SUMMARY
Mammalian barrier surfaces are constitutively colonized by numerous microorganisms. We explored how the microbiota was sensed by the immune system and the defining properties of such responses. Here we show that a skin commensal can induce T cell responses in a manner restricted to non-classical MHC Class I molecules. These responses are uncoupled from inflammation and highly distinct from pathogen-induced cells. Commensal-specific T cells express a defined gene signature characterized by expression of effector genes together with immunoregulatory and tissue repair signatures. As such, non-classically restricted commensal-specific immune responses not only promoted protection to pathogens but also accelerated skin wound closure. Thus, the microbiota can induce a highly physiological and pleiotropic form of adaptive immunity that couples antimicrobial function with tissue repair. Our work also reveals that non-classical MHC Class I molecules, an evolutionarily ancient arm of the immune system, can promote homeostatic immunity to the microbiota.
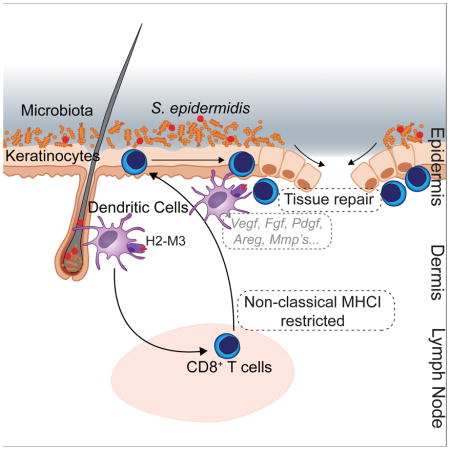
Graphical Abstract
Microbiota induce a form of adaptive immunity that couples antimicrobial function with tissue repair.
INTRODUCTION
The immune system acts as a formidable regulator of host homeostasis to sustain and restore tissue function in the context of microbial encounters and environmental challenges. The development of defined arms of the immune system and, more particularly, those associated with adaptive immunity has coincided with the acquisition of a complex microbiota suggesting that a large fraction of this machinery has evolved as a means to maintain symbiotic relationships with these highly diverse microbial communities (Belkaid and Hand, 2014).
Most of what we understand today about the function of the immune system has come from the exploration of inflammatory settings or responses to pathogenic microbes. However, the vast majority of immune system-microbial encounters are those resulting from the symbiotic relationship with the microbiota. The characteristics and properties of this class of immunity remain largely unknown. Far from being ignored by the immune system as originally perceived, microbes at all barrier surfaces are tonically educating tissues for antimicrobial functions and are actively recognized by both the innate and adaptive immune systems (Belkaid and Hand, 2014; Honda and Littman, 2016; Pamer, 2016). In the gastrointestinal tract, host-microbe communications are mediated by IgA, Th17, and Treg cell responses, a dialogue that can be in part explained by the unique requirement of the GI tract for absorption (Belkaid and Hand, 2014; Honda and Littman, 2016). However, it is now becoming clear that even at more stringent barrier sites such as the skin, the immune system is poised to sense and to respond to the microbiota (Naik et al., 2015; Scharschmidt et al., 2015). These commensal-specific responses not only control microbiota containment but also promote antimicrobial defenses via their action on both innate and epithelial cells (Ivanov et al., 2009; Naik et al., 2015; Yang et al., 2014). However, despite the extraordinary number of potential antigens expressed by the microbiota, only a handful of epitopes have been identified thus far (Cong et al., 2009; Yang et al., 2014), and the mechanisms underlying antigen presentation of microbiota-derived antigens remains poorly understood.
The skin is the body’s most exposed environmental interface and acts as a first line of physical and immunological defense. This organ is also a complex and dynamic ecosystem inhabited by a multitude of microorganisms including bacteria, fungi and viruses (Belkaid and Segre, 2014). Skin-specific microbiota control diverse aspects of tissue physiology, including innate and adaptive immunity to pathogens (Belkaid and Tamoutounour, 2016). Even in the context of an intact barrier, encounter of the skin immune system with a commensal or mutualistic microbe can drive cognate, long-lasting immune responses that promote broad antimicrobial responses (Naik et al., 2015). One striking feature of commensal-specific immunity is its uncoupling from inflammation and the maintenance of tissue homeostasis at both the induction and effector stages of the response (Naik et al., 2015). These observations raise several intriguing questions. Particularly, it remains unclear to what extent does adaptive immunity induced under these physiological settings obey conventional rules of adaptive immunity to pathogens, and what are the defining properties of such homeostatic immune responses. Here we show that commensal derived N-formyl methionine-containing (fMet) peptides can be presented by a member of an ancient and evolutionarily conserved arm of the immune system. These results provide direct evidence that non-classical MHC Class Ib (MHCIb) molecules can function to present commensal-derived antigens to T cells and are thereby direct mediators in the interaction between the host and its microbiota. This work also reveals an ancient form of immunity endowed with a pleiotropic arsenal of immune functions ranging from protective immunity to tissue repair.
RESULTS
Staphylococcus epidermidis elicits Tc17 and Tc1 cells to skin
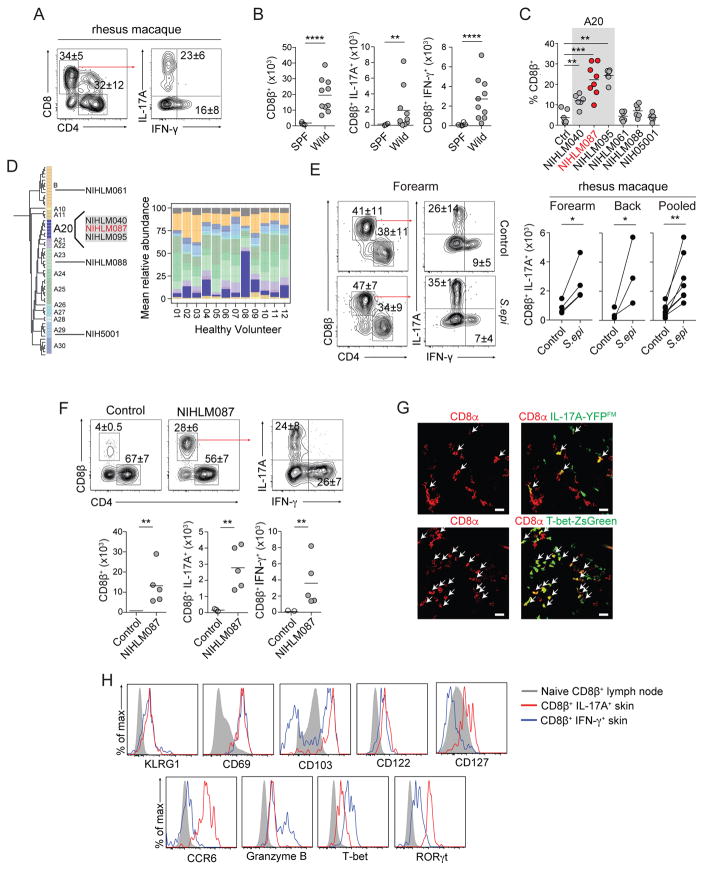
We previously demonstrated in mice that skin association with defined commensals that persist at low levels leads to the induction of CD8+ T cells (Naik et al., 2015). This response occurred in the absence of inflammation (Naik et al., 2015). The accumulation of CD8+ T cells able to express IL-17A (Tc17) or IFN-γ (Tc1) is also a common feature of the skin tissue in non-human primates and humans under steady state conditions (Naik et al., 2015) (Figure 1A and S1A). Further, wild-caught outbred mice, that are exposed to physiological microbial partners (Beura et al., 2016), contained a significantly increased number of IL-17A and IFN-γ producing CD8+ T cells in the skin compared to specific pathogen free (SPF) C57BL/6 mice (Figure 1B). Skin accumulation of CD8+ T cells can also be reproduced in SPF mice colonized with defined skin commensals such as S. epidermidis (Naik et al., 2015). Taken together, these observations point towards the physiological relevance of skin-resident Tc17 and Tc1 and suggest homeostatic functionality.
Figure 1. Tc17 cells are enriched in the skin of non-human primates and mice.
A) Frequencies of IL-17A+ or IFN-γ+ CD8+ T cells isolated from Rhesus macaque. All cells were isolated from glabella. Left plot is gated on live CD45+ CD3ε+ T cells.
B) Numbers of total, IL-17A+, or IFN-γ+ CD8β+ T cells in naïve SPF or wild-caught mice. All cells were isolated from ear pinnae. Plots are gated on live CD45+ TCRβ+ CD8β+ T cells. Each dot represents an individual mouse.
C) Frequencies of CD8β+ T cells in control mice or mice topically associated with the indicated S. epidermidis isolate. S. epidermidis isolate highlighted in red (NIHLM087) is the strain used for all remaining experiments unless specified otherwise. All cells were isolated from ear pinnae. Plots are gated on live CD45+ TCRβ+ CD8β+ T cells. Each dot represents an individual mouse.
D) Phylogenetic tree of S. epidermidis clades and relative abundance plot of S. epidermidis clades across healthy volunteers. Individual bacterial clades are differentiated by color.
E) Frequencies and numbers of IL-17A+ or IFN-γ+ CD8+ T cells in unassociated (control) or two weeks post S. epidermidis (NIHLM087) association (contralateral side) isolated from Rhesus macaques. All cells were isolated from dorsal forearm or upper back. Left plot is gated on live CD45+ CD3ε+ T cells. Paired points denote same animal.
F) Frequencies and numbers of total, IL-17A+, or IFN-γ+ CD8β+ T cells in naïve SPF mice (control) or two weeks post S. epidermidis (NIHLM087) association. All cells were isolated from ear pinnae. Plots are gated on live CD45+ TCRβ+ CD8β+ T cells. Each dot in graph represents an individual mouse.
G) Representative confocal imaging volume projected along the z-axis of epidermal skin from mice 14 days post S. epidermidis (NIHLM087) association. Arrows show CD8α+IL-17A-Cre+, Rosa26-YFP+ (IL-17A-YFPFM=fate map) (top), CD8α+ T-bet-ZsGreen+ (bottom) cells. Scale bars, 20μm.
H) Expression of: KLRG1, CD69, CD103, CD122, CD127, CCR6, Granzyme B, T-bet, or RORγt by CD8β+ T cells in ear pinnae of mice, two weeks post S. epidermidis association. Plots are gated on live CD45+ TCRβ+ CD8β+ T cells. Solid Gray=CD44low CD8β+ T cells in skin draining lymph nodes. Blue line=IFN-γ+ CD8β+ T cells in ear pinnae. Red line=IL-17A+ CD8β+ T cells in ear pinnae.
(A–C, E, G) cells were stimulated with PMA and Ionomycin.
(B–C, F) Student’s t test was used to measure significance. **P<0.005, *** P<0.0005, **** P<0.0001.
(E) Paired student’s t test was used to measure significance. * P<0.05, **P<0.005
Data are representative of 2–3 independent experiments.
Using a collection of fully sequenced S. epidermidis isolates from both healthy donors and patients with nosocomial infections (Conlan et al., 2012), we found that while several isolates tested promoted the accumulation of Th1 and Th17 within the skin (Figure S1B), the ability of S. epidermidis to induce CD8+ T cell responses were restricted to isolates belonging to a specific clade (A20) (Figures 1C and 1D). This clade is highly represented on human skin and is enriched in healthy adults compared to pediatric atopic dermatitis patients (Byrd et al., 2017) (Figure 1D). Topical association of NIHLM087 (from A20 clade) to the skin of non-human primates (Rhesus macaque) also significantly increased the absolute number of Tc17 cells but not CD4+T cells in all animals tested (Figure 1E and S1C). Thus, the ability of S. epidermidis to promote skin CD8+ T cell responses is conserved in non-human primates.
In mice, Tc17 and Tc1 cells induced by NIHLM087 association (Figure 1F and 1G) shared phenotypic features with previously described pathogen induced skin resident memory CD8+ T cells. Notably, both subsets expressed high levels of CD69 and CD103, and low levels of killer cell lectin-like receptor G1 (KLRG1) and the IL-2Rβ subunit (CD122) (Figure 1H). Tc17 selectively expressed CCR6, RORγt and the IL-7Rα subunit (CD127), a phenotype that distinguishes them from both commensal-induced Tc1 (expressing T-bet, and bimodal CD103 and Granzyme B) and previously described skin TRM (Schenkel and Masopust, 2014) (Figure 1H).
Staphylococcus epidermidis-specific CD8+ T cells are restricted to MHCIb
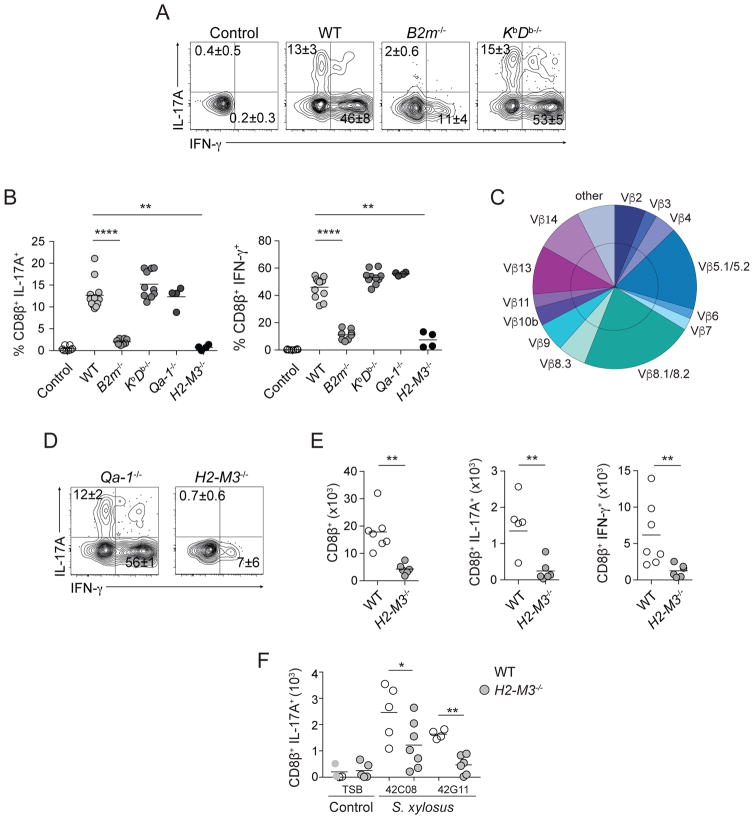
The homeostatic induction of CD8+ T cells in response to a non-invasive microbe led us to explore the possibility that unconventional mechanisms of antigen presentation may contribute to the initiation and/or maintenance of these responses. The S. epidermidis-specific CD8+ T cell response is β2-microglobulin (B2m)-dependent (Naik et al., 2015) (Figures 2A and 2B). The adaptor molecule B2m binds not only MHCIa molecules, but also to numerous MHCIb molecules (Rodgers and Cook, 2005). CD8+ T cells from previously associated wild-type (WT) mice were purified from the skin by cell sorting and incubated with KbDb−/− dendritic cells (DCs) loaded with heat-killed S. epidermidis. Both alleles of MHCIa are deleted in KbDb−/− DCs (Vugmeyster et al., 1998). Bacteria loaded KbDb−/− DCs recalled IL-17A and IFN-γ production in a manner comparable to WT DCs, indicating that S. epidermidis-induced CD8+ T cells were restricted to an MHCIb molecule (Figures 2A and B). Additionally, S. epidermidis loaded DCs isolated from several mouse strains (bearing non-complementary haplotypes) could recall CD8+ T cells similarly to DCs from C57BL/6 mice, formally demonstrating that CD8+ T cells isolated from the skin of S. epidermidis associated mice are restricted to a non-polymorphic B2m-dependent MHCIb molecule (Figure S2A).
Figure 2. S. epidermidis-specific CD8+ T cells are restricted to the MHCIb molecule H2-M3.
A,B,D) In vitro recall of commensal induced CD8+ T cells from skin with heat-killed S. epidermidis loaded splenic DCs. Frequencies of IFN-γ or IL-17A-producing CD8β+ T cells after overnight culture. Splenic dendritic cells were enriched from wild-type (WT), β 2m−/−, KbDb−/−, Qa-1−/−, or H2-M3−/− mice. Control=unloaded WT splenic DCs.
C) Frequencies of Vβ chain usage among total CD8β+ T cells from skin.
E) Numbers of total, IFN- γ, or IL-17A-producing CD8β+ T cells in S. epidermidis associated WT or H2-M3−/− mice. Cells were stimulated with PMA and Ionomycin.
F) Numbers of IL-17A+ CD8β+ T cells in WT or H2-M3−/− mice topically associated with TSB (control) or with the indicated S. xylosus isolate.
All cells were isolated from ear pinnae. Plots are gated on live CD45+ TCRβ+ CD8β+ T cells. Each dot represents an individual mouse. All cells were isolated from ear pinnae of S. epidermidis or S. xylosus associated mice 2 weeks after application. Plots are gated on live CD45+ TCRβ+ CD8β+ T cells. Each dot represents an individual mouse. Student’s t test was used to measure significance. *P<0.05, **P<0.005, *** P<0.0005. Data are representative of 2–4 independent experiments.
CD8+ T cell recall responses using S. epidermidis-conditioned media were equivalent to those obtained when DCs were loaded with heat-killed bacteria, supporting the idea that the antigen(s) sensed by the immune system were secreted (Figure S2B). Pre-incubation of the antigen preparation with the broad-spectrum serine protease Proteinase K (Pro.K) prior to DC loading abolished the response (Figure S2B). In addition, Tap1−/− DCs phenocopied WT DCs in our recall assay, consistent with studies showing that Tap1 is not necessary for certain MHCIb molecules (Rodgers and Cook, 2005) (Figure S2C). Together these results indicate that the S. epidermidis-derived antigen(s) recognized by CD8+ T cells come from secreted proteins, most likely peptides, processed and presented in a Tap1-independent manner.
CD8+ T cells induced by topical association with S. epidermidis utilized a diverse T cell receptor (TCR) Vβ usage pattern (Figure 2C). Among MHCIb molecules, Qa-1 and H2-M3 are known to bind to TCRs with diverse Vβ chains (Lindahl et al., 1997). Although these molecules are considered “middle-aged” compared to other MHCIb types, they emerged early in mammalian evolution (>65 million years ago), prior to the radiation of placental taxa (Doyle et al., 2003). Qa-1 and H2-M3 MHCIb each bind a limited repertoire of both self and microbe-derived peptides; Qa-1 is specific for MHCI leader peptides and viral peptides and H2-M3 is specific for mitochondrial and bacterial peptides (Godfrey et al., 2015; Lindahl et al., 1997). In vitro recall of WT CD8+ T cells with S. epidermidis-loaded Qa-1−/− DCs phenocopied the cytokine response elicited by WT loaded DCs (Figures 2B and D). However, IL-17A production by CD8+ T cells exposed to S. epidermidis-loaded H2-M3−/− DCs was abrogated and the IFN-γ response was severely reduced compared to S. epidermidis-loaded WT DCs (Figures 2B and D). To assess the relevance of this pathway in vivo, H2-M3−/− and WT mice were associated with S. epidermidis. Accumulation of both Tc17 and Tc1 was dramatically impaired in H2-M3−/− compared to WT associated mice (Figure 2E). Association of mice with isolates of a common murine skin commensal Staphylococcus xylosus also promoted CD8+ T cell accumulation in an H2-M3-dependent manner (Figure 2F). These results indicate that H2-M3 was required for the induction and/or function of CD8+ T cell responses to S. epidermidis and potentially to other dominant skin microbes.
S. epidermidis-specific CD8+ T cells respond to S. epidermidis-derived N-formyl methionine peptides
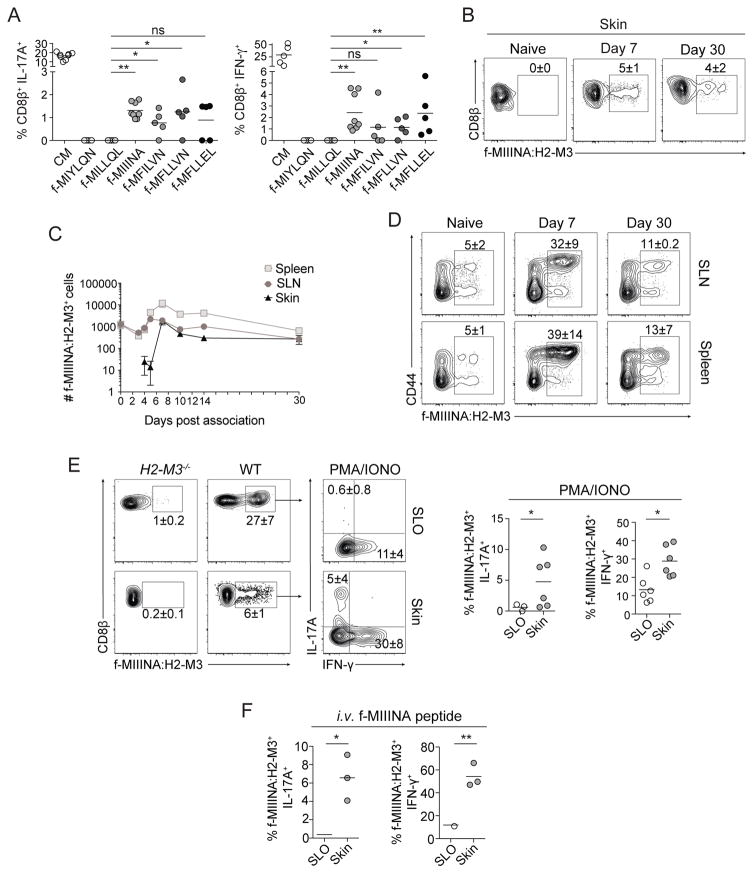
We next sought to identify the relevant bacterially-derived H2-M3-bound antigenic ligands. H2-M3 binds peptides that contain an N-formyl methionine (fMet), which is required to initiate protein translation in bacteria and mitochondria (Lindahl et al., 1997). We interrogated the S. epidermidis genome for fMet peptides able to bind H2-M3, using a previously described algorithm that identifies peptides likely to bind H2-M3 based on a known sequence motif (Chun et al., 2001). Analysis of the S. epidermidis genome revealed 30 fMet peptides bearing this motif (Figure S3A). Two fMet peptides (f-MIIINA and f-MFLLVN) could drive both IL-17A and IFN-γ production in vitro by S. epidermidis-elicited CD8+ T cells (Figures 3A and S3B).
Figure 3. H2-M3 restricted CD8+ T cells are specific to S. epidermidis derived fMet peptide ligands.
A) In vitro recall of S. epidermidis-induced CD8+ T cells from FACsorted skin with S. epidermidis conditioned medium (CM) or fMet peptide loaded splenic DCs. Frequencies of IFN-γ or IL-17A-producing CD8β+ T cells after overnight culture. Gated on live CD45+ TCRβ+ CD8β+ T cells. Splenic dendritic cells were enriched from wild-type mice.
B–F) Frequencies and numbers of f-MIIINA:H2-M3 tetramer positive cells or tetramer positive IFN-γ or IL-17A-producing CD8β+ T cells in (B,C,E,F) ear pinnae, (C–D) magnetic bead enriched spleen or skin draining lymph nodes (SLN), or (E–F) magnetic bead enriched combined spleen and skin draining lymph nodes (SLO) of (B–F) intact WT (SPF) or (E) H2-M3−/− mice.
B–D) cells were isolated from naïve mice or mice after indicated time-point following first S. epidermidis skin application.
C) Data are represented as mean ± SEM.
E) Cells were stimulated with PMA and Ionomycin.
F) Mice were injected with 100μg f-MIIINA peptide intravenously and tissues harvested after three hours.
E–F) All cells were isolated from S. epidermidis associated mice 2 weeks after application.
B–F) Plots are gated on live CD45+ Lineage (MHCII, CD11b, γδTCR)− CD90.2+ CD3ε+ CD8β+ T cells.
Student’s t test was used to measure significance. * P<0.05, ** P<0.005. Data are representative of 2–4 independent experiments.
In order to track S. epidermidis-specific CD8+ T cell responses in vivo, we generated an fMet peptide:H2-M3 tetramer reagent using f-MIIINA, the fMet peptide that triggered the strongest cytokine response during in vitro recall of S. epidermidis-specific CD8+ T cells (Figure 3A and S3B). In naïve mice, no tetramer positive cells were detected within the skin compartment. However, 7 days post-association with S. epidermidis, f-MIIINA:H2-M3+CD8+ cells began to accumulate in this tissue (Figure 3B and C). Accumulation within peripheral tissues was restricted to the skin and not detectable at other barrier sites such as the gastrointestinal tract (data not shown). From this time point and onward, ~5–10% of total CD8+ T cells within the skin were f-MIIINA:H2-M3-specific, indicating that the response against this specific peptide was dominant and sustained for at least one month post microbial colonization (Figures 3B and C).
We next assessed how systemic the response could be post skin association by using a previously described method of tetramer staining followed by magnetic bead enrichment to identify all f-MIIINA:H2-M3-specific cells in skin draining lymph nodes (SLN) and spleen (Moon et al., 2007). We confirmed enrichment efficiency (Figures 3D and S3D) and that the tetramer bound MHCI-restricted CD8+ T cells, but not MHCII-restricted CD4+ T cells (Figure S3C). As an additional control for specificity, we stained the SLO of naïve mice with two f-MIIINA:H2-M3 tetramers each containing either phycoerythrin or allophycocyanin fluorochromes (Obar et al., 2008). Approximately 80% of tetramer binding cells bound to both tetramers, indicating this reagent stains f-MIIINA:H2-M3-specific cells with a high degree of specificity (Figure S3E). Next, we enumerated f-MIIINA:H2-M3-specific T cells within skin draining lymph nodes and spleen and detected an expanded population at day 5 post-association, a time point that preceded skin accumulation (Figure 3C). f-MIIINA:H2-M3-specific cell activation was first detectable at day 2 after S. epidermidis skin association in the skin draining lymph nodes (but not in the spleen) as evidenced by upregulation of CD69 (Figure S3F). These data support the idea that activation and expansion of H2-M3-restricted T cells first occurs in lymphoid structures, similar to conventional T cell responses. Further, these results reveal that response to a skin commensal is not restricted to the skin draining lymph nodes and can be more systemic than previously appreciated.
In naïve mice, following tetramer staining and magnetic bead enrichment, we identified ~2500 f-MIIINA:H2-M3-specific cells in the pooled SLO (Figures 3C and D). Of note, the number of f-MIIINA:H2-M3-specific CD8+ cells in naïve mice is high compared to the average number of naïve CD8+ T cell precursors specific to foreign antigens presented by conventional MHCIa pathways (~200 cells) (Obar et al., 2008). About half of the f-MIIINA:H2-M3-specific cells in pooled SLO were CD44hi, supporting the idea that these T cell receptors may have previously responded to other peptide:MHCI complexes. To test the possibility that S. epidermidis f-MIIINA:H2-M3-specific cells in naïve SPF mice may be cross-reactive with pre-existing members of the microbiota, we assessed the frequency and phenotype of f-MIIINA:H2-M3-specific T cells in mice raised in the absence of live microbes (gnotobiotic). Surprisingly, the frequency of CD44hi f-MIIINA:H2-M3-specific CD8+ T cells was comparable between germ-free and SPF mice (Figure S3G).
Previous work investigating responses to Listeria monocytogenes infection identified that H2-M3-restricted TCRs can be cross-reactive (Ploss et al., 2003). To test whether f-MIIINA:H2-M3-specific cells were cross-reactive to a previously described Listeria-derived H2-M3-binding fMet peptide, we utilized a previously described method of double tetramer staining (Kerksiek et al., 2001) of SLO from naïve SPF mice with f-MIGWII:H2-M3 and f-MIIINA:H2-M3 tetramers in the same sample and observed that staining was exclusive (Figure S3H). To address the possibility that some of the CD8+ T cells elicited by S. epidermidis could promiscuously respond to the thirteen mitochondrial-encoded or other previously identified microbe-derived H2-M3-binding fMet peptides, we recalled S. epidermidis elicited CD8+ T cells in vitro with previously described fMet peptide epitopes derived from mitochondria or Listeria monocytogenes (Lindahl et al., 1997). Mitochondrial derived peptides did not significantly recall CD8+ T cells above background (Figure S3I). Thus, CD8+ T cells induced by S. epidermidis may not be broadly promiscuous when induced under homeostatic conditions. While we cannot exclude the possibility of cross-reactivity with other bacterial-derived fMet peptides or with mitochondrial-derived peptides during development, these results further support the idea that CD8+ T cells elicited by S. epidermidis skin association are specific to S. epidermidis-derived fMet peptides and that CD44hi f-MIIINA:H2-M3-specific cells may exist in naïve mice as a result of positive selection on hematopoietic cells in the thymus as previously reported (Cho et al., 2011).
Stimulation of f-MIIINA:H2-M3-specific cells promoted IFN-γ production in all compartments analyzed at 14 days post-association (Figure 3E). Conversely, and consistent with the role of skin-derived IL-1 in licensing cytokine production in this model (Naik et al., 2012), IL-17A was only detected within the skin and IFN-γ production was significantly enhanced in both cell frequency and intensity in this compartment (Figure 3E). While IL-17A and IFN-γ production by f-MIIINA:H2-M3-specific CD8+ T cells mirrored that of the total CD8+ T cell population, tetramer staining allowed a significantly higher f-MIIINA:H2-M3-specific CD8+ T cell detection frequency (Figure 3B) than detectable via in vitro peptide recall (Figure 3A). As expected, f-MIIINA:H2-M3-specific cells were absent in S. epidermidis associated H2-M3−/− mice (Figure 3E). When f-MIIINA:H2-M3-specific CD8+ T cells were re-stimulated in vivo via intravenous peptide injection, we detected a similarly enhanced IFN-γ and IL-17A production within the skin compartment compared to SLO (Figure 3F). This indicates that tissue regulation of cytokine production by commensal-specific CD8+ T cells occurs proximal to T cell receptor signaling machinery.
Together these results uncover a canonical class of bacterial antigens (fMet peptides) as potent inducers of immunity to a non-invasive microbe. Further, these results show that even in the absence of inflammation, the response to a skin microbe can be broad and systemic.
Commensal-specific CD8+ T cells express a immunoregulatory/tissue repair signature
Having determined that S. epidermidis-induced CD8+ T cells have a distinct cytokine profile and an unconventional MHCI restriction, we next assessed if these cells expressed a defined transcriptional profile. To this end, we compared S. epidermidis-induced CD8+ T cells to distinct CD8+ T cell subsets (Tc17 and Tc1) isolated from the skin in the context of distinct infections or inflammatory states (Figure S4A). S. epidermidis-induced CD8+ T cells were isolated as CD8+ CCR6+ (Tc17) or CD8+ CCR6- (Tc1) populations (Figure 1H). The same strategy was utilized for isolation of Tc17 and Tc1 cells from inflammatory contexts. Notably, Tc1 and Tc17 induced by topical association with S. epidermidis were compared to: 1) CD8+ T cells induced by S. epidermidis in its pathogenic form via intradermal (i.d.) injection, consisting of exclusively Tc1 of which 50% are H2-M3-restricted (Figure S4B); 2) Tc1 cells elicited upon acute skin infection with Herpes simplex virus (HSV)(Gebhardt et al., 2009); 3) Tc1 cells elicited to skin during chronic infection with Leishmania major (Belkaid et al., 2002); 4) Skin Tc17 induced in the context of experimental psoriasis triggered by treatment with the TLR7 agonist Imiquimod (Figure S4C). These multiple comparisons allowed us to control for location (skin), subset (CD8+ T cells), differentiation (Tc1/Tc17), inflammation (homeostasis/acute/chronic) and mode of interaction with a microbe (colonization versus barrier breach and infection). Dendritic epidermal T cells (DETC), a population of epidermal-resident innate lymphocytes, TEM, and naïve CD8+ T cells from SLOs were also used as reference populations (Figure S4A).
We normalized isolation of cell populations post-S. epidermidis association and S. epidermidis or HSV infection at two weeks post treatment. At this time point, the non-inflammatory CD8+ T cell response to topical S. epidermidis was stable (Figure 3C) and inflammation induced by HSV (van Lint et al., 2004) and intradermal S. epidermidis injection has resolved (Naik et al., 2015). Tc17 cells induced by Imiquimod treatment were purified at 10 days post-initial treatment and Tc1 cells induced by L. major skin infection were purified during the chronic stage of the infection (30 days post-infection).
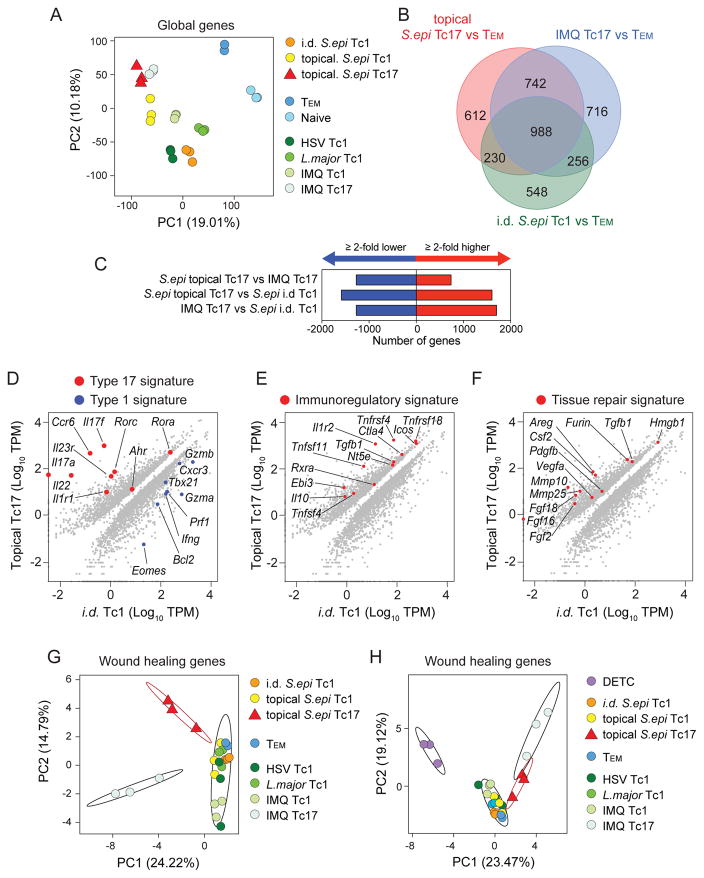
Based upon global transcriptome signatures from RNA-seq, principle component analysis recapitulated the distinct nature of these T cell populations, (Figure 4A, Figure S4D). After comparison to TEM cells, we focused on transcripts either common or unique to S. epidermidis-induced Tc17, Imiquimod-induced Tc17, and Tc1-elicited by i.d. infection with S. epidermidis (Figure 4B). We identified a core signature of ~1000 genes shared amongst these populations (Figure 4B). When compared to Tc17 cells induced by Imiquimod, S. epidermidis-induced Tc17 cells differentially expressed over 2000 genes (Figure 4C).
Figure 4. Commensal-specific Tc17 cells are a distinct population and enriched in transcripts associated with immune regulation and tissue repair.
A) Principle component analysis of global gene expression from FACsorted populations identified in (Figure S4A).
B) Venn diagram from RNA-seq analysis of genes differentially expressed in pairwise comparisons between topical S. epidermidis-elicited Tc17, imiquimod (IMQ)-elicited Tc17, and intradermal (i.d.) S. epidermidis-elicited Tc1 cells in skin relative to skin draining lymph node Tem cells in naïve mice. Numbers of genes with greater >2-fold increase relative to Tem are shown.
C) Numbers of genes with a >2-fold increase (red) or decrease (blue) in expression levels between the indicated populations.
D–F) Diagonal plots showing differentially expressed genes (>2-fold, Padj<0.05) comparing topical S. epidermidis-elicited Tc17 to i.d. S. epidermidis-elicited Tc1 cells in skin.
G–H) Principle component analysis of curated wound healing gene expression from FACsorted populations identified in (Figure S4A). Ellipses denote 95% confidence intervals of the mean of selected cell populations: topical S. epidermidis-elicited Tc17, imiquimod (IMQ)-elicited Tc17, and all others.
We next compared cells generated by the same microbe under steady state or inflammatory conditions. Tc17 induced by topical association with S. epidermidis and Tc1 elicited by infection with S. epidermidis demonstrated distinct gene expression profiles, underlined by Type 17 (Rorc, Il17a, Il17f, Il22, Csf2, Ahr) and Type 1 (Eomes, Tbx21, Gzma, Gzmb, Prf1, Cxcr3) signatures, respectively (Figure 4D). Of note, many of the genes elevated in S. epidermidis-specific Tc17 cells compared to other populations were associated with immune regulation and regulatory T cells including the Tumor Necrosis Factor family members: Tnfsf4 (OX40L), Tnfsf11 (RANKL), Tnfrsf4 (OX40), Tnfrsf11a (RANK), the transcription factor: Rxra (Retinoid X receptor alpha), as well as transcripts for the cytokine Il10 and the cytokine subunit Ebi3 (IL-27 beta) (Figure 4E). Strikingly, S. epidermidis-elicited Tc17 cells also expressed higher levels of multiple transcripts associated with tissue repair relative to Tc1 (Figure 4F). Utilizing a curated list of tissue repair genes (Werner and Grose, 2003), we identified that S. epidermidis-elicited Tc17 cells were enriched in expression of a range of pleiotropic factors involved in various aspects of wound healing, such as angiogenesis (Csf2, Fgf2, Vegfa, Pdgfb), chemotaxis (Pdgfb, Csf2, Tgfb1, Furin), tissue remodeling (Mmp10, Mmp25), and extracellular matrix production (Fgf2, Pdgfb, Tgfb1, Furin)(Werner and Grose, 2003) (Figure 4F). A large number of these transcripts are factors known to affect keratinocyte proliferation: Areg, Fgf2, Fgf16, Fgf18, Tgfb1, Furin, and Pdgfb (Werner and Grose, 2003). Type 17, immunoregulatory and tissue repair signatures were also identified when S. epidermidis-specific Tc17 cells were compared to either HSV-specific Tc1 cells or topical S. epidermidis-specific Tc1 cells (Figure S4E).
Given the tissue repair gene signature of Tc17 cells elicited by skin colonization with S. epidermidis, we sought to comprehensively compare the expression of wound healing genes among all cell populations. Using principle component analysis to reduce data dimensionality, we found that the wound healing gene expression profile of Tc17 cells elicited by S. epidermidis was indeed distinct from all CD8+ T cell populations (including Imiquimod Tc17) (Figure 4G). At the global gene level as well as based on our tissue repair gene list, DETC, a population previously shown to contribute to tissue repair (Jameson et al., 2002) was also highly distinct from all the other cell subsets (Figure 4H and S4D). Focused analysis of tissue repair genes expressed by S. epidermidis-specific Tc17 cells and DETC identified distinct expression for genes previously associated with DETC function (Figure S4F) (Fgf7, Fgf10 and Igf1) (Nielsen et al., 2017). Additionally, we identified Fgf2, Csf2, Mmp10, Mmp25 as transcripts selectively expressed by S. epidermidis induced Tc17 (Figure S4G) and Fgf16, Fgf18, Tgfb1, Pdgfb, Hmgb1 and Furin as highly expressed by both S. epidermidis Tc17 and DETC populations (Figure S4H). Together, these results support the idea that S. epidermidis-elicited Tc17 cells represent a unique population and that T cells induced by commensals may contribute to tissue repair.
Commensal-specific CD8+ T cells accelerate wound healing
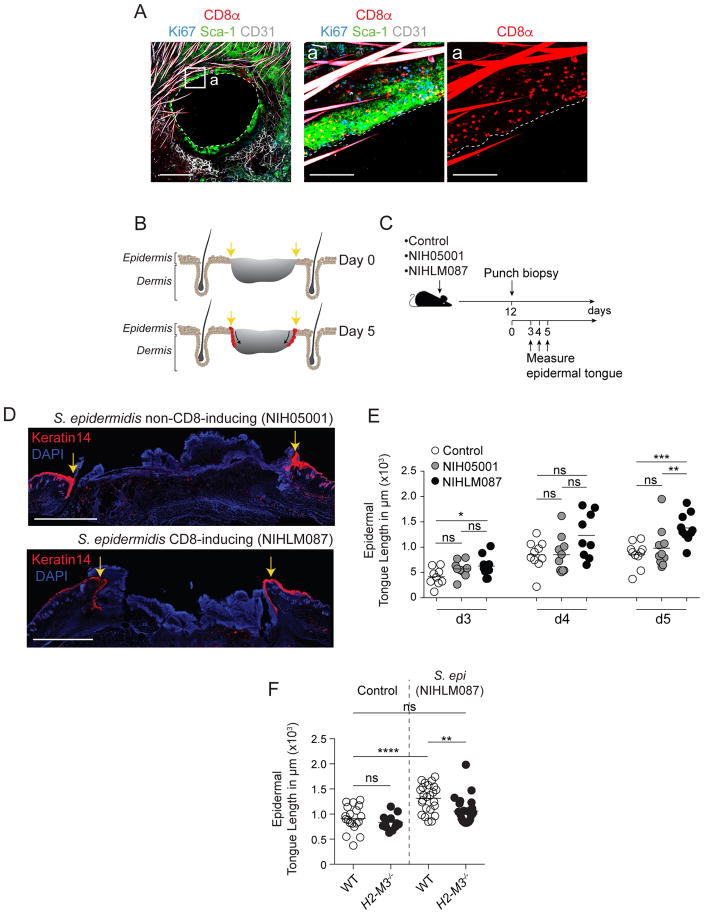
The skin is the most exposed body surface and provides protection from mechanical injury, chemical hazard and microbial invasion and therefore, tissue repair is of the utmost importance at this site. The tissue repair gene expression signature of CD8+ T cells induced by commensals prompted us to address the possibility that, in addition to their protective properties, these cells may contribute to tissue repair. A model of skin wounding induced by a punch biopsy in the ear pinnae of mice previously associated with S. epidermidis revealed a striking accumulation of the CD8+ T cells at the wound edge as shown at 3 days post injury (Figure 5A). We next utilized an approach of backskin punch biopsy that allows precise quantification of repair (Keyes et al., 2016). In this model, progression of the healing wound can be measured via the length of the proliferating epidermal keratinocyte tongue that migrates into the wound bed over time, eventually resulting in wound closure (Figure 5B). Wounds were performed at 12 days post-association with S. epidermidis (NIHLM087), a time-point near the peak of the CD8+ T cell response with sustained low bacterial burden (Figure S5A). To control for bacterial exposure and potential innate immunity contribution (Lai et al., 2010), we included a group associated with a non-CD8-inducing S. epidermidis strain (NIH05001) (Figures 1C and 5C). Wounds were measured 3, 4, and 5 days post-wounding, a time span that was previously shown to have the highest re-epithelialization activity (Keyes et al., 2016) (Figure 5C). We identified significant differences in basal keratinocyte epidermal tongue length on day 3 and 5 between wounds in mice that were previously associated with the CD8+ T cell inducing S. epidermidis strain as compared to one or more control groups (Figures 5D and 5E). Thus, S. epidermidis-induced CD8+ T cells promoted more rapid epidermal keratinocyte tongue progression and therefore accelerated wound healing. This response was independent of the presence of DETC (Figure S5B). Accelerated healing was not associated with changes in inflammatory infiltrates between mice associated with CD8-inducing or non-inducing strains (Figure S5C). To assess the contribution of S. epidermidis-induced CD8+ T cells to this process, we measured epidermal tongue length in wounds of WT and H2-M3 deficient mice that were previously topically associated with S. epidermidis. The ability of S. epidermidis to accelerate this process was significantly reduced in H2-M3−/− mice formally linking non-classical MHCI restricted cells, the microbiota, and tissue repair (Figure 5F). Differences in tissue repair between WT and H2-M3−/− mice was not associated with changes in skin hematopoietic cell infiltrate (Figure S5D), but with a transient and discrete change in tissue gene expression associated with muscle responses (Figure S5E and F) supporting the idea that at least one of the targets of this process may be the arrector pili muscles (Torkamani et al., 2017). Together, these results identify an important role for commensal-specific, H2-M3-restricted CD8+ T cells in driving enhanced tissue repair.
Figure 5. Commensal-specific CD8+ T cells accelerate wound healing.
A) Representative imaging of whole mount ear pinnae at 3 days after 2.5mm ear punch biopsy from mice (15 days post initial S. epidermidis association). Whole mounts were stained for CD8+ T cells (CD8α), keratinocytes (Sca-1), blood vessels (CD31), cycling cells (Ki67) and imaged by confocal microscopy. Scale bars: 150μm, 1000μm, and 1000μm, respectively. Hair is autofluorescent in CD8α and CD31 channels.
B) Schematic depicting progress of re-epithelialization over time after wounding. Yellow arrows denote wound edge. Red indicates advancing epidermal tongue of basal keratinocytes that stains brightly for Keratin 14.
C) Mice were topically associated with growth medium alone (control), non-CD8+ T cell inducing S. epidermidis isolate (NIH05001), or CD8+ T cell inducing S. epidermidis isolate (NIHLM087) followed by a backskin punch biopsy twelve days later. Epidermal keratinocyte tongue length was measured at days 3,4, and 5 post wounding.
D) Immunofluorescence images of backskin wounds at day 5 after punch biopsy. Sections are immunolabeled for basal epidermal keratinocytes (K14) and co-stained with DAPI. Scale bars, 1000μm. Yellow arrows denote wound edge.
E) Quantification of the length of the tongue of epidermal keratinocytes that migrate in from the wound edges during re-epithelialization of the wound bed in WT mice. Mice were topically skin associated with growth medium (control), a non-CD8+ T cell inducing S. epidermidis (NIH5001) strain, or a CD8+ T cell inducing S. epidermidis (NIHLM087) strain. Tongue lengths were measured at 3–5 days after backskin punch biopsy.
F) Quantification of the length of the tongue of epidermal keratinocytes that migrate in from the wound edges during re-epithelialization of the wound bed in WT or H2-M3−/− mice. Mice were topically skin associated with growth medium (control) or NIHLM087. Tongue lengths were measured at 5 days after backskin punch biopsy.
Student’s t test was used to measure significance. * P<0.05, ** P<0.005, *** P<0.0005, **** P<0.0001. Data are representative of 2–4 independent experiments.
DISCUSSION
Most of what we understand today about the function of the immune system comes from the exploration of responses to pathogenic microbes. However, the vast majority of immune system-microbial encounters are those resulting from the symbiotic relationship with the microbiota (Belkaid and Hand, 2014). Here we show that this fundamental class of immunity can promote specific immune-microbial antigen dialogue and is endowed with pleiotropic functions.
Our present results propose that the symbiotic dialogue between the microbiota and the immune system could be dominantly mediated by non-classical MHCI molecules. Non-polymorphic MHC representatives, referred to as non-classical MHC for those encoded by genes located within the MHC region and MHC-like molecules for those outside the MHC, exist for both class I and class II proteins, the most diverse being those related to the MHC class I molecules (Adams and Luoma, 2013). While these molecules have been shown to contribute to the recognition of pathogenic microbes, a protective role for these responses has only recently been addressed (Seaman et al., 2000; Shang et al., 2016; Van Rhijn et al., 2015). Our present results propose that non-classical MHCI molecules may have been dominantly maintained as a means to engage in a dialogue with the microbiota. In support of this, barrier sites are enriched in cells restricted to non-classical MHCI presentation including NK cells, NKT cells, γδ T cells, MAIT cells (Fan and Rudensky, 2016), and as shown here, H2-M3-restricted CD8+ T cells. Although previous work has identified that H2-M3-restricted TCRs can be promiscuous (Ploss et al., 2003), our work supports the idea that H2-M3-restricted CD8+ T cells to a commensal, at least under conditions of homeostatic immunity, may not be as cross-reactive.
While our present results highlight a role for H2-M3 in mice, it has been suggested that an analog may exist in humans. Precedence for this comes from the identification of the analog to the mouse MHCIb molecule Qa-1 in humans, which was identified as HLA-E. These two molecules have little amino acid sequence homology, yet bind similar peptides (Rodgers and Cook, 2005). Further, many non-classical MHCI molecules exist in humans that may serve a similar purpose. Intriguingly, association of non-human primate skin with S. epidermidis was sufficient to significantly enhance skin Tc17. The nature of the potential MHC I involved in such setting remains to be identified.
Of particular relevance to our present observation, MHCIb-restricted immune cell populations have been shown to be enriched at barrier sites during early life (Gensollen et al., 2016), a time of extraordinary and dynamic exposure to the microbiota. For instance, bacteria derived sphingolipids can interact via non-classical CD1d molecule with NKT cells early in life, a response that shapes subsequent susceptibility to inflammation (An et al., 2014). The ability of MHCIb molecules to present antigens with specific motifs, which can be derived from a large constituency of the microbiota, place immune cells that recognize them as ideal candidates for the constitutive sensing and recognition of microbiota-derived antigens or metabolites.
One remarkable feature of the CD8+ T cells induced by S. epidermidis is their unusual and complex signature that distinguishes them from canonical CD8+ T cells. Whether this unique phenotype results from the engagement of MHCIb molecules or the non-inflammatory setting surrounding these responses remains to be addressed. Nonetheless, we can speculate that induction of immunity under homeostatic conditions may poise cells to develop a less polarized phenotype and may sustain the development of pleiotropic programs. Notably, CD8+ T cells induced by S. epidermidis express a high number of transcripts associated with tissue repair compared to CD8+ T cells induced by pathogenic encounter and were able to promote skin repair. In the skin, tissue repair is of the utmost importance and complex and redundant pathways converge to promote this vital process. For instance, DETC were also previously shown to promote tissue repair (Nielsen et al., 2017) but were not required for the tissue repair effect of S. epidermidis, supporting the idea that these two cell subsets play distinct roles in this process. A link between the microbiota and tissue repair was previously provided by the observation that, in the gut, TLR activation by commensals was required to promote tissue repair and host survival following acute injury (Rakoff-Nahoum et al., 2004). In the skin, a defined product of S. epidermidis, Lipoteichoic Acid (LTA), can also mitigate inflammation via its capacity to bind to Toll Like receptor (TLR) 2, thereby promoting wound healing (Lai et al., 2009). While we do not exclude a synergistic role for this molecule, in our setting we found that S. epidermidis (NIH5001) decorated with LTA, but unable to induce CD8+ T cells failed to promote tissue repair. Of interest, no differences in cellular recruitment were observed in the presence or absence of H2-M3 molecules post injury in the context of S. epidermidis suggesting that T cells induced by this commensal may target the non-hematopoietic compartment. The impact of CD8+ T cells induced by S. epidermidis was detectable during the proliferation phase of wound healing, perhaps through an effect on keratinocyte proliferation. Notably, CD8+ T cells induced by S. epidermidis express high levels of amphiregulin, a molecule described for its mitogenic role on keratinocytes through binding the epidermal growth factor receptor (EGFR) (Werner and Grose, 2003). Additionally, these cells expressed fibroblast growth factor (FGF) family members with previously identified roles as keratinocyte mitogens (Kawano et al., 2005; Werner and Grose, 2003). This production of an EGFR ligand and FGFs by commensal-specific CD8+ T cells supports the idea that these cells may utilize redundant mechanisms to promote tissue repair. We also found that CD8+ T cells induced by S. epidermidis localized to the edge of the wound post-injury, a tropism that may serve the dual purpose of enhancing keratinocyte proliferation while at the same time promoting antimicrobial defense at a site of high vulnerability. In addition to keratinocytes, our results support the idea that S. epidermidis-induced CD8+ T cells may also impact the skin muscle compartment. The skin is enriched in arrector pili muscles that are attached to hair follicles and these muscles have been proposed to contribute to the maintenance of hair follicle integrity (Torkamani et al., 2017). How commensal-specific T cells restore the function of these structures and how such process contribute to tissue repair remains to be addressed.
There are at least two possible mechanisms to explain how commensal-specific CD8+ T cells are able to sense and respond to tissue injury. The first could be in an innate manner, through binding of signal molecules to receptors other than the TCR, the other could be through TCR binding to S. epidermidis fMet peptide:H2-M3 ligands made available via barrier breach. Such phenomenon may be applicable to a large fraction of cells residing at barrier sites. Indeed, because of the extraordinary number of potential antigens expressed by the microbiota, a significant fraction of resident lymphocytes including non-conventional lymphocytes are expected to be microbiota specific. Consequently, any barrier breach mediated by infection or injury is likely to occur in the context of a much broader recall response against diverse microbial antigens (Hand et al., 2012), a response that as we show here could also contribute to tissue repair.
Together, our work proposes that the rules and properties of immunity to the microbiota are distinct from those induced by pathogenic encounters. Here, we uncover some defining features of commensal-specific immunity, a pleiotropic class of immunity that we show to be highly entwined with tissue physiology. These findings may have important clinical implications. Indeed, despite the importance of healing for host survival, mechanisms underpinning tissue repair and its failure to heal are still poorly understood and current therapies are limited (Eming et al., 2014). As such, poor wound healing after trauma, surgery, acute illness, or chronic disease conditions remains a serious public health concern (Eming et al., 2014). In this context, by uncovering a link between commensal-specific immunity/non-classical MHC and tissue repair, our work may open the door to novel therapeutic interventions based on the physiological integration of host microbiota interactions.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yasmine Belkaid (YBelkaid@niaid.nih.gov).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6NTac and SJL/JCrNTac (SJL) specific pathogen-free mice were purchased from Taconic Farms. Germ-free C57BL/6 mice were bred and maintained in the NIAID Microbiome Program gnotobiotic animal facility. B6.SJL (B6.SJL-Cd45a(Ly5a)/Nai), B10.A (B10.A-Cd45a(Ly5a)/NAi N5), T-bet-ZsGreen (C57BL/6-Tbet-ZsGreen[Tg]), and B2m−/− (B6.129-B2mtm1Jae N12) mice were obtained through the NIAID-Taconic exchange program. BALB/c, FVB, KbDb−/− (B6.129P2-H2-Kbtm1 H2-Dbtm1 N12), R26-stop-EYFP (B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J), Tap1−/− (B6.129S2-Tap1tm1Arp/J) and TCRδ−/− (B6.129P2-Tcrdtm1Mom/J) mice were purchase from the Jackson Laboratory. IL-17A-Cre mice were kindly provided by Dr. B. Stockinger (The Francis Crick Institute). H2-M3−/− mice were kindly provided by Dr. C. Wang (Northwestern University) and bred in house with WT C57BL/6. Qa-1−/− mouse spleens were kindly provided by Dr. H Cantor (Harvard University). Congenic β2m−/− (B6.129-B2mtm1Jae N12 x B6-LY5.2/Cr) were kindly provided by Dr. R. Bosselut (National Cancer Institute, NIH). All mice were bred and maintained under pathogen-free conditions at an American Association for the Accreditation of Laboratory Animal Care (AAALAC)-accredited animal facility at the NIAID and housed in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals. All experiments were performed at the NIAID under an animal study proposal (LPD11E) approved by the NIAID Animal Care and Use Committee. Sex and age-matched mice between 6 and 12 weeks of age were used for each experiment. Wild Mus musculus domesticus were trapped in Maryland. Trapping was carried out in concordance with local laws and was carried out with the approval of the relevant regulatory bodies under protocol approved by the NIDDK Animal Care and Use Committee.
Non-human primates
Non-human primates were housed at the NIH animal center. All procedures were carried out under Ketamine anesthesia by trained personnel under the supervision of veterinary staff and all efforts were made to ameliorate the welfare and to minimize animal suffering in accordance with the “Weatherall report for the use of non-human primates” recommendations under animal protocol LPD26. Animals were housed in adjoining individual primate cages allowing social interactions, under controlled conditions of humidity, temperature and light (12-hour light/12-hour dark cycles). Food and water were available ad libitum. Animals were monitored twice daily and fed commercial monkey chow, treats and fruit twice daily by trained personnel. The study included 3 adult rhesus macaques (2 males and 1 female). All monkeys were housed and cared in accordance with AAALAC standards in AAALAC-accredited facilities, and all animal procedures performed according to protocols approved by the NIAID Animal Care and Use Committee.
METHODS DETAILS
Topical association and intradermal infection of mice with Staphylococcus epidermidis
Staphylococcus epidermidis strains (NIHLM040, NIHLM095, NIHLM061, NIHLM088, NIH05001, and NIHLM087) and Staphylococcus xylosus strains (42C08 and 42G11) were cultured for 18 hours in tryptic soy broth at 37°C. For topical association of bacteria, each mouse was associated by placing 5 ml of the bacterial suspension (approximately 109 CFU/ml) across the entire skin surface (approximately 36 cm2) using a sterile cotton swab. Application of bacterial suspension was repeated every other day a total of four times. In experiments involving topical application of various bacterial species or strains, 18-hour cultures were normalized using OD600 to achieve similar bacterial density (approximately 109 CFU/ml). For some experiments, mice were infected intradermally in the ear pinnae with 107 c.f.u. of S. epidermidis strain NIHLM087.
Leishmania major and HSV-1 skin infections
C57BL/6 mice were infected intradermally in the ear pinnae with L. major (5×103 metacyclic promastigotes) using a 29 gauge needle in a 10μl volume. For HSV infection, C57BL/6 mice were anaesthetized with isofluorane and areas of the flank were depilated with Nair. The next day mice were anaesthetized with Avertin and a 2 cm2 area of the flank was abraded for 5 s with a Dremel 4000 variable speed tool (setting 10) containing a 1-cm width medium grit sanding bit. 1×106 plaque forming units (PFU) of HSV-1 (F) in DMEM containing 10% FBS was applied and gently rubbed into the abraded area with a pipette tip. Mice were maintained with the flank exposed until recovery. Mice that received a productive skin infection were sacrificed 14 days later and flanks were processed for cell sorting.
Imiquimod induced psoriasis-like dermatitis
Mice between 6 and 8 weeks of age were treated daily for 7 days on ear pinnae with 10 mg of 5% imiquimod (IMQ) cream (Aldara Cream 5%; 3M Health Care, UK) (adapted from (van der Fits et al., 2009)).
Non-human primate skin and association with Staphylococcus. epidermidis
For characterization of CD8+ T cells at steady state, non-human primate skin tissue was obtained from the glabella of 4 healthy rhesus macaques (Macaca mulatta). For association with S. epidermidis: rhesus macaques (RM) were anesthetized with ketamine and a 5-cm x 5-cm area of each dorsal forearm and each side of upper back was shaved with a hair clipper (4 sites total per RM). As a control, two 6-mm biopsies were taken from forearm and upper back of one side of each RM for T cell analysis prior to S. epidermidis association. This was done to prevent contamination of the control site with bacteria. Next, contralateral forearm and upper back sites were topically associated with a culture of S. epidermidis soaked onto a 5-cm x 5-cm piece of gauze dressing and taped to the site for one hour prior to removal. Topical application was repeated every other day for a total of 4 applications. Fourteen days later two 6-mm punch biopsies were taken from each contralateral site and analyzed for T cell responses. Values from the two biopsies from each site were averaged when computing frequencies and total numbers of T cells.
Staphylococcus epidermidis conditioned medium preparation
NIHLM087 was cultured for 24 hours in IMDM (Invitrogen) supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate and nonessential amino acids, 20 mM HEPES with shaking at 37°C. Bacterial culture was centrifuged at 3200 g for 30 minutes. Supernatant was then filtered through 0.22 μm filter (Millipore). For experiments that involved proteinase K digestion (Sigma), 5×109 cfu heat-killed NIHLM087 in PBS or 1mL of NIHLM087 conditioned medium was incubated with β-mercaptoethanol (1mM final concentration, Sigma) and 300μg Proteinase K. The solutions were incubated for 24 hours at 56°C followed by a 95°C, 30 minutes denatu ration step.
Tissue processing
Murine tissues
Cells from lymph nodes and spleen were mashed through a 70-micron cell strainer to generate single cell suspensions. Ears were excised and separated into ventral and dorsal sheets. Flank (dorsal) skin was shaved with chrom mini (Wahl), subcutaneous fat tissue was removed with a number 10 scalpel, and skin was cut into 1cm by 1cm pieces. Ear pinnae and flank tissues were digested in digestion media (RPMI 1640 media supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate and nonessential amino acids, 55 μM β-mercaptoethanol, 20 mM HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin and 0.25 mg/ml Liberase TL purified enzyme blend, Roche), and incubated for 2 hours at 37°C and 5% CO2. Digested skin sheets were homogenized using the Medicon/Medimachine tissue homogenizer system (Becton Dickinson).
Non-human primate skin
Subcutaneous fat tissue was scraped off with a number 10 scalpel blade. Skin tissue was then weighted and perfused with 100–500 μl of digestion media (RPMI 1640 media supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate and nonessential amino acids, 20 mM HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin and 0.25 mg/ml Liberase TL purified enzyme blend, Roche). Perfused skin tissue was placed in 5 ml of digestion media dermal side down and incubated for 1 hour at 4°C. Tissue was then minced with scissors in 5 m l of fresh digestion media and incubated at 37°C with shaking. After 1 hour, digestion was stopped by adding 100 μl of 0.5 M EDTA and 1 ml of fetal bovine serum. Digested tissue was then mashed through a 70-micron cell strainer to obtain a single cell suspension.
Wounding study
C57BL/6 mice in the telogen phase of the hair cycle were anaesthetized and given punch biopsies on the ear or back. For ear wounds, a 2.5mm diameter corneal trephine blade (Ambler Surgical) was used to punch a through and through wound (hole) in the center of each ear. Tissue was collected at 3d after wounding for microscopy. For backskin wounds, dorsal hairs were cut with a clipper and a 6mm biopsy punch was used to partially perforate skin. An iris scissors was then used to cut epidermis and dermis along outline to create a full thickness wound in the shape of a circle.
Immunofluorescence/confocal microscopy of ear pinnae
Ear pinnae were split with forceps, fixed in 1% paraformaldehyde solution (Electron Microscopy Sciences) overnight at 4°C and blocked in 1% BSA, 0.25% Triton X blocking buffer for 2 hours at room temperature. Tissues were first stained with anti-CD8α (clone 53-6.7, eBioscience), anti-Sca-1 (D7, eBioscience) and/or rabbit anti-CD31 (390, eBioscience), and anti-Ki67 (solA15, eBioscience) antibodies overnight at 4°C, washed three time with PBS and then stained with 4′, 6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich) for 5 min at room temperature and before being mounted with ProLong Gold (Molecular Probes) antifade reagent. Ear pinnae images were captured on a Leica TCS SP8 confocal microscope with a 40X oil objective (HC PL APO 40X/1.3 oil). Images were analyzed using Imaris Bitplane software.
Immunofluorescence/epifluorescence microscopy of backskin wounds
Wounded backskin tissue was incubated in 30% sucrose at 4°C overnight, fixed in 4% paraformaldehyde, embedded in OCT compound (Tissue-Tek), frozen on dry ice, and cryo-sectioned (20 μm section thickness). Sections were fixed in 4% paraformaldehyde, rinsed with PBS, permeabilized 10 min with 0.1% Triton X-100 (Sigma) in PBS, then blocked for 1 hour in blocking buffer (2.5% normal goat serum (Jackson ImmunoResearch), 1% BSA, 0.3% Triton X-100). Primary antibody to K14 (chicken, 1:400, Biolegend) was used. Primary antibody was diluted in blocking buffer with Rat gamma globulin (Jackson ImmunoResearch) and anti-CD16/32 (ebioscience) and incubated at 4°C overnight. After washing with PBS, a secondary antibody, conjugated with Alexa647 (goat, Jackson ImmunoResearch) was added for 2 hours at room temperature. Slides were washed with PBS, counterstained with DAPI and mounted in Prolong Gold. Wound images were captured with a Leica DMI 6000 widefield epifluorescence microscope equipped with a Leica DFC360X monochrome camera. Tiled and stitched images of wounds were collected using a 20X/0.4NA dry objective. Images were analyzed using Imaris Bitplane software.
In vitro re-stimulation
For detection of basal cytokine potential, single cell suspensions from various tissues were cultured directly ex vivo in a 96-well U-bottom plate in complete medium (cRPMI-RPMI 1640 supplemented with 10% fetal bovine serum [FBS], 2 mM L-glutamine, 1 mM sodium pyruvate and nonessential amino acids, 20 mM HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin, 50 mM β-mercaptoethanol) and stimulated with 50 ng/ml phorbol myristate acetate (PMA) (Sigma-Aldrich) and 5 μg/ml (mouse) or 1 μg/ml (non-human primate) ionomycin (Sigma-Aldrich) in the presence of brefeldin A (GolgiPlug, BD Biosciences) for 2.5 hours at 37°C in 5% CO 2. After stimulation, cells were assessed for intracellular cytokine production as described below.
Phenotypic analysis
Murine single cell suspensions were incubated with fluorochrome-conjugated antibodies against surface markers: CCR6 (29-2L17), CD3ε (145-2C11), CD4 (clone RM4-5), CD8β (eBioH35-17.2), CD11b (M1/70), CD11c (N418), CD19 (6D5), CD24 (M1/69), CD44 (IM7), CD45 (30-F11), CD45.1 (A20), CD45.2 (104), CD64 (X54-5/7.1), CD69 (H1.2F3), CD103 (2E7), CD122 (TM-b1), CD127 (A7R34), KLRG1 (2F1), Ly-6C (HK1.4), Ly-6G (1A8), MHCII (M5/114.15.2), TCRβ (H57-597), TCRvγ3+ (536), and mouse Vβ screening panel (Becton Dickinson) in Hank’s buffered salt solution (HBSS) for 20 min at 4°C and then washed. LIVE/DEAD Fixable Blue Dead Cell Stain Kit (Invitrogen Life Technologies) was used to exclude dead cells. Cells were then fixed for 20 min at 4°C using BD Cytofix/Cytoperm (Becton Dickinson) and washed twice. For intracellular cytokine staining, cells were stained with fluorochrome-conjugated antibodies against IFN-γ (XMG-1.2), IL-17A (eBio17B7), and/or Granzyme B (human, GB11) in BD Perm/Wash Buffer (Becton Dickinson) for 45 min at 4°C. For transcription factor staining, cells were fixed and permeabilized with the FoxP3/Transcription Factor staining buffer set (eBioscience) and stained with fluorochrome-conjugated antibodies against T-bet (ebio4B10) or RORγt (B2D) for 45 min at 4°C. Each staining was perfor med in the presence of purified anti-mouse CD16/32 (2.4G2), 0.2 mg/ml purified rat gamma globulin (Jackson Immunoresearch). Staining of cells from non-human primate skin tissue was performed using a similar protocol and the following antibodies against human proteins: CD3ε (SP34-2), CD4 (L200), CD8α (RPA-T8), CD45 (D058-1283), IFN-γ (B27) and IL-17A (BL168). All antibodies were purchased from eBioscience, Biolegend, BD Biosciences, or Miltenyi Biotec. Cell acquisition was performed on an LSRII flow cytometer using FACSDiVa software (BD Biosciences) and data were analyzed using FlowJo software (TreeStar).
In vivo recall of f-MIIINA:H2-M3-specific CD8+ T cells via intravenous peptide injection
Cytokine production was elicited by i.v. injection of 100 μg of f-MIIINA peptide in PBS via the tail vein of mice that were previously topically associated with S. epidermidis strain NIHLM087. Mice were euthanized after 3 hours, and single-cell suspensions were made from pooled skin draining lymph nodes and spleen or ear pinnae in cRPMI medium supplemented with brefeldin A (10 μg/mL; Sigma). Cytokine production by f-MIIINA:H2-M3-specific CD8+ T cells from mice injected with f-MIIINA peptide was determined before (ear pinnae) or after tetramer-based cell enrichment (spleen and lymph nodes) as described below.
Cell enrichment
Spleen and lymph node cells were prepared and stained for 1 hour at room temperature with f-MIIINA:H2-M3-streptavidin-phycoerythrin, f-MIIINA:H2-M3-streptavidin-allophycocyanin, and/or f-MIGWII:H2-M3-streptavidin-allophycocyanin tetramers. Samples were then enriched for bead-bound cells on magnetized columns, and a portion was removed for counting. The rest of the sample underwent surface staining on ice with a mixture of antibodies described above.
DC-T cell co-culture assay
CD45+ CD90.2+ CD8β+ T cells (>95% purity) were FACsorted from the ear skin tissue of C57BL/6 mice two weeks after topical association with S. epidermidis strain NIHLM087 using a FACSAria cell sorter. For splenic DC purification, single-cell suspensions from the spleen of congenic wild-type, B2m−/−, KbDb−/−, Qa-1−/−, or H2-M3−/− mice were magnetically enriched for CD11c+ cells by positive selection using CD11c MicroBeads and MACS separation columns (Miltenyi Biotec). Purified splenic dendritic cells (SpDC) and CD8β+ T cells were co-cultured at a 15:1 ratio (5×103 CD8β+ T cells) in a 96-well U-bottom plate in complete medium for 16 hours at 37°C in 5% CO2. Brefeldin A (GolgiPlug) was added for the final 4 hours of culture. SpDC were previously incubated for 2 hours with or without heat-killed S. epidermidis (bacteria:SpDC ratio, 500:1) and washed before co-culture with CD8β+ T cells. For samples in which S. epidermidis conditioned medium (CM) was added, 40 μl of CM was added directly to co-cultured cells and incubated for 16 hours. For samples in which fMet peptides were added, peptides were added at a final concentration of 15 μg/ml. CD8β+ T cell cytokine production following co-culture was assessed by flow cytometry after surface and intracellular cytokine staining using the following antibodies: CD4, CD8β, CD45.1, CD45.2, TCR-β, IFN-γ and IL-17A.
T cell isolation and RNA extraction for RNA-sequencing
T cells were FACsorted from the skin draining lymph nodes, ear or flank skin tissue of C57BL/6 mice. T cells were isolated from naïve mice or those topically-associated with S. epidermidis NIHLM087, infected via intradermal route with S. epidermidis NIHLM087, or Leishmania major, topically administered with Imiquimod, or infected with Herpes Simplex Virus via skin scarification. Groups included: TEM (lymph nodes; CD45+ CD90.2+ TCRβ+ CD8β+ CD44high CD62Llow), Tc17 (skin; Lineage- CD45+ CD90.2+ TCRγδ− TCRβ+ CD8β+ CCR6+), Tc1 (skin; Lineage− CD45+ CD90.2+ TCRγδ− TCRβ+ CD8β+ CCR6−), DETC (skin; Lineage− CD45+ CD90.2+ TCRvγ3+ TCRβ− CD4− CD8β−). The lineage cocktail consisted of antibodies to CD11b, CD11c, MHC-II, NK1.1, CD49f and CD19. RNA was extracted from T cell populations using the Arcturus PicoPure RNA Isolation Kit (Applied Biosystems), as per manufacturer’s instructions.
RNA library preparation and sequencing
mRNA libraries were constructed using SMARTer Ultra Low Input RNA kit (Clonetech) + Nextera XT DNA library prep kit (Illumina). mRNA samples were pooled and sequenced on 3 lanes of HiSeq40000 with Illumina Hiseq3000/4000 chemistry. The reads from the Illumina Hi-seq sequencer in fastq format were verified for quality control using fastqc software package. The low-quality segments of the read along with adaptors were trimmed with the tool Trimmomatic (Ewels et al., 2016). The quality filtered reads were aligned to mouse genome GRCM38 using RSEM package (Li and Dewey, 2011) with default parameters and STAR aligner (Dobin et al., 2013). The expected RSEM counts were rounded to the nearest integer value and the transcripts with zero counts across all the samples are filtered out. T cell populations were compared using DESeq2 package (Anders and Huber, 2010).
Whole tissue RNA-sequencing and transcriptome analysis
A ~1mm skin region surrounding the wound site of backskin wounds was micro-dissected at each time point after initial wounding described above and submerged in RNAlater (Sigma) and stored at −20C. Total tissue RNA was isolated from skin tissue using the RNeasy Fibrous Tissue Mini Kit (Qiagen), as per manufacturer’s instructions. A 3′end mRNA sequencing library was prepared using 100ng of total input RNA with the QuantSeq 3′ mRNA-Seq Library Prep Kit FWD for Illumina (Lexogen) as per manufacturer’s instructions. Libraries were quantified using an Agilent Tapestation (High Sensitivity D1000 ScreenTape) and Qubit (Thermo Fisher Scientific). Libraries (n=24) were then pooled at equimolar concentrations and sequenced on an Illumina Nextseq 500 using the High Output v2 kit (75 cycles). Resultant data was demultiplexed on Illumina Basespace server using blc2fastq tool. The reads from the Illumina Next-seq sequencer in fastq format were verified for quality control using FastQC software package and summarized using multi-qc (Ewels et al., 2016). The low-quality segments of the read are trimmed with the tool Trimmomatic (Bolger et al., 2014) using: seed mismatches:3, palindrome clip threshold:50, simple clip threshold:10, sliding window:4, required quality:20, minimum length after trimming: 40 and maximum read length to error rate of 50:0.8. The quality filtered reads are aligned to mouse genome GRCM38 using RSEM package (Li and Dewey, 2011) with default parameters and STAR aligner (Dobin et al., 2013). The expected RSEM counts are rounded to the nearest integer value and the transcripts with zero counts across all the samples are filtered out. Differential expression analysis was performed using DESeq package (Anders and Huber, 2010) between two cohorts of mice (WT and H2-M3−/−) at different time points after wounding.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Groups were compared with Prism software (GraphPad) using the two-tailed unpaired or paired Student’s t-test. Flow cytometry plots are presented as mean ± standard deviation. All other data are presented as mean only or mean ± standard error of the mean. P<0.05 was considered significant unless otherwise stated.
DATA AND SOFTWARE AVAILABILITY
The RNA-Seq data sets have been deposited in NCBI BioProject under ID code BioProject ID: PRJNA419368.
Highlights.
Non-classical MHC Class I molecules promote homeostatic immunity to the microbiota
Commensal-specific T cells express immunoregulatory and tissue repair signatures
Commensal-specific T cells accelerate wound closure
Acknowledgments
This work was supported by the Division of Intramural Research of the NIAID, NCI, NIAMS and NIDDK. J.L. was supported by the NIGMS Postdoctoral Research Associate (PRAT) fellowship program. O.J.H. was supported by a National Psoriasis Foundation Early Career Research Grant. S.T. was supported by the Long Term EMBO Fellowship. We thank the NIAID animal facility staff; the NIAID Microbiome Program gnotobiotic animal facility for the use of germ-free mice; K. Holmes, C. Eigsti, and E. Stregevsky (NIAID Flow Cytometry facility); O. Schwartz (NIAID Biological Imaging facility) and K. Beacht, J. LeGrand, and J. Davis for technical assistance; Dr. R. Bosselut for the B2m−/− mice; Dr. H. Cantor for the Qa-1−/− mice; We thank all the members of the Belkaid laboratory for critical reading of the manuscript and helpful discussions.
Footnotes
AUTHOR CONTRIBUTIONS
J.L.L. and Y.B. designed the studies and wrote the manuscript. J.L.L performed experiments and analyzed the data. O.J.H., S.H., and N.B. participated in performing experiments, provided intellectual expertise, and helped to interpret experimental results. I.V., A.L.B., A.V.V., J.J.O., S.K.S., J.S., A.D., G.T., and T.M.K. assisted with the RNAseq study. S.P.R. and B.R. assisted with the wild mouse study. S.H. and S.T. performed whole tissue mount imaging studies. M.S. and N.C. assisted with wound imaging studies. O.J.H., S.H., C.W., and N.B. assisted with manuscript preparation. J.M.B. provided primate tissues and guidance on the preparation and staining. All authors read and commented on the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams EJ, Luoma AM. The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu Rev Immunol. 2013;31:529–561. doi: 10.1146/annurev-immunol-032712-095912. [DOI] [PubMed] [Google Scholar]
- An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, Kasper DL. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science. 2014;346:954–959. doi: 10.1126/science.1260144. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, Tamoutounour S. The influence of skin microorganisms on cutaneous immunity. Nat Rev Immunol. 2016;16:353–366. doi: 10.1038/nri.2016.48. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, Von Stebut E, Mendez S, Lira R, Caler E, Bertholet S, Udey MC, Sacks D. CD8+ T cells are required for primary immunity in C57BL/6 mice following low-dose, intradermal challenge with Leishmania major. J Immunol. 2002;168:3992–4000. doi: 10.4049/jimmunol.168.8.3992. [DOI] [PubMed] [Google Scholar]
- Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, Thompson EA, Fraser KA, Rosato PC, Filali-Mouhim A, et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 2016;532:512–516. doi: 10.1038/nature17655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd AL, Deming C, Cassidy KB, Harrison OJ, Conlan S, Belkaid Y, Segre JA, Kong HH NISC Comparative Sequencing Program. Staphylococcus aureus and S. epidermidis strain diversity underlying human atopic dermatitis. Science Translational Medicine. 2017 doi: 10.1126/scitranslmed.aal4651. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Bediako Y, Xu H, Choi HJ, Wang CR. Positive selecting cell type determines the phenotype of MHC class Ib-restricted CD8+ T cells. Proc Natl Acad Sci U S A. 2011;108:13241–13246. doi: 10.1073/pnas.1105118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T, Serbina NV, Nolt D, Wang B, Chiu NM, Flynn JL, Wang CR. Induction of M3-restricted cytotoxic T lymphocyte responses by N-formylated peptides derived from Mycobacterium tuberculosis. J Exp Med. 2001;193:1213–1220. doi: 10.1084/jem.193.10.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci U S A. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan S, Mijares LA, Program NCS, Becker J, Blakesley RW, Bouffard GG, Brooks S, Coleman H, Gupta J, Gurson N, et al. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol. 2012;13:R64. doi: 10.1186/gb-2012-13-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle CK, Davis BK, Cook RG, Rich RR, Rodgers JR. Hyperconservation of the N-formyl peptide binding site of M3: evidence that M3 is an old eutherian molecule with conserved recognition of a pathogen-associated molecular pattern. J Immunol. 2003;171:836–844. doi: 10.4049/jimmunol.171.2.836. [DOI] [PubMed] [Google Scholar]
- Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: mechanisms, signaling, and translation. Sci Transl Med. 2014;6:265sr266. doi: 10.1126/scitranslmed.3009337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Rudensky AY. Hallmarks of Tissue-Resident Lymphocytes. Cell. 2016;164:1198–1211. doi: 10.1016/j.cell.2016.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, Carbone FR. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. 2009;10:524–530. doi: 10.1038/ni.1718. [DOI] [PubMed] [Google Scholar]
- Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, Moody DB. The burgeoning family of unconventional T cells. Nat Immunol. 2015;16:1114–1123. doi: 10.1038/ni.3298. [DOI] [PubMed] [Google Scholar]
- Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagan AJ, Pepper M, Maynard CL, Elson CO, 3rd, Belkaid Y. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553– 1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson J, Ugarte K, Chen N, Yachi P, Fuchs E, Boismenu R, Havran WL. A role for skin gammadelta T cells in wound repair. Science. 2002;296:747–749. doi: 10.1126/science.1069639. [DOI] [PubMed] [Google Scholar]
- Kawano M, Komi-Kuramochi A, Asada M, Suzuki M, Oki J, Jiang J, Imamura T. Comprehensive analysis of FGF and FGFR expression in skin: FGF18 is highly expressed in hair follicles and capable of inducing anagen from telogen stage hair follicles. J Invest Dermatol. 2005;124:877–885. doi: 10.1111/j.0022-202X.2005.23693.x. [DOI] [PubMed] [Google Scholar]
- Kerksiek KM, Busch DH, Pamer EG. Variable immunodominance hierarchies for H2-M3-restricted N-formyl peptides following bacterial infection. J Immunol. 2001;166:1132–1140. doi: 10.4049/jimmunol.166.2.1132. [DOI] [PubMed] [Google Scholar]
- Keyes BE, Liu S, Asare A, Naik S, Levorse J, Polak L, Lu CP, Nikolova M, Pasolli HA, Fuchs E. Impaired Epidermal to Dendritic T Cell Signaling Slows Wound Repair in Aged Skin. Cell. 2016;167:1323–1338. e1314. doi: 10.1016/j.cell.2016.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Cogen AL, Radek KA, Park HJ, Macleod DT, Leichtle A, Ryan AF, Di Nardo A, Gallo RL. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J Invest Dermatol. 2010;130:2211–2221. doi: 10.1038/jid.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, Wu ZR, Hooper LV, Schmidt RR, von Aulock S, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009;15:1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl KF, Byers DE, Dabhi VM, Hovik R, Jones EP, Smith GP, Wang CR, Xiao H, Yoshino M. H2-M3, a full-service class Ib histocompatibility antigen. Annu Rev Immunol. 1997;15:851–879. doi: 10.1146/annurev.immunol.15.1.851. [DOI] [PubMed] [Google Scholar]
- Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, Wilhelm C, Conlan S, Himmelfarb S, Byrd AL, Deming C, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520:104– 108. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MM, Witherden DA, Havran WL. gammadelta T cells in homeostasis and host defence of epithelial barrier tissues. Nat Rev Immunol. 2017 doi: 10.1038/nri.2017.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obar JJ, Khanna KM, Lefrancois L. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 2008;28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamer EG. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science. 2016;352:535–538. doi: 10.1126/science.aad9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploss A, Lauvau G, Contos B, Kerksiek KM, Guirnalda PD, Leiner I, Lenz LL, Bevan MJ, Pamer EG. Promiscuity of MHC class Ib-restricted T cell responses. J Immunol. 2003;171:5948–5955. doi: 10.4049/jimmunol.171.11.5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol. 2005;5:459–471. doi: 10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- Scharschmidt TC, Vasquez KS, Truong HA, Gearty SV, Pauli ML, Nosbaum A, Gratz IK, Otto M, Moon JJ, Liese J, et al. A Wave of Regulatory T Cells into Neonatal Skin Mediates Tolerance to Commensal Microbes. Immunity. 2015;43:1011– 1021. doi: 10.1016/j.immuni.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity. 2014;41:886–897. doi: 10.1016/j.immuni.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MS, Wang CR, Forman J. MHC class Ib-restricted CTL provide protection against primary and secondary Listeria monocytogenes infection. J Immunol. 2000;165:5192–5201. doi: 10.4049/jimmunol.165.9.5192. [DOI] [PubMed] [Google Scholar]
- Shang S, Siddiqui S, Bian Y, Zhao J, Wang CR. Nonclassical MHC Ib-restricted CD8+ T Cells Recognize Mycobacterium tuberculosis-Derived Protein Antigens and Contribute to Protection Against Infection. PLoS Pathog. 2016;12:e1005688. doi: 10.1371/journal.ppat.1005688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamani N, Rufaut N, Jones L, Sinclair R. The arrector pili muscle, the bridge between the follicular stem cell niche and the interfollicular epidermis. Anat Sci Int. 2017;92:151–158. doi: 10.1007/s12565-016-0359-5. [DOI] [PubMed] [Google Scholar]
- van der Fits L, Mourits S, Voerman JS, Kant M, Boon L, Laman JD, Cornelissen F, Mus AM, Florencia E, Prens EP, et al. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J Immunol. 2009;182:5836–5845. doi: 10.4049/jimmunol.0802999. [DOI] [PubMed] [Google Scholar]
- van Lint A, Ayers M, Brooks AG, Coles RM, Heath WR, Carbone FR. Herpes simplex virus-specific CD8+ T cells can clear established lytic infections from skin and nerves and can partially limit the early spread of virus after cutaneous inoculation. J Immunol. 2004;172:392–397. doi: 10.4049/jimmunol.172.1.392. [DOI] [PubMed] [Google Scholar]
- Van Rhijn I, Godfrey DI, Rossjohn J, Moody DB. Lipid and small-molecule display by CD1 and MR1. Nat Rev Immunol. 2015;15:643–654. doi: 10.1038/nri3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vugmeyster Y, Glas R, Perarnau B, Lemonnier FA, Eisen H, Ploegh H. Major histocompatibility complex (MHC) class I KbDb −/− deficient mice possess functional CD8+ T cells and natural killer cells. Proc Natl Acad Sci U S A. 1998;95:12492–12497. doi: 10.1073/pnas.95.21.12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol Rev. 2003;83:835–870. doi: 10.1152/physrev.2003.83.3.835. [DOI] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]