In this report, we describe a hybrid MinION whole-genome sequencing pipeline and the genomic characteristics of the first eight Fusobacterium strains deposited in the FusoPortal database. This collection of highly accurate and complete genomes drastically improves upon previous multicontig assemblies by correcting and newly identifying a significant number of open reading frames. We believe that the availability of this resource will result in the discovery of proteins and molecular mechanisms used by an oral pathogen, with the potential to further our understanding of how Fusobacterium nucleatum contributes to a repertoire of diseases, including periodontitis, preterm birth, and colorectal cancer.
KEYWORDS: Fusobacterium, Fusobacterium nucleatum, pathogenesis, colon cancer, colorectal cancer, genomics, host-pathogen
ABSTRACT
Here we present FusoPortal, an interactive repository of Fusobacterium genomes that were sequenced using a hybrid MinION long-read sequencing pipeline, followed by assembly and annotation using a diverse portfolio of predominantly open-source software. Significant efforts were made to provide genomic and bioinformatic data as downloadable files, including raw sequencing reads, genome maps, gene annotations, protein functional analysis and classifications, and a custom BLAST server for FusoPortal genomes. FusoPortal has been initiated with eight complete genomes, of which seven were previously only drafts that ranged from 24 to 67 contigs. We have showcased that the genomes in FusoPortal provide accurate open reading frame annotations and have corrected a number of large (>3-kb) genes that were previously misannotated due to contig boundaries. In summary, FusoPortal (http://fusoportal.org) is the first database of MinION-sequenced and completely assembled Fusobacterium genomes, and this central Fusobacterium genomic and bioinformatic resource will aid the scientific community in developing a deeper understanding of how this human pathogen contributes to an array of diseases, including periodontitis and colorectal cancer.
IMPORTANCE In this report, we describe a hybrid MinION whole-genome sequencing pipeline and the genomic characteristics of the first eight Fusobacterium strains deposited in the FusoPortal database. This collection of highly accurate and complete genomes drastically improves upon previous multicontig assemblies by correcting and newly identifying a significant number of open reading frames. We believe that the availability of this resource will result in the discovery of proteins and molecular mechanisms used by an oral pathogen, with the potential to further our understanding of how Fusobacterium nucleatum contributes to a repertoire of diseases, including periodontitis, preterm birth, and colorectal cancer.
INTRODUCTION
Multiple Fusobacterium species are oral pathogens that infect a broad range of human organ and tissue niches (1, 2). Fusobacterium nucleatum has recently been connected with colorectal cancer (CRC) (3, 4), with studies showing that this bacterium induces a proinflammatory microenvironment and chemoresistance against drugs used to treat CRC (5–7). Despite the importance of Fusobacterium in human diseases, a lack of complete genomes of biomedically relevant isolates has hindered protein cataloging and virulence factor identification. Many bacterial draft genomes have been sequenced and partially assembled using short-read technologies (Illumina, 454 Life Sciences), leaving complete genome assembly difficult due to the presence of repeat regions. The reference genome of F. nucleatum subsp. nucleatum ATCC 25586 was completed using cosmid and λ phage technologies to achieve long (10-to-35-kb) cosmid insertions and whole-genome assembly (8). However, we show in a parallel report that whereas this genome was assembled into one complete chromosome, we uncovered a 452-kb inversion using our sequencing and assembly methods. This inversion does not appear to change many open reading frames, but it could be useful in whole-genome comparisons to analyze detailed gene orientations. With the emergence of next-generation long-read sequencing (Pacific Biosciences, Oxford Nanopore MinION), assembling whole genomes has become standard practice and affordable for academic research settings. The recent combination of MinION long-read and Illumina short-read technologies to scaffold and polish DNA sequencing data, respectively, has created a robust pipeline for bacterial genome completion and subsequent gene identification and characterization (9, 10).
The motivation for complete sequencing and assembly of Fusobacterium genomes came from our discovery that bioinformatic analysis identified a high percentage of large genes (~3,000 to 12,000 bp) in the F. nucleatum subsp. nucleatum ATCC 23726 genome that appeared to correspond to proteins missing critical domains at either the N or C terminus (e.g., >2,000-amino acid deletions). We showed that these genomic discrepancies are not isolated to F. nucleatum subsp. nucleatum ATCC 23726 by correcting a substantial number of large proteins (up to 5,300-amino-acid corrections) encoded in all eight genomes. Since the largest proteins in Fusobacterium are autotransporters of the type 5 secretion system, these new genomes will be important to reevaluate the virulence factor landscape of these pathogenic bacteria.
To provide ease of use and data accessibility to the community, we have used this study to launch the FusoPortal repository (http://fusoportal.org), which provides the first eight completely sequenced, assembled, and annotated Fusobacterium genomes using MinION and Illumina technology. While databases such as KEGG, NCBI, and UniProt are crucial for researchers to find open reading frames, our goal was to create a central database in which researchers interested in Fusobacterium biology could obtain high-quality data in an easy-to-navigate platform. The FusoPortal repository framework has been developed to allow additional genomes to efficiently be added, with the goal of assembling 25 previously incomplete genomes spanning a broad range of Fusobacterium species. In summary, genomes and bioinformatic analyses available in the FusoPortal repository provide key resources to further determine how this understudied pathogen contributes to a variety of human infections and diseases. Here we highlight not only how users can interact with the FusoPortal website but also additional bioinformatic analysis that was made possible by improved genome sequencing and assembly.
RESULTS AND DISCUSSION
Genome sequencing, assembly, and annotation.
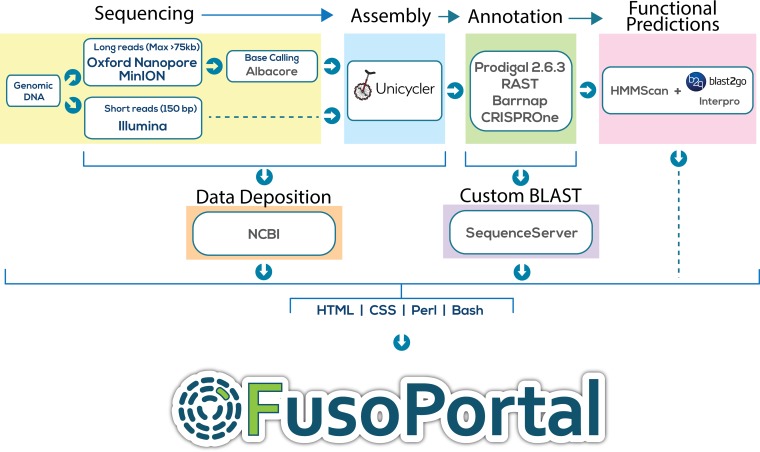
In this study, we successfully completed seven new Fusobacterium genomes and resequenced the widely referenced strain F. nucleatum subsp. nucleatum 25586. Each genome now consists of a single circular chromosome, and in the case of F. varium 27725, we report the discovery of a 42-kb circular plasmid that contains 70 genes encoding proteins ranging in size from 38 to 1,678 amino acids. For each genome, we have provided access to all protein- and RNA-encoding genes and have highlighted the previously unidentified CRISPR systems. In Fig. 1, we describe the workflow used to complete genome sequencing, assembly, annotation, functional prediction, and implementation of the interactive FusoPortal repository. We made a concerted effort to use open-source software when possible, to make this workflow reproducible and accessible to the scientific community.
FIG 1 .
Schematic of all genomic, bioinformatic, and scripting workflows used to create FusoPortal.
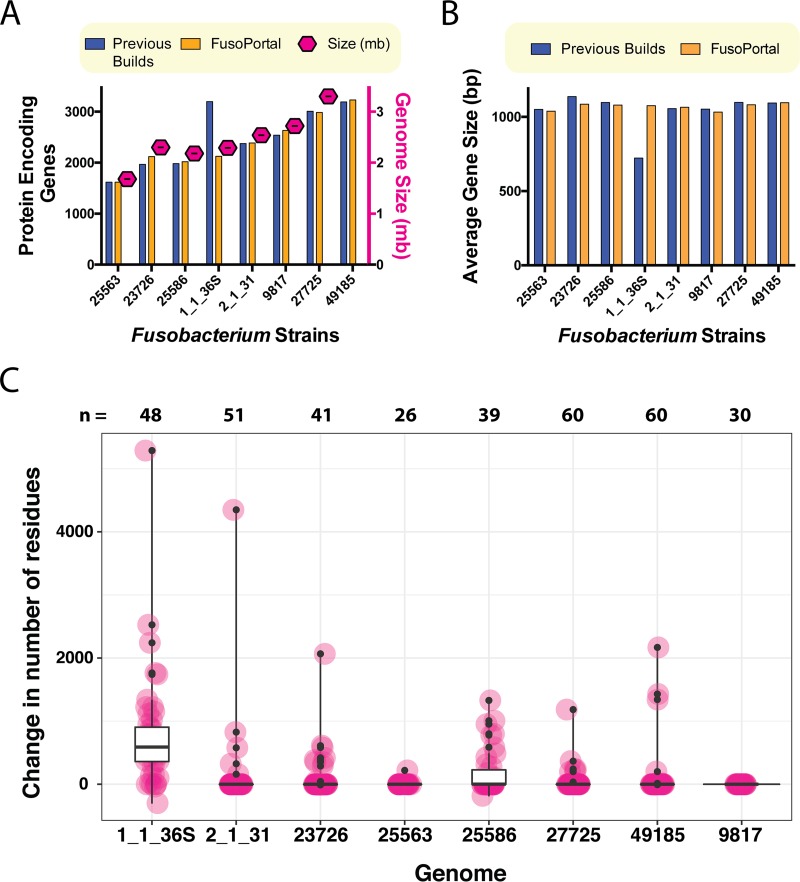
We highlight that the strain with the largest change in gene number from a reference genome to our single-chromosome build was F. necrophorum funduliforme 1_1_36S, which was previously reported with 3,197 protein-encoding genes. We analyzed our new prediction of 2,125 genes and show that this number is much more consistent with the genome size seen in comparisons of all Fusobacterium genomes (Fig. 2A) (Table 1). These data show that the previous annotation contained an abundance of short open reading frames and that the complete genome increases the average gene length by more than 250 bp; the average gene size of ~1,000 bp agrees well with the data for the remaining seven Fusobacterium genomes (Fig. 2B).
FIG 2 .
Correction of open reading frames in Fusobacterium genomes. (A) A comparison of the genome size to the number of protein-encoding genes per genome in both the NCBI and FusoPortal genomes. (B) A comparison of all proteins for average gene size in the NCBI and FusoPortal genomes. (C) Analysis of all proteins 1,000 residues in size and above in annotated FusoPortal proteomes and of how many residues were added (or, in rare cases, removed) compared to the previous annotations present in the NCBI database. n = number of proteins 1,000 residues in size or greater in each genome.
TABLE 1 .
Statistics and NCBI accession numbers for FusoPortal genomes
| Species | Strain | NCBI GenBank sequence accession no. |
No. of NCBI contigsa |
Size (Mb) |
No. of protein genes |
FusoPortal GenBank sequence accession no. |
No. of FusoPortal contigs |
Size (Mb) |
No. of protein genes |
|---|---|---|---|---|---|---|---|---|---|
| F. nucleatum | 23726 | GCF_000178895.1 | 67 | 2.23 | 1,983 | GCA_003019785.1 | 1 | 2.30 | 2,111 |
| F. nucleatum | 25586 | GCA_000007325.1 | 1 | 2.17 | 1,968 | GCA_003019295.1 | 1 | 2.18 | 2,019 |
| F. varium | 27725 | GCA_000159915.2 | 39 | 3.30 | 3,008 | GCA_003019655.1 | 1 | 3.35 | 3,054b |
| F. ulcerans | 49185 | GCA_000158315.2 | 49 | 3.49 | 3,191 | GCA_003019675.1 | 1 | 3.54 | 3,230 |
| F. mortiferum | 9817 | GCA_000158195.2 | 44 | 2.67 | 2,538 | GCA_003019315.1 | 1 | 2.72 | 2,631 |
| F. gonidiaformans | 25563 | GCA_000158835.2 | 24 | 1.70 | 1,618 | GCA_003019695.1 | 1 | 1.68 | 1,617 |
| F. periodonticum | 2_1_31 | GCA_000158215.3 | 61 | 2.55 | 2,375 | GCA_003019755.1 | 1 | 2.54 | 2,388 |
| F. necrophorum | 1_1_36S | GCA_000242215.1 | 40 | 2.31 | 3,197 | GCA_003019715.1 | 1 | 2.29 | 21,215 |
Contig, continuous stretch of genomic DNA with breaks.
Data include 70 genes from a 42-kb plasmid (GCA_003019655.1).
Correcting large protein-encoding reading frames.
As expected, the largest change in the F. necrophorum funduliforme 1_1_36S genome is in the number of genes that are now annotated with 1,000 amino acids or more. In fact, the combined analyses of all eight genomes show that we have corrected a significant percentage of proteins that were previously annotated with <1,000 amino acids and are now annotated with >1,000 amino acids or of proteins that were already annotated with >1,000 amino acids and are now annotated with expanded protein open reading frames (Fig. 2C). The most extreme example was a previously annotated protein encoded in the F. necrophorum funduliforme 1_1_36S genome that was 960 amino acids in size (EHO18576.1) and that has now been annotated as a 6,248-amino acid protein in FusoPortal. F. mortiferum 9817 did not have any amino acid additions in proteins over 1,000 residues in size, but we note that the largest protein encoded in this genome consists of 1,602 residues (EEO35225.2), and as expected, proteins in this range have fewer open reading frame errors than the larger proteins frequently found in Fusobacterium genomes. We highlight that in the biomedically significant and genetically tractable strain F. nucleatum subsp. nucleatum ATCC 23726, several large stretches of amino acids were added to proteins that are homologous to the previously characterized virulence protein Fap2 (11–13). In addition, we corrected an abnormally large number of genes in the reference strain F. nucleatum subsp. nucleatum ATCC 25586, which we attribute to improved bacterial annotation software and not to genomic errors in the previous complete genome.
Whole-genome phylogenetic analysis of a diverse group of Fusobacterium spp.
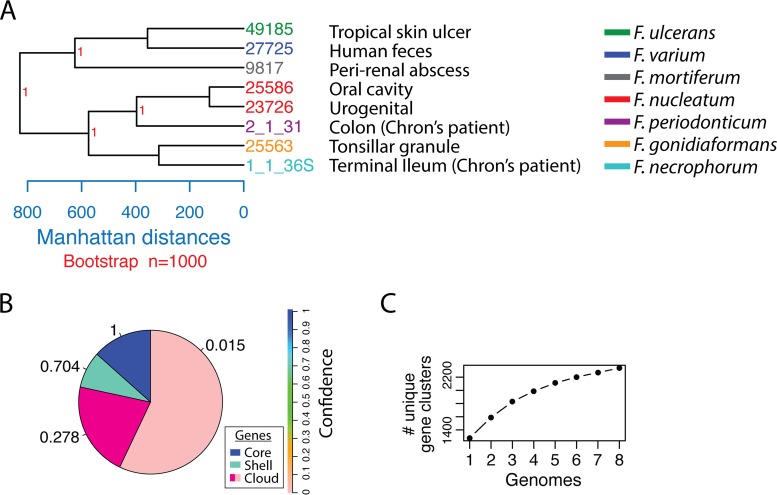
With newly identified genes in our reported Fusobacterium genomes, we set out to create a phylogenetic tree to compare to previous reports. By using all open reading frames from each genome to build a tree, we show that while F. nucleatum subsp. nucleatum ATCC 23726 and F. nucleatum subsp. nucleatum ATCC 25586 are quite close phylogenetically, the remaining genomes are quite diverse in their genetic makeup (Fig. 3). Because of the diversity of these eight genomes, there are a large number of unique gene clusters which had been analyzed previously and were hypothesized to govern differences in intracellular invasion and virulence potential (14). We highlight the site of isolation of each strain in Fig. 3A as described by Manson-McGuire et al. (14) and anticipate that as we complete more genomes to expand the phylogenetic analysis this tree may provide more extensively detailed insight into genotype-specific populations associated with tissue distribution and human diseases.
FIG 3 .
Phylogenetic analysis of complete Fusobacterium genomes. (A) Tree created using complete proteomes in the microPan package in R. Bootstrap values are indicated in red. Locations of where strains were isolated in the human body are highlighted. (B) Micropan analysis of pan-genome gene families with models predicting the percentages of core genes, shell genes (present in most genomes), and cloud genes (found in few genomes). (C) Analysis of the number of unique gene clusters found among the eight complete FusoPortal genomes.
Features of the FusoPortal repository.
FusoPortal was built on an HTML5 framework and is therefore functional on all full-size computers and mobile devices. The home page gives a description of features available in FusoPortal and provides links to all genomes and bioinformatic data. Links to all raw (Illumina and MinION DNA reads) and processed genomes and annotations are available as shown in Fig. 4A. In Fig. 4B, we highlight a genome map in which directional arrows represent clickable open reading frame designations that send the user to individual pages containing DNA and protein coding sequences in FASTA format. We have also analyzed all proteins on FusoPortal using HMMer (15) (downloadable .out files) and have produced custom linked InterPro pages with full bioinformatic analyses (Fig. 4C). These resources alleviate the need for users to exit the site to acquire bioinformatic data and provide functional predictions for entire proteomes.
FIG 4 .
Navigating the FusoPortal Web interface for genome analysis. (A) Links are provided for downloading all genomic data (raw and analyzed), with Bandage plots to show the completeness of each genome build. (B) Whole-genome maps were produced with links to custom webpages for each gene that provide access to all genomic and bioinformatic analysis. (C) Each gene contains a full InterPro analysis page with links to multiple functional prediction databases (e.g., PFAM, TIGR, Gene3D, CDD, GO). No IPR, no InterPro accession number.
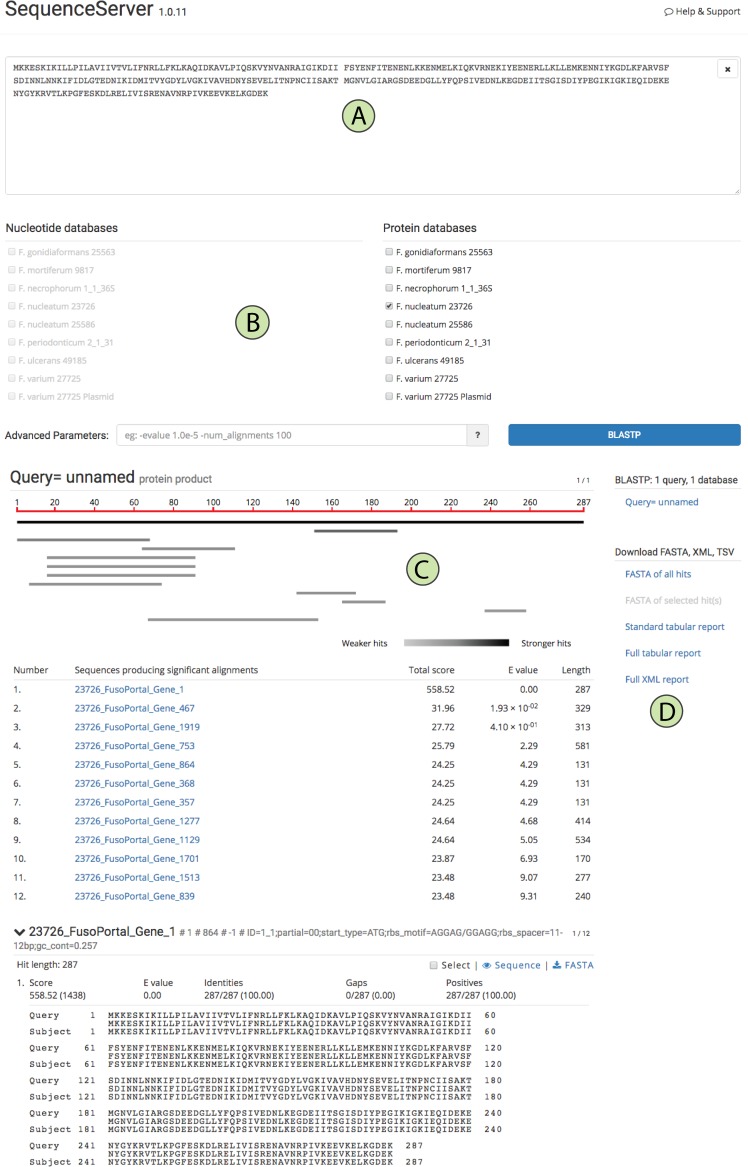
A custom BLAST database to search FusoPortal.
To additionally aid in virulence factor identification, we built a custom BLAST server using the open source software Sequenceserver (16). All eight genomes, including the F. varium 27725 plasmid, can be searched using a DNA or protein sequence input as shown in Fig. 5A and B. Results are provided as alignments with E values (customizable inputs for thresholds), and all acquired sequences and alignments can be downloaded in various formats (Fig. 5C and D).
FIG 5 .
A custom BLAST database to search FusoPortal genomes built with SequenceServer. (A) Data entry panel that autodetects protein or DNA sequences. (B) The available genomes that users can choose to search. (C) BLAST results for identified genes. (D) Links to downloadable files.
Conclusion.
FusoPortal is a database of fully sequenced, annotated, and bioinformatically characterized Fusobacterium genomes that provides a central location to increase our understanding of these virulent bacteria, which contribute to a wide range of diseases. At this time, we are keeping FusoPortal as a resource that is populated only with genomes sequenced and assembled in the laboratory of D. J. Slade. However, we are open to accepting requests to sequence and deposit additional Fusobacterium strains that are of value to the scientific community. Currently, FusoPortal excels at providing genomes that are accurate in both their sequences and their gene annotations. Our goal was to create a clean user interface that allows users to find data in an efficient manner compared with larger databases with steeper learning curves. The introduction of a custom BLAST server provides an interactive environment to detect specific proteins in Fusobacterium genomes. At this time, FusoPortal does not contain as many features as other websites such as the BioCyc database collection, EcoGene, and KEGG websites, but we aim to expand the capabilities provided on FusoPortal with both internal and external contributions. In summary, we have provided FusoPortal as a Fusobacterium-specific resource and aim to expand this database for increased efficiency in performing detailed comparative genomics and virulence factor predictions for Fusobacterium species.
MATERIALS AND METHODS
Data to populate FusoPortal.
Detailed sequencing statistics and assembly methods are reported in a concurrent publication by Todd and colleagues (17). Briefly, genomic DNA was isolated from Fusobacterium cultures and sequenced using MinION (Oxford Nanopore Technologies) and Illumina platforms. Genomes were assembled using the open source software package Unicycler version 0.4.3 (9). Gene annotations were obtained using Prodigal version 2.6.3 (18), RAST-SDK version 0.0.12 (via KBase) (19), Barrnap version 0.8 (20), and CRISPRone (21). Prediction of protein functions was achieved with the stand-alone HMMer version 3.1 program (15) and with InterPro (22) from within the Blast2GO software platform (23). A graphical representation of methods for DNA sequencing, genome assembly, bioinformatics, and construction of FusoPortal is presented in Fig. 1. As this resource could require expansion or correction of sequencing or reading frame annotation, we note that are implementing versioning and state on the FusoPortal website that all genomes have been initiated at version 1.
Development of interactive genome maps.
Protein open reading frames predicted by Prodigal (18) were used to identify boundaries that were used in the R package gggenes to create complete genome maps. For each genome, DNA gene coordinates were placed in a .csv file and imported into the gggenes package in RStudio to create linear genome maps. Maps were then manually loaded into Adobe Illustrator for custom FusoPortal formatting, followed by adding links to genes using Adobe Acrobat Pro. Final genome maps were exported as .pdf files for incorporation into the FusoPortal website. All .csv files with gene coordinates and a template R file are provided on our Open Science Framework database (http://osf.io/2c8pv).
Construction of FusoPortal using HTML and automated field-filling scripts.
FusoPortal was built using HTML5 and CSS, and automation of HTML page filling with genomic and bioinformatic data was implemented using custom Bash and Perl scripts. All scripts are available on an Open Science Framework database (http://osf.io/2c8pv) run by D. J. Slade. Scripts were developed to automate the acquisition of custom HMMer models for all genomes and to produce linear genome maps using gggenes in the Studio package. For populating FusoPortal HTML pages, scripts were created to extract Prodigal gene coordinates from .gbk files, extract gene and protein sequences, and populate webpages with InterPro functional annotations from .csv files for population into HTML pages. InterPro analysis pages were produced by Blast2GO, and the FusoPortal logo was added to each page through scripting.
Construction of a FusoPortal custom BLAST server.
The FusoPortal custom BLAST server was built using the Sequenceserver software package (16). Briefly, a custom Apache server was implemented on an Amazon Light Sail private server, and all FusoPortal genomes containing protein open reading frames in DNA (.fna) and amino acid (.faa) formats were implemented as guided by the Sequenceserver advanced setup and configuration documentation. This BLAST server can be accessed at http://18.216.121.101/blast/.
Accession number(s).
All accession numbers for data are provided in Table 1. In addition, all raw sequencing data have been deposited in the NCBI sequence read archive (SRA) as documented in a companion manuscript accompanying this report (17).
ACKNOWLEDGMENTS
This work was supported by the USDA National Institute of Food and Agriculture and by startup funding from the Department of Biochemistry at Virginia Tech to D.J.S. We thank Virginia Tech's Open Access Subvention Fund for publication funding.
B.E.S. was responsible for data curation, code writing, and review and editing of the writing; A.U. was responsible for data curation, code writing, and review and editing of the writing; J.A.L. was responsible for code writing and review and editing of the writing; D.J.S. was responsible for conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, methodology, writing of the original draft, project administration, and review and editing of the writing.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/mSphere.00269-18.
REFERENCES
- 1.Dahya V, Patel J, Wheeler M, Ketsela G. 2015. Fusobacterium nucleatum endocarditis presenting as liver and brain abscesses in an immunocompetent patient. Am J Med Sci 349:284–285. doi: 10.1097/MAJ.0000000000000388. [DOI] [PubMed] [Google Scholar]
- 2.Signat B, Roques C, Poulet P, Duffaut D. 2011. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol 13:25–36. [PubMed] [Google Scholar]
- 3.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. 2012. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. 2012. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. 2013. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. 2013. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang JY. 2017. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170:548–563.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapatral V, Anderson I, Ivanova N, Reznik G, Los T, Lykidis A, Bhattacharyya A, Bartman A, Gardner W, Grechkin G, Zhu L, Vasieva O, Chu L, Kogan Y, Chaga O, Goltsman E, Bernal A, Larsen N, D’Souza M, Walunas T, Pusch G, Haselkorn R, Fonstein M, Kyrpides N, Overbeek R. 2002. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol 184:2005–2018. doi: 10.1128/JB.184.7.2005-2018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb Genom 3:e000132. doi: 10.1099/mgen.0.000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abed J, Emgård JEM, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, Mellul A, Chaushu S, Manson AL, Earl AM, Ou N, Brennan CA, Garrett WS, Bachrach G. 2016. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe 20:215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklić K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. 2015. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan CW, Ma X, Paranjpe A, Jewett A, Lux R, Kinder-Haake S, Shi W. 2010. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect Immun 78:4773–4778. doi: 10.1128/IAI.00567-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manson McGuire A, Cochrane K, Griggs AD, Haas BJ, Abeel T, Zeng Q, Nice JB, MacDonald H, Birren BW, Berger BW, Allen-Vercoe E, Earl AM. 2014. Evolution of invasion in a diverse set of Fusobacterium species. MBio 5:e01864-14. doi: 10.1128/mBio.01864-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 16.Priyam A, Woodcroft BJ, Rai V, Munagala A, Moghul I, Ter F, Gibbins MA, Moon H, Leonard G, Rumpf W, Wurm Y. 2015. Sequenceserver: a modern graphical user interface for custom BLAST databases. BioRvix https://www.biorxiv.org/content/early/2015/11/27/033142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Todd SM, Settlage RE, Lahmers KK, Slade DJ. 2018. Fusobacterium genomics using MinION and Illumina sequencing enables genome completion and correction. mSphere 3:e00269-18 10.1128/mSphere.00269-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seemann T. Bacterial ribosomal RNA predictor. https://github.com/tseemann/barrnap. [Google Scholar]
- 21.Zhang Q, Ye Y. 2017. Not all predicted CRISPR-Cas systems are equal: isolated cas genes and classes of CRISPR like elements. BMC Bioinformatics 18:92. doi: 10.1186/s12859-017-1512-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztányi Z, El-Gebali S, Fraser M, Gough J, Haft D, Holliday GL, Huang H, Huang X, Letunic I, Lopez R, Lu S, Marchler-Bauer A, Mi H, Mistry J, Natale DA, Necci M, Nuka G, Orengo CA, Park Y, Pesseat S, Piovesan D, Potter SC, Rawlings ND, Redaschi N, Richardson L, Rivoire C, Sangrador-Vegas A, Sigrist C, Sillitoe I, Smithers B, Squizzato S, Sutton G, Thanki N, Thomas PD, Tosatto SC, Wu CH, Xenarios I, Yeh LS, Young SY, Mitchell AL. 2017. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res 45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]