Abstract
The glycine cleavage system (GCS) is a complex of four enzymes enabling glycine to serve as a source of one-carbon units to the cell. We asked whether concentrations of glycine, dimethylglycine, formate, and serine in blood are influenced by variation within GCS genes in a sample of young, healthy individuals. Fifty-two variants tagging (r2 < 0.9) the four GCS genes were tested; one variant, GLDC rs2297442-G, was significantly associated (p = .0007) with decreased glycine concentrations in serum.
Keywords: Glycine cleavage system (GCS), One-carbon metabolism, Glycine, Folate, Neural tube defects
1. Introduction
The glycine cleavage system (GCS) is a crucial component of mitochondrial one-carbon metabolism (OCM). The GCS involves four distinct enzymes: glycine decarboxylase (GLDC), aminomethyltransferase (AMT), glycine cleavage system protein H (GCSH) and dihydrolipoamide dehydrogenase (DLD). These enzymes act sequentially in the mitochondrion to cleave glycine obtained from the cytoplasm or from the breakdown of serine in the mitochondrion to produce N,N-methylene tetrahydrofolate (CH2-THF), an active folate derivative that contributes to the transfer of one‑carbon units in cellular reactions [1].
Loss-of-function mutations in two GCS genes (GLDC and AMT [2]) cause non-ketotic hyperglycinemia (NKH), a rare recessive disease. Additionally, two NKH patients have been observed to have a complex rearrangement of GCSH [3]. Affected patients suffer from neurological impairments, seizures, and developmental delay, suggesting that the GCS is important for normal brain development and function [4]. In a study population of patients from the UK, Sweden and Japan, two of the genes encoding GCS enzymes, AMT and GLDC, were found to have missense variants associated with neural tube defects (NTDs) [5]. Similarly, variants in these genes were identified in a study of American patients with myelomeningocele, a type of NTD [6]. Finally, GLDC rs14742391 (p.Ser951Tyr) has been observed in a patient with NKH [2] and a patient with anencephaly, a type of NTD [7].
Although genome-wide association studies (GWASs) can identify gene variants that influence traits, they are often underpowered to detect signals [8] from real but small effects of common alleles, or large effects of rare alleles. GWASs have been carried out for serum glycine, identifiying a single nucleotide polymorphism (SNP) in carbamoyl-phosphate synthase 1 (rs715) [[9], [10], [11], [12]]. Blood serine has been associated with a SNP in phosphoglycerate dehydrogenase (rs477992) [9, 12, 13]). To test for other genetic modifiers of OCM that may have been missed by the GWAS method, we examined whether common variants in the genes of the GCS influence relevant metabolites in a healthy population.
2. Materials and methods
2.1. The Trinity Student Study (TSS) cohort
The TSS cohort comprises a population of 2232 healthy Irish students, as described [[14], [15], [16]]. Informed consent and ethical approval were obtained from all participants. Briefly, blood samples were collected into EDTA and clotting tubes, processed within 3 h, and stored at −80 °C before assaying. Glycine (interassay CV = 3.3%) and serine (interassay CV = 5.7%) in serum were measured with gas-chromatography tandem mass spectrometry (GC–MS/MS [17]) and plasma dimethylglycine (interassay CV <10%) by liquid-chromatography tandem mass spectrometry (LC-MS/MS [18]) by the laboratory of Bevital, Norway (www.bevital.no). Formate was measured using a newly developed GC–MS method [19]. A single control sample was frozen in aliquots for daily analysis with formate assays of participant serum samples. The day-to-day variation for this sample was 7.4% (n = 38). Genome-wide SNP genotyping was conducted at the Center for Inherited Disease Research (CIDR, USA) using Illumina 1 M HumanOmni1-Quad_v1-0_B chips.
2.2. Metabolite association analyses
Conservative (r2 < 0.9) tagging variants in the four GCS genes including 10 kb flanking regions were selected to test for association with levels of serum glycine, plasma dimethylglycine, serum serine, and serum formate. Log10 transformations of metabolite concentrations were used to meet normality assumptions. Linkage disequilibrium (LD) analyses were performed with Haploview; a total of 52 tag SNPs covering 93 variants were selected for association testing [20, 21]. For each SNP, association was tested using linear regression with a 1-df test (additive genetic model) or a 2-df test (genotypic model) (R statistical language and environment (version 3.3.2) [22], R package snpStats [23]). Bonferroni correction for 52 tests was applied to the nominal significance threshold (p < .05) to obtain a corrected significance threshold (p < .00096). Associations between log10 transformed metabolite data were assessed using Pearson's correlation coefficient (r).
2.3. GLDC expression association analyses
The public Gene-Tissue Expression resource (https://www.gtexportal.org/home) was used to search for associations between SNPs and GLDC mRNA levels (mRNA expression quantitative trait locus, eQTLs).
3. Results
The 52 tag SNPs in the four glycine cleavage genes (GLDC, AMT, GCSH, and DLD) were tested for association with blood levels of glycine, dimethylglycine, formate, and serine in 2232 young healthy Irish adults (Table 1). Serum glycine concentrations were significantly correlated with all the other metabolites measured. The strength of these associations varied from r = 0.52; p < .001 with serine to −0.07; p = .004 with formate. Only one GCS SNP is significantly associated with a metabolite. GLDC rs2297442 returns a p-value of 0.0007 following 1-df association testing with log10 transformed glycine concentration. The 2-df test of association is also significant (p = .0031), nominally.
Table 1.
Metabolite characteristics in the TSS and by genotype group of its most associated GCS variant.
| Serine (μM) | Glycine (μM) | Dimethylglycine (μM) | Formate (μM) | |
|---|---|---|---|---|
| Mean +/− SD | 147.2 +/− 23.9 | 293.7 +/− 63.9 | 4.17 +/− 1.22 | 25.9 +/− 7.8 |
| Median | 145.6 | 288.1 | 3.99 | 24.8 |
| Number⁎ | 2210 | 2210 | 2227 | 1535 |
| Top SNP | rs2297442 | rs2297442 | rs16924717 | rs7031325 |
| p-value⁎⁎ | 0.0225 | 0.0007 | 0.0047 | 0.0133 |
| Genotype (No.) | AA (1198) | AA (1198) | AA (2130) | TT (647) |
| Mean +/− SD | 148.1 +/− 24.5 | 297.6 +/− 65.8 | 4.2 +/− 1.2 | 26.1 +/− 8.2 |
| Genotype (No.) | AG (848) | AG (848) | AG (95) | TC (701) |
| Mean +/− SD | 146.9 +/− 23.7 | 290.6 +/− 63.2 | 4.6 +/− 2 | 25.7 +/− 7.1 |
| Genotype (No.) | GG (164) | GG (164) | GG (2) | CC (187) |
| Mean +/− SD | 143.1 +/− 23.4 | 281.1 +/− 53.7 | 3.8 +/− 2.7 | 25.9 +/− 9 |
Number of participants with metabolite and genotype values.
The Bonferroni-corrected threshold for significance is p < .00096.
We then sought to replicate the association between rs2297442-G and decreased levels of circulating glycine by examining results from two recent GWASs in which glycine was measured. Our observation was replicated in two studies of Finnish individuals (n = 16,506, ß = −0.039, p = .0014 [24]; n = 8545, ß = −0.039, p = .028 [25]).
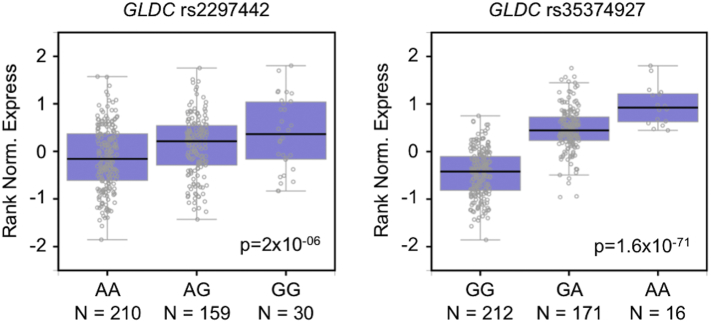
We also asked if GLDC mRNA expression levels are influenced by rs2297442 genotype. A Gene-Tissue Expression (GTEx) query of rs2297442 did not reveal a strong or consistent effect on GLDC mRNA levels across multiple tissues. In contrast, several other GLDC SNPs strongly influenced GLDC mRNA levels in several tissues (p < 1 × 10−40). For example, in thyroid tissue, GLDC rs35374927 is estimated to have a large impact on GLDC mRNA levels compared to GLDC rs2297442 (Fig. 1). These eQTLs of strong effect reside in LD blocks near the 5′ end of the gene, as opposed to the SNPs in the LD block at the 3′ end of the gene where rs2297442 is located.
Fig. 1.
Association of rs2297442 (left) or rs35374927 (right) with GLDC mRNA levels in human thyroid. In these graphs generated by GTEx, participants are grouped by genotype and plotted for ranked normalized expression of GLDC mRNA in thyroid. Unadjusted p-values for linear regression analyses are shown. GLDC rs2297442 effect size (i.e., slope of the linear regression) = 0.3. GLDC rs35374927 effect size =1.0.
4. Discussion
The results of these association tests confirm that variation in GLDC can influence serum glycine concentrations. The variant reported here, GLDC rs2297442, is a noncoding polymorphism found in the seventh of the ten introns in this gene. The GTEx data revealed that other, unlinked SNPs in the gene have a larger impact on GLDC mRNA levels, albeit in limited, specific tissues. We conclude that rs2297442 does not influence serum glycine levels via modulation of GLDC transcript levels. It is possible that rs2297442 is simply linked to the causal SNP. A search for a linked, exonic SNP in a population of European ancestry [26] failed to identify any strongly related variants (r2 < 0.2).
These results are consistent with the presence of a common SNP in GLDC that may increase the activity or alter the tissue distributions of this enzyme, causing reduced levels of circulating glycine. We predict that this effect on serum glycine concentrations may not be clinically relevant in this healthy population; however, it may contribute to diseases with a complex mode of inheritance, where combinations of many environmental and genetic factors of small individual effect must converge. The known link between folic acid supplementation and neural tube defect (NTD) prevention implicates perturbations of OCM as a key risk factor. In a study of 258 NTD patients, 27 single-base substitutions were discovered in GLDC, six of which influenced enzyme activity in an in vitro system [5]. The association between GLDC rs2297442 and serum glycine levels in the healthy populations in this study and others identifies a candidate locus for studies investigating the genetic basis of NKH, NTDs or any pathophysiology involving OCM.
Acknowledgments
Acknowledgements
The authors acknowledge the contributions made by the study participants in the Trinity Student Study and the replication cohorts. The authors are grateful to Narisu Narisu and colleagues for providing summary statistics for the METSIM study. The authors acknowledge genotyping performed by the Center for Inherited Disease Research (CIDR) under contract HHSN268201200008I.
Data statement
The data upon which these analyses are based have been deposited in and are available from the dbGaP database under dbGaP accession phs000789.v1.p1.
Funding
This work was supported by funding from the Intramural Research Programs of the National Human Genome Research Institute.
Conflict of interest statement
The authors have no conflicts of interest to declare.
References
- 1.Tibbetts A.S., Appling D.R. Compartmentalization of mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- 2.Coughlin C.R., II The genetic basis of classic nonketotic hyperglycinemia due to mutations in GLDC and AMT. Genet. Med. 2017;19(1):104–111. doi: 10.1038/gim.2016.74. [DOI] [PubMed] [Google Scholar]
- 3.Koyata H., Hiraga K. The glycine cleavage system: structure of a cDNA encoding human H-protein, and partial characterization of its gene in patients with hyperglycinemias. Am. J. Hum. Genet. 1991;48(2):351–361. [PMC free article] [PubMed] [Google Scholar]
- 4.Pai Y.J. Glycine decarboxylase deficiency causes neural tube defects and features of non-ketotic hyperglycinemia in mice. Nat. Commun. 2015;6:6388. doi: 10.1038/ncomms7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narisawa A. Mutations in genes encoding the glycine cleavage system predispose to neural tube defects in mice and humans. Hum. Mol. Genet. 2012;21(7):1496–1503. doi: 10.1093/hmg/ddr585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah R.H. Genetic association of the glycine cleavage system genes and myelomeningocele. Birth Defects Res. A Clin. Mol. Teratol. 2016;106(10):847–853. doi: 10.1002/bdra.23552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishida M. A targeted sequencing panel identifies rare damaging variants in multiple genes in the cranial neural tube defect, anencephaly. Clin. Genet. 2018;93(4):870–879. doi: 10.1111/cge.13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W.Y. Genome-wide association studies: theoretical and practical concerns. Nat. Rev. Genet. 2005;6(2):109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 9.Draisma H.H.M. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nat. Commun. 2015;6:7208. doi: 10.1038/ncomms8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartiala J.A. Genome-wide association study and targeted metabolomics identifies sex-specific association of CPS1 with coronary artery disease. Nat. Commun. 2016;7 doi: 10.1038/ncomms10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin S.Y. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014;46(6):543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie W. Genetic variants associated with glycine metabolism and their role in insulin sensitivity and type 2 diabetes. Diabetes. 2013;62(6):2141–2150. doi: 10.2337/db12-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suhre K. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477(7362):54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molloy A.M. A common polymorphism in HIBCH influences Methylmalonic acid concentrations in blood independently of cobalamin. Am. J. Hum. Genet. 2016;98(5):869–882. doi: 10.1016/j.ajhg.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stone N. Bioinformatic and genetic association analysis of microRNA target sites in one-carbon metabolism genes. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0021851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills J.L. Do high blood folate concentrations exacerbate metabolic abnormalities in people with low vitamin B-12 status? Am. J. Clin. Nutr. 2011;94(2):495–500. doi: 10.3945/ajcn.111.014621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Midttun O. Combined measurement of 6 fat-soluble vitamins and 26 water-soluble functional vitamin markers and amino acids in 50 muL of serum or plasma by high-throughput mass spectrometry. Anal. Chem. 2016;88(21):10427–10436. doi: 10.1021/acs.analchem.6b02325. [DOI] [PubMed] [Google Scholar]
- 18.Midttun O., Kvalheim G., Ueland P.M. High-throughput, low-volume, multianalyte quantification of plasma metabolites related to one-carbon metabolism using HPLC-MS/MS. Anal. Bioanal. Chem. 2013;405(6):2009–2017. doi: 10.1007/s00216-012-6602-6. [DOI] [PubMed] [Google Scholar]
- 19.Lamarre S.G. An isotope-dilution, GC-MS assay for formate and its application to human and animal metabolism. Amino Acids. 2014;46(8):1885–1891. doi: 10.1007/s00726-014-1738-7. [DOI] [PubMed] [Google Scholar]
- 20.Barrett J.C. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 21.de Bakker P.I. Efficiency and power in genetic association studies. Nat. Genet. 2005;37(11):1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 22.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2016. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 23.Clayton D. 2015. snpStats: SnpMatrix and XSnpMatrix Classes and Methods. [Google Scholar]
- 24.Teslovich T.M. Identification of seven novel loci associated with amino acid levels using single variant and gene-based tests in 8,545 Finnish men from the METSIM study. Hum. Mol. Genet. 2018;27(9):1664–1674. doi: 10.1093/hmg/ddy067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kettunen J. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat. Commun. 2016;7:11122. doi: 10.1038/ncomms11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genomes Project C. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]