Abstract
Background
Abnormal microRNAs (miRNAs) were reported to be involved in the mechanism of Graves’ disease (GD). Dysregulated miRNAs may be overlapping in different cells and can be secreted to circulation. We chose miRNAs which were previously reported to be differentially expressed in peripheral blood mononuclear cells (PBMCs) in patients with GD with different disease stage, detected the expression of those miRNAs in serum, corroborated the findings in thyroid tissue, and validated the target gene in vitro to investigate the possible role of circulating miRNAs in GD.
Methods
A total of 54 individuals with untreated GD, 12 individuals with GD in remission and 14 disease-free controls were enrolled. The expression of miR-142-3p, miR-154-3p, miR-431-3p, miR-590-5p, and let-7b was detected in the serum. Ten thyroid tissue samples from patients with GD and six disease-free thyroid samples were used for further validation. The potential target genes were identified and validated in vitro.
Results
miR-142-3p, miR-154-3p, miR-431-3p, miR-590-5p, and let-7b were present in serum and two of them (miR-142-3p and let-7b) were significantly increased in serum of patients with untreated GD (for serum miR-142-3p, P = 0.033, for serum let-7b, P = 0.026) and gradually decreased to normal levels in patients with GD in remission. Correlation analysis showed that let-7b level was strongly correlated with TRAb level (r = 0.305, P = 0.001). let-7b directly inhibited promyelocytic leukemia zinc finger (PLZF) expression and increased the expression of TSHR in thyroid cells in vitro. Furthermore, let-7b levels in GD thyroid tissue were found to be inversely correlated with PLZF levels (r = − 0.849, P = 0.033). Decreased PLZF and increased TSHR was validated in thyroid tissue in patients with GD.
Conclusions
The present study confirmed that a portion of miRNAs in PBMCs were also presented and differentially expressed in serum and thyroid tissue. Upregulated in all these three compartments, let-7b may be used as a disease biomarker and therapeutic targets in patients with GD. Circulating let-7b had a strong correlation with disease severity and let-7b may participate in the production of TRAb via targeting PLZF in patients with GD.
Electronic supplementary material
The online version of this article (10.1186/s12967-018-1565-9) contains supplementary material, which is available to authorized users.
Keywords: Circulating miRNAs, Let-7b, Graves’ disease, Thyrotropin receptor antibody
Background
Graves’ disease (GD) is the most common cause of hyperthyroidism in the clinic, affecting about 1% of the general population [1, 2]. GD is an antibody-mediated organ-specific autoimmune disease and is initiated by the interplay of susceptibility genes, environment, and immune disorders [3, 4]. It is well established that human thyrotropin receptor (hTSHR) is the primary autoantigen of Graves’ disease. The circulating autoantibodies to TSHR are called thyrotropin receptor antibodies (TRAb) which can mimic the role of TSH, binding to TSHR and leading to overproduction of thyroid hormones and thyroid hyperplasia [5]. TRAb is detectable in 95–100% untreated GD patients and decreased by treatment and, when they persist, may predict recurrence [6]. Thus, TRAb is disease specific and plays a central role in the etiology of GD. However, the mechanism of the production of TRAb remained unclear.
MicroRNAs (miRNAs) are noncoding RNAs that interact with their target RNAs in a sequence-dependent manner [7]. miRNAs are involved in several important biological processes including immunity, apoptosis, cell differentiation and development, proliferation, and metabolism [8]. Similar to other autoimmune diseases, recently studies concentrated on the specific miRNAs profiles in GD. The importance of miRNAs in the pathogenesis of GD was highlighted in many reports [9–12]. Originating from the active or passive release by cells, circulating miRNAs were an emerging area in recent years. Evidence suggested that circulating miRNAs can be used as biological markers for diseases such as tumors and inflammatory diseases [13, 14]. Studies demonstrated that circulating miRNAs had a strong correlation with disease severity in patients with GD. Yamada et al. explored circulating miRNA expression profiles in 17 patients with GD through microarray. miR-16, miR-22, miR-375, and miR-451 were significantly increased in circulation in patients with GD [15]. Increased circulating miR-23b-5p and miR-92a-39, as well as decreased circulating let-7g-3p and miR-339-5p were associated with intractable GD [16]. Interestingly, circulating miRNAs were reported to be disseminated through the extracellular fluid to reach target cells and participated in the intercellular communication. Although the specific mechanism remained unknown, miRNA can be transferred between different cells through apoptotic body or extracellular vesicles were observed in vitro systems. Circulating miRNAs might have hormone-like effects leading to widespread responses within different types of cells [17]. Thus, circulating miRNAs might dysregulated in different compartments and paly crucial roles in the pathogenesis of the disease. In multiple sclerosis, miR-21, miR-142-3p, miR-146a, miR-155 and miR-326 was found to be upregulated in both PBMCs and brain white matter lesions from patients and mouse model as well [18]. Marazuela et al. investigated the differential expression profiles of miRNAs in thyroid tissue in GD samples, and then detected these miRNAs in serum. The results showed that some miRNAs that were highly expressed in thyroid tissues were also highly expressed in the serum and serum let-7d was strongly correlated with disease severity [19].
The differential expression of miRNAs in peripheral blood mononuclear cells (PBMCs) in untreated GD patients was reported in our previous study, where miR-142-3p, miR-154-3p, miR-431-3p, miR-590-5p, and let-7b were significantly differential expressed [10]. Those miRNAs have been reported in several autoimmune diseases such as rheumatoid arthritis [20], systemic lupus erythematosus [21], multiple sclerosis [18, 22] and psoriasis [23] (shown in Additional file 1: Table S1). We hypothesized that those miRNAs, which were involved in various inflammatory signaling pathways, may be presented and differentially expressed in serum and tissue and may participate in the pathogenesis mechanism of GD and the production of TRAb. However, very few studies focused on the expression of those miRNAs candidates in serum or thyroid tissue of patients with GD. Therefore, we detected the expression of these miRNAs in serum in GD patients with different stage, correlated their expression with GD patients’ clinical data, identified the target genes and validated in thyroid tissue to investigate the possible pathogenesis role of circulating miRNAs in patients with GD.
Methods
Enrollment of patients and disease-free controls
Patients and disease-free controls were enrolled from the outpatient department of Endocrinology of Ruijin Hospital affiliated to Shanghai JiaoTong University Medicine School. The diagnosis of GD was based on clinical manifestation and laboratory examination. Typical manifestations included heat intolerance, fatigue, increased appetite, increased sweating, weight loss, muscle weakness, tremors, and diffusely enlarged thyroid gland. Laboratory examinations included increased serum concentration of free triiodothyronine (FT3), free thyroxine (FT4), decreased serum concentration of sensitive thyrotropin (sTSH), and high tilters of TRAb. Patients with untreated GD were all newly diagnosed and untreated in our study. Patients were considered to be in remission when thyroid function returned to normal (TSH, FT3, and FT4 within the reference range and negative TRAb) under the treatment of anti-thyroid drugs. The diagnosis of Graves’ disease was confirmed by the experienced physician or endocrinologist in Ruijin Hospital affiliated to Shanghai JiaoTong University. Further, a group of subjects without thyroid disease as disease-free controls were also recruited. Disease-free controls were subjects who had no personal and family history of thyroid disease, with normal thyroid ultrasound imaging, and serum TSH, FT3/FT4 within reference range and negative thyroid antibody.
Serum microRNA extraction
Three to 5 mL peripheral blood were collected and centrifuged at 1900g (3000 rpm) for 10 min at 4 °C to isolate serum. Serum isolation was completed within 4 h after collection. miRNA was isolated from 200 μL of serum using the miRNeasy Serum/Plasma Kit (Qiagen) following the manufacturer’s protocol. miRNeasy Serum/Plasma Spike-In Control (C. elegans miR-39) was added to serum as a control before miRNA extraction.
Thyroid tissue miRNA extraction
We collected 10 thyroid tissue samples from patients with GD and six disease-free thyroid tissue samples confirmed by pathological examination from thyroid nodule patients who had undergone thyroidectomy between April 2017 and October 2017 at Ruijin Hospital. miRNA extraction from thyroid tissue samples was performed using an miRNeasy Mini Kit (Qiagen) by following the manufacturer’s protocol.
Real-time reverse transcription polymerase chain reaction
cDNAs were synthesized using a miScript Reverse Transcription Kit (Qiagen, Hilden, Germany). The expression levels of miRNAs were confirmed with a miScript SYBR Green PCR kit and miRNA-specific primers (Qiagen). Commercially available primers purchased from Qiagen were used as follows: Hs_miR-142-3p miScript Primer Assay, Hs_miR-154-3p miScript Primer Assay, Hs_ miR-431-3p miScript Primer Assay, Hs_miR-590-5p miScript Primer Assay, and Hs_let-7b miScript Primer Assay. RNU6-2 served as the internal normalization control for the expression in thyroid tissue. C. elegans mir-39 served as the internal normalization control for the expression in serum. Light Cycler 480 software (Roche Applied Science, Indianapolis, IN, USA) was used to analyze data. All reactions were performed in triplicate and the results were calculated by determining the ΔΔCt value.
Cell isolation and cell culture
Primary thyroid cells were isolated from fresh thyroid tissue from normal thyroid tissue. Primary thyroid cells and Nthy-ori3-1 cells were maintained in RMPI 1640 supplemented with 10% FBS and 1% penicillin–streptomycin at 37 °C in 5% CO2. 293T cells were maintained in DMEM supplemented with 10% FBS and 1% penicillin–streptomycin at 37 °C in 5% CO2. The cells were passaged every 3 days.
The transient transfection
Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) was used as a transfection reagent. let-7b mimics, let-7b inhibitors, and mimic control or negative control were mixed with the transfection reagent and then added to the cells, followed by incubation for 24 h at 37 °C in 5% CO2.
Western blot analysis
Cell lysates were subjected to western blot analysis according to standard protocols. After blocking, the membranes were incubated overnight at 4 °C with primary antibodies to PLZF (1:1000; SAB, USA) or TSHR (1:1000; Proteintech, USA). GAPDH (1:3000; Cell Signaling Technology, USA) was used as a normalization control. Next, the membranes were incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (1:2000; Cell Signaling Technology, USA). Protein bands were illuminated using ECL Prime Western Blotting Detection Reagent (GE Healthcare, Little Chalfont, Buckinghamshire, UK).
Luciferase activity assay
The pmiR-RB-REPORT vector (RIBOBIO) was used to clone the PLZF 3′UTR sequence containing the putative miR-4443 binding sites, designated as wild type 1 (binding sites: 1889–1895; PLZF-WT1) and wild type 2 (binding sites: 4766–4773; PLZF-WT2). Reporter plasmids of the corresponding mutation (Mut1 and Mut2) were also constructed. The PLZF-WT1 (1889–1895), PLZF-WT2 (4766–4773), and mutant type (PLZF-mut1 and PLZF mut2) constructs were amplified and cloned downstream of a luciferase reporter gene in the pmiR-RB-REPORT vector. HEK293T cells were seeded in 24-well plates the day before transfection. For each well, 100 ng of wild-type or mutant reporter plasmid was transiently co-transfected with miRNA mimics or negative control using Lipofectamine 3000 (Invitrogen). Cell lysates were harvested 48 h after transfection, and the cells were subjected to a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Renilla luciferase activity was normalized to firefly luciferase activity to control for transfection efficiency.
Statistical analysis
SPSS version 19.0 were used for statistical calculations. All figures were completed with Graph Pad Prism 6.0. Data are presented as mean ± SD. P < 0.05 was considered significant. Statistical difference was evaluated using Mann–Whitney U-test for continuous variables among three groups. The Chi square test was used to test for categorical variables. Correlations between the different variables were analyzed by Spearman correlation.
Results
The general clinical characteristics of the study population
A total of 54 patients with untreated GD, 12 patients with GD in remission, and 14 disease-free controls were enrolled in our study. The general characteristics of patients and disease-free controls are summarized in Table 1. There was no significance in age and gender composition between untreated GD patients and disease-free controls. FT3, FT4, TRAb, TPOAb, and TgAb were highest and TSH was lowest in patients with untreated GD. FT3, FT4, TSH and TRAb were returned to normal in patients with GD in remission.
Table 1.
Clinical characteristics of the study population
| Disease-free controls | Untreated GD | GD in remission | |
|---|---|---|---|
| N (F/M) | 14 (9/5) | 54 (38/16) | 12 (9/3) |
| Age (years) | 31.57±6.81 | 38.87±11.30 | 47.55±11.61* |
| FT3 (pmol/L) | 4.62±0.51# | 26.90±14.17* | 4.13±0.49# |
| FT4 (pmol/L) | 13.76±1.57# | 41.44±28.87* | 13.47±1.10# |
| TSH (mIU/L) | 1.93±1.17# | 0.32±2.33* | 2.38±0.95# |
| TRAb (IU/L) | 0.46±0.24# | 17.15±12.88* | 0.75±0.13# |
| TPOAb (IU/mL) | 19.02±68.15# | 435.77±366.92* | 274.38±157.71 |
| TgAb (IU/mL) | 6.01±13.57# | 209.71±275.97* | 74.25±61.31 |
* Compared to disease-free controls, P < 0.001
# Compared to untreated GD, P < 0.001
The expression of miR-142-3p, miR-154-3p, miR-431-3p, miR-590-5p, and let-7b in serum and correlation with TRAb in Graves’ disease
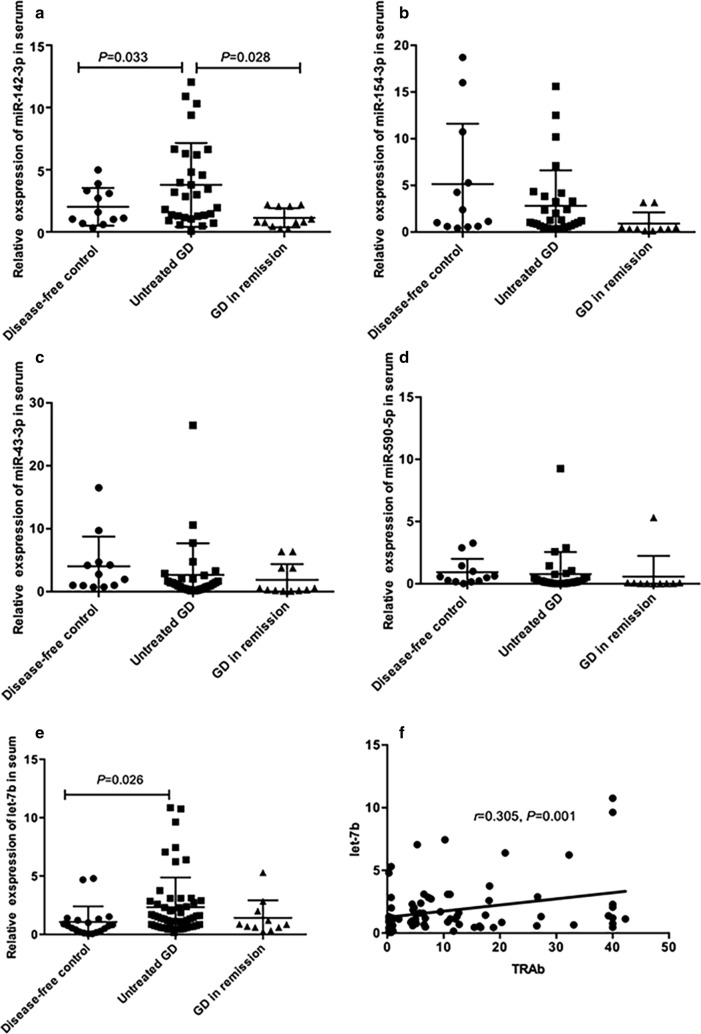
The expression of miR-142-3p, miR-154-3p, miR-431-3p, miR-590-5p and let-7b were detected in serum. Compared to disease-free controls, serum miR-142-3p and let-7b expression was significantly increased in patients with untreated GD (for serum miR-142-3p, P = 0.033; for serum let-7b, P = 0.026), and gradually decreased to normal under the anti-thyroid treatment in patients with GD in remission (Fig. 1a, e). There was no significance difference in serum miR-154-3p, miR-431-3p, and miR-590-5p levels among the three groups (Fig. 1b–d). Correlation analysis showed that only serum let-7b level was positively correlated with TRAb level (r = 0.305, P = 0.001, Fig. 1f). Serum miR-142-3p level was not correlated with TRAb (r = 0.231, P = 0.147).
Fig. 1.
Relative expression of miR-142-3p, miR-154, miR-431-3p, miR-590-5p and let-7b in serum and correlation with TRAb. Relative expression of miR-142-3p (a), miR-154 (b), miR-431-3p (c), miR-590-5p (d) and let-7b (e) expression in patients with hyperthyroidism with untreated Graves’ disease (GD), patients in remission with TRAb-negative GD, and disease-free controls. f The correlation between the expression levels of let-7b in serum and TRAb levels
PLZF is a direct target gene of let-7b
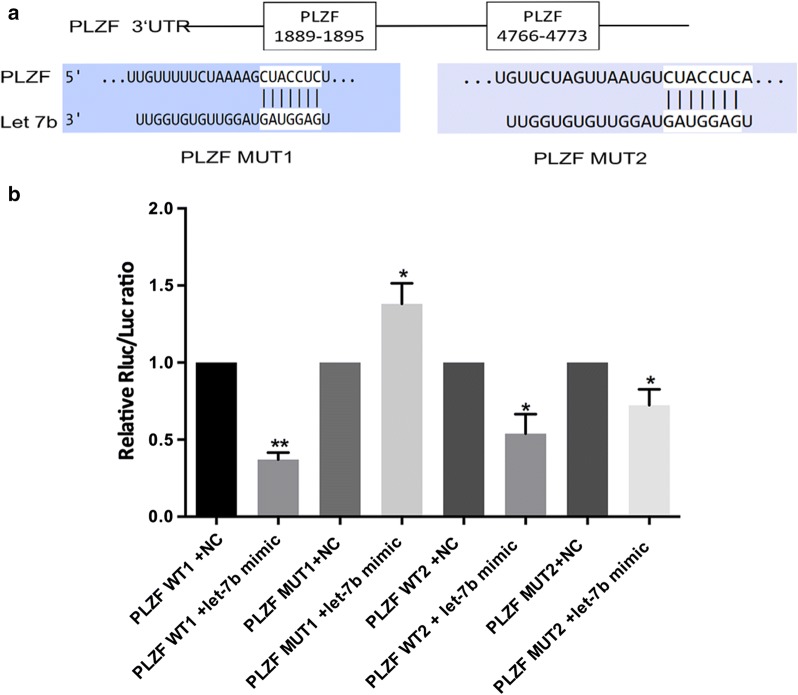
It is well known that miRNAs function by regulating their target genes. In the present study, the putative let-7b target genes were predicted using miRbase and TargetScan. PLZF possesses two potential complementary sites for the let-7b seed region in its 3′UTR (Fig. 2a). To further explore the direct binding and repression effect between let-7b and PLZF, a luciferase activity assay was performed. Co-transfection of let-7b mimics and PLZF-WT1 in HEK293T cells markedly reduced luciferase activity compared with activity in the negative control (P < 0.001, Fig. 2b). However, co-transfection of let-7b mimics and PLZF-mut1 in HEK293T cells did not reduce luciferase activity. Additionally, no difference in luciferase activity was observed between PLZF-WT2 and PLZF-mut2. The luciferase reporter assay showed that let-7b binds more at the putative binding sites of 1889–1895.
Fig. 2.
PLZF is a direct target gene of let-7b. a 2 potential binding sites between let-7b and PLZF. 293T cells were co-transfected with the PLZF gene sequence with wild-type and mutated let-7b. Fluorescence intensity was measured 24 h later using the Dual-Luciferase Reporter Assay System and the results are shown in (b)
Expression of let-7b, PLZF, and TSHR in thyroid tissue
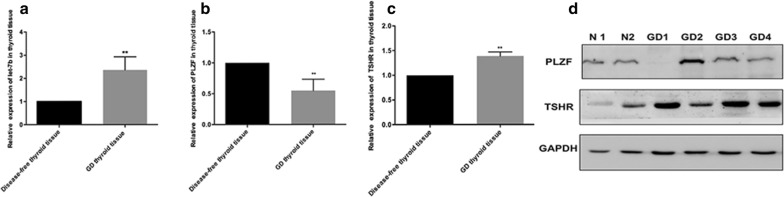
let-7b level was strongly correlated with TRAb level, and PLZF, which inhibits TSHR expression in the thyroid, was its target gene. Thus, we validated the expression of let-7b, PLZF, and TSHR in thyroid tissue. Compared to levels in normal thyroid tissue from patients with thyroid nodules, the expression of let-7b was significantly increased in thyroid tissue in patients with GD (P < 0.001, Fig. 3a). PLZF expression significantly decreased while TSHR level significantly increased in GD thyroid tissue as determined by Real-time PCR and western blotting (Fig. 3b–d). Additionally, correlation analysis showed that the expression of let-7b was significantly inversely correlated with PLZF level in thyroid tissue (r = − 0.849, P = 0.033).
Fig. 3.
Expression of let-7b, PLZF, and TSHR in thyroid tissue. Relative expression of let-7b, PLZF, and TSHR in thyroid tissue. The expression of let-7b (a), PLZF mRNA (b), and TSHR mRNA (c) in thyroid tissues from individuals with Graves’ disease (GD) and normal thyroid tissues by real-time PCR. d Protein levels of PLZF and TSHR in GD thyroid tissue. **P < 0.001
Let-7b increases TSHR expression in thyroid tissue via inhibiting PLZF
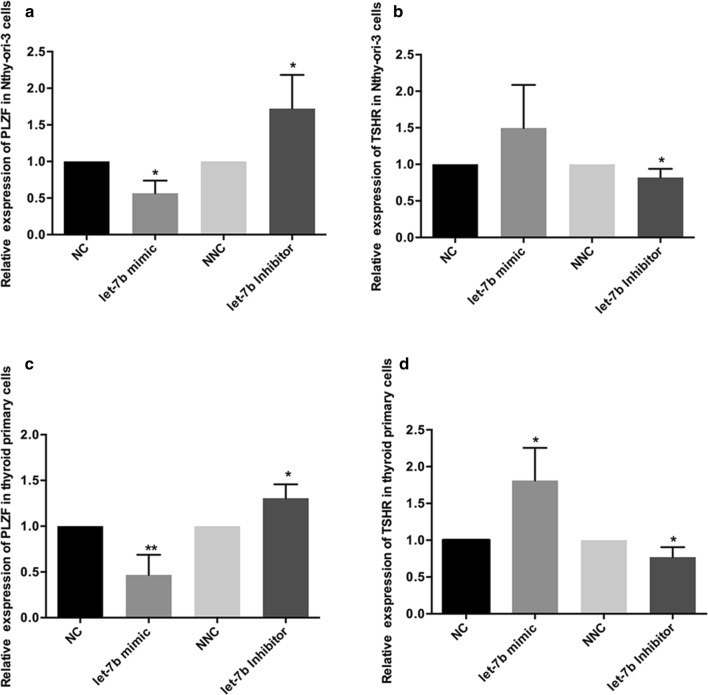
To explore whether let-7b increased the expression of TSHR via inhibiting PLZF in thyroid cells, Nthy-ori3-1 and primary thyroid cells were transfected with let-7b mimics or let-7b inhibitors in vitro. After transfection with let-7b mimics, reduced PLZF mRNA and increased TSHR mRNA levels were found in both Nthy-ori3-1 and primary thyroid cells, whereas let-7b inhibition resulted in increased PLZF mRNA and decreased TSHR mRNA expression (Fig. 4).
Fig. 4.
let-7b regulates TSHR expression via targeting PLZF. The expression of PLZF and TSHR in thyroid tissue. Thyroid primary cells were isolated from fresh thyroid tissue and cultured in vitro. Negative control or let-7b mimics, Inhibitor negative control, or let-7b inhibitors were transfected into primary thyroid cells (a, c) and human normal thyroid primary cells (b, d) and PLZF mRNA (a, b) and TSHR mRNA (c, d) was detected by real-time PCR. Results were calculated by three independent experiments, *P < 0.05, **P < 0.001
Discussion
Circulating miRNAs are emerging area in autoimmune disease and attracted our attention. Because of their highly stable and reproducible characteristics, circulating miRNAs are reliable biological markers for diseases such as tumors and inflammatory diseases [13, 14]. Overexpression of circulating miR-191-5p, miR-24-3p, miR-128-3p, and miR-376c-3 was detected in patients with multiple sclerosis [24]. In addition, miR-24-3p and miR-128-3p tended to associate with dysfunction accumulation and disease activity, respectively [25]. Limited research reported the circulating miRNAs in patients with GD. Yamada et al. [15] found that miR-16, miR-22, miR-375, and miR-451 were significantly increased in circulation in patients with GD. Hiratsuka et al. demonstrated that let-7g and miR-339-5p levels were decreased in patients with GD in remission compared with levels in patients with intractable GD [16].
It was thought circulating miRNAs have hormone-like effects leading to widespread responses within different types of cells [17]. Circulating miRNAs were originated from the passive released from cells and could reach to remote target cells to communicate. Differential expression of miRNAs in specific cells might lead to differential expression in circulation. And, differential expression of miRNAs may be overlapping in different cells. In multiple sclerosis, miR-21, miR-142-3p, miR-146a, miR-155 and miR-326 were found to be upregulated in both PBMCs and brain white matter lesions from patients and mouse model [18]. Nagarkatti et al. demonstrated miR-155 could serve as diagnostic markers and therapeutic targets in multiple sclerosis since miR-155 was all upregulated in plasma, PBMCs and brain white matter lesions from patients with multiple sclerosis. Besides, evidence from animal studies suggested that suppression miR-155 could inhibit the development of Th1 and Th17 cells and anti-miR-155 could inhibit EAE development [26, 27]. Thus, some overlapping in the differentially expression miRNA profiles in disease may play crucial roles in disease and could be used as potential therapeutic target.
Based on our previous study, miR-142-3p, miR-154-3p, miR-431-3p, miR-590-5p, and let-7b were differentially expressed in PBMCs in untreated GD patients [10]. Those miRNAs were involved in various pathways such as NF-κB pathways, TNF pathways, Toll-like receptor pathways which were closely related with the pathogenesis of GD and were reported in previous literatures to be related with autoimmune diseases (shown in Additional file 1: Table S1) [28, 29]. In this regard, we investigated the expression of miR-142-3p, miR-154-3p, miR-431-3p, miR-590-5p, and let-7b in serum of patients with GD in this study. The results showed that serum miR-142-3p and serum let-7b levels were significantly increased in patients with untreated GD and recovered in GD patients with remission.
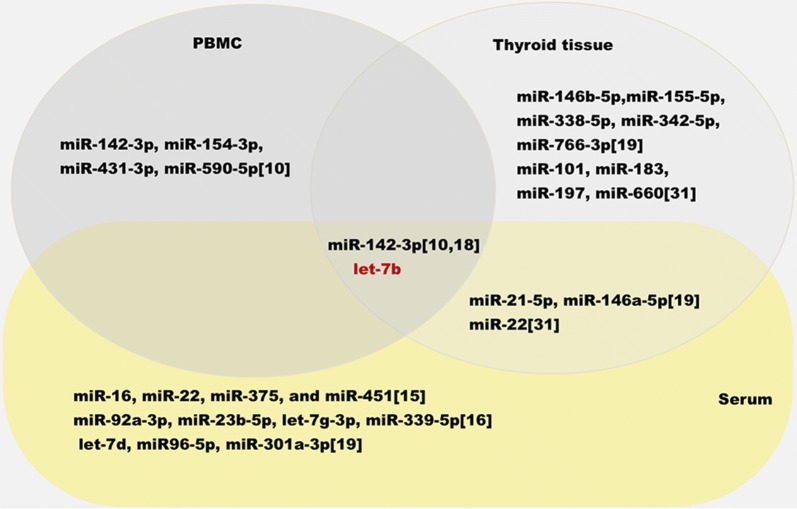
In addition, we summarized dysregulated profiles of miRNAs in patients with GD based on previous literatures (Fig. 5). Coupled with our results, we found some overlapping of these miRNAs in patients with GD. miR-22 was reported by Qin in lesioned tissue of GD and reported by Yamada et al. to be increased significantly in circulation in patients with GD [15, 30]. Marazuela et al. demonstrated that miR-142-3p, miR-21-5p, miR-146a-5p were dysregulated in both thyroid tissue and serum [19].
Fig. 5.
Overlapping between dysregulated miRNAs in serum and peripheral blood mononuclear cells and thyroid tissue. The data of dysregulated miRNAs was compiled from miRNAs expression profiling studies in serum [15, 16, 19], in peripheral blood mononuclear cells [10] and in thyroid tissue [19, 30]. Identical dysregulated miRNAs were shown in overlapping area. Non-identical dysregulated miRNAs were shown in single area
Consistent with the results of their study, miR-142-3p and let-7b level was significantly increased in serum in our study. Dysregulated in three compartments. miR-142-3p was upregulated in serum and thyroid tissue and downregulated in PBMCs in patients with GD. let-7b was upregulated in serum, thyroid tissue and PBMCs in patients with GD. miR-142-3p is abundant in cells of hematopoietic origin and has gained considerable attention for its role in regulating immune response [31]. Software predicted that miR-142-3p was involved in inflammtory pathways such as B cell receptor signaling pathway and TNF signaling pathway, which was responsible for the production of antibody. Especially, evidence suggested that miR-142-3p impaired the inhibitory Treg cells on the proliferative response and cytokine production of CD4+ CD25− T cells [32]. The impaired inhibitory function of Treg cells in patients with GD were confirmed in previous literatures [33, 34]. Thus, dysregulated miR-142-3p might exacerbated the immune process via impairing Treg cell function in patients with GD. However, there was no correlation between miR-142-3p and thyroid function parameters in our study, and we did not validate furtherly. Instead, we found that let-7b level strongly correlated with TRAb level in this study, which was used as an indicator of GD diagnosis and disease severity, suggesting a possible role of let-7b in the mechanism of GD and TRAb production.
let-7b belongs to the let-7 miRNA family, which is one of the important families of miRNAs. The members of let-7 miRNA family include let-7a-1, let-7a-2, let-7a-3, let-7b, let-7c, let-7d, let-7d, let-7e, let-7f-1, let-7f-2, let-7g, let-7i, mir-98, and mir-202 [35]. Circulating let-7 was reported in various other autoimmune diseases. let-7b levels were increased in the serum of patients with immunoglobulin A nephropathy and were a reliable predictor of the probability of having immunoglobulin A nephropathy [36]. Gandhi et al. [37] identified circulating miRNAs (let-7 and miR-92) that were differentially expressed in relapsing-remitting MS patients compared to secondary progressive MS patients. Mature let-7 miRNAs are highly conserved among different species and regulate multiple target genes affecting cell cycle and cell proliferation and differentiation [35, 38]. However, few reports have investigated circulating let-7b and its target gene in GD.
Promyelocytic leukemia zinc finger (PLZF), the target gene of let-7b, was validated in this study. There are two putative binding sites in PLZF (positions 1889–1895 and positions 4766–4773 in the 3′UTR of PLZF). As determined by a dual-luciferase reporter assay, let-7b mainly targeted the first binding site (positions 1889–1895 in the 3′UTR of PLZF). One recent study, whose results were consistent with ours, showed that let-7 miRNAs target the 3′UTR of zbtb mRNA to inhibit the expression of PLZF protein, which is required for innate-lineage T cell generation in the thymus. During the development of natural killer T (NKT) cells, the expression of let-7b is dynamically upregulated, directing NKT cells to eventually differentiate into IFN-producing NKT1 cells rather than IL-4-producing NKT2 cells or NKT17 cells that produce IL-17 [39]. PLZF is a transcription factor belonging to the BTB/POZ family, and functions by recruiting multiprotein complexes to regulatory gene elements [40, 41]. Findings of Stefan et al. suggest that PLZF can act as a transcriptional repressor of TSHR and downregulate TSHR expression [42]. TSHR is the primary autoantigen of GD. An enhanced presentation of TSHR on thyrocytes was reported in patients with GD [43], but the mechanism remains to be elucidated. We found upregulation of let-7b reduced the expression of PLZF and increased the TSHR expression in both primary normal thyroid cells and normal human thyroid cell lines. The opposite effect was observed when let-7b was downregulated in primary normal thyroid cells and normal human thyroid cell lines. We corroborated the expression of PLZF in thyroid tissue in patients with GD and found that PLZF was significantly decreased and inversely correlated with let-7b expression in the thyroid. These results suggested that changes in let-7b levels affect the expression of TSHR via PLZF. Therefore, we demonstrated that let-7b was involved in the production of TRAb by targeting PLZF.
The strengthen of our study was that we detected some miRNAs in serum and thyroid tissue in patients with GD. For the first time, we found that miR-142-3p and let-7b were dysregulated in three compartments. miR-142-3p was significantly upregulated in serum in patients with GD and could be used in the discrimination of GD patients from healthy control. let-7b was all upregulated in serum, thyroid tissue and PBMCs in patients with GD. let-7b significantly correlated with TRAb level in GD and let-7b may participate in the production of TRAb via PLZF. Therefore, let-7b may be a potential therapeutic target for patients with GD. One major limitation of this study was its relatively small sample size. Further studies need to confirm the utility of let-7b in patients with GD, which could help us further understand the pathogenesis of circulating miRNAs in GD and contribute to the development of potential therapeutic strategies in patients with GD.
Conclusions
In the present study, we detected the expression of miRNAs in serum of patients with GD which was the differentially expressed in PBMCs. For the first time, we demonstrated that let-7b was upregulated in PBMCs, serum and thyroid tissue of patients with GD. Serum let-7b significantly correlated with TRAb level in GD and let-7b may participate in the production of TRAb via PLZF.
Additional file
Additional file 1: Table S1. The predicted candidate gene, pathway and related other inflammatory disease of the selected microRNA candidates.
Authors’ contributions
XC, FH and YQ designed the study, performed the laboratory work, analyzed the data. XC drafted the manuscript. MZ, QY, YP and YZ performed the recruitment. GN and SW were responsible for the grants. All authors read and approved the final manuscript.
Acknowledgements
All thanks go to the patients and disease-free controls in our study. Special thanks go to Dr. Jiqi Yan in the department of thyroid surgery in Ruijin Hospital.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All procedures performed in this study are in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Funding
The research was supported by National Natural Science Foundation of China (No. 81570707).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- GD
Graves’ disease
- TSHR
thyrotropin receptor
- TRAb
thyrotropin receptor antibody
- miRNAs
microRNA
- PBMCs
peripheral blood mononuclear cells
- TNF
tumor necrosis factor
- TLRs
Toll-like receptors
- PLZF
promyelocytic leukemia zinc finger
Footnotes
Xinxin Chen and Fengjiao Huang contributed equally to this manuscript
Electronic supplementary material
The online version of this article (10.1186/s12967-018-1565-9) contains supplementary material, which is available to authorized users.
Contributor Information
Xinxin Chen, Email: creammonkey@sjtu.edu.cn.
Fengjiao Huang, Email: huang_fengjiao@163.com.
Yicheng Qi, Email: qionecheng@163.com.
Mengxi Zhou, Email: mency0510@163.com.
Qinglei Yin, Email: yinqinglei128@163.com.
Ying Peng, Email: pennyppp@126.com.
Yulin Zhou, Email: yulinzhou6@163.com.
Guang Ning, Email: gning@sibs.ac.cn.
Shu Wang, Phone: +86-021-64370045, Email: shuwang999@hotmail.com.
References
- 1.Weetman AP. Graves’ disease. N Engl J Med. 2000;343:1236–1248. doi: 10.1056/NEJM200010263431707. [DOI] [PubMed] [Google Scholar]
- 2.Shan Z, Chen L, Lian X, Liu C, Shi B, Shi L, et al. Iodine status and prevalence of thyroid disorders after introduction of mandatory universal salt iodization for 16 years in China: a cross-sectional study in 10 cities. Thyroid. 2016;26:1125–1130. doi: 10.1089/thy.2015.0613. [DOI] [PubMed] [Google Scholar]
- 3.Wang L, Wang FS, Gershwin ME. Human autoimmune diseases: a comprehensive update. J Intern Med. 2015;278:369–395. doi: 10.1111/joim.12395. [DOI] [PubMed] [Google Scholar]
- 4.Shukla SK, Singh G, Ahmad S, Pant P. Infections, genetic and environmental factors in pathogenesis of autoimmune thyroid diseases. Microb Pathog. 2018;116:279–288. doi: 10.1016/j.micpath.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Morshed SA, Davies TF. Graves’ disease mechanisms: the role of stimulating, blocking, and cleavage region TSH receptor antibodies. Horm Metab Res. 2015;47:727–734. doi: 10.1055/s-0035-1559633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross DS, Burch HB, Cooper DS, Greenlee MC, Laurberg P, Maia AL, et al. American Thyroid Association guidelines for diagnosis and management of hyperthyroidism and other causes of thyrotoxicosis. Thyroid. 2016;26:1343–1421. doi: 10.1089/thy.2016.0229. [DOI] [PubMed] [Google Scholar]
- 7.Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun. 2009;32:189–194. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 9.Bernecker C, Lenz L, Ostapczuk MS, Schinner S, Willenberg H, Ehlers M, et al. MicroRNAs miR-146a1, miR-155-2, and miR-200a1 are regulated in autoimmune thyroid diseases. Thyroid. 2012;22:1294–1295. doi: 10.1089/thy.2012.0277. [DOI] [PubMed] [Google Scholar]
- 10.Liu R, Ma X, Xu L, Wang D, Jiang X, Zhu W, et al. Differential microRNA expression in peripheral blood mononuclear cells from Graves’ disease patients. J Clin Endocrinol Metab. 2012;97:968–972. doi: 10.1210/jc.2011-2982. [DOI] [PubMed] [Google Scholar]
- 11.Shen L, Huang F, Ye L, Zhu W, Zhang X, Wang S, et al. Circulating microRNA predicts insensitivity to glucocorticoid therapy in Graves’ ophthalmopathy. Endocrine. 2015;49:445–456. doi: 10.1007/s12020-014-0487-4. [DOI] [PubMed] [Google Scholar]
- 12.Qi Y, Zhou Y, Chen X, Ye L, Zhang Q, Huang F, et al. MicroRNA-4443 causes CD4+ T cell dysfunction by targeting TNFR-associated factor 4 in Graves’ disease. Front Immunol. 2017;8:1440. doi: 10.3389/fimmu.2017.01440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada H, Itoh M, Hiratsuka I, Hashimoto S. Circulating microRNAs in autoimmune thyroid diseases. Clin Endocrinol (Oxf) 2014;81:276–281. doi: 10.1111/cen.12432. [DOI] [PubMed] [Google Scholar]
- 16.Hiratsuka I, Yamada H, Munetsuna E, Hashimoto S, Itoh M. Circulating microRNAs in Graves’ disease in relation to clinical activity. Thyroid. 2016;26:1431–1440. doi: 10.1089/thy.2016.0062. [DOI] [PubMed] [Google Scholar]
- 17.Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating miRNAs: cell-cell communication function? Front Genet. 2013;4:119. doi: 10.3389/fgene.2013.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma X, Zhou J, Zhong Y, Jiang L, Mu P, Li Y, Singh N, Nagarkatti M, Nagarkatti P. Expression, regulation and function of microRNAs in multiple sclerosis. Int J Med Sci. 2014;11:810–818. doi: 10.7150/ijms.8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez-Hernández R, Sampedro-Núñez M, Serrano-Somavilla A, Ramos-Leví AM, de la Fuente H, Triviño JC, et al. A microRNA signature for evaluation of risk and severity of autoimmune thyroid diseases. J Clin Endocrinol Metab. 2018;103:1139–1150. doi: 10.1210/jc.2017-02318. [DOI] [PubMed] [Google Scholar]
- 20.Kim SJ, Chen Z, Essani AB, Elshabrawy HA, Volin MV, Volkov S, et al. Identification of a Novel Toll-like Receptor 7 endogenous ligand in rheumatoid arthritis synovial fluid that can provoke arthritic joint inflammation. Arthritis Rheumatol. 2016;68:1099–1110. doi: 10.1002/art.39544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Zhu L, Xie GL, Bao JF, Yu Q. Let-7 miRNAs modulate the activation of NF-κB by targeting TNFAIP3 and are involved in the pathogenesis of lupus nephritis. PLoS ONE. 2015;10:e0121256. doi: 10.1371/journal.pone.0121256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller A, Leidinger P, Lange J, Borries A, Schroers H, Scheffler M, et al. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS ONE. 2009;4:e7440. doi: 10.1371/journal.pone.0007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joyce CE, Zhou X, Xia J, Ryan C, Thrash B, Menter A, et al. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet. 2011;20:4025–4040. doi: 10.1093/hmg/ddr331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vistbakka J, Elovaara I, Lehtimaki T, Hagman S. Circulating microRNAs as biomarkers in progressive multiple sclerosis. Mult Scler. 2017;23:403–412. doi: 10.1177/1352458516651141. [DOI] [PubMed] [Google Scholar]
- 25.Vistbakka J, Sumelahti ML, Lehtimäki T, Elovaara I, Hagman S. Evaluation of serum miR-191-5p, miR-24-3p, miR-128-3p, and miR-376c-3 in multiple sclerosis patients. Acta Neurol Scand. 2018 doi: 10.1111/ane.12921. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Cheng Y, Cui W, Li M, Li B, Guo L. MicroRNA-155 modulates Th1 and Th17 cell differentiation and is associated with multiple sclerosis and experimental autoimmune encephalomyelitis. J Neuroimmunol. 2014;266:56–63. doi: 10.1016/j.jneuroim.2013.09.019. [DOI] [PubMed] [Google Scholar]
- 27.Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J Immunol. 2011;187:2213–2221. doi: 10.4049/jimmunol.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vergoulis T, Kanellos I, Kostoulas N, Georgakilas G, Sellis T, Hatzigeorgiou A, Dalamagas T. mirPub: a database for searching microRNA publications. Bioinformatics. 2015. http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=mirpub/index. Accessed 20 June 2018. [DOI] [PMC free article] [PubMed]
- 29.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015: gkv403.http://snf-515788.vm.okeanos.grnet.gr. Accessed 20 June 2018. [DOI] [PMC free article] [PubMed]
- 30.Qin Q, Wang X, Yan N, Song RH, Cai TT, Zhang W, et al. Aberrant expression of miRNA and mRNAs in lesioned tissues of Graves’ disease. Cell Physiol Biochem. 2015;35:1934–1942. doi: 10.1159/000374002. [DOI] [PubMed] [Google Scholar]
- 31.Sharma S. Immunomodulation: a definitive role of microRNA-142. Dev Comp Immunol. 2017;77:150–156. doi: 10.1016/j.dci.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Huang B, Zhao J, Lei Z, Shen S, Li D, Shen GX, et al. miR-142-3p restricts cAMP production in CD4+ CD25− T cells and CD4+ CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep. 2009;10:180–185. doi: 10.1038/embor.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marazuela M, Garcia-Lopez MA, Figueroa-Vega N, de la Fuente H, Alvarado-Sanchez B, Monsivais-Urenda A, et al. Regulatory T cells in human autoimmune thyroid disease. J Clin Endocrinol Metab. 2006;91:3639–3646. doi: 10.1210/jc.2005-2337. [DOI] [PubMed] [Google Scholar]
- 34.Mao C, Wang S, Xiao Y, Xu J, Jiang Q, Jin M, et al. Impairment of regulatory capacity of CD4+ CD25+ regulatory T cells mediated by dendritic cell polarization and hyperthyroidism in Graves’ disease. J Immunol. 2011;186:4734–4743. doi: 10.4049/jimmunol.0904135. [DOI] [PubMed] [Google Scholar]
- 35.Roush S, Slack FJ. The let-7 family of micrornas. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Serino G, Pesce F, Sallustio F, De Palma G, Cox SN, Curci C, et al. In a retrospective international study, circulating miR-148b and let-7b were found to be serum markers for detecting primary IgA nephropathy. Kidney Int. 2016;89:683–692. doi: 10.1038/ki.2015.333. [DOI] [PubMed] [Google Scholar]
- 37.Gandhi R, Healy B, Gholipour T, Egorova S, Musallam A, Hussain MS, et al. Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann Neurol. 2013;73:729–740. doi: 10.1002/ana.23880. [DOI] [PubMed] [Google Scholar]
- 38.Bussing I, Slack FJ, Grosshans H. Let-7 micrornas in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Pobezinsky LA, Etzensperger R, Jeurling S, Alag S, Kadakia T, McCaughtry TM, et al. Let-7 microRNAs target the lineage-specific transcription factor PLZF to regulate terminal NKT cell differentiation and effector function. Nat Immunol. 2015;16:517–524. doi: 10.1038/ni.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suliman BA, Xu D, Williams BR. The promyelocytic leukemia zinc finger protein: two decades of molecular oncology. Front Oncol. 2012;2:74. doi: 10.3389/fonc.2012.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ono R, Masuya M, Nakajima H, Enomoto Y, Miyata E, Nakamura A, et al. Plzf drives MLL-fusion-mediated leukemogenesis specifically in long-term hematopoietic stem cells. Blood. 2013;122:1271–1283. doi: 10.1182/blood-2012-09-456665. [DOI] [PubMed] [Google Scholar]
- 42.Stefan M, Wei C, Lombardi A, Li CW, Concepcion ES, Inabnet WB, 3rd, et al. Genetic–epigenetic dysregulation of thymic TSH receptor gene expression triggers thyroid autoimmunity. Proc Natl Acad Sci USA. 2014;111:12562–12567. doi: 10.1073/pnas.1408821111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuang M, Wang S, Wu M, Ning G, Yao Z, Li L. Expression of IFNalpha-inducible genes and modulation of HLA-DR and thyroid stimulating hormone receptors in Graves’ disease. Mol Cell Endocrinol. 2010;319:23–29. doi: 10.1016/j.mce.2009.12.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The predicted candidate gene, pathway and related other inflammatory disease of the selected microRNA candidates.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.