Summary
The traditional paradigm of cardiovascular disease research derives insight from large-scale, broadly inclusive clinical studies of well-characterized pathologies. These insights are then put into practice according to standardized clinical guidelines. However, stagnation in the development of new cardiovascular therapies and variability in therapeutic response implies that this paradigm is insufficient for reducing the cardiovascular disease burden. In this state-of-the-art review, we examine 3 interconnected ideas we put forth as key concepts for enabling a transition to precision cardiology: 1) precision characterization of cardiovascular disease with machine learning methods; 2) the application of network models of disease to embrace disease complexity; and 3) using insights from the previous 2 ideas to enable pharmacology and polypharmacology systems for more precise drug-to-patient matching and patient-disease stratification. We conclude by exploring the challenges of applying a precision approach to cardiology, which arise from a deficit of the required resources and infrastructure, and emerging evidence for the clinical effectiveness of this nascent approach.
Key Words: cardiology, clinical informatics, multi-omics, precision medicine, translational bioinformatics
Abbreviations and Acronyms: CAD, coronary artery disease; EHR, electronic health record; GWAS, genome-wide association studies; HF, heart failure
Central Illustration
The assumption of precision medicine is that insight into pathophysiological mechanisms of cardiovascular disease enables rational development of targeted treatments and procedures. Improved understanding of cardiovascular pathophysiologies comes at multiple scales. On the level of gross anatomy, understanding that occlusion of the coronary arteries typically leads to myocardial infarction or ventricular dysfunction led to the development of angiography and bypass surgery in the 1960s and 1970s, and later to percutaneous coronary intervention (1).
The arrival of molecular biology techniques in the 1970s and 1980s enabled the discovery of biological pathways such as the renin-angiotensin-aldosterone system. Such advances enabled the creation of drugs inhibiting specific targets such as angiotensin-converting enzyme (2). Angiotensin-converting enzyme inhibitors and other analogously designed drug classes, such as beta-blockers, 3-hydroxy-3-methylglutaryl-CoA reductase inhibitors, and glycoprotein IIb/IIIa inhibitors, have led to significant decreases in cardiovascular disease morbidity and mortality for millions of people around the world 3, 4, 5.
Although targeting single molecules has worked for some cardiovascular diseases in the past, future success will require the adoption of new paradigms. Chronic cardiovascular disease encompasses a wide variety of pathological processes whose etiologies are genetic, environmental, and idiopathic. Even determining precise etiologies is often challenging. Although genome-wide association studies (GWAS) have revealed several highly significant loci 6, 7, 8, 9 associated with cardiovascular disease (Figure 1), the overall contribution of these loci to heritability in complex disease is often <10% (10). This “missing heritability” poses a significant problem for drug discovery: it implies that the strategy of targeting genetic regions discovered via GWAS, phenome-wide association studies, or loss-of-function studies will not provide clear-cut improvements for managing complex cardiovascular diseases moving forward 11, 12. The productivity of drug discovery pipelines has declined despite accumulating demand for new therapies 13, 14, 15. For example, highly targeted therapies such as the “-trapib” class of cholesterol esterase transfer protein inhibitors have repeatedly failed clinical trials 16, 17, 18, 19.
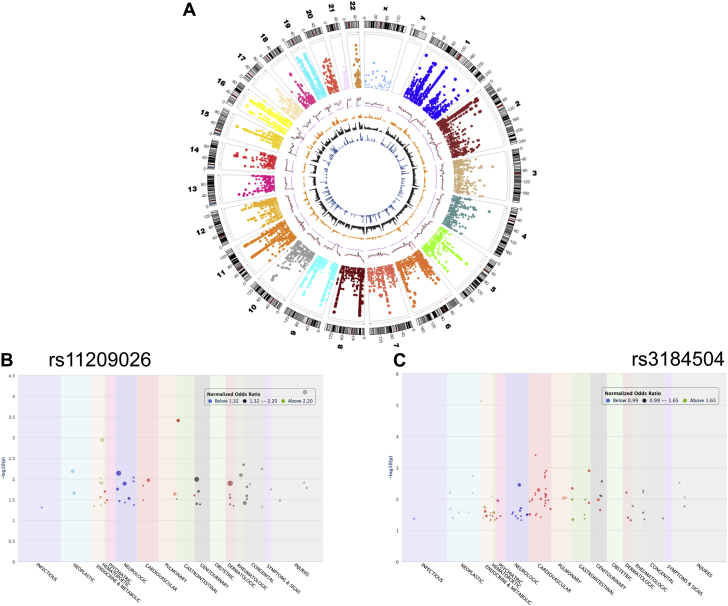
Figure 1.
Genome and Phenome-Wide Associations of Coronary Atherosclerosis, a Fundamental Mechanism Driving Several Cardiovascular Diseases
(A) Circos plot representing genome-wide associations of coronary atherosclerosis with each section representing human chromosomes. Phenome-wide associations of cardiovascular disease variants across other disease categories are represented in different colors. (B) rs11209026, a coding variant (c.G>A: R381Q) localized on interleukin-23 receptor (IL-23R); and (C) rs3184504 coding variant (c.784T>C: W262R) localized to PH domain of the SH2B3 protein.
In this state-of-the-art review, we identify 3 interconnected areas for new therapeutic opportunities in cardiovascular disease (20). First, we discuss the incorporation of precision medicine concepts into cardiology, or precision cardiology. We use the term precision cardiology to mean more accurate and refined characterization and stratification of disease states and individual patient pathologies using multiple molecular and clinical features (21). Precision characterization of cardiovascular disease consolidates heterogeneous sources of information into disease-related features. Until now, disease classification has relied upon experiential knowledge to decide a priori what information should be used to determine disease status. Instead, we propose to use multiscale data in combination with computational methods to better delineate boundaries between disease states, with the ultimate aim of choosing more precise therapies. Second, we generate and utilize disease networks to uncover and treat comorbidities associated with chronic cardiovascular diseases. Improved understanding of disease comorbidities will allow for new therapeutic opportunities. Third, we investigate the cardiovascular drug space in the frame of systems pharmacology, including drug repurposing and the identification of treatments that may act on multiple targets (polypharmacology). We conclude with a discussion on the potential role of precision cardiology in improving health care delivery through cost optimization, care coordination, and value-based standards of care.
Defining Precision Cardiology
Despite enormous public interest and federal investment into precision medicine as epitomized by the recent establishment of the Precision Medicine Initiative 22, 23, 24, 25, there are several competing definitions of precision medicine. The term is currently most often associated with the field of oncology, where rapid disease progression in cancer results from a series of somatic mutational events, which often clearly define a before- and after-disease state. This dichotomy provides a clear avenue to target treatments to an individual patient’s mutational profile 26, 27, 28, 29. The term is also used to define the application of genomic profiling and pharmacogenomics in a public health setting 30, 31, 32, 33. Although genomic medicine 34, 35 utilizes genetic information, we envision going further by incorporating information from the transcriptome, proteome, and metabolome with longitudinal health care data, such as disease diagnoses, procedures, medications, and environmental exposure data (36). We thus define precision cardiology as the application of multidimensional data to delineate subsets of the heterogeneous cardiovascular disease space. The ultimate aim of this approach is to enable patient stratification that can be used to better guide therapeutic interventions.
Many concepts from precision medicine in oncology are not directly applicable to cardiovascular diseases because there are substantial differences between heart disease and cancer. Somatic hypermutation is a central feature of cancer, but is not paramount in cardiology. Most cardiovascular diseases are chronic processes where the pathoetiology may begin decades before there are any symptomatic manifestations of the disease. Cardiovascular diseases are highly heterogeneous and present as comorbid or multimorbid with other conditions, whereas, for a given affected individual, cancer often presents as a more uniform pathological process (although an expressed malignancy in an individual can exhibit appreciable molecular and pathophysiological diversity due to clonal heterogeneity). Clinically, cardiology often uses broad, inclusive disease definitions that may conceal subtle disease variance. Symptoms are encountered late in disease progression. Finally, there is a strong temporal effect in cardiovascular disease—that is, the same disease encountered at different time points may require completely different interventions for prevention or treatment.
Traditional Quantitative Approaches Are Inadequate for Precision Cardiology
Several important factors drive the need to develop new quantitative approaches for precision cardiology. First, biological systems are inherently complex and display dynamic, emergent properties resulting from myriad potential interactions between individual molecules and coordinated pathways (37). In humans, vital functionality occurs at scales ranging from cellular genomics to gross anatomy, with numerous layers of molecular and cellular physiology in between. Second, there are challenges to interpreting data for several reasons. Data collected from patients during clinical encounters is often limited. When this information is entered into electronic health records (EHRs), limitations of this format can make later analysis more difficult. Because collecting data is expensive and time-consuming in clinical settings, sample sizes are often small. Collectively, these challenges hinder our attempts to build comprehensive deterministic models of complex disease that could be used to better guide patient treatment.
Because of the issues with deterministic models, clinical researchers often utilize traditional statistical approaches such as logistic or Cox regression models. These techniques allow investigators to draw conclusions about associations between a limited number of predictor variables without complete characterization of the system.
However, tested hypotheses must be specified ahead of time, and these models do not easily fit data that may have underlying hidden structure (38). Instead, the implementation of more advanced computational and informatics approaches is an integral requirement for precision cardiology. Specifically, machine learning techniques can be used to model and explore data in an unsupervised fashion (Central Illustration).
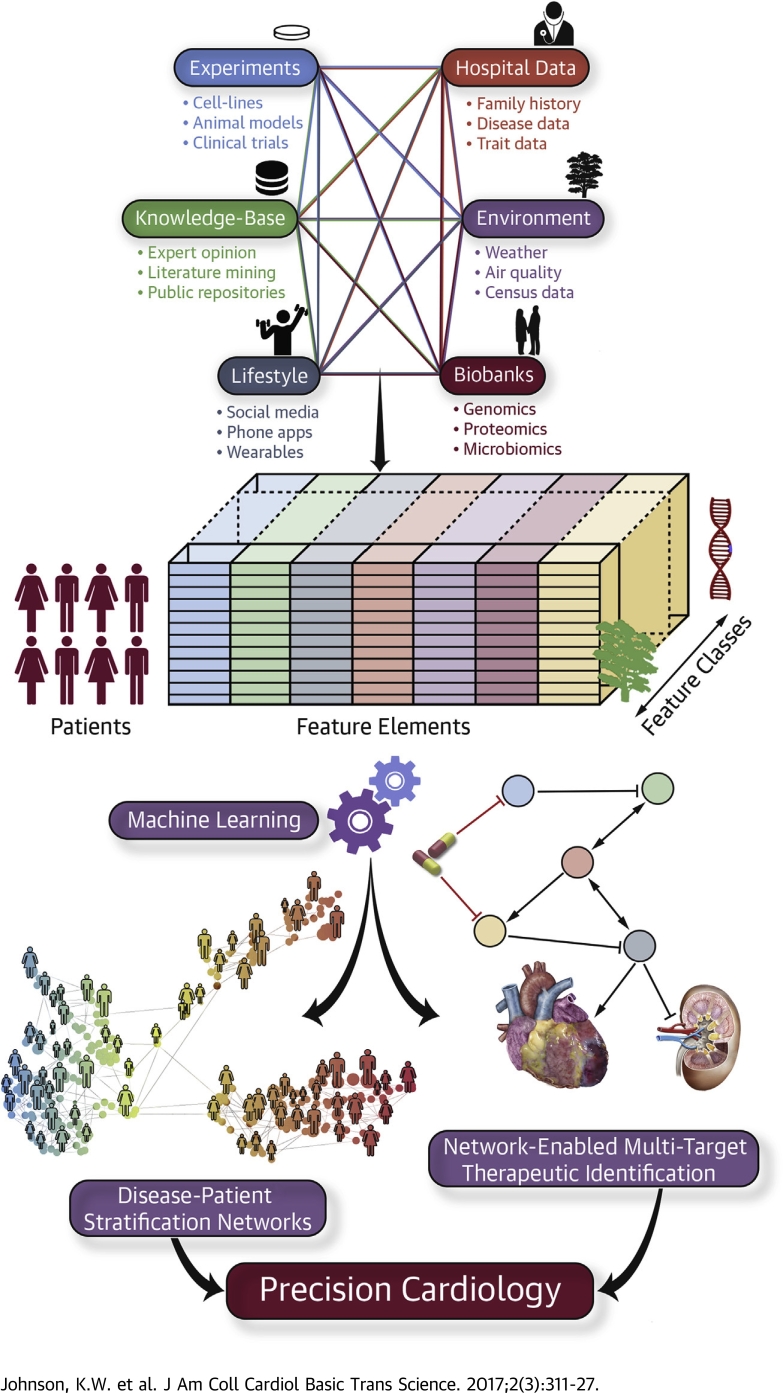
Central Illustration.
Machine Learning-Driven Precision Cardiology
Multiple sources of heterogeneous data, including experimental evidence, bioinformatics databases, lifestyle measurements, electronic health records, environmental influences, and biobank findings, can be incorporated together using machine learning algorithms to identify causal disease networks, stratify patients, and ultimately predict more efficacious therapies.
Medicine lags behind other industries with regard to incorporating dynamic, longitudinal data into event detection and decision-making processes (39). For example, credit card companies collect large, longitudinal datasets containing customer information and individual transactions, but these datasets often do not include labels for whether particular transactions are fraudulent. Legitimate transactions generally far outnumber fraudulent transactions, and this unbalanced dependent variable problem has made the application of traditional statistical approaches unreliable. Instead, machine learning methods such as neural networks have been applied to solve this problem through separation of the entirety of the collected dataset into different strata representing different transaction classes. These classes are then tested for enrichment of different features, such as fraudulent transactions. This strategy has proven to be highly effective for fraud detection, and we propose the adoption of similar strategies to deal with medical data in cardiology (40). The practice of medicine and health care delivery is unique in that it must concern itself with patient safety and privacy concerns unparalleled in other industries. Nonetheless, we believe that medicine and, in particular, precision cardiology have much to gain by looking out into other industries and their use of data-driven methods. As an early example of the success of machine-learning in cardiology, data from devices such as the mobile electrocardiographic device Kardia (AliveCor Inc., San Francisco, California) have been successfully used in combination with machine learning to detect atrial fibrillation.
A great number of resources are available for incorporation into precision cardiology efforts. Clinical data traditionally collected from patients, such as vital signs, imaging results, laboratory tests, and patient histories, can be processed and incorporated into predictive models. Furthermore, machine learning methods have already been successfully applied to echocardiography results 41, 42. We can also add patient descriptors, such as medications, diagnoses, procedures, and billing codes, to the set of clinical information. Other clinical data like medications can be linked to external adverse-event and drug-drug interaction databases. We also envision including information from emerging biosensors and consumer devices, such as smart watches or mobile health applications, into the data to be analyzed. Genomic information can be utilized in predictive models in the form of single nucleotide polymorphisms, copy number variants, and structural variants discovered from genome-wide association studies or other techniques. Associations from epigenetic, transcriptomic, and proteomic studies can also be added to represent genomic function. Metabolomics results (i.e., small molecules in fluids or tissues) present a tightly coupled representation of biological status related to clinical traits and may also be incorporated. The previous tools can then stratify phenotypes identified in phenome-wide association studies. We collectively call this set of multiscale data multi-omics.
Analyzing Cardiovascular Multiscale Data With Machine Learning Methods
In multi-omics studies, we do not know the exact mechanistic model that connects the different types of data. To address such uncertainties, statistical machine learning methods can be used to interrogate, model, and learn from complex multi-omics data.
The mathematical strategies underlying machine learning can be broadly defined as either supervised or unsupervised learning (Table 1). Supervised learning requires pre-defined labeling of the dataset (e.g., “cases” and “controls”). The labeled data is typically divided into training and testing datasets to reduce overfitting and produce better predictive models. Supervised learning algorithms (e.g., support vector machines, random forests, naïve Bayes classifiers, as well as ensembles thereof) can be selected to perform a particular machine learning task depending on the nature of the data (continuous or categorical), sparseness or completeness of the data, or nature of the machine learning task (prediction, classification, and so on). Many classification and clustering problems in biology and medicine, including cardiovascular disease classification, precision phenotyping, and clinical diagnostic support systems, have leveraged machine learning methods 41, 43, 44.
Table 1.
Statistical Learning Approach to Precision Cardiology
| Type of Learning | Problem Tasks | Algorithms | Example Application in Cardiology | PMID |
|---|---|---|---|---|
| Supervised learning | ||||
| Regression | Ordinary least squares regression | Many | ||
| Classification | Logistic regression | Many | ||
| K nearest neighbors | Many | |||
| Predictive modeling | Lasso regression | Sex-dependent risk factors for myocardial infarction | 25515680 | |
| Survival analysis | Ridge regression | Discovery of biomarkers associated with CAD prognosis | 27224515 | |
| Elastic net regression | Discovery of biomarkers associated with CAD prognosis | 27224515 | ||
| Naïve Bayes | Classification of cardiovascular risk level | 27525161 | ||
| Support vector machines | Diagnosis of acute coronary syndrome | 26815338 | ||
| Information maximizing component analysis | Feature extraction of left ventricle shape changes following myocardial infarction | 26531126 | ||
| Bayesian networks | Meta-analysis of stroke prevention treatments; real-time prediction of cardiovascular events | 27570467; 26456181 | ||
| Decision trees | Estimating risk in congenital heart surgery | 26774238 | ||
| Random forests | Predictive modeling of chemoreflex sensitivity; predictive modeling of pediatric dilated cardiomyopathy from miRNAs | 27099934 | ||
| AdaBoost classifiers | Myocardial perfusion analysis from CT imaging | 26073787 | ||
| Neural networks | Length of hospital stay prediction; automated detection of stent failure; pharmacokinetics of losartan; prediction of heart failure outcomes | 27195660; 27036565; 26885213; 24387896 | ||
| Ensemble methods | All-cause mortality prediction | 27252451 | ||
| Unsupervised learning | ||||
| Dimensionality reduction | Hierarchical clustering | Many | ||
| Clustering | K means | Many | ||
| Principal components | Many | |||
| Self-organizing map neural network | Clustering ECG complexes | 10916254 | ||
| Linear discriminant analysis | Quantifying self-similarity of multimodal signals in the ICU; evaluation of atherosclerosis from multimodal imaging | 27454256; 25722204 | ||
| Topological data analysis | Pulmonary embolism diagnosis; few other examples | 26515513 | ||
| Deep learning | Ultrasound image processing; causal phenotype discovery | 21947526; 26958203 | ||
Table of selected statistical learning approaches with previous example applications in cardiology.
CAD = coronary artery disease; CT = computed tomography; ECG = electrocardiogram; ICU = intensive care unit; miRNA = microribonucleic acid; PMID = PubMed identifier number.
Methods drawn from machine learning known collectively as “unsupervised learning” can be used to model and learn from multi-omics data. Unsupervised learning can broadly be described as an approach to machine learning to discover hidden structure in datasets without comparing the data to an external reference label, such as disease status, mortality rate, or other commonly studied dependent variables. This strategy is often used to reduce the dimension of noisy, highly multivariate datasets—essentially distilling complex sources of information into smaller representative datasets, which can then be more meaningfully used for downstream analysis. Many traditional approaches to unsupervised learning such as principal components analysis, singular value decomposition, and various methods of clustering remain robust and applicable despite their maturity. However, as a highly active area of interest in the computer science and bioinformatics communities, machine learning has been revolutionized in the context of medicine by a significant number of new, emerging techniques such as advanced algorithms for matrix factorization 45, 46, 47; topological data analysis 48, 49, 50, 51; autoencoder artificial neural networks (52); and, in particular, deep learning, or artificial neural networks with multiple layers of hidden neurons 53, 54, 55, 56, 57, 58, 59, 60, 61, 62. To highlight 1 recent example of unsupervised learning, Li et al. (63) used topological data analysis to cluster patients with a diagnosis of type 2 diabetes mellitus into 3 separate subtypes of patients using data from a health system biobank, which included multiscale information from EHRs, clinical observations, and genetic data. In another example, deep learning-based denoising autoencoders were used successfully to build new aggregates of breast cancer gene expression data, which could then be used to predict mortality better than previous methods (52). Deep learning is perhaps the area of machine learning that has made the most rapid advance in recent years: in situations with large enough available datasets, deep learning has been demonstrated to produce best-of-class results and has thus been widely applied in areas such as drug discovery 58, 59 and digital pathology 54, 55. Briefly, deep learning strategies employ many layers of stacked models to represent data in hierarchies of concepts. Each layer is comprised of many different models, which interpret data provided to them and then pass their results to another higher-level model. This process is repeated until output results are obtained from the top-level model. In a recent study, Miotto et al. (36) demonstrated the promise of deep learning by applying the technique to model sparse, repetitive, and layered data found in the EHR to classify patient disease status in an unsupervised fashion. They termed this concept “deep patient” (36).
Accurate determination of a patient’s disease phenotypes can be a difficult problem. EHRs were established primarily to support health care provision and billing, and not for research. Moreover, different physicians, health systems, and scientists may use different criteria to diagnose a particular disease (64). These factors may lead to different conclusions and study results if not accounted for. The desire to consistently assign case and control status from EHRs has led to the development of standardized algorithms such as those from the eMERGE consortium 65, 66. These electronic phenotyping algorithms serve to standardize patient populations across studies. Many such algorithms are deposited in the Phenotype KnowledgeBase (PheKB) (67), a centralized repository of validated E-phenotyping algorithms for high-quality phenotyping using EHR data (68). For each disease algorithm, case and control cohorts are defined using standardized criteria that can be consistently applied across different settings. However, due to the rigorous nature of these algorithms, only a limited amount has been developed to date. These algorithmic disease classification practices have an important role in building a foundation for more precise approaches. However, it is important to note that “E-phenotyping” approaches such as those by the eMERGE consortium are not a precision approach by themselves; instead, they generally propose to define the entire complexity of disease processes into simple “present” or “absent” disease classifications.
In contrast to the algorithmic “E-phenotyping” approach described in the previous text, a differing approach called computational phenotyping has emerged (69). The goal of computational phenotyping is to embrace the complexity inherent in disease mechanisms through machine learning approaches to define accurate phenotypes.
Often, the goal is to use these phenotypes to model future responses in a dynamic fashion. For example, 1 group of investigators sought to discover new genetic diseases in patients with suspected genetic diseases. The investigators combined genetic sequencing with clinical trait data to build predictive scoring models for different putative mutations. They found that their model could correctly predict the genetic etiology for 28% of new cases for which there existed no previous diagnosis (70). In another example, the investigators developed a new machine-learning technique to predict antibiotic resistance phenotypes in a variety of bacterial species (71). Here, we envision a particular type of computational phenotyping that we call precision subtyping, which may use insights from both E-phenotyping (to establish baseline monolithic disease case-control cohorts) as well as from computational phenotyping (delineating new subsets of these cohorts).
Criteria for Cardiovascular Diseases Amenable to a Precision Approach
The diseases with the most potential to be treated in a more precise manner through multiscale approaches are those that meet several of the following criteria:
-
1.
Diseases that are classified symptomatically or according to their clinical phenotype instead of according to pathoetiology. Such diseases may often arise from subtly distinct pathways that could benefit from data-driven differential treatment. Many cardiovascular diseases fit this paradigm. For example, essential hypertension is a disease defined purely phenotypically (systolic or diastolic hypertension). Using machine-learning strategies to stratify essential hypertension may allow for differential treatment on the basis of etiology. We believe there is a compelling need for more personalized approaches to hypertension treatment, as current therapies are unsatisfactory. It is estimated that 44% of patients with essential hypertension were unable to achieve blood pressure control despite pharmacological therapy (72).
-
2.
Diseases that are treated according to nonspecific guidelines or treatment algorithms established as the best practice by clinical trials that focus on the mean response to an intervention instead of examining variability in response (21). Precision approaches have an opportunity to capture variability and suggest possibilities for differential treatment. Again, many cardiovascular diseases fit this paradigm. Hypertension clinical trials, for example, generally examine outcome effects as the sample mean blood pressure is decreased. Stent clinical trials generally look at mean revascularization rates or other outcomes by device instead of allowing for more precise patient-to-device matching.
-
3.
Diseases that are prominently characterized by biomarkers that do not reveal the underlying complexity of the disease. For example, atherosclerosis is strongly clinically associated with elevated low-density lipoprotein levels. However, the underlying biology is more complex, as suggested by the clinical failure of evacetrapib despite significant effects on low-density lipoprotein, and the accumulating evidence of the importance of the pleiotropic effects of statin therapy 73, 74, 75, 76, 77.
-
4.
Diseases that manifest variably over an extended time frame, providing an axis upon which patients may be stratified and treated differentially. Coronary artery disease is a pertinent example: atherosclerosis is a progressive disease in which the first pathology typically emerges in one’s 20s, yet it takes decades to manifest symptomatically. Even when it does finally manifest, reasons for differential outcomes are complex and are not fully understood.
-
5.
Diseases that are comorbid or multimorbid with other disease phenotypes. These diseases may in many cases have a specific molecular signature that could be corroborated with a pathophysiological mechanism. The majority of chronic cardiovascular pathologies have strong associations with comorbidities such as diabetes, obesity and metabolic syndrome, cancer (78), rheumatologic disease (79), as well as other cardiovascular diseases.
Examples of Cardiovascular Diseases Amenable to Precision Subtyping
Profiling patient populations using multi-omics approaches could help us to perform precision phenotyping of several cardiovascular diseases (Figure 2). As noted by Antman and Loscalzo (21), 1 example of a cardiovascular phenotype amenable to more precise treatment is essential hypertension. Essential hypertension’s definition is based upon a single physiological finding; yet, it likely has a complex and varied pathoetiology that is not well accounted for in current clinical treatment guidelines 21, 80. For example, a recent large meta-analysis of 18 studies containing 350,000 patients of varied ancestry with hypertension identified several rare single nucleotide variants with larger effect sizes than many common genetic variants associated with hypertension (81). The presence of substantial effect sizes with rare variants implies that there may be different or complementary molecular hypertension pathways. These different pathways may benefit from differential pharmacological treatment. Indeed, there are at least 10 different classes of drugs that can be used to treat hypertension (82); yet, standard guidelines and clinical trials generally emphasize a rote treatment algorithm in which drugs are used sequentially guided only by mean blood pressure measurements, and there are few attempts to tailor treatment to patients’ specific hypertension profiles 83, 84.
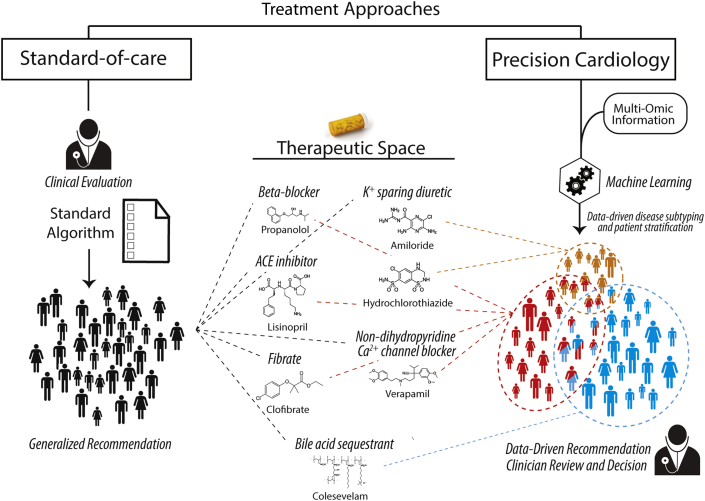
Figure 2.
Conceptualization of a Personalized Medicine Approach to Cardiology Contrasted With the Current Standard of Care
In the precision approach to cardiology, multi-omic information is incorporated to identify subtle strata of patients which can be differentially treated within the existing therapeutic space. ACE = angiotensin-converting enzyme.
Similarly, heart failure (HF) is another highly heterogeneous disease that results from complex interactions of dynamic physiological systems that are currently defined primarily symptomatically 85, 86. For example, the New York Heart Association functional classes are completely symptomatic, although more recent classification systems now take into account pre-symptomatic and at-risk subjects (87). Even more objective measures such as left ventricular ejection fraction also only crudely predict the degree of disease, let alone underlying disturbances in physiology. The lack of proper measures that could be used for subset identification of HF patients has been implicated in the failure of recent HF trials 88, 89. There have already been successful attempts at defining clusters of HF patients: for example, Ahmad et al. (90) used an unsupervised clustering approach to isolate 4 distinct clusters of HF patients: from clinical variables and biomarkers and found that the clusters were significantly associated with differential hospitalization and mortality risk. However, there remains an identified need for further efforts at HF patient stratification 91, 92. To our knowledge, only a single paper published to date has attempted to apply deep learning methods to HF patients (57).
Finally, coronary artery disease (CAD) is the third example of cardiovascular pathology amenable to a more precision-oriented approach. CAD fits all of our criteria for precision medicine application: it develops over an extended period of decades, beginning with atherosclerosis and manifesting variably along a spectrum from asymptomatic to stable ischemic heart disease, acute coronary syndrome, and sudden cardiac death.
Furthermore, despite a plethora of imaging techniques and biomarkers, CAD is still primarily diagnosed and managed symptomatically (93). Recent advances in characterizing CAD gene networks 94, 95 could be coupled with advanced imaging 96, 97 and a wide variety of novel biomarkers 98, 99, 100, 101, 102, 103, 104 to stratify patients with this heterogeneous disease and make more informed treatment decisions.
Disease Networks for Patient Stratification and Population Health Intelligence
Network biology offers a powerful paradigm for representing and learning from complex biomedical data. In general, networks are defined as objects (called nodes or vertices) linked to each other by edges (called links or edges). Edges may be either directed or undirected. In biological applications, many different types of networks can be constructed on the basis of the availability of data types to define nodes (e.g., gene-gene network, gene-drug network, or gene-disease network). Edge relationships can be inferred based upon biological or clinical contexts (e.g., coexpression level, availability of drug-target information, and gene-disease linkage information) (Figure 3). The resulting networks can be analyzed using a set of algorithms to prioritize individual nodes, to prioritize network hubs, or to highlight subsets of nodes (subnetworks or cliques).
Figure 3.
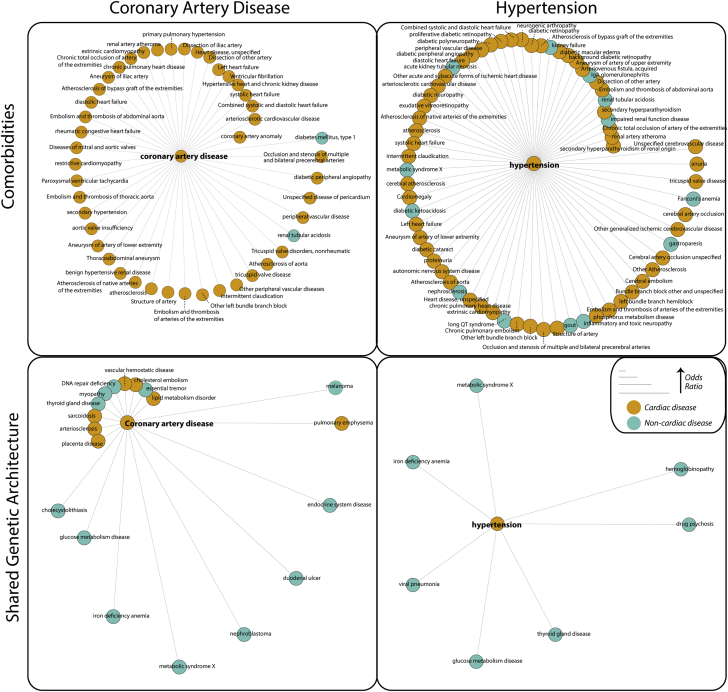
Comorbidity and Shared Genetic Architecture Between Hypertension and Coronary Artery Disease
(Top) Example of disease comorbidity networks for coronary artery disease (CAD) and hypertension (HTN), with comorbid diseases ascertained from Mount Sinai Hospital’s electronic health record data arranged around the central node. Distance from the central node is proportional to comorbidity odds ratio. We calculated comorbidity from ICD-9 codes using a logistic regression model controlling for age, sex, and self-reported ethnicity. Due to space limitations, we only show disease comorbidities with odds ratio ≥2. (Bottom) Networks of shared genetic architecture between CAD and HTN and other diseases, with shared genetic architecture defined as shared genome-wide association studies (GWAS) loci (gene level) between the 2 diseases. We compiled all data from GWASdb version 2 (August 2015) (159) and associated genes to a disease if they were GWAS threshold significant (p < 5 × 10−6) and conferred an increased risk. We calculated shared genetic architecture using a 1-sided Fisher exact test. Distance from central node is proportional to odds ratio. DNA = deoxyribonucleic acid.
Network analysis metrics such as the clustering coefficient, centrality measures, and connectivity scores can then be used to characterize network properties and differences between disease states 105, 106, 107, 108, 109. Additionally, groups of highly related nodes can be tested for statistical enrichment of various properties, such as ontology or functional classification. To highlight a recent example of a biomedical network analysis, Glicksberg et al. (110) built a large-scale human disease network by compiling phenomic data from the EHR. The organized network used 1,988 disease conditions and 37,282 disease pairs from 1.02 million patients of 3 different ancestries: Caucasian, African American, and Hispanic/Latino. Using this network, we identified several interesting pairwise disease comorbidities that may not be captured using traditional epidemiological studies. For example, we discovered 51 key “hub” diseases along with 2,158 significantly comorbid conditions for Caucasian patients, 3,265 for African-American patients, and 672 for Hispanic/Latino patients. By integrating network-level information with demographic information and genomic data, the sequelae of diseases can be used to identify the subsets of at-risk populations and stratify patients for optimized care delivery. In another recent example, we used a network approach to define cis and trans gene regulation of several hundred risk single nucleotide polymorphisms identified by GWAS for cardiometabolic diseases, providing strong evidence for extensive sharing of causal disease genes across tissues and different cardiometabolic diseases (95).
Drug Repositioning as an Avenue for Directed Interventions
Drug repositioning is a drug-discovery strategy that relies on a priori knowledge to recommend an existing drug for a new indication based on evidence from clinical data (e.g., off-label prescriptions) or biological experiments (e.g., gene expression profiling after drug perturbations) 111, 112. Briefly, drug repositioning (Figure 4) relies on the concept that biological processes are mediated by a finite set of genes or gene-products and several biological entities (e.g., genes, proteins, and transcripts) have pleiotropic effects and mediate similar functions. We outline a detailed overview of drug repositioning and various repositioning strategies in our recent review exploring experimental and computational approaches in the drug repositioning space 20, 113. We recently developed a reference database for drug repositioning investigations using primary indications and secondary indications of rare, common, and chronic diseases (RepurposeDB) (114). By analyzing the RepurposeDB, we identified biological, chemical, and epidemiological factors driving successful drug repositioning. For example, 1 of the most prominent successes of drug repositioning is the example of beta-blockers for HF 115, 116, 117. Originally used as antihypertensive agents, they were once considered to be contraindicated in systolic HF, but are now the cornerstone of pharmacological management for this disease.
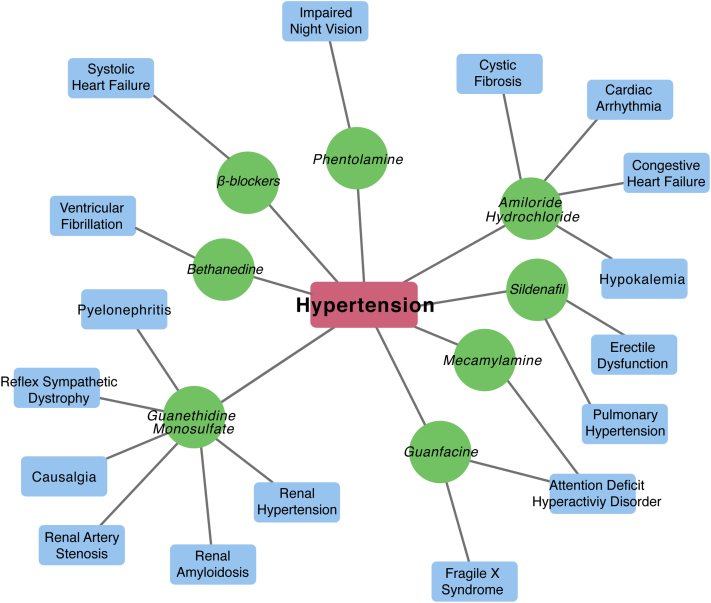
Figure 4.
Hypertension Drug Repositioning Bipartite Network From RepurposeDB
Example of drugs originally developed or used for hypertension that have been repurposed for other indications.
Another prominent example of repositioning is sildenafil. This compound was originally developed for hypertension, targeting the phosphodiesterase family of enzymes with the hope of augmenting atrial natriuretic peptide function by antagonizing breakdown of the second messenger molecule cyclic guanosine monophosphate. Pre-clinical trials showed that the compound’s actions lead to vasodilation, and the focus was then changed to angina. In clinical trials, it was noted that male patients reported the surprising side effect of penile erection after taking the drug. This observation allowed it to be eventually repositioned for erectile dysfunction. Later, it was observed that PDE-5 (a major target of sildenafil) was up-regulated in the lungs of patients with pulmonary hypertension (118). Sildenafil, as a known inhibitor of PDE-5, was then successfully repurposed for the indication of pulmonary hypertension 119, 120. Similarly, several cardiovascular therapies have beneficial effects on other disease processes (e.g., aspirin for cancer) (121). Drugs like colchicine (122) that have systemic action are also particularly eligible for repurposing. It should be noted that no therapeutic interventions are devoid of side effects: onco-cardiology effects and the effect of chemotherapy are growing concerns of cancer patients in remission (123). Thus, building personalized prescription recommendation models to optimize therapeutic efficacy and limit side effects is a prominent goal of precision cardiology for both patient outcomes and cost-effectiveness. In addition to small molecules, recently, biological drugs (see the comprehensive list of small molecule and repurposed biological drugs compiled in RepurposeDB [124]) including monoclonal antibodies, peptide inhibitors, and microribonucleic acid–based therapeutics have shown potential for drug repositioning. Thus, exploring such emerging therapeutic interventions and their effect on cardiovascular disease could help accelerate repositioning discoveries in clinical trials 125, 126, 127.
Opportunities Through Polypharmacology
Polypharmacological therapeutic development refers to the goal of developing therapies targeting complex diseases like cardiovascular diseases and their known comorbidities, utilizing a multitarget approach 128, 129, 130, 131. Briefly, polypharmacology aims to find a therapy or combination of therapies that would fit the patient more precisely than a generic therapy. For example, highly active antiretroviral therapy is a combination therapy used in human immunodeficiency virus treatment, sulfamethoxazole/trimethoprim is a combination drug targeting folate biosynthesis pathways in bacteria (132), and dalfopristin/quinupristin is another combination agent that targets the bacterial 50S ribosome (133). An extension of this approach called systems polypharmacology utilizes an entire network view rather than a single or a few targets, and is evolving as a new area of active research in pharmacology (134). Although both multitarget therapeutics and systems pharmacology have led to several rational drug discovery candidates, their application in drug repositioning has thus far been limited. Performing drug repositioning studies on multitarget therapies could help to find new potential candidates for drug repositioning for cardiovascular disease.
Challenges in Integrating Precision Cardiology With Other Medical Specialties
The incidence rate of cardiovascular disease and the cost of management in the United States are increasing, which presents another potential application and opportunity for precision cardiology. For example, a recent estimate suggests that by 2030, 40.5% of patients will have at least 1 cardiovascular illness, which will raise the total cost of care to over $818 billion (135) worldwide. As cardiovascular-related conditions are often associated with life-style choices and have modifiable risk factors and well-characterized biological profiles, precision medicine approaches could help to reduce both the incidence rate and societal expense 12, 136, 137, 138. However, unlike precision oncology, precision cardiology is missing several important building blocks to implement workflows in a clinical setting. For example, genomic profiling data are often siloed under individual investigator databases, and raw datasets available in resources like dbGAP are not directly usable by cardiologists. The successful emergence of precision oncology can be attributed to the availability of several user-friendly bioinformatics resources, customized bioinformatics workflows, and unique datasets (e.g., genomics, transcriptomics, methylation, and proteomics) built as part of projects like The Cancer Genome Atlas. This raw, patient-level data is further used to create user-friendly applications like CBioPortal (139), which are fueling the genomic medicine revolution in oncology and could eventually lead to a standardized precision oncology practice 140, 141. The recent announcement of the U.S. Cancer Moonshot initiative also offers continuous federal funding for innovative oncology projects that would further broaden the applications, reference databases, and resources required to implement precision oncology 142, 143. Cardiology lacks both a centralized resource like The Cancer Genome Atlas as well as innovative funding programs that are a prerequisite for implementing precision practices. For example, building a Cardiovascular Disease Atlas by collecting longitudinal phenomic data integrated with multi-omics profiling using genomics, transcriptomics, proteomics, and metabolomics would be the foundation for enabling precision cardiology. A recent collaboration between the American Heart Association and Google proposed a moonshot grant, but unlike the 2020 Cancer Moonshot, the funding is restricted to a single project as part of a “1 team, 1 vision” approach (144). Furthermore, cardiologists and cardiovascular researchers should embrace practices including data sharing and other data commons strategies for faster knowledge distribution and dissemination of data for immediate replication, validation, reuse, and implementation into practice.
Precision Cardiology: Moving Forward From Computers to Patient Communities
Implementing precision cardiology in the clinical setting will require concerted efforts from undergraduate medical education, residency and fellowship training programs, medical school faculty, and practicing clinicians. Students must be trained in several interdisciplinary areas, including data science, mathematical modeling, systems biology, and bioinformatics, to become proficient practitioners 21, 22, 145, 146. Cardiology clinicians and physician-scientists will need to collaborate with mathematicians, engineers, and bioinformatics scientists to build and apply complex models of disease. Furthermore, health care delivery experts and physician leadership will need to rapidly adopt new guidelines, personalized clinical workflows, and emerging regulatory frameworks. Patients and patient advocates will be a central component of implementing precision cardiology in the clinical setting 147, 148. For example, communicating various aspects of precision medicine and including patients in clinical decisions using methods like shared-decision making is a vital step. Improvements in EHRs and clinical decision support tools will also be necessary. Design, development, and deployment of informatics applications and tools for visual analytics, predictive modeling, and risk analytics at the point-of-care are other key elements 149, 150, 151.
Integration with existing clinical infrastructure should promote cost-effectiveness as various longitudinal clinical data and some deep profiling data for large patient populations already exist 36, 152, 153. As the value of next-generation health care is a moving target and the current utility of precision medicine has been questioned, the success of precision cardiology will ultimately depend upon its effectiveness at reducing morbidity and mortality and increasing the quality of life for the cardiovascular patient population.
Precision Cardiology: Future Outlook
Physicians and scientists have a pressing need for a new paradigm to address deficiencies in the treatment of cardiovascular disease. The prevalence and incidence of cardiovascular diseases are increasing, and precision cardiology methods may offer a strategy to achieve improved outcomes while simultaneously decreasing health care expenditure (135). There are many early indications of the potential for precision cardiology through multi-omics methods, disease networks, and polypharmacology.
Application of these methods will become commonplace as data sources expand and it becomes easier to implement advanced algorithms. The current challenge with precision cardiology is determining how it can be most immediately translated into clinical practice to improve patient outcomes. The most ambitious precision cardiology plans require extensive data collection and analysis for which cost-effectiveness evidence does not currently exist. Such “translational-delays” from “bench-to-bedside” are not unexpected; consider the example of the implementation of pharmacogenetic testing for warfarin metabolism. Challenges such as the high cost of demonstrating utility in clinical trials and weaker intellectual property protections have delayed its uptake nationally (154). However, after years of doubt, recent studies have confirmed the cost-effectiveness of this approach 155, 156, 157, 158. It is likely that advances in precision cardiology will follow a similar path to clinical implementation. After demonstrating success through the integration of multidimensional and heterogeneous sources of data, there will be enough justification to collect, support, and maintain the next generation of clinical care and information gathering as well as to inform clinical trials for disease subtypes. As scientists and clinicians who care foremost about improving patient outcomes and delivering optimized patient care, enabling precision cardiology is an important step we can take.
Footnotes
Dr. Sengupta is a consultant for TeleHealthRobotics, Heart Test Labs, and Hitachi-Aloka Ltd. Dr. Björkegren is a member of the AstraZeneca Translational Science Centre-Karolinska Institutet. Dr. Kovacic has received financial compensation from AstraZeneca as a lecturer in 2014. Dr. Dudley has received consulting fees or honoraria from Janssen Pharmaceuticals, GlaxoSmithKline, AstraZeneca, and Hoffman-La Roche; is a scientific advisor to LAM Therapeutics; and holds equity in NuMedii Inc., Ayasdi Inc., and Ontomics, Inc. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
All authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the JACC: Basic to Translational Scienceauthor instructions page.
References
- 1.Iqbal J., Gunn J., Serruys P.W. Coronary stents: historical development, current status and future directions. Br Med Bull. 2013;106:193–211. doi: 10.1093/bmb/ldt009. [DOI] [PubMed] [Google Scholar]
- 2.Cushman D.W., Ondetti M.A. History of the design of captopril and related inhibitors of angiotensin converting enzyme. Hypertension. 1991;17:589–592. doi: 10.1161/01.hyp.17.4.589. [DOI] [PubMed] [Google Scholar]
- 3.Fox K.A., Steg P.G., Eagle K.A. Decline in rates of death and heart failure in acute coronary syndromes, 1999–2006. JAMA. 2007;297:1892–1900. doi: 10.1001/jama.297.17.1892. [DOI] [PubMed] [Google Scholar]
- 4.Ho P.M., Magid D.J., Shetterly S.M. Medication nonadherence is associated with a broad range of adverse outcomes in patients with coronary artery disease. Am Heart J. 2008;155:772–779. doi: 10.1016/j.ahj.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Sidney S., Quesenberry C.P., Jr., Jaffe M.G. Recent trends in cardiovascular mortality in the United States and public health goals. JAMA Cardiol. 2016;1:594–599. doi: 10.1001/jamacardio.2016.1326. [DOI] [PubMed] [Google Scholar]
- 6.Smith J.G., Newton-Cheh C. Genome-wide association studies of late-onset cardiovascular disease. J Mol Cell Cardiol. 2015;83:131–141. doi: 10.1016/j.yjmcc.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kessler T., Vilne B., Schunkert H. The impact of genome-wide association studies on the pathophysiology and therapy of cardiovascular disease. EMBO Mol Med. 2016;8:688–701. doi: 10.15252/emmm.201506174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pjanic M., Miller C.L., Wirka R., Kim J.B., DiRenzo D.M., Quertermous T. Genetics and genomics of coronary artery disease. Curr Cardiol Rep. 2016;18:102. doi: 10.1007/s11886-016-0777-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen N.B., Lloyd-Jones D., Hwang S.J. Genetic loci associated with ideal cardiovascular health: a meta-analysis of genome-wide association studies. Am Heart J. 2016;175:112–120. doi: 10.1016/j.ahj.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Visscher P.M., Brown M.A., McCarthy M.I., Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjorkegren J.L., Kovacic J.C., Dudley J.T., Schadt E.E. Genome-wide significant loci: how important are they? Systems genetics to understand heritability of coronary artery disease and other common complex disorders. J Am Coll Cardiol. 2015;65:830–845. doi: 10.1016/j.jacc.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shameer K., Denny J.C., Ding K. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum Genet. 2014;133:95–109. doi: 10.1007/s00439-013-1355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horrobin D.F. Realism in drug discovery—could Cassandra be right? Nat Biotechnol. 2001;19:1099–1100. doi: 10.1038/nbt1201-1099. [DOI] [PubMed] [Google Scholar]
- 14.Pammolli F., Magazzini L., Riccaboni M. The productivity crisis in pharmaceutical R&D. Nat Rev Drug Discov. 2011;10:428–438. doi: 10.1038/nrd3405. [DOI] [PubMed] [Google Scholar]
- 15.Fordyce C.B., Roe M.T., Ahmad T. Cardiovascular drug development: is it dead or just hibernating? J Am Coll Cardiol. 2015;65:1567–1582. doi: 10.1016/j.jacc.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 16.Joy T.R., Hegele R.A. The failure of torcetrapib: what have we learned? Br J Pharmacol. 2008;154:1379–1381. doi: 10.1038/bjp.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLain J.H., Alsterda A.J., Arora R.R. Cholesteryl ester transfer protein inhibitors: trials and tribulations. J Cardiovasc Pharmacol Ther. 2016 Aug 10 doi: 10.1177/1074248416662349. [E-pub ahead of print] [DOI] [PubMed] [Google Scholar]
- 18.Barter P.J., Rye K.A. Cholesteryl ester transfer protein inhibition is not yet dead—pro. Arterioscler Thromb Vasc Biol. 2016;36:439–441. doi: 10.1161/ATVBAHA.115.306879. [DOI] [PubMed] [Google Scholar]
- 19.Chen B., Butte A.J. Network medicine in disease analysis and therapeutics. Clin Pharmacol Ther. 2013;94:627–629. doi: 10.1038/clpt.2013.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodos R.A., Kidd B.A., Shameer K., Readhead B.P., Dudley J.T. In silico methods for drug repurposing and pharmacology. Wiley Interdiscip Rev Syst Biol Med. 2016;8:186–210. doi: 10.1002/wsbm.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antman E.M., Loscalzo J. Precision medicine in cardiology. Nat Rev Cardiol. 2016;13:591–602. doi: 10.1038/nrcardio.2016.101. [DOI] [PubMed] [Google Scholar]
- 22.Shah S.H., Arnett D., Houser S.R. Opportunities for the cardiovascular community in the precision medicine initiative. Circulation. 2016;133:226–231. doi: 10.1161/CIRCULATIONAHA.115.019475. [DOI] [PubMed] [Google Scholar]
- 23.Jaffe S. Planning for US Precision Medicine Initiative underway. Lancet. 2015;385:2448–2449. doi: 10.1016/S0140-6736(15)61124-2. [DOI] [PubMed] [Google Scholar]
- 24.Ashley E.A. The precision medicine initiative: a new national effort. JAMA. 2015;313:2119–2120. doi: 10.1001/jama.2015.3595. [DOI] [PubMed] [Google Scholar]
- 25.Collins F.S., Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H.M., Liao C.T., Yen T.C. Clues toward precision medicine in oral squamous cell carcinoma: utility of next-generation sequencing for the prognostic stratification of high- risk patients harboring neck lymph node extracapsular extension. Oncotarget. 2016;7:63082–63092. doi: 10.18632/oncotarget.11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirsch F.R., Scagliotti G.V., Mulshine J.L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 28.Kerr K.M., Lopez-Rios F. Precision medicine in NSCLC and pathology: how does ALK fit in the pathway? Ann Oncol. 2016;27(Suppl 3):iii16–iii24. doi: 10.1093/annonc/mdw302. [DOI] [PubMed] [Google Scholar]
- 29.Stella G.M., Gentile A., Baderacchi A., Meloni F., Milan M., Benvenuti S. Ockham's razor for the MET-driven invasive growth linking idiopathic pulmonary fibrosis and cancer. J Transl Med. 2016;14:256. doi: 10.1186/s12967-016-1008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruderfer D.M., Charney A.W., Readhead B. Polygenic overlap between schizophrenia risk and antipsychotic response: a genomic medicine approach. Lancet Psychiatry. 2016;3:350–357. doi: 10.1016/S2215-0366(15)00553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashley E.A., Butte A.J., Wheeler M.T. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kullo I.J., Jouni H., Austin E.E. Incorporating a genetic risk score into coronary heart disease risk estimates: effect on low-density lipoprotein cholesterol levels (the MI-GENES Clinical Trial) Circulation. 2016;133:1181–1188. doi: 10.1161/CIRCULATIONAHA.115.020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shameer K., Klee E.W., Dalenberg A.K., Kullo I.J. Whole exome sequencing implicates an INO80D mutation in a syndrome of aortic hypoplasia, premature atherosclerosis, and arterial stiffness. Circ Cardiovasc Genet. 2014;7:607–614. doi: 10.1161/CIRCGENETICS.113.000233. [DOI] [PubMed] [Google Scholar]
- 34.Feero W.G., Guttmacher A.E., Collins F.S. Genomic medicine—an updated primer. N Engl J Med. 2010;362:2001–2011. doi: 10.1056/NEJMra0907175. [DOI] [PubMed] [Google Scholar]
- 35.Guttmacher A.E., Collins F.S. Genomic medicine—a primer. N Engl J Med. 2002;347:1512–1520. doi: 10.1056/NEJMra012240. [DOI] [PubMed] [Google Scholar]
- 36.Miotto R., Li L., Kidd B.A., Dudley J.T. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep. 2016 May 17;6:26094. doi: 10.1038/srep26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adami C., Ofria C., Collier T.C. Evolution of biological complexity. Proceedings of the National Academy of Sciences. 2000;97:4463–4468. doi: 10.1073/pnas.97.9.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstein B.A., Navar A.M., Carter R.E. Moving beyond regression techniques in cardiovascular risk prediction: applying machine learning to address analytic challenges. Eur Heart J. 2016 Jul 19 doi: 10.1093/eurheartj/ehw302. [E-pub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandl K.D., Kohane I.S. Escaping the EHR trap—the future of health IT. N Engl J Med. 2012;366:2240–2242. doi: 10.1056/NEJMp1203102. [DOI] [PubMed] [Google Scholar]
- 40.Bolton R.J., Hand D.J. Unsupervised profiling methods for fraud detection. Proc Credit Scoring and Credit Control VII. 2001:5–7. [Google Scholar]
- 41.Sengupta P.P., Huang Y.M., Bansal M. Cognitive machine-learning algorithm for cardiac imaging: a pilot study for differentiating constrictive pericarditis from restrictive cardiomyopathy. Circ Cardiovasc Imaging. 2016;9:e004330. doi: 10.1161/CIRCIMAGING.115.004330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narula S., Shameer K., Salem Omar A.M., Dudley J.T., Sengupta P.P. Machine-learning algorithms to automate morphological and functional assessments in 2D echocardiography. J Am Coll Cardiol. 2016;68:2287–2295. doi: 10.1016/j.jacc.2016.08.062. [DOI] [PubMed] [Google Scholar]
- 43.Shameer K., Pugalenthi G., Kandaswamy K.K., Sowdhamini R. 3dswap-pred: prediction of 3D domain swapping from protein sequence using Random Forest approach. Protein Pept Lett. 2011;18:1010–1020. doi: 10.2174/092986611796378729. [DOI] [PubMed] [Google Scholar]
- 44.Shameer K., Pugalenthi G., Kandaswamy K.K., Suganthan P.N., Archunan G., Sowdhamini R. Insights into protein sequence and structure-derived features mediating 3D domain swapping mechanism using support vector machine based approach. Bioinform Biol Insights. 2010;4:33–42. doi: 10.4137/bbi.s4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ammad-Ud-Din M., Khan S.A., Malani D. Drug response prediction by inferring pathway-response associations with kernelized Bayesian matrix factorization. Bioinformatics. 2016;32:i455–i463. doi: 10.1093/bioinformatics/btw433. [DOI] [PubMed] [Google Scholar]
- 46.Vural S., Wang X., Guda C. Classification of breast cancer patients using somatic mutation profiles and machine learning approaches. BMC Syst Biol. 2016;10(Suppl 3):62. doi: 10.1186/s12918-016-0306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stamile C., Kocevar G., Cotton F., Maes F., Sappey-Marinier D., Huffel S.V. Multi-parametric non-negative matrix factorization for longitudinal variations detection in white matter fiber-bundles. IEEE J Biomed Health Inform. 2016 Aug 3 doi: 10.1109/JBHI.2016.2597963. [E-pub ahead of print] [DOI] [PubMed] [Google Scholar]
- 48.Camara P.G., Rosenbloom D.I., Emmett K.J., Levine A.J., Rabadan R. Topological data analysis generates high-resolution, genome-wide maps of human recombination. Cell Syst. 2016;3:83–94. doi: 10.1016/j.cels.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alagappan M., Jiang D., Denko N., Koong A.C. A multimodal data analysis approach for targeted drug discovery involving topological data analysis (TDA) Adv Exp Med Biol. 2016;899:253–268. doi: 10.1007/978-3-319-26666-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nielson J.L., Paquette J., Liu A.W. Topological data analysis for discovery in preclinical spinal cord injury and traumatic brain injury. Nat Commun. 2015;6:8581. doi: 10.1038/ncomms9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hinks T., Zhou X., Staples K. Multidimensional endotypes of asthma: topological data analysis of cross-sectional clinical, pathological, and immunological data. Lancet. 2015;385(Suppl 1):S42. doi: 10.1016/S0140-6736(15)60357-9. [DOI] [PubMed] [Google Scholar]
- 52.Tan J., Ung M., Cheng C., Greene C.S. Unsupervised feature construction and knowledge extraction from genome-wide assays of breast cancer with denoising autoencoders. Pac Symp Biocomput. 2015;20:132–143. [PMC free article] [PubMed] [Google Scholar]
- 53.Singh R., Lanchantin J., Robins G., Qi Y. DeepChrome: deep-learning for predicting gene expression from histone modifications. Bioinformatics. 2016;32:i639–i648. doi: 10.1093/bioinformatics/btw427. [DOI] [PubMed] [Google Scholar]
- 54.Huynh B.Q., Li H., Giger M.L. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J Med Imaging (Bellingham) 2016;3:034501. doi: 10.1117/1.JMI.3.3.034501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Janowczyk A., Madabhushi A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J Pathol Inform. 2016;7:29. doi: 10.4103/2153-3539.186902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liang Z., Huang J.X., Zeng X., Zhang G. DL-ADR: a novel deep learning model for classifying genomic variants into adverse drug reactions. BMC Med Genomics. 2016;9(Suppl 2):48. doi: 10.1186/s12920-016-0207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi E., Schuetz A., Stewart W.F., Sun J. Using recurrent neural network models for early detection of heart failure onset. J Am Med Inform Assoc. 2017;24:361–370. doi: 10.1093/jamia/ocw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ekins S. The Next Era: Deep learning in pharmaceutical research. Pharm Res. 2016;33:2594–2603. doi: 10.1007/s11095-016-2029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gawehn E., Hiss J.A., Schneider G. Deep learning in drug discovery. Mol Inform. 2016;35:3–14. doi: 10.1002/minf.201501008. [DOI] [PubMed] [Google Scholar]
- 60.Ortiz A., Munilla J., Gorriz J.M., Ramirez J. Ensembles of deep learning architectures for the early diagnosis of the Alzheimer's disease. Int J Neural Syst. 2016;26:1650025. doi: 10.1142/S0129065716500258. [DOI] [PubMed] [Google Scholar]
- 61.Ngo T.A., Lu Z., Carneiro G. Combining deep learning and level set for the automated segmentation of the left ventricle of the heart from cardiac cine magnetic resonance. Med Image Anal. 2016;35:159–171. doi: 10.1016/j.media.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 62.Aliper A., Plis S., Artemov A., Ulloa A., Mamoshina P., Zhavoronkov A. Deep learning applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data. Mol Pharm. 2016;13:2524–2530. doi: 10.1021/acs.molpharmaceut.6b00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li L., Cheng W.Y., Glicksberg B.S. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci Transl Med. 2015;7:311ra174. doi: 10.1126/scitranslmed.aaa9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Houle D., Govindaraju D.R., Omholt S. Phenomics: the next challenge. Nat Rev Genet. 2010;11:855–866. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- 65.Gottesman O., Kuivaniemi H., Tromp G. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15:761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.National Human Genome Research Institute. Electronic Medical Records and Genomics (eMERGE) Network. Available at: https://www.genome.gov/27540473/electronic-medical-records-and-genomics-emerge-network/. Accessed April 23, 2017.
- 67.PheKB. What is the Phenotype KnowledgeBase? Available at: https://phekb.org. Accessed April 23, 2017.
- 68.Kirby J.C., Speltz P., Rasmussen L.V. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc. 2016;23:1046–1052. doi: 10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oellrich A., Collier N., Groza T. The digital revolution in phenotyping. Brief Bioinform. 2016;17:819–830. doi: 10.1093/bib/bbv083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zemojtel T., Kohler S., Mackenroth L. Effective diagnosis of genetic disease by computational phenotype analysis of the disease-associated genome. Sci Transl Med. 2014;6:252ra123. doi: 10.1126/scitranslmed.3009262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Drouin A., Giguere S., Deraspe M. Predictive computational phenotyping and biomarker discovery using reference-free genome comparisons. BMC Genomics. 2016;17:754. doi: 10.1186/s12864-016-2889-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cost-effectiveness of hypertension therapy according to 2014 guidelines. N Engl J Med. 2015;372:1677. doi: 10.1056/NEJMx150016. [DOI] [PubMed] [Google Scholar]
- 73.Bedi O., Dhawan V., Sharma P.L., Kumar P. Pleiotropic effects of statins: new therapeutic targets in drug design. Naunyn Schmiedebergs Arch Pharmacol. 2016;389:695–712. doi: 10.1007/s00210-016-1252-4. [DOI] [PubMed] [Google Scholar]
- 74.Alfonsi J.E., Hegele R.A., Gryn S.E. Pharmacogenetics of lipid-lowering agents: precision or indecision medicine? Curr Atheroscler Rep. 2016;18:24. doi: 10.1007/s11883-016-0573-6. [DOI] [PubMed] [Google Scholar]
- 75.Birnbaum Y., Ye Y. Pleiotropic effects of statins: the role of eicosanoid production. Curr Atheroscler Rep. 2012;14:135–139. doi: 10.1007/s11883-012-0232-5. [DOI] [PubMed] [Google Scholar]
- 76.Antonopoulos A.S., Margaritis M., Shirodaria C., Antoniades C. Translating the effects of statins: from redox regulation to suppression of vascular wall inflammation. Thromb Haemost. 2012;108:840–848. doi: 10.1160/TH12-05-0337. [DOI] [PubMed] [Google Scholar]
- 77.Mihos C.G., Salas M.J., Santana O. The pleiotropic effects of the hydroxy-methyl-glutaryl-CoA reductase inhibitors in cardiovascular disease: a comprehensive review. Cardiol Rev. 2010;18:298–304. doi: 10.1097/CRD.0b013e3181f52a7f. [DOI] [PubMed] [Google Scholar]
- 78.Tashakkor A.Y., Moghaddamjou A., Chen L., Cheung W.Y. Predicting the risk of cardiovascular comorbidities in adult cancer survivors. Curr Oncol. 2013;20:e360–e370. doi: 10.3747/co.20.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Crepaldi G., Scire C.A., Carrara G. Cardiovascular comorbidities relate more than others with disease activity in rheumatoid arthritis. PLoS One. 2016;11:e0146991. doi: 10.1371/journal.pone.0146991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson R.J., Feig D.I., Nakagawa T., Sanchez-Lozada L.G., Rodriguez-Iturbe B. Pathogenesis of essential hypertension: historical paradigms and modern insights. J Hypertens. 2008;26:381–391. doi: 10.1097/HJH.0b013e3282f29876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Surendran P., Drenos F., Young R. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat Genet. 2016;48:1151–1161. doi: 10.1038/ng.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kent S.T., Shimbo D., Huang L. Antihypertensive medication classes used among medicare beneficiaries initiating treatment in 2007–2010. PLoS One. 2014;9:e105888. doi: 10.1371/journal.pone.0105888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ambrosius W.T., Sink K.M., Foy C.G. The design and rationale of a multicenter clinical trial comparing two strategies for control of systolic blood pressure: the Systolic Blood Pressure Intervention Trial (SPRINT) Clin Trials. 2014;11:532–546. doi: 10.1177/1740774514537404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.James P.A., Oparil S., Carter B.L. 2014 evidence-based guideline for the management of high blood pressure in adults: report from the panel members appointed to the Eighth Joint National Committee (JNC 8) JAMA. 2014;311:507–520. doi: 10.1001/jama.2013.284427. [DOI] [PubMed] [Google Scholar]
- 85.Francis G.S., Cogswell R., Thenappan T. The heterogeneity of heart failure: will enhanced phenotyping be necessary for future clinical trial success? J Am Coll Cardiol. 2014;64:1775–1776. doi: 10.1016/j.jacc.2014.07.978. [DOI] [PubMed] [Google Scholar]
- 86.Louridas G.E., Lourida K.G. Systems biology and clinical phenotypes of heart failure syndrome. J Am Coll Cardiol. 2015;65:1269–1270. doi: 10.1016/j.jacc.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 87.Eckstein D., Korabathina R. Heart failure update: diagnosis and classification. FP Essent. 2016;442:11–17. [PubMed] [Google Scholar]
- 88.Vaduganathan M., Greene S.J., Ambrosy A.P., Gheorghiade M., Butler J. The disconnect between phase II and phase III trials of drugs for heart failure. Nat Rev Cardiol. 2013;10:85–97. doi: 10.1038/nrcardio.2012.181. [DOI] [PubMed] [Google Scholar]
- 89.Gheorghiade M., Larson C.J., Shah S.J. Developing new treatments for heart failure: focus on the heart. Circ Heart Fail. 2016;9 doi: 10.1161/CIRCHEARTFAILURE.115.002727. [DOI] [PubMed] [Google Scholar]
- 90.Ahmad T., Pencina M.J., Schulte P.J. Clinical implications of chronic heart failure phenotypes defined by cluster analysis. J Am Coll Cardiol. 2014;64:1765–1774. doi: 10.1016/j.jacc.2014.07.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Madias J.E. Applying cluster analysis to data of previously published chronic heart failure trials. J Am Coll Cardiol. 2015;65:1268–1269. doi: 10.1016/j.jacc.2014.11.070. [DOI] [PubMed] [Google Scholar]
- 92.Ahmad T., Felker G.M. Reply: applying cluster analysis to data of previously published chronic heart failure trials: systems biology and clinical phenotypes of heart failure syndrome. J Am Coll Cardiol. 2015;65:1270–1271. doi: 10.1016/j.jacc.2014.12.050. [DOI] [PubMed] [Google Scholar]
- 93.Welch T.D., Yang E.H., Reeder G.S., Gersh B.J. Modern management of acute myocardial infarction. Curr Probl Cardiol. 2012;37:237–310. doi: 10.1016/j.cpcardiol.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 94.Talukdar H.A., Foroughi Asl H., Jain R.K. Cross-tissue regulatory gene networks in coronary artery disease. Cell Syst. 2016;2:196–208. doi: 10.1016/j.cels.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Franzen O., Ermel R., Cohain A. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science. 2016;353:827–830. doi: 10.1126/science.aad6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rymer J.A., Newby L.K. Back to the future: improving the use of guidelines-recommended coronary disease secondary prevention at the dawn of the precision medicine era. Circulation. 2015;131:1234–1235. doi: 10.1161/CIRCULATIONAHA.115.015707. [DOI] [PubMed] [Google Scholar]
- 97.Shurlock B. The emergence of cardiovascular imaging as a subspecialty and individualized treatment of coronary artery disease: two related trends. Eur Heart J. 2014;35:3392–3393. doi: 10.1093/eurheartj/ehu434. [DOI] [PubMed] [Google Scholar]
- 98.Gijsberts C.M., den Ruijter H.M., Asselbergs F.W., Chan M.Y., de Kleijn D.P., Hoefer I.E. Biomarkers of coronary artery disease differ between Asians and Caucasians in the general population. Glob Heart. 2015;10:301–311.e11. doi: 10.1016/j.gheart.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 99.Kleber M.E., Goliasch G., Grammer T.B. Evolving biomarkers improve prediction of long-term mortality in patients with stable coronary artery disease: the BIO-VILCAD score. J Intern Med. 2014;276:184–194. doi: 10.1111/joim.12189. [DOI] [PubMed] [Google Scholar]
- 100.Darabi F., Aghaei M., Movahedian A., Pourmoghadas A., Sarrafzadegan N. The role of serum levels of microRNA-21 and matrix metalloproteinase-9 in patients with acute coronary syndrome. Mol Cell Biochem. 2016;422:51–60. doi: 10.1007/s11010-016-2805-z. [DOI] [PubMed] [Google Scholar]
- 101.Cheng J., Chen Y., Xu B., Wu J., He F. Association of soluble fibrinogen-like protein 2 with the severity of coronary artery disease. Intern Med. 2016;55:2343–2350. doi: 10.2169/internalmedicine.55.6149. [DOI] [PubMed] [Google Scholar]
- 102.Genoux A., Lichtenstein L., Ferrieres J. Serum levels of mitochondrial inhibitory factor 1 are independently associated with long-term prognosis in coronary artery disease: the GENES Study. BMC Med. 2016;14:125. doi: 10.1186/s12916-016-0672-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stone P.A., Thompson S.N., Khan M., Northfield E., Schillinger R., Skaff P. The impact of biochemical markers on major adverse cardiovascular events and contralateral carotid artery stenosis progression following carotid interventions. Ann Vasc Surg. 2017;38:144–150. doi: 10.1016/j.avsg.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 104.Li H.Y., Zhao X., Liu Y.Z. Plasma microRNA-126-5p is associated with the complexity and severity of coronary artery disease in patients with stable angina pectoris. Cell Physiol Biochem. 2016;39:837–846. doi: 10.1159/000447794. [DOI] [PubMed] [Google Scholar]
- 105.Barabasi A.L., Oltvai Z.N. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 106.Giri K., Shameer K., Zimmermann M.T. Understanding protein-nanoparticle interaction: a new gateway to disease therapeutics. Bioconjug Chem. 2014;25:1078–1090. doi: 10.1021/bc500084f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Burt R.S., Kilduff M., Tasselli S. Social network analysis: foundations and frontiers on advantage. Annu Rev Psychol. 2013;64:527–547. doi: 10.1146/annurev-psych-113011-143828. [DOI] [PubMed] [Google Scholar]
- 108.Vidal M., Cusick M.E., Barabasi A.L. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Glicksberg B.S., Li L., Cheng W.Y. An integrative pipeline for multi-modal discovery of disease relationships. Pac Symp Biocomput. 2015:407–418. [PMC free article] [PubMed] [Google Scholar]
- 110.Glicksberg B.S., Li L., Badgeley M.A. Comparative analyses of population-scale phenomic data in electronic medical records reveal race-specific disease networks. Bioinformatics. 2016;32:i101–i110. doi: 10.1093/bioinformatics/btw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Novac N. Challenges and opportunities of drug repositioning. Trends Pharmacol Sci. 2013;34:267–272. doi: 10.1016/j.tips.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 112.Sirota M., Dudley J.T., Kim J. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3:96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shameer K., Readhead B., Dudley J.T. Computational and experimental advances in drug repositioning for accelerated therapeutic stratification. Curr Top Med Chem. 2015;15:5–20. doi: 10.2174/1568026615666150112103510. [DOI] [PubMed] [Google Scholar]
- 114.RepurposeDB. Repositioning investigations. Available at: http://repurposedb.dudleylab.org/. Accessed April 23, 2017.
- 115.Packer M., Bristow M.R., Cohn J.N., for the U.S. Carvedilol Heart Failure Study Group The effect of carvedilol on morbidity and mortality in patients with chronic heart failure. N Engl J Med. 1996;334:1349–1355. doi: 10.1056/NEJM199605233342101. [DOI] [PubMed] [Google Scholar]
- 116.Swedberg K., Hjalmarson A., Waagstein F., Wallentin I. Prolongation of survival in congestive cardiomyopathy by beta-receptor blockade. Lancet. 1979;1:1374–1376. doi: 10.1016/s0140-6736(79)92010-5. [DOI] [PubMed] [Google Scholar]
- 117.Epstein S.E., Braunwald E. The effect of beta adrenergic blockade on patterns of urinary sodium excretion. Studies in normal subjects and in patients with heart disease. Ann Intern Med. 1966;65:20–27. doi: 10.7326/0003-4819-65-1-20. [DOI] [PubMed] [Google Scholar]
- 118.Sanchez L.S., de la Monte S.M., Filippov G., Jones R.C., Zapol W.M., Bloch K.D. Cyclic-GMP-binding, cyclic-GMP-specific phosphodiesterase (PDE5) gene expression is regulated during rat pulmonary development. Pediatr Res. 1998;43:163–168. doi: 10.1203/00006450-199802000-00002. [DOI] [PubMed] [Google Scholar]
- 119.Barnett C.F., Machado R.F. Sildenafil in the treatment of pulmonary hypertension. Vasc Health Risk Manag. 2006;2:411–422. doi: 10.2147/vhrm.2006.2.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ghofrani H.A., Osterloh I.H., Grimminger F. Sildenafil: from angina to erectile dysfunction to pulmonary hypertension and beyond. Nat Rev Drug Discov. 2006;5:689–702. doi: 10.1038/nrd2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bertolini F., Sukhatme V.P., Bouche G. Drug repurposing in oncology—patient and health systems opportunities. Nat Rev Clin Oncol. 2015;12:732–742. doi: 10.1038/nrclinonc.2015.169. [DOI] [PubMed] [Google Scholar]
- 122.Deftereos S., Giannopoulos G., Papoutsidakis N. Colchicine and the heart: pushing the envelope. J Am Coll Cardiol. 2013;62:1817–1825. doi: 10.1016/j.jacc.2013.08.726. [DOI] [PubMed] [Google Scholar]
- 123.Brown S.A., Sandhu N., Herrmann J. Systems biology approaches to adverse drug effects: the example of cardio-oncology. Nat Rev Clin Oncol. 2015;12:718–731. doi: 10.1038/nrclinonc.2015.168. [DOI] [PubMed] [Google Scholar]
- 124.RepurposeDB. Drugs. Available at: http://repurposedb.dudleylab.org/browseDrugs. Accessed April 23, 2017.
- 125.Bharadwaj U., Eckols T.K., Kolosov M. Drug-repositioning screening identified piperlongumine as a direct STAT3 inhibitor with potent activity against breast cancer. Oncogene. 2015;34:1341–1353. doi: 10.1038/onc.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Corbett A., Pickett J., Burns A. Drug repositioning for Alzheimer's disease. Nat Rev Drug Discov. 2012;11:833–846. doi: 10.1038/nrd3869. [DOI] [PubMed] [Google Scholar]
- 127.Rukov J.L., Wilentzik R., Jaffe I., Vinther J., Shomron N. Pharmaco-miR: linking microRNAs and drug effects. Brief Bioinform. 2014;15:648–659. doi: 10.1093/bib/bbs082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Millan M.J. Multi-target strategies for the improved treatment of depressive states: Conceptual foundations and neuronal substrates, drug discovery and therapeutic application. Pharmacol Ther. 2006;110:135–370. doi: 10.1016/j.pharmthera.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 129.Lu J.J., Pan W., Hu Y.J., Wang Y.T. Multi-target drugs: the trend of drug research and development. PLoS One. 2012;7:e40262. doi: 10.1371/journal.pone.0040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zimmermann G.R., Lehar J., Keith C.T. Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov Today. 2007;12:34–42. doi: 10.1016/j.drudis.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 131.Medina-Franco J.L., Giulianotti M.A., Welmaker G.S., Houghten R.A. Shifting from the single to the multitarget paradigm in drug discovery. Drug Discov Today. 2013;18:495–501. doi: 10.1016/j.drudis.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Masters P.A., O'Bryan T.A., Zurlo J., Miller D.Q., Joshi N. Trimethoprim-sulfamethoxazole revisited. Arch Intern Med. 2003;163:402–410. doi: 10.1001/archinte.163.4.402. [DOI] [PubMed] [Google Scholar]
- 133.Allington D.R., Rivey M.P. Quinupristin/dalfopristin: a therapeutic review. Clin Ther. 2001;23:24–44. doi: 10.1016/s0149-2918(01)80028-x. [DOI] [PubMed] [Google Scholar]
- 134.Hopkins A.L. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 135.Heidenreich P.A., Trogdon J.G., Khavjou O.A. Forecasting the future of cardiovascular disease in the United States: a policy statement from the American Heart Association. Circulation. 2011;123:933–944. doi: 10.1161/CIR.0b013e31820a55f5. [DOI] [PubMed] [Google Scholar]
- 136.Kullo I.J., Shameer K., Jouni H. The ATXN2-SH2B3 locus is associated with peripheral arterial disease: an electronic medical record-based genome-wide association study. Front Genet. 2014;5:166. doi: 10.3389/fgene.2014.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yusuf S., Hawken S., Ounpuu S. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case- control study. Lancet. 2004;364:937–952. doi: 10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
- 138.Pearson T.A., Blair S.N., Daniels S.R. AHA guidelines for primary prevention of cardiovascular disease and stroke: 2002 update: consensus panel guide to comprehensive risk reduction for adult patients without coronary or other atherosclerotic vascular diseases. American Heart Association Science Advisory and Coordinating Committee. Circulation. 2002;106:388–391. doi: 10.1161/01.cir.0000020190.45892.75. [DOI] [PubMed] [Google Scholar]
- 139.cBioPortal for Cancer Genomics. Available at: http://www.cbioportal.org/. Accessed April 23, 2017.
- 140.Cerami E., Gao J., Dogrusoz U. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tang X., Baheti S., Shameer K. The eSNV-detect: a computational system to identify expressed single nucleotide variants from transcriptome sequencing data. Nucleic Acids Res. 2014;42:e172. doi: 10.1093/nar/gku1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Aelion C.M., Airhihenbuwa C.O., Alemagno S. The US Cancer Moonshot initiative. Lancet Oncol. 2016;17:e178–e180. doi: 10.1016/S1470-2045(16)30054-7. [DOI] [PubMed] [Google Scholar]
- 143.Kennedy Sheldon L. Oncology nurses and the Cancer Moonshot 2020. Clin J Oncol Nurs. 2016;20:355–356. doi: 10.1188/16.CJON.355-356. [DOI] [PubMed] [Google Scholar]
- 144.American Heart Association News. ‘1 Team, 1 Vision’—AHA, Google Life Sciences launch $50 million project. Available at: http://news.heart.org/american-heart-association-google-life-sciences-seeking-novel-strategies-to-prevent-heart-disease/. Accessed April 23, 2017.
- 145.Pitt G.S. Cardiovascular precision medicine: hope or hype? Eur Heart J. 2015;36:1842–1843. doi: 10.1093/eurheartj/ehv226. [DOI] [PubMed] [Google Scholar]
- 146.Eden C., Johnson K.W., Gottesman O., Bottinger E.P., Abul-Husn N.S. Medical student preparedness for an era of personalized medicine: findings from one US medical school. Per Med. 2016;13:129–141. doi: 10.2217/pme.15.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Maojo V., Kulikowski C.A. Bioinformatics and medical informatics: collaborations on the road to genomic medicine? J Am Med Inform Assoc. 2003;10:515–522. doi: 10.1197/jamia.M1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Carlile S., Sefton A.J. Healthcare and the information age: implications for medical education. Med J Aust. 1998;168:340–343. doi: 10.5694/j.1326-5377.1998.tb138963.x. [DOI] [PubMed] [Google Scholar]
- 149.Badgeley M.A., Shameer K., Glicksberg B.S. EHDViz: clinical dashboard development using open-source technologies. BMJ Open. 2016;6:e010579. doi: 10.1136/bmjopen-2015-010579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vaitsis C., Nilsson G., Zary N. Visual analytics in healthcare education: exploring novel ways to analyze and represent big data in undergraduate medical education. PeerJ. 2014;2:e683. doi: 10.7717/peerj.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Charles C., Gafni A., Whelan T. Shared decision-making in the medical encounter: what does it mean? (or it takes at least two to tango) Soc Sci Med. 1997;44:681–692. doi: 10.1016/s0277-9536(96)00221-3. [DOI] [PubMed] [Google Scholar]
- 152.Rumsfeld J.S., Joynt K.E., Maddox T.M. Big data analytics to improve cardiovascular care: promise and challenges. Nat Rev Cardiol. 2016;13:350–359. doi: 10.1038/nrcardio.2016.42. [DOI] [PubMed] [Google Scholar]
- 153.Peiris D.P., Joshi R., Webster R.J. An electronic clinical decision support tool to assist primary care providers in cardiovascular disease risk management: development and mixed methods evaluation. J Med Internet Res. 2009;11:e51. doi: 10.2196/jmir.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chalkidou K., Rawlins M. Pharmacogenetics and cost-effectiveness analysis: a two-way street. Drug Discov Today. 2011;16:873–877. doi: 10.1016/j.drudis.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 155.Pink J., Pirmohamed M., Lane S., Hughes D.A. Cost-effectiveness of pharmacogenetics- guided warfarin therapy vs. alternative anticoagulation in atrial fibrillation. Clin Pharmacol Ther. 2014;95:199–207. doi: 10.1038/clpt.2013.190. [DOI] [PubMed] [Google Scholar]
- 156.Verhoef T.I., Redekop W.K., Langenskiold S. Cost-effectiveness of pharmacogenetic- guided dosing of warfarin in the United Kingdom and Sweden. Pharmacogenomics J. 2016;16:478–484. doi: 10.1038/tpj.2016.41. [DOI] [PubMed] [Google Scholar]
- 157.Gor D., Kim K., Chumnumwat S. Cost-effectiveness of a novel pharmacist guided warfarin pharmacogenetic service. Value Health. 2015;18:A390. [Google Scholar]
- 158.You J.H. Pharmacogenetic-guided selection of warfarin versus novel oral anticoagulants for stroke prevention in patients with atrial fibrillation: a cost-effectiveness analysis. Pharmacogenet Genomics. 2014;24:6–14. doi: 10.1097/FPC.0000000000000014. [DOI] [PubMed] [Google Scholar]
- 159.Li M.J., Liu Z., Wang P. GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2016;44:D869–D876. doi: 10.1093/nar/gkv1317. [DOI] [PMC free article] [PubMed] [Google Scholar]