and DNA methylation changes to influence transcriptional activity after fertilization. In addition to protamine- and histone-bound DNA, specific sperm DNA regions are attached to the nuclear matrix, in so-called matrix attachment regions (MARs), which constitute an additional layer of chromatin structure information in sperm [62], [63]. At the functional levels, MARs are essential to normal embryonic development, and implicated in DNA replication and formation of the male pronucleus after fertilization [62]. Thus, the distribution of protamines along sperm DNA drives not only histone-related epigenetic signals but, in parallel, defines the specific DNA regions engaged in MARs and, subsequently, may tune various early developmental events post-fertilization. Yet, the influence of lifestyle and environmental factors on protamine positioning remains vastly unknown. Concentration of factors contained in seminal fluid such as zinc may modulate protamine formation and subsequently chromatin structure in a very dynamic fashion [64]. In addition to their positively-charged groups, which neutralize the negative charge of the DNA backbone, protamines contain thiol groups that are engaged into zinc-stabilized bridges, allowing high degree of compaction [65]. In vitro evidence suggests that variations in extracellular zinc concentration may affect zinc content in the sperm head and influence DNA compaction [65]. In vivo, protamine structure may be modulated when entering the ooplasm, thereby allowing decompaction of paternal DNA. In addition, environmentally-induced changes in zinc concentration in seminal fluid, various time of exposure to seminal fluid during assisted reproduction techniques, or various time in the female tract may influence zinc concentration in the sperm head and chromatin structure. However, additional studies are warranted to determine the influence of the seminal fluid and the female tract on protamine structure and organization in sperm.
More evidence support that nutritional factors change histone marks in sperm in various model organisms. Male Drosophila fed a high sugar diet showed transcriptional derepression in sperm in a chromatin-state-specific manner [48]. Response to paternal diet was dependent on sperm chromatin plasticity, notably through H3K9me3-and H3K27me3-dependent silencing [48]. Another study using mice as a model demonstrated the role of chromatin marks in sperm on epigenetic inheritance [66]. Remodeling of sperm H3K4me2 marks using a transgenic model of KDM1A histone lysine 4 demethylase overexpression during mouse spermatogenesis showed reduced survival and developmental abnormalities in three subsequent generations [66]. Increased KDM1A activity during sperm development was associated with altered histone methylation and sperm RNA expression, which suggests that histone modification could participate, at least in part, in the observed transgenerational effects [66]. Interestingly, the fact that remodeling of the sperm H3K4me2 marks was associated with expression of small non-coding RNA suggest cooperation between the different epigenetic marks in epigenetic inheritance [66]. The notion of epigenetic cooperation is further supported by a previous study showing that early traumatic stress induces transgenerational alteration of behavioral and metabolic phenotype [18]. In this investigation, early traumatic stress induced remodeling of sperm-borne miRNA expression in F0 but not F1 sperm, despite propagation of the behavioral and metabolic phenotype to the second (F2) generation [18]. Since microinjection experiments demonstrated the role of sperm RNA in the transgenerational response to early traumatic stress [18], this study suggests that other epigenetic signals than RNAs cooperate to propagate transgenerational effects.
The role of histone positioning and modification on epigenetic inheritance is not established. Evidence that some histone marks in sperm are erased post-fertilization [67] indicates that, if histone signals carried by paternal DNA are involved in epigenetic inheritance, these signals are not propagated to adult tissues. Still, histone marks in sperm may participate in intergenerational effects, but more likely through altering the very early stage of developmental reprogramming.
4. Sperm-borne small RNAs
Small RNAs (sRNAs) function as epigenetic regulators of gene expression either by interaction with the translation machinery and/or by inducing degradation of their complementary mRNA targets. During spermatogenesis, the cytoplasm along with most of its transcripts is depleted as a residual body, leaving the spermatozoa inert at the translational level, as supported by the absence of intact rRNA [68]. However, some transcripts are left as a pool, consisting of both coding and non-coding RNAs in a quantity of about 200 times less than in somatic cells [69]. In the past, the remaining RNAs were thought to be non-functional, remnant molecules from earlier steps of spermatogenesis, or simply contamination from somatic cells. With development of new methods and analysis tools, it is now clear that the remaining transcripts are in fact selectively retained, suggesting that sperm-borne RNAs are functional. Among these are fragmented and non-degraded mRNAs, pi (piwi-interacting)RNAs, t (transfer)RNA fragments (tRFs), si (small-interfering)RNAs, mi (micro)RNAs and lnc (long non-coding)RNAs. A role for sperm transcripts in embryonic development was suggested by Krawetz and colleagues, who showed that paternal messenger RNA is delivered to the oocytes at fertilization [70]. Another study showed that, by injecting RNA coding for phospholipase C-zeta into mouse oocytes, a portion of these transcripts were translated into functional proteins [71]. A seminal study later implicated sperm RNAs in the transmission of non-Mendelian phenotypes [72]. Together, these studies have demonstrated a role for sperm-borne transcripts in the early development of the embryo and suggested a role in epigenetic inheritance.
Piwi-interacting RNAs (26–31 nucleotides) are specifically expressed in the gonads, and are thought to silence transposable elements, especially in the germline, protecting the integrity of the genome. PiRNAs mediate their effect through PIWI-proteins, a subfamily of the argonaute family of proteins. In addition to their role in male fertility [73] piRNAs are involved in epigenetic inheritance in Drosophila and C. Elegans [74], [75]. Spermatozoa from obese humans and rats on a high-fat diet have an altered piRNA signature compared to their lean counterparts, which could point to a role for piRNAs in epigenetic inheritance of metabolic dysfunction [6], [8], [9]. In otherwise healthy obese humans of the fertile age, expression of 37 piRNAs was altered in spermatozoa, as compared to spermatozoa from lean males [8]. Computational target prediction of these piRNA-based complementary seeding sequences to mRNA, returned that the Cocaine and Amphetamine Regulated Transcript (CART) gene, a negative regulator of food intake involved in obesity, was a putative target (reviewed in [76]). Data exist showing that lifestyle factors can modulate piRNA in sperm. Six weeks of exercise training alters the expression of 6 different piRNAs in spermatozoa from lean and healthy young individuals [9]. Importantly, and as discussed earlier, altered piRNA expression was not due to a simple effect of collection time between the untrained and the trained state, since piRNA expression changes were reverted following a 3-month detraining period. These data indicate that lifestyle effects are indeed specific and that sperm piRNA expression is dynamic. Target prediction of piR-hsa-28160, one of the differentially expressed piRNAs in response to exercise, returned the ILF3/NF90-interacting RNA Small ILF3/NF90-associated RNA (SNAR), a regulator of the let7 family member let7a, which is a miRNA family known to be involved in inflammation, glucose metabolism [77], [78], [79] and, more recently, epigenetic inheritance as mentioned in this review [6], [20].
Other important molecules belonging to small RNAs and thought to contribute to epigenetic inheritance are tRNA fragments, or tRFs. tRFs vary in length (10–45 nucleotides) and are mainly derived from the 5′ end of either mature tRNA or pre-tRNA. A role of sperm-borne tRFs in epigenetic inheritance was suggested by the observation that tRFs participate in piRNAs synthesis in mouse gametes and zygotes [80]. More recently, several groups have provided compelling evidence that tRFs play a role in epigenetic inheritance [17], [20]. A low-protein diet remodels the expression of several fragments of tRFs in spermatozoa from founders at the origin of parental effects [20]. The potential role of sperm tRFs was suggested via the intracytoplasmic sperm injection (ICSI) of sperm from founders on a low-protein diet. These experiments identified tRF-Gly-GCC as a repressor for genes linked to an endogenous retroelement active in the preimplantation embryo [20]. A similar approach was used that discovered altered tRNA fragments in sperm from mice fed a high-fat diet. This study further demonstrated that injection of tRNA fragments isolated from HFD-sperm into control zygotes resulted in metabolic disorders of the offspring alongside altered gene expression of metabolic pathways in the early embryos [17]. Together, these results convincingly indicate that sperm tsRNAs could play a role in the transmission of dietary-induced epigenetic signals from father to offspring, through modulation of embryonic development.
Micro RNAs (miRNA) are the most well-characterized subtype of sRNAs. MiRNAs are constituted of a 22 nucleotide-long, hairpin-structured RNA and are well described in regulation of gene expression. In a canonical pathway, miRNAs downregulate gene expression by targeting mRNAs on their 3′ UTRs, which results in either translational repression or degradation of the mRNA [81]. One miRNA can target several mRNAs; one mRNA can be targeted by several miRNA. This means that miRNA-mRNA networks can be complex to resolve and the function of individual miRNAs difficult to identify. Several studies have identified the function of sperm-borne miRNAs. For example, miR-34c is the most abundant miRNA in human sperm and was shown to be required for the first cell divisions in mouse embryos [82]. Moreover, several groups have provided evidence that miRNAs play an important role in the provision of signals for early embryonic development [18], [19], [72], [83], [84], [85]. Mice who received a zygotic injection of a combination of nine miRNAs that were altered in paternal sperm after exposure to chronic stress had offspring that developed altered stress-responses closely resembling the phenotype of the father [86]. More recently, miRNA let-7c was identified as a potential carrier of transgenerational epigenetic inheritance induced by paternal high-fat diet [6]. The let-7 miRNA family is known to control lipid and glucose metabolism, and was found to be differentially expressed in spermatozoa of HFD-fed rats as well as in the spermatozoa of their offspring [6]. As a response to paternal obesity, offspring developed disturbances in glucose and lipid metabolism, together with observed altered expression of let-7c in liver, white adipose, and muscle tissue of the adult offspring [6], [87]. Interestingly, the let-7 family of miRNAs appears to be implicated in other models of diet-induced transgenerational inheritance as well. When exposing mice to a low-protein diet in the period up to conception, several let-7 species are down-regulated in spermatozoa associated with an altered phenotype of the offspring [20]. These discoveries, made by distinguished laboratories, indicate that let-7 family members are diet-responsive sperm sRNAs that mediate intergenerational inheritance.
While some sRNA molecules appear to respond to a variety of stimuli, for example nutritional stress, it is noteworthy that the composition of the different subgroups of sRNAs is found to vary among animal species. In humans, piRNAs, followed by tRNAs and miRNAs, constitute the most abundant sncRNA subgroups [8], [88], whereas in rodents, tRNAs is by far the most abundant group when considering the content of fully mature spermatozoa [20], [85]. This composition changes, however, throughout maturation of the spermatozoon, with piRNAs as the most abundant form in the first processes of spermatogenesis then slowly decreasing in quantity whereas tRNAs increase in abundance as maturation proceeds [17]. While stage-dependent cleavage of specific tRNAs may be at the origin of sperm tRFs, evidence exists supporting that epididymal epithelial cells provide tRNAs from epididymosomes, during the travel of spermatozoa in the epididymis [20]. The difference in sRNA composition between species may of course be evolutionarily established, but it is possible that differences in methods for the collection of mature spermatozoa are involved. In human studies, semen cells are collected and isolated from sperm ejaculates, whereas most rodent studies harvest spermatozoa directly from the epididymis. As mentioned, epithelial cells can deliver sRNAs to the mature spermatozoa [20]; therefore, it is possible that methodological differences when collecting spermatozoa may account for composition disparities. Indeed, spermatozoa in ejaculates have been through the entire male genital tract, while collection of spermatozoa in various places of the epididymis (cauda, corpus and caput) may result in immature spermatozoa, in terms of RNA content. Yet, several groups have succeeded in partly replicating phenotypic alterations obtained by natural mating through generations using IVF/ICSI techniques, or microinjection of sperm-isolated RNA, where sperm was collected in the epididymis [17], [18], [20], [84]. However, among other more obvious confounders, method dissimilarities make it difficult to deduct findings from one species to the next. A way to account for this could be semen collection by rectal electric stimulation in rodents, but this method is ethically and methodologically challenging to establish, due to its stressful and painful nature.
Despite the above-mentioned evidence that sperm-borne sRNAs contribute to epigenetic inheritance, the actual mechanisms of action, i.e. in the developing embryo, are still insufficiently defined. The ongoing development of new technologies such as transcriptomic profiling at the single- or few cell level, has however greatly facilitated the investigations about the transcriptomic role of sRNAs. In addition, accumulating evidence suggests sperm sRNAs influence other epigenetic marks. For instance, in gene inactivation, where sRNAs contribute to the recruitment of histone methylases, such as during establishment of X chromosome inactivation and at imprinted regions [89], [90]. Additionally, sRNAs can affect DNA methylation levels at specific loci through interaction with DNA methyltransferases [91], [92]. Argonaute proteins and different histone modifications co-immunoprecipitate, which leads to the speculation that sRNAs alter chromatin structure at specific target sites (reviewed in [93]). A cross-talk between epigenetic marks would imply that investigations focusing on the transmission of epigenetic signals from the gamete to the somatic cells of the offspring should be exhaustive to all the main epigenetics marks, DNA methylation, histone modification and sRNA expression, which represents a very time-consuming and financially demanding strategy at the present date.
5. Evolutionary considerations
The high CpG density of genes referred as “control of neurological processes” in ontology databases raises the additional question of a possible role of epigenetic variation in genetic variation. As suggested earlier [94], methylated cytosines are thought to constitute hot spots for deamination (where methylated cytosines are converted into thymines) and mutation. The notion that methylated cytosines are mutagenic has been at the origin of a theory that hypomethylation is a mechanism protecting DNA sequence integrity [94]. This theory is supported by the computational observation that SNPs in the human genome occur at a higher frequency in CpG sites [95]. To our knowledge, only one study, performed in E. Coli, has experimentally addressed the mutagenicity of methylated cytosines, whereby deamination rates of 5-methylcytosine within double stranded DNA was higher than that of cytosine [96]. The mutagenicity of cytosines is further supported by an in silico study, in which comparison of orthologous CpG-rich sequences across primates suggests that an active process of CpG conservation exists at specific genomic regions [95]. Notably, the imprinted regions H19 and GTL2/DLK1 in the male germ line show lower CpG loss rates than expected [95]. Moreover, CpG islands located in exonic regions also show low cytosine divergence rates [95]. These observations could altogether indicate that some genomic regions are prone to epigenetic variation and subsequent genetic mutations, while other regions are safe-guarded for methylation-induced mutagenicity. Our data shows that epigenetically variable regions in obese men overlap with variants previously identified in genome-wide association studies of obesity. This would favor that epigenetic variation induced by one's lifestyle also occurs at hotspots for genetic variation [8]. This process has been described by the developmental biologist Conrad Waddington as “genetic assimilation” [97]. The evolutionary implication of epigenetic variation as a source of genetic variation is appealing, since it would reconcile two theoretical models of species evolution that are often opposed; evolution through Lamarckian inheritance, where the genome is directly modified by environmental stressors and then transmitted transgenerationally, and evolution through Darwinian selection, where continuous genetic variation exists within a population and leads to selection at the extremes of phenotype. Mathematical modeling, however, indicates that epigenetic variation may be genetically selected [41], which is in contradiction with the opposite model in which epigenetic variation is a cause of genetic variation. While both models can cooperate, more experimental evidence, other than computationally-based, should be provided to address the mutagenicity of regions subjected to environmentally-induced epigenetic variation and the evolutionary implications.
6. Concluding remarks
While current research points to epigenetic regulation as a feasible control mechanism for paternal inheritance, it is challenging to rule out confounding (non-gametic) factors that could be at play in parental effects. For example, exchange of microbiota at time of mating may participate in an altered developmental milieu for the embryo. Also, factors of the paternal seminal fluid may theoretically affect fetal environment before or at the fusion of gametes, as it is known to contain potential signaling factors such as hormones. Interestingly, elevated seminal levels of insulin and leptin have been detected in obese men [98], and mass spectrometry analyses has revealed more than 20 proteins differentially expressed in seminal plasma of smokers [99]. Spermatozoa are not in contact with the full fraction of seminal fluid before ejaculation, but as it often takes few hours before the spermatozoa reach the oocyte, the spermatozoa are susceptible to be exposed to these factors many hours before fertilization. The common assumption is that DNA methylation changes in sperm occur exclusively prior to ejaculation. The presence of DNA methylation-modifying enzymes in mature sperm [100], [101] and the fact that dynamic DNA methylation can occur in non-dividing cells [102], would however support the provocative hypothesis that sperm DNA methylation can be altered in mature sperm. In addition, somatic cells may deliver sRNA-content directly to the mature spermatozoon as suggested [20]. Thus, mature sperm cells, even after ejaculation, may be epigenetically remodeled by the seminal fluid, or by factors or cells of the female genital tract.
6.1. Are diet-induced epigenetic marks transmitted to the offspring?
Understanding the mechanisms by which environmentally-driven changes of the sperm epigenome affect the metabolism of the offspring represents a very fundamental gap to fill. Given the importance of the sperm epigenome in embryonic development, it is very plausible that any modulation of the gametic epigenetic signal will alter the developmental programming of the embryo. Such alteration would be amplified through the so-called Waddington model, according to which small epigenetic changes that occur early during cell differentiation drive tissue-specific gene expression [97]. The characterization of the environmentally-driven changes passed down to the stem cells of the offspring would be highly informative.
It is tempting to postulate that chromatin and DNA methylation marks that vary under environmental stress serve a function post-fertilization and are not simply decorative. This assumption may be hard to test, however, since examination the effect of specific DNA methylation patterns in sperm on the developmental programming of the embryo remains technically challenging. Editing tools using the nuclease-free CRISPR-Cas9 system, for example the CRISPR-Cas9 fused to DNA methyltransferase 3A [103] or the demethylation-participating enzyme TET1 [104] may be used. Yet, if DNA methylation signals localized in a plethora of loci are differentially methylated, these tools may not be able to target all genomic sites in the same sperm cell.
On the other hand, microinjection experiments using sRNA have convincingly provided evidence that sRNA can carry parental effects to the next generation [10], [17], [18], [19], [20]. However, the fact that many of these experiments led to altered metabolism in the offspring does not indicate high-specificity [17], [19], [20]. As discussed earlier in this review, metabolic features like glucose and insulin tolerance constitute readouts that are likely to be affected by every slight alteration in embryo programming and therefore, microinjection of any sRNA species is likely to affect metabolism in one way or another. Using a sRNA type that does not vary under the specific environmental insult as a negative control for microinjection experiment, rather than scrambled molecules for example, would help determine the specificity of sRNA in epigenetic inheritance.
At present, in the field of epigenetic inheritance, many questions are still left unanswered. The mechanism of how epigenetic factors are established and altered in the germline, as well as in somatic cells, are not well understood. Causality is yet to be explained, and it is still highly debated to what extent genetic and epigenetic factors interplay in the environmentally influenced manipulation of gene expression and phenotype. An overview of the diversity of epigenetic responses to lifestyle or environmental insults (Table 1) is prompting researchers in the field to better understand the contribution of the timing and duration of exposure, sex, species, and the technology used on epigenetic changes in sperm (Figure 1). Future research efforts may be able to identify a unified epigenetic remodeling response to lifestyle stress across species. Understanding the role of environmentally-driven epigenetic changes in gametes on the phenotype of the offspring constitutes not only a fascinating biological question on its own but also represents a moral obligation for the health of future generations.
Table 1.
Selection of studies providing evidence of transgenerational epigenetic inheritance in murine models.
| Author, year | Intervention | Species, generations investigated | Epigenetic marks studied | Technique(s) used |
|---|---|---|---|---|
| Carone et al., 2010 | Paternal low-protein diet before conception | Mouse, F0 + F1 | Sperm, F0: DNA methylation and small ncRNA Liver, F1: DNA methylation, small ncRNA |
Sperm, F0: MeDIP-seq, bisulfite seq, RNA microarray Liver, F1: DNA microarray, RRBS, bisulfite seq, HT-seq of miRNA |
| Radford et al., 2014 | Maternal caloric restriction during gestation (F0) | Mouse, (F0) + F1 + F2 | Sperm, F1: DNA Methylation Liver/brain, F2: DNA methylation |
Sperm, F1: MeDIP-seq, bisulfite pyrovateseq F2, liver/brain: Bisulfite pyrovateseq |
| de castro Barbosa et al., 2016 | Paternal high-fat diet before conception | Rat, F0+F1+F2 | Sperm, F0+F1+F2: DNA methylation, small ncRNA Metabolic tissues, F1+F2: small ncRNA |
Sperm, F0+F1+F2: MBD-seq, RNA-seq Metabolic tissue, F1+F2: RNA-seq |
| Dias & Ressler, 2014 | Paternal odor fear conditioning | Mouse, F0+F1+F2 Cross-fostering, IVF |
Sperm, F0+F1: DNA methylation, chromatin | Sperm, F0+F1: Bisulfite seq Sperm, F0: Native-ChIP Main olfactory epithelium, F1+F2: Bisulfite seq |
| Manikkam et al., 2013 | Maternal exposure to endocrine disruptors during gestation (F0) | Rat, F0+F1+F3 | Sperm, F3: DNA methylation | Sperm, F3: MeDIP-Chip |
| McPherson et al., 2015 | Paternal exposure to high-fat diet and/or exercise before conception | Mouse, F0+F1 | Sperm, F0: microRNA | Sperm, F0: MicroRNA Array |
| Wei et al., 2014 | Paternal high-fat diet + low-dose streptozodocin before conception | Mouse, F0+F1+F2 | Sperm, F0: DNA methylation Pancreas, F1: DNA methylation Pancreas, F2: DNA methylation |
Sperm, F0: MeDIP-Seq, bisulfite seq Pancreas, F1: MeDIP-Seq, bisulfite seq Pancreas, F2: MeDIP-qPCR |
| Gapp et al., 2014 | Paternal traumatic stress in early life | Mouse, F0+F1+F2 Microinjection of sperm RNA into ctrl zygotes |
Sperm, F0+F1+F2: Small ncRNA Brain + serum, F0+F1+F2: Small ncRNA |
RNA-seq (all tissues) |
| Sharma et al., 2016 | Paternal low-protein diet before ceonception | Mouse, F0+F1 IVF, ICSI/microinjection of specific RNA into ctrl zygote |
Sperm, epididymis, testis, F0: ncRNA Single embryo, F1: ncRNA |
Sperm, epididymis, testis F0: RNA-seq Single embryo, F1: RNA-seq |
| Chen et al., 2016 | Paternal high-fat diet before conception | Mouse, F0+F1 Microinjection of sperm head/total RNA/specific RNA into ctrl zygotes |
Sperm, F0: ncRNA Pancreas, embryo/blastocyst, F1: ncRNA, DNA methylation |
Sperm, F0: RNA-seq Pancreas, F1: RNA-seq, RRBS Single embryo/blastocyst: RNA-seq |
| Grandjean et al., 2009 | Microinjection of specific microRNA into ctrl zygotes | Mouse, Microinjection of specific microRNA into ctrl zygotes (F1)+F2+F3 | Embryo + adult tissue, F1: Specific microRNA, chromatin | Embryo + adult tissue, F1: RT-qPCR, ChIP |
| Wagner et al., 2008 | Microinjection of specific microRNA into ctrl zygotes | Mouse, Microinjection of specific microRNA into ctrl zygotes (F1) | F1, heart: Specific microRNA | Heart, F1: RT-qPCR |
| Cropley et al., 2016 | Paternal congenic obesity/pre-diabetes mouse model | Mouse, F0+F1+F2+F3 | Sperm, F1: ncRNA | Sperm, F1: RNA-seq |
| Grandjean et al., 2015 | Paternal high-fat/high-sugar diet before conception | Microinjection of testis and sperm total RNA/specific microRNA into ctrl zygotes (F1) | Testis, sperm, F0: ncRNA | Testis, F0: RNA-seq Testis, sperm, F0: RT-qPCR |
Seq = sequencing; MeDIP = Methylated DNA immunoprecipitation; RRBS = Reduced representative bisulfite sequencing; (RT-)qPCR: (reverse transcription) quantitative polymerase chain reaction; MBD = Methyl binding domain; ChIP: Chromatin immunoprecipitation.
Acknowledgements
This work has been funded by the Novo Nordisk Foundation Endocrinology Nordic Grant 21448. The Novo Nordisk Foundation Centre for Basic Metabolic Research is an independent research Centre at the University of Copenhagen partially funded by an unrestricted donation from the Novo Nordisk Foundation.
Conflict of interest
None to declare.
References
- 1.Noble D. Evolution evolves: physiology returns to centre stage. The Journal of Physiology. 2014;592(11):2237–2244. doi: 10.1113/jphysiol.2014.273151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pembrey M.E. Sex-specific, male-line transgenerational responses in humans. European Journal of Human Genetics. 2006;14(2):159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- 3.Carone B.R. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143(7):1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng S.F. Paternal high-fat diet consumption induces common changes in the transcriptomes of retroperitoneal adipose and pancreatic islet tissues in female rat offspring. FASEB Journal. 2014;28(4):1830–1841. doi: 10.1096/fj.13-244046. [DOI] [PubMed] [Google Scholar]
- 5.Radford E.J. In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science. 2014;345(6198):1255903. doi: 10.1126/science.1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Castro Barbosa T. High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Molecular Metabolism. 2016;5(3):184–197. doi: 10.1016/j.molmet.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denham J. Genome-wide sperm DNA methylation changes after 3 months of exercise training in humans. Epigenomics. 2015;7(5):717–731. doi: 10.2217/epi.15.29. [DOI] [PubMed] [Google Scholar]
- 8.Donkin I. Obesity and bariatric surgery drive epigenetic variation of spermatozoa in humans. Cell Metabolism. 2016;23(2):369–378. doi: 10.1016/j.cmet.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Ingerslev L.R. Endurance training remodels sperm-borne small RNA expression and methylation at neurological gene hotspots. Clinical Epigenetics. 2018 doi: 10.1186/s13148-018-0446-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dias B.G. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nature Neuroscience. 2014;17(1):89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manikkam M. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One. 2013;8(1):e55387. doi: 10.1371/journal.pone.0055387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McPherson N.O. Preconception diet or exercise intervention in obese fathers normalizes sperm microRNA profile and metabolic syndrome in female offspring. American Journal of Physiology-Endocrinology and Metabolism. 2015;308(9):E805–E821. doi: 10.1152/ajpendo.00013.2015. [DOI] [PubMed] [Google Scholar]
- 13.Ge Z.J. Maternal diabetes causes alterations of DNA methylation statuses of some imprinted genes in murine oocytes. Biology of Reproduction. 2013;88(5):117. doi: 10.1095/biolreprod.112.105981. [DOI] [PubMed] [Google Scholar]
- 14.Ge Z.J. DNA methylation in oocytes and liver of female mice and their offspring: effects of high-fat-diet-induced obesity. Environmental Health Perspectives. 2014;122(2):159–164. doi: 10.1289/ehp.1307047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stilling R.M. Microbial genes, brain & behaviour - epigenetic regulation of the gut-brain axis. Genes, Brain and Behavior. 2014;13(1):69–86. doi: 10.1111/gbb.12109. [DOI] [PubMed] [Google Scholar]
- 16.Curley J.P. Epigenetics and the origins of paternal effects. Hormones and Behavior. 2011;59(3):306–314. doi: 10.1016/j.yhbeh.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Q. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351(6271):397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 18.Gapp K. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nature Neuroscience. 2014;17(5):667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grandjean V. RNA-mediated paternal heredity of diet-induced obesity and metabolic disorders. Scientific Reports. 2015;5:18193. doi: 10.1038/srep18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma U. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351(6271):391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ventura-Junca P. In vitro fertilization (IVF) in mammals: epigenetic and developmental alterations. Scientific and bioethical implications for IVF in humans. Biological Research. 2015;48:68. doi: 10.1186/s40659-015-0059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaenisch R. DNA methylation and imprinting: why bother? Trends in Genetics. 1997;13(8):323–329. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- 23.Bird A.P. Methylation-induced repression–belts, braces, and chromatin. Cell. 1999;99(5):451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 24.Ito S. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tahiliani M. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodcock D.M. The majority of methylated deoxycytidines in human DNA are not in the CpG dinucleotide. Biochemical and Biophysical Research Communications. 1987;145(2):888–894. doi: 10.1016/0006-291x(87)91048-5. [DOI] [PubMed] [Google Scholar]
- 27.Lister R. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan J. Evidence for non-CpG methylation in mammals. Experimental Cell Research. 2011;317(18):2555–2561. doi: 10.1016/j.yexcr.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 29.Barres R. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metabolism. 2009;10(3):189–198. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Ichiyanagi T. Accumulation and loss of asymmetric non-CpG methylation during male germ-cell development. Nucleic Acids Research. 2013;41(2):738–745. doi: 10.1093/nar/gks1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Messerschmidt D.M. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes and Development. 2014;28(8):812–828. doi: 10.1101/gad.234294.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seisenberger S. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Molecular Cell. 2012;48(6):849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang W.W. Specification and epigenetic programming of the human germ line. Nature Reviews Genetics. 2016;17(10):585–600. doi: 10.1038/nrg.2016.88. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi H. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Research. 2013;23(4):616–627. doi: 10.1101/gr.148023.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gkountela S. DNA demethylation dynamics in the human prenatal germline. Cell. 2015;161(6):1425–1436. doi: 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X. Ultra-performance liquid chromatography/tandem mass spectrometry for accurate quantification of global DNA methylation in human sperms. Journal of Chromatography. B. 2011;879(19):1647–1652. doi: 10.1016/j.jchromb.2011.04.002. Analytical Technologies in the Biomedical and Life Sciences. [DOI] [PubMed] [Google Scholar]
- 37.Molaro A. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146(6):1029–1041. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ehrlich M. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Research. 1982;10(8):2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simar D. DNA methylation is altered in B and NK lymphocytes in obese and type 2 diabetic human. Metabolism. 2014;63(9):1188–1197. doi: 10.1016/j.metabol.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 40.Tang W.W. A unique gene regulatory network resets the human germline epigenome for development. Cell. 2015;161(6):1453–1467. doi: 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feinberg A.P. Evolution in health and medicine Sackler colloquium: stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proceedings of the National Academy of Sciences of the United States. 2010;107(Suppl 1):1757–1764. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feinberg J.I. Paternal sperm DNA methylation associated with early signs of autism risk in an autism-enriched cohort. International Journal of Epidemiology. 2015;44(4):1199–1210. doi: 10.1093/ije/dyv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kerkel K. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nature Genetics. 2008;40(7):904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 44.Gertz J. Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS Genetics. 2011;7(8):e1002228. doi: 10.1371/journal.pgen.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.You J.S. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22(1):9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei Y. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proceedings of the National Academy of Sciences of the United States. 2014;111(5):1873–1878. doi: 10.1073/pnas.1321195111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shea J.M. Genetic and epigenetic variation, but not diet, shape the sperm methylome. Developmental Cell. 2015;35(6):750–758. doi: 10.1016/j.devcel.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ost A. Paternal diet defines offspring chromatin state and intergenerational obesity. Cell. 2014;159(6):1352–1364. doi: 10.1016/j.cell.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 49.Ward W.S. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biology of Reproduction. 1991;44(4):569–574. doi: 10.1095/biolreprod44.4.569. [DOI] [PubMed] [Google Scholar]
- 50.Barral S. Histone variant H2A.L.2 guides transition protein-dependent protamine assembly in male germ cells. Molecules and Cells. 2017;66(1):89–101. doi: 10.1016/j.molcel.2017.02.025. e108. [DOI] [PubMed] [Google Scholar]
- 51.Rousseaux S. Histone acylation beyond acetylation: terra incognita in chromatin biology. Cell Journal. 2015;17(1):1–6. doi: 10.22074/cellj.2015.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goudarzi A. Dynamic competing histone H4 K5K8 acetylation and butyrylation are hallmarks of highly active gene promoters. Molecules and Cells. 2016;62(2):169–180. doi: 10.1016/j.molcel.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaucher J. From meiosis to postmeiotic events: the secrets of histone disappearance. FEBS Journal. 2010;277(3):599–604. doi: 10.1111/j.1742-4658.2009.07504.x. [DOI] [PubMed] [Google Scholar]
- 54.Arpanahi A. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Research. 2009;19(8):1338–1349. doi: 10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erkek S. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nature Structural and Molecular Biology. 2013;20(7):868–875. doi: 10.1038/nsmb.2599. [DOI] [PubMed] [Google Scholar]
- 56.Hammoud S.S. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Human Reproduction. 2011;26(9):2558–2569. doi: 10.1093/humrep/der192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carone B.R. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Developmental Cell. 2014;30(1):11–22. doi: 10.1016/j.devcel.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samans B. Uniformity of nucleosome preservation pattern in Mammalian sperm and its connection to repetitive DNA elements. Developmental Cell. 2014;30(1):23–35. doi: 10.1016/j.devcel.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 59.Kurimoto K. Mechanism and reconstitution in vitro of germ cell development in mammals. Cold Spring Harbor Symposia on Quantitative Biology. 2015;80:147–154. doi: 10.1101/sqb.2015.80.027425. [DOI] [PubMed] [Google Scholar]
- 60.van der Heijden G.W. Sperm-derived histones contribute to zygotic chromatin in humans. BMC Developmental Biology. 2008;8:34. doi: 10.1186/1471-213X-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teperek M. Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Research. 2016;26(8):1034–1046. doi: 10.1101/gr.201541.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamauchi Y. Paternal pronuclear DNA degradation is functionally linked to DNA replication in mouse oocytes. Biology of Reproduction. 2007;77(3):407–415. doi: 10.1095/biolreprod.107.061473. [DOI] [PubMed] [Google Scholar]
- 63.Ward W.S. Function of sperm chromatin structural elements in fertilization and development. Molecular Human Reproduction. 2010;16(1):30–36. doi: 10.1093/molehr/gap080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bjorndahl L. Sequence of ejaculation affects the spermatozoon as a Carrier and its message. Reproductive BioMedicine Online. 2003;7(4):440–448. doi: 10.1016/s1472-6483(10)61888-3. [DOI] [PubMed] [Google Scholar]
- 65.Bjorndahl L. Human sperm chromatin stabilization: a proposed model including zinc bridges. Molecular Human Reproduction. 2010;16(1):23–29. doi: 10.1093/molehr/gap099. [DOI] [PubMed] [Google Scholar]
- 66.Siklenka K. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science. 2015;350(6261):aab2006. doi: 10.1126/science.aab2006. [DOI] [PubMed] [Google Scholar]
- 67.Zheng H. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Molecules and Cells. 2016;63(6):1066–1079. doi: 10.1016/j.molcel.2016.08.032. [DOI] [PubMed] [Google Scholar]
- 68.Johnson G.D. Cleavage of rRNA ensures translational cessation in sperm at fertilization. Molecular Human Reproduction. 2011;17(12):721–726. doi: 10.1093/molehr/gar054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goodrich R.J. Isolating mRNA and small noncoding RNAs from human sperm. Methods in Molecular Biology. 2013;927:385–396. doi: 10.1007/978-1-62703-038-0_33. [DOI] [PubMed] [Google Scholar]
- 70.Ostermeier G.C. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature. 2004;429(6988):154. doi: 10.1038/429154a. [DOI] [PubMed] [Google Scholar]
- 71.Sone Y. Nuclear translocation of phospholipase C-zeta, an egg-activating factor, during early embryonic development. Biochemical and Biophysical Research Communications. 2005;330(3):690–694. doi: 10.1016/j.bbrc.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 72.Rassoulzadegan M. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441(7092):469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 73.Deng W. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Developmental Cell. 2002;2(6):819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 74.Ashe A. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150(1):88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brennecke J. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322(5906):1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lau J. CART in the regulation of appetite and energy homeostasis. Frontiers in Neuroscience. 2014;8:313. doi: 10.3389/fnins.2014.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castella S. Ilf3 and NF90 functions in RNA biology. Wiley Interdisciplinary Reviews: RNA. 2015;6(2):243–256. doi: 10.1002/wrna.1270. [DOI] [PubMed] [Google Scholar]
- 78.Jiang L.Q. Autocrine role of interleukin-13 on skeletal muscle glucose metabolism in type 2 diabetic patients involves microRNA let-7. American Journal of Physiology-Endocrinology and Metabolism. 2013;305(11):E1359–E1366. doi: 10.1152/ajpendo.00236.2013. [DOI] [PubMed] [Google Scholar]
- 79.Zhu H. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147(1):81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garcia-Lopez J. Global characterization and target identification of piRNAs and endo-siRNAs in mouse gametes and zygotes. Biochimica et Biophysica Acta. 2014;1839(6):463–475. doi: 10.1016/j.bbagrm.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 81.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 82.Liu W.M. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proceedings of the National Academy of Sciences of the United States. 2012;109(2):490–494. doi: 10.1073/pnas.1110368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grandjean V. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development. 2009;136(21):3647–3655. doi: 10.1242/dev.041061. [DOI] [PubMed] [Google Scholar]
- 84.Wagner K.D. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Developmental Cell. 2008;14(6):962–969. doi: 10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 85.Cropley J.E. Male-lineage transmission of an acquired metabolic phenotype induced by grand-paternal obesity. Molecular Metabolism. 2016;5(8):699–708. doi: 10.1016/j.molmet.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rodgers A.B. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proceedings of the National Academy of Sciences of the United States. 2015;112(44):13699–13704. doi: 10.1073/pnas.1508347112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alm P.S. Grandpaternal-induced transgenerational dietary reprogramming of the unfolded protein response in skeletal muscle. Molecular Metabolism. 2017;6(7):621–630. doi: 10.1016/j.molmet.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krawetz S.A. A survey of small RNAs in human sperm. Human Reproduction. 2011;26(12):3401–3412. doi: 10.1093/humrep/der329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nagano T. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322(5908):1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 90.Pandey R.R. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Molecular Cell. 2008;32(2):232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 91.Benetti R. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nature Structural and Molecular Biology. 2008;15(9):998. doi: 10.1038/nsmb0908-998b. [DOI] [PubMed] [Google Scholar]
- 92.Lai F. Where long noncoding RNAs meet DNA methylation. Cell Research. 2014;24(3):263–264. doi: 10.1038/cr.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li L.C. Chromatin remodeling by the small RNA machinery in mammalian cells. Epigenetics. 2014;9(1):45–52. doi: 10.4161/epi.26830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bird A. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40(1):91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 95.Cohen N.M. Primate CpG islands are maintained by heterogeneous evolutionary regimes involving minimal selection. Cell. 2011;145(5):773–786. doi: 10.1016/j.cell.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 96.Shen J.C. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Research. 1994;22(6):972–976. doi: 10.1093/nar/22.6.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Waddington C.H. The epigenotype. 1942. International Journal of Epidemiology. 2012;41(1):10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- 98.Leisegang K. Obesity is associated with increased seminal insulin and leptin alongside reduced fertility parameters in a controlled male cohort. Reproductive Biology and Endocrinology. 2014;12:34. doi: 10.1186/1477-7827-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fariello R.M. Effect of smoking on the functional aspects of sperm and seminal plasma protein profiles in patients with varicocele. Human Reproduction. 2012;27(11):3140–3149. doi: 10.1093/humrep/des287. [DOI] [PubMed] [Google Scholar]
- 100.Marques C.J. DNA methylation imprinting marks and DNA methyltransferase expression in human spermatogenic cell stages. Epigenetics. 2011;6(11):1354–1361. doi: 10.4161/epi.6.11.17993. [DOI] [PubMed] [Google Scholar]
- 101.Ni K. TET enzymes are successively expressed during human spermatogenesis and their expression level is pivotal for male fertility. Human Reproduction. 2016;31(7):1411–1424. doi: 10.1093/humrep/dew096. [DOI] [PubMed] [Google Scholar]
- 102.Pattamaprapanont P. Muscle contraction induces acute hydroxymethylation of the exercise-responsive gene Nr4a3. Frontiers in Endocrinology. 2016;7:165. doi: 10.3389/fendo.2016.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vojta A. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Research. 2016;44(12):5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu X.S. Editing DNA methylation in the mammalian genome. Cell. 2016;167(1):233–247. doi: 10.1016/j.cell.2016.08.056. e217. [DOI] [PMC free article] [PubMed] [Google Scholar]