Abstract
Lipid-based micellar nanoparticles promote aggregation of huntingtin exon-1 peptides. Here we characterize the interaction of two such peptides, httNTQ7 and httNTQ10 comprising the N-terminal amphiphilic domain of huntingtin followed by 7 and 10 glutamine repeats, respectively, with 8 nm lipid micelles using NMR chemical exchange saturation transfer (CEST), circular dichroism and pulsed Q-band EPR. Exchange between free and micelle-bound httNTQn peptides occurs on the millisecond time scale with a KD ~ 0.5–1 mM. Upon binding micelles, residues 1–15 adopt a helical conformation. Oxidation of Met7 to a sulfoxide reduces the binding affinity for micelles ~3–4-fold and increases the length of the helix by a further two residues. A structure of the bound monomer unit is calculated from the backbone chemical shifts of the micelle-bound state obtained from CEST. Pulsed Q-band EPR shows that a monomer–dimer equilibrium exists on the surface of the micelles and that the two helices of the dimer adopt a parallel orientation, thereby bringing two disordered polyQ tails into close proximity which may promote aggregation upon dissociation from the micelle surface.
Huntington’s disease is a fatal neurodegenerative disease arising from the presence of 36 or more CAG repeats within exon 1 of the Huntingtin (htt) gene, resulting in expansion of the polyQ domain that lies immediately downstream of the 16-residue N-terminal amphiphilic sequence (httNT) of the huntingtin protein.1 The presence of a long polyQ stretch results in the rapid formation of polymorphic fibrils,2 the rate of which is modulated by the presence of flanking regions (httNT and the proline rich domain C-terminal to the polyQ sequence), as well as additional factors including binding to lipid membranes and the presence of reactive oxygen species.3 HttNT is known to target a variety of membrane-containing structures within cells,4 undergoes a random coil to α-helix transition upon binding lipid membranes,5 and mediates aggregation of polyQ on membrane surfaces.3a,6 Here we explore the interaction of two httNTQn (n = 7 and 10) constructs with lipid-based micellar nanoparticles7 using a combination of NMR chemical exchange saturation transfer (CEST) and pulsed Q-band EPR spectroscopy.
The shorter httNTQ 7 construct remains soluble over a prolonged period of time (~15 days at 150 μM and 10 °C) permitting solution NMR studies, whereas the longer httNTQ 10 construct aggregates with a t1/2 of ~150 h (Figure S1), in agreement with previous findings.2a Aggregation of httNTQ 10 is significantly enhanced in the presence of lipid micelles (t1/2 ~ 70 h) (Figure S1). Upon oxidation of Met7 to a sulfoxide (Met7O), no aggregation of httNTQ 10 in the presence or absence of micelles is observed, consistent with previous observations.3c Micellar nanoparticles, comprising a 3:1 molar ratio of 1-palmitoyl-2-hydroxy-sn-glycero-3-phosphocholine (LPC) and 1-palmitoyl-1-hydroxy-sn-glycero-3-phospho-(1-rac-glycerol) (LPG), were used as a membrane mimetic (see SI). The micelle size distribution is homogeneous (diameter ~ 8 nm; polydispersity index ~ 0.15), corresponding to a molecular mass of ~91 ± 3 kDa (Figure S2).
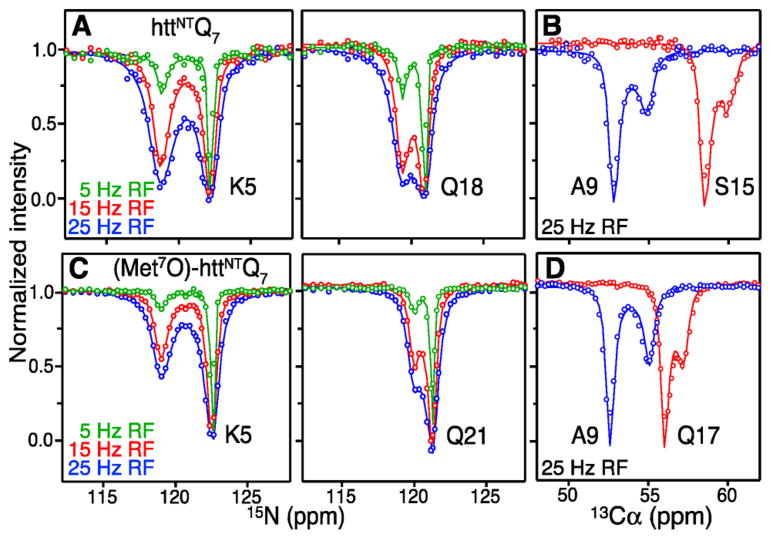
Exchange between free and micelle-bound native and (Met7O)-httNTQ7 was probed using CEST. CEST permits one to obtain the chemical shifts of the sparsely populated micelle-bound species in a system undergoing exchange on the millisecond time scale.8 Examples of 15N and 13Cα CEST8,9 profiles are shown in Figure 1 and display two well separated intensity dips, with the smaller one corresponding to the micelle-bound state. Fitting all the 15N CEST profiles to a two-state exchange model using the Bloch–McConnell Equations (see SI) yields values for the pseudo-first-order association rate constant ( ), the dissociation rate constant (koff) and the micelle-bound population (pB) of 15.0 ± 0.2 s −1, 56.1 ± 0.8 s−1 and 21.2 ± 0.3%, respectively, for httNTQ7, and 6.7 ± 0.1 s−1, 82.8 ± 1.6 s−1 and 7.8 ± 0.2%, respectively, for (Met7O)-httNTQ7.
Figure 1.
Kinetics of binding of httNTQ 7 and (Met7O)-httNTQ 7 to lipid-based micellar nanoparticles using CEST. Examples of 15N and 13Cα CEST profiles for native (panels A and B) and (Met7O) (panels C and D) httNTQ 7 in the presence of LPC/LPG micelles recorded at 700 MHz and 10 °C. The experimental data are shown as circles and the best-fit profiles for a two-state exchange model as continuous lines (see main text and SI). Panels A and C: 150 μM peptide in the presence of micelles at a 1:2 peptide to lipid molar ratio. Panels B and D: 0.96 mM peptide in the presence of micelles at 1:0.5 and 1:2 peptide to lipid molar ratios, respectively.
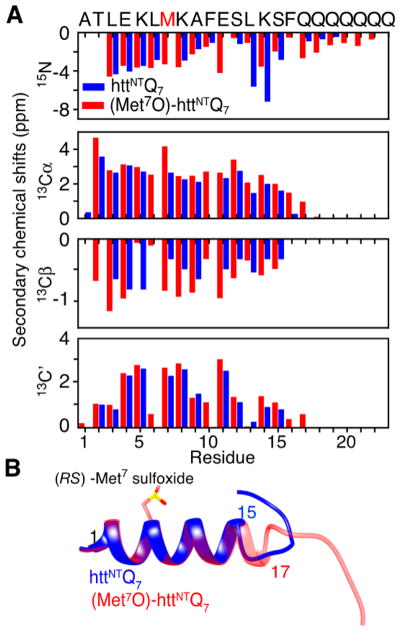
Analysis of 15N, 13Cα, 13Cβ9 and 13C′10 CEST profiles (Figure S3) yields a complete set of backbone 15N/13C shifts for the micelle-bound states of native and (Met7O)-httNTQ 7 (Figure 2A and Table S1), from which it can be deduced11 that residues 1–15 adopt an α-helical conformation that is extended by a further two residues upon oxidation of Met7. This may be due to an increase in the strength of the interaction between Met7 and Phe10 upon oxidation.12 The backbone chemical shifts were used to compute structures (Figure 2B) using the program CS-ROSETTA.13 The average transverse relaxation rate, <15N-R2B>helix, for the helical residues in the bound state obtained from the CEST fits is ~79 s−1, approximately 30% lower than the value predicted for a 91 kDa particle at 10 °C (~110 s−1 at 700 MHz), consistent with the existence of whole-body rocking motion about an axis lying in the plane of the micelle surface.14
Figure 2.
Structural characterization of micelle-bound native and Met7-sulfoxide httNTQ 7. (A) Secondary backbone chemical shifts derived from the CEST data with the sequence shown above the panel and Met7 indicated in red, and (B) structures calculated from the backbone shifts using CS-ROSETTA13 for micelle-bound native (blue) and Met7O (red) httNTQ 7.
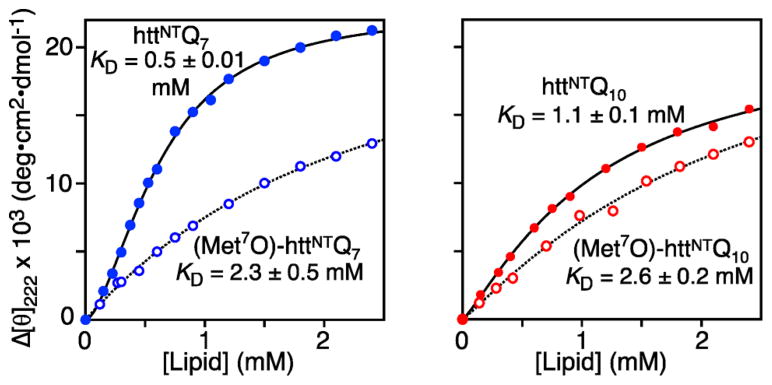
We next investigated the binding of native and (Met7O)-httNTQ 7 and httNTQ 10 to micelles by monitoring the change in circular dichroism (CD) ellipticity at 222 nm (Figure 3 and Figure S4). The number of helical residues for the micelle-bound state obtained from CD data is in excellent agreement with the CEST data (see SI). The equilibrium dissociation constant for httNTQ 7 (KD ~ 0.5 mM) is about 2-fold lower than that for the longer httNTQ 10 construct (KD ~ 1 mM). Moreover, the affinity is further reduced upon oxidation of Met7 (KD ~ 2.3–2.6 mM for both constructs) presumably due to unfavorable electrostatic interactions between Met7-sulfoxide and the lipid head groups. At low lipid concentrations the binding curve for httNTQ 7 deviates from a hyperbolic binding isotherm indicative of some type of binding cooperativity, possibly arising from exclusion of micelle surface at low lipid concentrations and/or repulsive interactions between surface-bound peptide molecules, both effects expected to be more pronounced with increasing saturation of the micelle surface (at low lipid concentrations). The data for httNTQ 7 can be fit phenomenologically with a Hill coefficient15 n = 1.7 (see SI). Minimal cooperativity (n = 1.12) is observed for httNTQ 10 and no cooperativity for the Met7O forms (n = 1).
Figure 3.
Binding of native and Met7-sulfoxide httNTQ 7 and httNTQ 10 to lipid micelles by CD. The experimental data are shown as circles and the continuous lines are best-fit curves to the Hill equation (see text and eq S3).
Neither the NMR nor CD data provide any information on the oligomeric state of httNTQn peptides on the micelle surface. We used pulsed Q-band EPR spectroscopy (see SI and Figures S5–S8 for experimental details) with httNTQn nitroxide spin-labeled at three sites individually: S15C-R1, S12C-R1 and A1C-R1, where R1 is (1-oxyl-2,2,5,5-tetramethyl-δ3-pyrroline-3-methyl)methanethiosulfonate (MTSL) conjugated to engineered cysteine residues.
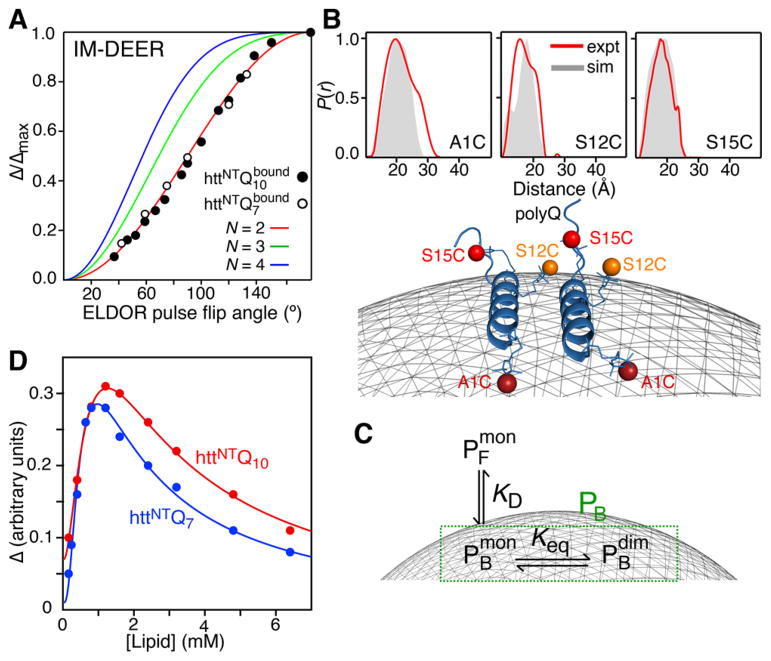
The multimeric state of httNTQn S15C-R1 was assessed using inversion-modulated double electron–electron resonance (IM-DEER) at 50 K in which the normalized modulation depth is measured as a function of ELDOR pulse flip angle from 0 to 180°, and the resulting curve (Figure 4A) is sensitive to the number of neighboring spins (eq S2).16 A comparison of the experimental IM-DEER data measured on flash-frozen samples of httNTQ 7 or httNTQ 10 S15C-R1 in the presence of micelles with that calculated for a dimer, trimer and tetramer indicates that dimers are formed on the micelle surface (Figure 4A). Note that the order of the oligomer for the Met7O forms could not be established unequivocally by IM-DEER owing to lower absolute modulation depths, and hence decreased signal-to-noise (Figure S12).
Figure 4.
Dimerization of httNTQn on the micelle surface characterized by Q-band pulsed EPR. (A) Normalized modulation depth (Δ/Δmax) as a function of ELDOR pulse flip angle in IM-DEER experiments measured on httNTQn (S15C-R1) in the presence of micelles. The experimental data are shown as circles, and the theoretical curves for a dimer (two spins), trimer (three spins) and tetramer (four spins) are shown in red, green and blue, respectively (see SI and eq S2). (B) Interspin label distance distributions, P(r), derived from the experimental DEER echo curves (Figure S7) by validated Tikhonov regularization using the program DEERAnalysis18 (red) for A1C-R1, S12C-R1 and S15C-R1 (top), and the dimer model calculated by docking of the CS-Rosetta monomer structure (cf. Figure 2B) using the DEER-derived mean distances as restraints with the membrane displayed as a gray mesh (bottom). The gray filled-in P(r) distributions shown in the top panels are calculated from the dimer model using MMM2013.219 (see Figure S9A). (C) Three-state model for the binding of httNTQn to the micelle surface. (D) Modulation depth of DEER echo curves as a function of lipid concentration. The experimental data are shown as circles, and the best-fit curves using the model shown in panel C (see eqs S7 and S8) are displayed as lines. All data were recorded on 80 μM httNTQ n; the peptide to lipid molar ratio was 1:30 and 1:80 in panels A and B, respectively.
DEER measurements (see SI and Figures S5–S8) on httNTQ 10 A1C-R1, S12C-R1 and S15C-R1 yield mean distances of 18.6, 17.1 and 19.7 Å, respectively, between spin labels in the dimer (Figure 4B, top), consistent with a parallel orientation of subunits. These distances allowed us to use Xplor-NIH17 to generate an approximate model of the dimer by docking the structure of the httNTQ 7 helical monomer (Figure 2B) employing the ansatz that helix–helix interactions are secondary to peptide–micelle interactions (see SI). The dimer model shown in Figure 4B (bottom) displays good agreement between experimental and calculated P(r) distributions (Figure 4B, top). Hydrophobic residues form a contiguous surface that interacts with the membrane (Figure S9B), and interhelical contacts comprise a mixture of hydrophobic and electrostatic interactions (Figure S9C). The side chains of Phe16 and Phe10 play an important role in binding to the micelle surface and in interhelical interactions, respectively. Interestingly, mutations of these two residues (F10A and F16A) inhibit aggregation even in the presence of 20 Q repeats.3c
To characterize the monomer–dimer equilibrium (Figure 4C) on the micelle surface, we monitored the modulation depth Δ of DEER echo curves recorded on S15C-R1 httNTQ 7 and httNTQ 10 as a function of lipid concentration (Figure 4D). Δ is a quantitative reporter of dipolar interactions between proximal electron spins and hence proportional to the population of dimeric micelle-bound peptide ( ) in a system characterized by a three-state equilibrium between free peptide ( ), bound monomer ( ) and bound dimer ( ) (Figure 4C). Δ initially increases with increasing lipid concentration until a maximum is reached, after which the value of Δ decreases with further increases in lipid concentration. This observation can be explained qualitatively as follows: initially, as the concentration of lipids is increased, the number of bound peptide particles (including dimers) per micelle increases; however, as the number of micelles grows, redistribution of bound peptide particles occurs such that ultimately the micelles will be occupied by either one or no peptide particles (making the formation of dimers impossible).
The lipid concentration at which Δ reaches its maximum value is dependent on the dimerization constant , where the curly brackets refer to average number of bound particles per micelle. Bearing in mind that , where {PB} is the average total number of bound particles per micelle, and that the population for each type of particle in Figure 4C can be expressed as (where [Ltot] and [Ptot] are the total lipid and peptide concentrations, respectively, and N the number of lipid molecules in a micelle), we derive an expression (eqs S7 and S8) for Δ = f(Keq, [Ltot], KD, n) (see SI for details; note KD and the Hill coefficient n are determined from analysis of the CD binding curves in Figure 3). The resulting best-fit curves are shown in Figure 4D and yield Keq values of 4.4(±1.7) × 10−3 and 7.0(±2.3) × 10−3 in units of (peptide particles/micelle)−1 for httNTQ 7 and httNTQ 10, respectively. The corresponding calculated dependence of { } and { } on lipid concentration is shown in Figure S10.
In summary, lipid-based micellar nanoparticles accelerate aggregation of httNT-polyQn peptides with as few as 10 glutamine repeats (Figure S1), confirming the existence of additional nucleation sites associated with membrane binding. The amphiphilic N-terminal segment of both httNTQ 7 and httNTQ 10 forms a 15-residue helix on the micelle surface (Figure 2B), which self-associates at high surface densities to form a parallel dimer (Figure 4B). The latter minimizes the intermolecular distance between the C-terminal polyQ tracts, which may in turn facilitate aggregation upon dissociation from the micelle surface through high local concentration effects.
Supplementary Material
Acknowledgments
We thank J. Baber, D. Garrett, J. M. Louis and J. Ying for technical support, L. E. Kay for the 13C CEST pulse code, and A. Szabo for useful suggestions. S.A.K. was supported by the Postdoctoral Research Associate Program of NIGMS (Award 1FiGM117609-01). This work was supported by the Intramural Programs of the National Institute of Diabetes and Digestive and Kidney Diseases (to G.M.C.) and the Center for Information Technology (to C.D.S.), National Institutes of Health.
Footnotes
Notes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b02619.
Experimental details, 1 Table and 12 Figures (PDF)
References
- 1.(a) Andresen JM, Gayan J, Djousse L, Roberts S, Brocklebank D, Cherny SS, Cardon LR, Gusella JF, MacDonald ME, Myers RH, Housman DE, Wexler NS. Ann Hum Genet. 2007;71:295–301. doi: 10.1111/j.1469-1809.2006.00335.x. [DOI] [PubMed] [Google Scholar]; (b) Urbanek A, Morato A, Allemand F, Delaforge E, Fournet A, Popovic M, Delbecq S, Sibille N, Bernado P. Angew Chem, Int Ed. 2018;57:3598–3601. doi: 10.1002/anie.201711530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Jayaraman M, Kodali R, Sahoo B, Thakur AK, Mayasundari A, Mishra R, Peterson CB, Wetzel R. J Mol Biol. 2012;415:881–899. doi: 10.1016/j.jmb.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Duim WC, Jiang Y, Shen K, Frydman J, Moerner WE. ACS Chem Biol. 2014;9:2767–2778. doi: 10.1021/cb500335w. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Lin HK, Boatz JC, Krabbendam IE, Kodali R, Hou Z, Wetzel R, Dolga AM, Poirier MA, van der Wel PCA. Nat Commun. 2017;8:15462. doi: 10.1038/ncomms15462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Burke KA, Hensal KM, Umbaugh CS, Chaibva M, Legleiter J. Biochim Biophys Acta, Biomembr. 2013;1828:1953–1961. doi: 10.1016/j.bbamem.2013.04.025. [DOI] [PubMed] [Google Scholar]; (b) Mitomi Y, Nomura T, Kurosawa M, Nukina N, Furukawa Y. J Biol Chem. 2012;287:34764–34775. doi: 10.1074/jbc.M112.387035. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Thakur AK, Jayaraman M, Mishra R, Thakur M, Chellgren VM, Byeon IJ, Anjum DH, Kodali R, Creamer TP, Conway JF, Gronenborn AM, Wetzel R. Nat Struct Mol Biol. 2009;16:380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Pandey NK, Isas JM, Rawat A, Lee RV, Langen J, Pandey P, Langen R. J Biol Chem. 2018;293:2597–2605. doi: 10.1074/jbc.M117.813667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Atwal RS, Truant R. Autophagy. 2008;4:91–93. doi: 10.4161/auto.5201. [DOI] [PubMed] [Google Scholar]; (b) Suopanki J, Gotz C, Lutsch G, Schiller J, Harjes P, Herrmann A, Wanker EE. J Neurochem. 2006;96:870–884. doi: 10.1111/j.1471-4159.2005.03620.x. [DOI] [PubMed] [Google Scholar]
- 5.(a) Michalek M, Salnikov ES, Bechinger B. Biophys J. 2013;105:699–710. doi: 10.1016/j.bpj.2013.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Michalek M, Salnikov ES, Werten S, Bechinger B. Biochemistry. 2013;52:847–858. doi: 10.1021/bi301325q. [DOI] [PubMed] [Google Scholar]
- 6.(a) Chaibva M, Burke KA, Legleiter J. Biochemistry. 2014;53:2355–2365. doi: 10.1021/bi401619q. [DOI] [PubMed] [Google Scholar]; (b) Gao X, Campbell WAT, Chaibva M, Jain P, Leslie AE, Frey SL, Legleiter J. Biochemistry. 2016;55:92–102. doi: 10.1021/acs.biochem.5b00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulder WJ, Strijkers GJ, van Tilborg GA, Griffioen AW, Nicolay K. NMR Biomed. 2006;19:142–164. doi: 10.1002/nbm.1011. [DOI] [PubMed] [Google Scholar]
- 8.Vallurupalli P, Bouvignies G, Kay LE. J Am Chem Soc. 2012;134:8148–8161. doi: 10.1021/ja3001419. [DOI] [PubMed] [Google Scholar]
- 9.Long D, Sekhar A, Kay LE. J Biomol NMR. 2014;60:203–208. doi: 10.1007/s10858-014-9868-5. [DOI] [PubMed] [Google Scholar]
- 10.Vallurupalli P, Kay LE. Angew Chem, Int Ed. 2013;52:4156–4159. doi: 10.1002/anie.201209118. [DOI] [PubMed] [Google Scholar]
- 11.Shen Y, Bax A. Methods Mol Biol. 2015;1260:17–32. doi: 10.1007/978-1-4939-2239-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis AK, Dunleavy KM, Senkow TL, Her C, Horn BT, Jersett MA, Mahling R, McCarthy MR, Perell GT, Valley CC, Karim CB, Gao J, Pomerantz WC, Thomas DD, Cembran A, Hinderliter A, Sachs JN. Nat Chem Biol. 2016;12:860–866. doi: 10.1038/nchembio.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Proc Natl Acad Sci U S A. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorieau JL, Louis JM, Bax A. J Am Chem Soc. 2011;133:14184–14187. doi: 10.1021/ja2045309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill ASV. J Physiol (Oxford, U K) 1910;40:190–224. doi: 10.1113/jphysiol.1910.sp001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt T, Ghirlando R, Baber J, Clore GM. ChemPhysChem. 2016;17:2987–2991. doi: 10.1002/cphc.201600726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwieters CD, Bermejo GA, Clore GM. Protein Sci. 2018;27:26–40. doi: 10.1002/pro.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. Appl Magn Reson. 2006;30:473–498. [Google Scholar]
- 19.Polyhach Y, Bordignon E, Jeschke G. Phys Chem Chem Phys. 2011;13:2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.