Abstract
HIV employs multiple means to evade the humoral immune response, particularly the elicitation of and recognition by broadly neutralizing antibodies (bnAbs). Such antibodies can act antivirally against a wide spectrum of viruses by targeting relatively conserved regions on the surface HIV envelope trimer spike. Elicitation of and recognition by bnAbs are hindered by the arrangement of spikes on virions and the relatively difficult access to bnAb epitopes on spikes, including the proximity of variable regions and a high density of glycans. Yet, in a small proportion of HIV-infected individuals, potent bnAb responses do develop, and isolation of the corresponding monoclonal antibodies has been facilitated by identification of favorable donors with potent bnAb sera and by development of improved methods for human antibody generation. Molecular studies of recombinant Env trimers, alone and in interaction with bnAbs, are providing new insights that are fueling the development and testing of promising immunogens aimed at the elicitation of bnAbs.
Keywords: vaccination, rational vaccine design, passive immunotherapy, conserved epitopes, immunization strategies
INTRODUCTION
Broadly neutralizing antibodies (bnAbs), i.e., antibodies capable of neutralizing the majority of strains of a given highly antigenically variable pathogen, have attracted considerable attention (1, 2). This is because they hold out the promise of helping to guide vaccine and drug design and serving themselves as prophylactics or therapeutics against some of the most difficult pathogens encountered in modern medicine. In the case of HIV, progress in isolating, characterizing, and understanding bnAbs has been dramatic, to the point that the vaccine field has received considerable stimulus and bnAbs are entering into clinical trials for prophylaxis and therapy. In HIV bnAb-based vaccine design (Figure 1), a number of advances have come together to energize the field, including the discovery of potent bnAbs; the description of the structure of the Env trimer, which is the sole target of bnAbs; the determination of the course of antibody-virus coevolution in a number of individuals who develop bnAb responses; the design of promising immunogens based on structural understanding; and the development of animal models to inform on how immunogens interact with the human antibody system. In this review, we report on recent developments in bnAbs, their interaction with Env trimers, and bnAb-based vaccine design. We link these developments to earlier findings to provide context to this rapidly expanding area of research.
Figure 1.
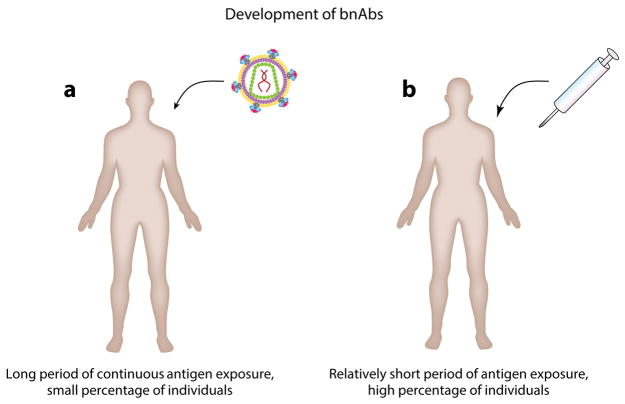
The essence of the problem of bnAbs in HIV vaccine design. (a) Potent bnAb responses develop in a small proportion of HIV-infected individuals, but this occurs over a long period (years) during which the individual is exposed to multiple viral strains. (b) A vaccine should induce such responses in the majority of individuals in a much shorter period with a manageable number of immunogens.
STRUCTURE OF ENV TRIMERS: LIGAND OF BROADLY NEUTRALIZING ANTIBODIES
Although the antibody response to HIV is diverse, only antibodies binding to the envelope surface spike are able to directly neutralize virions. On each HIV virion, there are only about 8–14 envelope spikes (3, 4). There is debate about how many spikes must be engaged by CD4 and CCR5 receptors for viral entry to proceed (5–8). The most detailed study suggests a range of 2–3, depending on the virus isolate (5). The low spike density is peculiar to the immunodeficiency viruses HIV and SIV and is enforced by the intracellular domain of gp41; in HIV or SIV in which the intracellular domain has been deleted, over 70 spikes can be found per virion (3, 4). Wide spacing of the envelope spikes is unfavorable for the activation of B cells, which are most effectively triggered by repetitive and rigid structures (9, 10). Yet, the wide spacing of envelope spikes on HIV probably leaves all sides of the spikes accessible (3, 4), in contrast to virions that have densely packed surfaces and on which only the apical tip of the spike is accessible to antibodies (e.g., rhabdoviruses) (10).
The envelope spike proteins are encoded by the env gene as a gp160 precursor protein that is posttranslationally processed by furin proteases (11) into the globular gp120 head that is responsible for receptor and coreceptor attachment, and the gp41 subunit that contains the fusion machinery and that anchors the envelope spikes into the viral membrane. These proteins form trimers of noncovalently linked dimers and are transported to the plasma membrane from where progeny viruses bud.
During transport, a median of 25 N-linked glycans (12) are attached to each gp120 subunit, making up to 50% of its mass (13). In many glycoproteins, high-mannose glycans are matured into complex sugars during transition through the Golgi compartment. However, in the case of native HIV envelope proteins, a large portion of the glycosylation is of the high-mannose type (14), indicating incomplete processing. This feature appears to be dictated by the compact native Env trimer structure, given that prolonged incubation with recombinant α-mannosidase I fails to trim the high-mannose chains (13). In contrast, monomeric gp120 exhibits more complex glycosylation, indicating that in the monomer, high-mannose glycans are successfully trimmed and then modified to yield complex glycans (15). In contrast to native trimers and recombinant trimers, such as SOSIP (below), that mimic their native counterparts, most recombinant gp120 and gp140 molecules possess a much higher proportion of complex glycans, which reflects their relatively open structures.
It is proposed that native HIV envelope spikes are generally rather unstable, decaying to nonfunctional configurations (16, 17). For example, in some cases, gp120 subunits dissociate from gp41, leaving the latter as stumps on the virion surface (17). An alternative view is that the spike is not unstable but rather emerges from the infected cell plasma membrane with a proportion of nonnative surface Env (18).
Five regions of considerable sequence variability are often described as hypervariable loops on gp120; some of these are quite disordered on monomeric gp120. Indeed, the overall conformation of gp120 is somewhat flexible. This proved to be a major obstacle in solving its structure in the late 1990s, requiring deletion of hypervariable loops 1 through 3, complexing with stabilizing antibodies, and binding to truncated CD4 receptor molecules to sufficiently stabilize the gp120 core so as to allow crystallization and solution of a core gp120 structure (19). Several core gp120 structures could be solved thereafter, but information on the native gp120/gp41 trimer complex and the conformations of the V1/V2 and V3 loops was lacking. Cryo-electron microscopy (cryo-EM) and cryotomography provided the first low-resolution view of the quaternary structure of Env spikes (3, 4). A much-higher-resolution picture emerged using cryo-EM and crystallography to solve the structure of a recombinant stabilized soluble envelope trimer molecule derived from clade A isolate BG505 (reviewed in Reference 20). BG505 SOSIP trimers (21–24) were stabilized by (a) a truncation of gp41 so that only a part of the extracellular domain remains, (b) a set of mutations referred to as SOS that connects the gp120 C-terminal domain to the ectodomain of gp41 via a disulfide bridge (25), and (c) a modification referred to as IP in which an isoleucine in the N-terminal heptad repeat domain is replaced with a proline residue to disfavor trimer disassembly (26). BG505 SOSIP structures were solved in complex with PGT122 [crystallography (22)], PGT122 and 35O22 [crystallography (24)], PGV04 [cryo-EM (23)], or VRC03 [cryo-EM (21)]. The BG505 SOSIP molecule exhibits a number of features indicating that it is a good structural mimic of the native functional trimer; most importantly, it binds neutralizing but not non-neutralizing antibodies, thereby reflecting the antigenic profile of native trimers (27–29). The high-resolution structures of the SOSIP trimer revealed the structural disposition of the protomers and the structures of the hypervariable loops (Figure 2). It became apparent that the envelope spikes form a propeller-like structure in which the gp120 outer domain (19) makes up the propeller’s blades and the inner domain forms the interface between the protomers and interacts with the gp41 subunit. Hyper-variable loops 1 through 3 assist the formation of the envelope trimers and provide additional protomer interactions (22, 23). In particular, the V1 and V2 loops of gp120 protomers interact and form the apex of the trimer, and the V3 loops are largely sequestered underneath the V1/V2 loop structure. The gp41 subunits form a perpendicular central three-helix bundle with the heptad 1 repeat region of each gp41 protomer (22), and a pedestal base that is formed by the respective heptad 2 repeat region (23). A higher-resolution crystal structure of the BG505 SOSIP trimer in complex with bnAbs PGT122 and 35O22 solved by crystallography revealed that prefusion gp41 encircles the N- and C-termini of gp120 with four helices that form a membrane-proximal collar, fastened by insertion of a methionine into a gp41-tryptophan clasp (24). It is believed that after gp120 has engaged the cellular receptors CD4 and CCR5, the clasp is opened, resulting in the exposure of the fusion peptide and its subsequent insertion into the host cell membrane. The two heptad repeats then align with each other and fold into a compact six-helix bundle. This transition pulls the host cell membrane into fusion distance of the viral membrane (21, 30).
Figure 2.
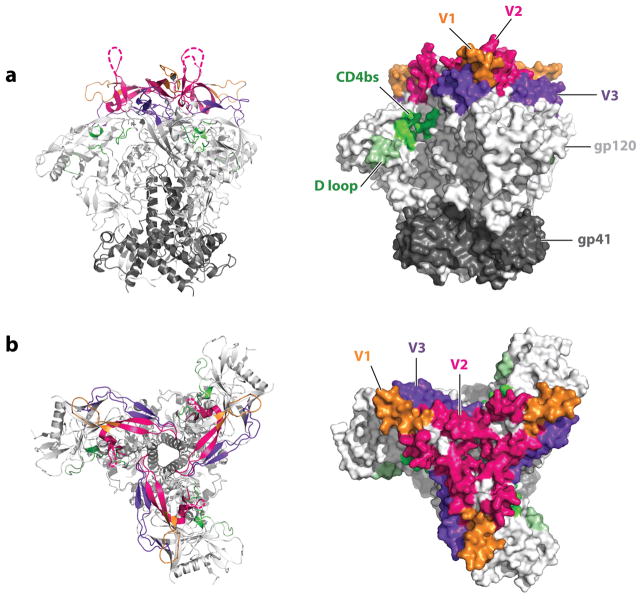
Structure of the HIV envelope spike. In a crystal structure of the HIV strain BG505 expressed as SOSIP.664 near-native trimers [PDB 4zmj (31)], the location of some of the most important Env trimer domains is indicated as viewed (a) from the side or (b) from the top. The gp120 subunits form the blades of a propeller-like structure, whereas the gp41 subunit forms a central stalk and a membrane-proximal, pedestal-like structure. As predicted from other experimental data, hypervariable loops V1 through V3 are located at the apex of the trimer, with hypervariable loop V3 being partially buried under hypervariable loops V1 and V2. These loops are also in proximity to the CD4 receptor binding site that is recessed left in the cleft formed by two propeller blades. Loops that did not resolve in the structure are indicated by a dashed line.
The above recombinant trimer structures were solved in complex with bnAbs that may, in principle, have altered the protein conformation. A recently solved structure of a ligand-free trimer further stabilized by the addition of a disulfide internal to gp120 argues against this, in that the structure is very similar to that of the liganded structures (31). Also, cryo-EM structures of the Env trimer from native virions are very much comparable with the structures found for SOSIP proteins (32, 32a).
Following the striking success of the SOSIP concept with a single isolate, BG505 (a clade A isolate), stable trimers have been generated for a number of other isolates of different clades (33, 34). Furthermore, alternative approaches to generate well-ordered trimers, generally defined as those showing a regular compact morphology in negative stain EM and an appropriate antigenic profile, are being explored. One approach uses a flexible linker to replace the gp120/gp41 cleavage site and maintain gp120-gp41 association together with an IP substitution, as for SOSIP. Trimers generated by this alternative approach are referred to as native, flexibly linked (or NFL) trimers (35). We can expect further well-ordered trimer designs to follow, building upon the experience with SOSIP.
FEATURES OF THE ENV SPIKE THAT RESIST ANTIBODY RECOGNITION
As a virus that can establish persistence without generally remaining completely latent for extended periods or being present only in immune privileged sites, HIV has evolved countermeasures to avoid elimination by antibodies. These countermeasures include both the prevention of immune recognition as well as escape from emerging neutralizing antibody responses.
Several structural properties of the virion surface and the envelope spike impair the generation of effective B cell responses. First, the large spacing between envelope spikes is unfavorable for B cell activation that requires antigen cross-linking of B cell receptors. Second, extremely extensive glycosylation of the envelope spike by the host cellular machinery cloaks the virus with a weakly immunogenic, potentially self, antigen and constrains antibody access to neutralizing epitopes. Moreover, the addition and loss of glycosylation sites is actively used by the virus to change the surface of the spike and thwart antibody recognition (36). Third, functionally essential structures that cannot easily be mutated in order to escape antibody recognition are presented in poorly accessible locations on the quaternary structure [e.g., the CD4 binding site (CD4bs), and the membrane-proximal external region (MPER)] and may be obstructed by structures that have no essential function and therefore can more readily be mutated in response to antibody pressure [e.g., V1/V2 (37)].
The CD4 receptor binding site, an important epitope for bnAbs, is a case in point. The site is located recessed on the left side of the cleft between two propeller blades of the trimer (Figure 2). It is surrounded by numerous glycans—a glycan fence (38)—that further conceal the conserved receptor binding site. In particular, glycosylation at position 276 was found to substantially restrict antibody access to the CD4bs (23). The glycan at N197 serves a similar function (38). The CD4bs is additionally shielded by the V2 loop that was found to be located above (22). In combination with the V1 loop, the V2 loop was also found to cover the V3 loop that is involved in coreceptor binding. The V1 and V2 loops are characterized by rapid evolution (changes in sequence, length, and glycosylation) in response to antibody pressure and facilitate escape from autologous antibody responses (39). Taken together, the location and surroundings of the CD4bs on the gp120 trimer only allow for a narrow range of approach angles for bnAbs.
An interesting and important feature of the Env spike related to antibody recognition is its ability to exist in different forms with varying degrees of openness (Figure 3). In the most compact closed conformation, conserved epitopes are best shielded. On binding of CD4, the spike opens out to permit coreceptor binding and coincidentally exposes additional neutralizing antibody epitopes (21, 28, 33, 40, 41). However, in addition to the fully open conformation, a tendency to breathe, i.e., to transition between a somewhat open conformation and the fully closed conformation, has been observed. This tendency appears to be dependent on the subtype and strain (42). While breathing, some antibody epitopes become available that are not very accessible on the closed conformation. For example, the CD4bs-specific antibody b12 preferentially binds to the relaxed conformation, which it then appears to maintain open (29, 33). In contrast, other bnAbs, such as VRC01, PG16, PGT128, PGT145, and 2G12 (see below), appear to favor the closed conformation, and their binding suppresses further breathing by locking down the trimer in its closed conformation (42).
Figure 3.
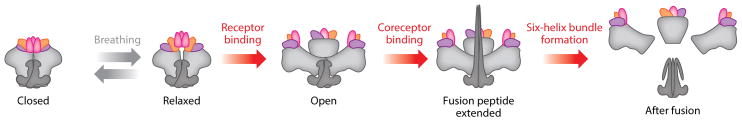
Conformations of the Env trimer. Several conformations have been described for the Env trimer. Before receptor engagement, and depending on the isolate, the Env trimers transit between closed, slightly more open, and relaxed conformations in a process referred to as breathing. Following CD4 receptor engagement, the trimers open out and the CCR5 (or CXCR4) coreceptor binding sites become available. Coreceptor binding then triggers insertion of the fusion peptide into the host membrane and formation of the six-helix bundle that pulls the host and the virus membrane into fusion proximity.
ANTIBODY RESPONSES TO THE HIV ENVELOPE GLYCOPROTEINS: THE FIRST GENERATION OF BROADLY NEUTRALIZING ANTIBODIES
Non-neutralizing antibody responses to envelope glycoproteins develop rapidly, within 8 days of infection. Initially only detectable in the form of immune complexes, free antibodies to gp41 and gp120 develop after about 2 and 4 weeks, respectively (43). Development of neutralizing antibodies, i.e., antibodies that can inactivate the virus directly, usually takes 3 to 12 months (36, 44). Such antibodies are typically directed against variable regions, e.g., the V1/V2 or V3 loops, and are of limited breadth (45). In contrast to the slow generation of neutralizing antibodies, the virus readily escapes selection pressure exerted by strain-specific antibody responses (36, 44), resulting in resistance of most circulating viruses to the concomitantly present neutralizing antibodies. Finally, as stated earlier, anywhere between 10% and 50% of individuals develop bnAb responses, depending upon the definition of potency and breadth (46, 47), but a much smaller fraction, a few percent (48), develop potent responses of the type that is associated with some of the best broadly neutralizing monoclonal antibodies. The reasons that some individuals develop much more potent responses than others is likely complex, involving a number of features, including viral load, features of infecting and evolving virus, and the quality of T cell help (46, 49).
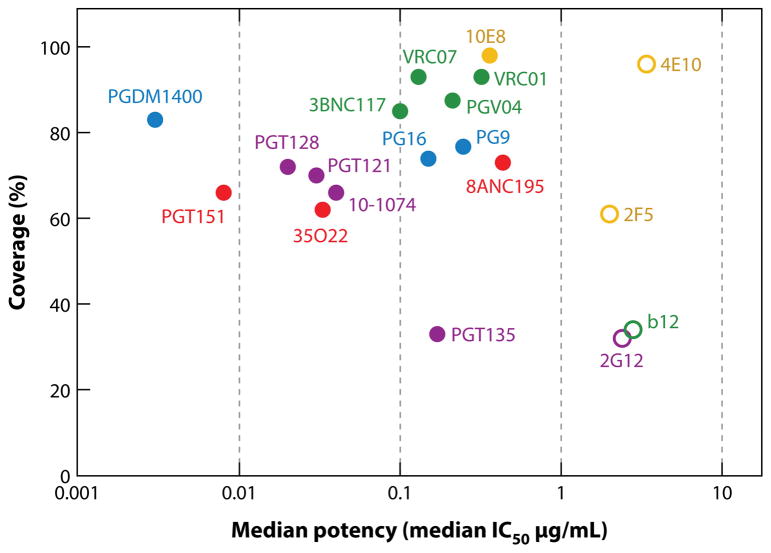
bnAb serum responses could in principle arise from a limited number of bnAb specificities or a very large number of strain-specific neutralizing antibodies. The isolation of a set of broadly neutralizing monoclonal antibodies by phage display and from human hybridomas in the 1990s was consistent with the former hypothesis. These antibodies led to the identification of conserved epitopes that are shared between subtypes and isolates and include the CD4bs on gp120 [bnAb b12 (50, 51)], the membrane-proximal external region (MPER) on gp41 [bnAbs 4E10 and 2F5 (52–54a)], and a glycan epitope on the outer domain of gp120 [bnAb 2G12 (55, 56)]. Since selection or screening for these antibodies was conducted using monomeric gp120, all of them recognize conserved epitopes that are expressed on a single gp120 protomer, and that are incompletely shielded on the trimer of the strains they neutralize. These first-generation monoclonal antibodies, particularly the gp120 monoclonal antibodies, were effective against clade B isolates, but compared to more recent bnAbs, they were of limited breadth and/or potency against global isolates (Figure 4). The first-generation monoclonal antibodies showed a number of features that have subsequently been described for bnAbs in general, including high levels of somatic hypermutation (SHM) compared to those typically observed in immune responses and a relatively long heavy chain complementarity determining region 3 (HCDR3) (Table 1). In the case of the glycan-specific bnAb 2G12, the antibody is mutated to such a high degree that it is domain exchanged (57, 58). This generates two additional potential antigen-binding sites and provides the antibody with sufficient avidity for neutralization by binding solely to glycans in the high-mannose patch (57). As depicted in Figure 4, the first generation of antibodies neutralizes between about 30% of global isolates (b12 and 2G12) and about 95% (4E10), but only at median IC50 concentrations greater than 1 μg/mL.
Figure 4.
Breadth and potency of bnAbs. The percentage of large panels of isolates neutralized at an IC50 < 50 μg/mL (coverage) in pseudovirus assays plotted against the median inhibitory concentration (IC50 in μg/mL) for all neutralization-sensitive viruses. First-generation antibodies are indicated by unfilled circles and second-generation antibodies by filled circles. The epitope specificity is indicated by color, with antibody specific for the CD4 binding site depicted in green, apex-specific antibodies in blue, antibodies binding to the high-mannose patch in purple, antibodies binding to the gp120-gp41 interface in red, and MPER-specific antibodies in light brown. Note that different panels of viruses have been used for many of the antibodies, so comparisons are approximate (74–79, 81, 84–86, 159). The locations of the epitopes recognized by the bnAbs are illustrated in Figure 5.
Table 1.
Overview of characteristics of representative broadly neutralizing antibodiesa
| Sequence ID | Chain | V gene and allele | D gene and allele | J gene and allele | CDR1 length | CDR2 length | CDR3 length | SHM, % nt | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| First generation | |||||||||
| 2G12 | Heavy | VH3-21*01 | D6-6*01 | JH3*01 | 8 | 8 | 16 | 20.3 | 55,56 |
| Light | VK1-5*03 | JK1*01 | 6 | 3 | 9 | 12.2 | |||
| b12 | Heavy | VH1-3*01 | D2-21*01 | JH6*03 | 8 | 8 | 20 | 13.8 | 50,51 |
| Light | VK3-20*01 | JK2*01 | 7 | 3 | 9 | 13.4 | |||
| 2F5 | Heavy | VH2-5*02 | D3-3*01 | JH6*02 | 10 | 7 | 24 | 12.1 | 53,54,56 |
| Light | VK1D-13*02 | JK4*01 | 6 | 3 | 9 | 11.8 | |||
| 4E10 | Heavy | VH1-69*06 | D6-19*06 | JH4*02 | 8 | 8 | 20 | 6.8 | 54a,56 |
| Light | VK3-20*01 | JK1*01 | 7 | 3 | 9 | 4.7 | |||
| Second generation | |||||||||
| PG9 | Heavy | VH3-33*05 | D3-3*01 | JH6*03 | 8 | 8 | 30 | 12.0 | 74 |
| Light | VL2-14*01 | JL3*02 | 9 | 3 | 10 | 9.2 | |||
| PG16 | Heavy | VH3-33*05 | D3-3*01 | JH6*03 | 8 | 8 | 30 | 13.1 | 74 |
| Light | VL2-14*01 | JL3*01 | 9 | 3 | 10 | 12.2 | |||
| PGT145 | Heavy | VH1-8*01 | D4-17*01 | JH6*02 | 8 | 8 | 33 | 16.2 | 75 |
| Light | VK2D-28*01 | JK1*01 | 11 | 3 | 9 | 15.5 | |||
| CAP256-VRC26.09 | Heavy | VH3-30*18 | D3-3*01 | JH3*02 | 8 | 8 | 39 | 12.8 | 77 |
| Light | VL1-51*02 | JL1*01 | 8 | 3 | 12 | 9.7 | |||
| PGDM1400 | Heavy | VH1-8*01 | D4-17*01 | JH6*02 | 8 | 8 | 34 | 24.3 | 81 |
| Light | VK2D-28*01 | JK1*01 | 11 | 3 | 9 | 11.3 | |||
| PGT121 | Heavy | VH4-59*07 | D3-3*02 | JH6*03 | 8 | 7 | 26 | 17.8 | 75 |
| Light | VL3-21*03 | JL3*02 | 6 | 3 | 12 | 15.5 | |||
| PGT124 | Heavy | VH4-59*03 | D3-3*01 | JH6*03 | 8 | 7 | 26 | 15.0 | 75 |
| Light | VL3-21*01 | JL3*02 | 6 | 3 | 12 | 16.6 | |||
| PGT128 | Heavy | VH4-39*07 | D3-3*01 | JH5*02 | 10 | 13 | 21 | 19.0 | 75 |
| Light | VL2-8*01 | JL3*01 | 4 | 3 | 10 | 8.7 | |||
| PGT135 | Heavy | VH4-39*07 | D2-21*01 | JH5*02 | 15 | 7 | 20 | 17.1 | 75 |
| Light | VK3-15*01 | JK1*01 | 6 | 3 | 9 | 15.9 | |||
| 10-1074 | Heavy | VH4-4*08 | D3-3*02 | JH6*03 | 8 | 7 | 26 | 26.7 | 83 |
| Light | VL3-9*02 | JL3*02 | 6 | 3 | 12 | 30.1 | |||
| VRC01 | Heavy | VH1-2*02 | D5-18*01 | JH2*01 | 8 | 8 | 14 | 29.7 | 79 |
| Light | VK3-NL1*01 | JK2*04 | 4 | 3 | 5 | 16.6 | |||
| 3BNC117 | Heavy | VH1-2*02 | D6-25*01 | JH2*01 | 8 | 8 | 12 | 23.5 | 158 |
| Light | VK1D-33*01 | JK3*01 | 6 | 3 | 5 | 16.6 | |||
| CH103 | Heavy | VH4-59*01 | D3-9*01 | JH4*02 | 8 | 7 | 15 | 18.0 | 132 |
| Light | VL3-1*01 | JL1*01 | 6 | 3 | 10 | 10.7 | |||
| PGT151 | Heavy | VH3-30-3*03 | D3-3*01 | JH6*02 | 8 | 8 | 28 | 18.5 | 85,98 |
| Light | VK2D-29*02 | JK4*02 | 11 | 3 | 9 | 12.6 | |||
| 35O22 | Heavy | VH1-18*03 | D3-10*01 | JH5*02 | 8 | 8 | 16 | 21.9 | 84 |
| Light | VL2-23*03 | JL1*01 | 9 | 3 | 10 | 22.4 | |||
| 8ANC195 | Heavy | VH1-69D*01 | D3-3-01 | JH4*02 | 9 | 6 | 22 | 27.2 | 86,87 |
| Light | VK1-5*03 | JK1*01 | 7 | 3 | 9 | 14.9 | |||
| 10E8 | Heavy | VH3-15*05 | D3-3*01 | JH1*01 | 8 | 10 | 22 | 19.4 | 78 |
| Light | VL3-19*01 | JL3*02 | 6 | 3 | 12 | 14.2 | |||
Abbreviation: SHM, somatic hypermutation.
V(D)J assignment was performed using the AbStar software (B. Briney et al., personal communication). The total number of somatic hypermutations corresponds to the sum of the mutations found in V and J elements. The epitope specificity is indicated by color: yellow for MPER (membrane-proximal external region), red for gp120-gp41 interface, purple for high-mannose patch, blue for apex, and green for CD4 binding site. First-generation bnAb sequences were obtained from personal communications; second-generation bnAb sequences were obtained from GenBank. Note that the short length of the D region often prevents an unequivocal identification of the encoding gene allele.
The first generation of bnAbs revealed much about the in vivo activity of such antibodies in humanized mice, monkeys, and humans. It became apparent that bnAbs have great potential to prevent infection, and to temporarily lower the viral load, although they failed to clear established infections (59, 60), at least in part because of rapid selection of escape variants in the treated subjects (60). Protection was observed in nonhuman primates against viral challenge via the intravenous (61, 62), rectal (63), and vaginal routes (64–67) when passive immunization achieved bnAb serum neutralizing titers generally of the very approximate order of 1:100, with some exceptions (62, 64, 68). Similar findings were made in humanized mice (69, 70). Of note, a CD4-IgG2 chimeric molecule (71) and 2G12 (72) both protected at unexpectedly low serum antibody neutralizing titers. The protective activity of bnAbs was partially attributed to Fc receptor–mediated effector functions as well as direct neutralization in monkey studies (66, 67). However, one study found no evidence that this effector function was antibody-dependent cellular cytotoxicity (ADCC) (73).
THE SECOND GENERATION OF BROADLY NEUTRALIZING ANTIBODIES
There are two main developments that lead to the discovery of a new generation of broadly neutralizing antibodies that are of greater breadth than the first generation. First, large cohorts of infected individuals, particularly from sub-Saharan Africa, were screened to identify those with potent broadly neutralizing sera to give the best opportunities to isolate bnAbs (48, 74). Second, improved approaches to human monoclonal antibody generation were adopted. The first approach takes advantage of improved methods to culture and activate individual memory B cells and then directly screens culture supernatants for neutralization (74–78). The antibody genes from positive wells are then rescued and cloned into a suitable antibody expression vector. The second approach relies on Env-specific B cell sorting, antibody gene rescue, and then screening of expressed antibodies for neutralization activity (79–82).
The first of the new generation of bnAbs, PG9 and PG16, were isolated by direct neutralization screening (74). Interestingly, these bnAbs defined a new epitope at the apex of the Env trimer, and they were shown to be strongly trimer preferring or trimer specific and therefore would be difficult to isolate by affinity selection using monomeric gp120. Next, a novel potent antibody, VRC01, was isolated to the CD4bs of gp120 (79). This antibody was isolated by affinity selection using a number of gp120s. The antibody was both notably more potent and broader than the first CD4bs antibody, b12 (Figure 4). The plethora of bnAbs that have been isolated since then are of ever increasing potency and considerable breadth, defining novel epitopes including a high-mannose patch overlapping the 2G12 epitope (75, 83) and a region at the interface between gp120 and gp41 (84–86). Many potent antibodies against the CD4bs have been isolated (51, 82, 87–90). Both direct neutralization screening and affinity selection of B cells have been used in isolating these antibodies. Of note, with the description of the well-ordered SOSIP trimer, this molecule has more recently been used to affinity select for B cells producing highly potent trimer-specific antibodies (81). It appears that a large fraction of the surface of the Env trimer is in fact accessible to bnAb recognition despite its dense glycan coating (Figure 5a).
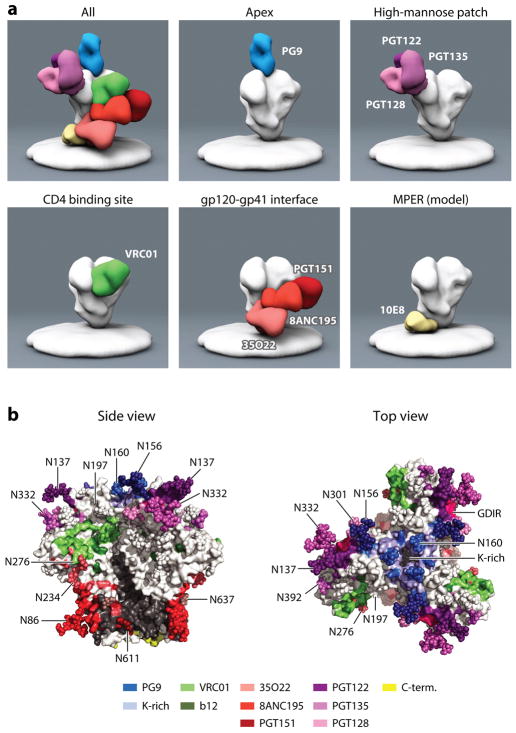
Figure 5.
(a) Negative-stain electron microscopy representations of bnAbs binding to the Env trimer. bnAbs can be divided according to the epitopes they recognize: those specific for the CD4 binding site, the apex, the high-mannose patch, the gp120-gp41 interface, and the MPER. Electron microscopy reconstructions, with the exception of the MPER panel, were kindly prepared by Jeong Hyun Lee, Gabriel Ozorowski, and Andrew Ward. The MPER antibody reconstruction has been very roughly modeled according to the location of the MPER epitope in the Env trimer structures. (b) The footprints of bnAbs on the Env trimer. The Env trimer coordinates from PDB 4TVP were extracted and stained in white and gray for gp120 and gp41. The contact residues for PG9 are blue (99); PGT122 dark purple (22); PGT128 light purple (96); PGT135 medium purple (136); VRC01 green (88); b12 olive green, from alanine scan (160); PGT151 dark red (98); 8ANC195 medium red (86, 87); and 35O22 light red (24). The MPER region is largely missing from the structure; the last available amino acids are in yellow. The glycan at position 156 of gp120 is recognized by both PGT9 and PGT122, which is reflected by the intermediate color. The glycan at position 332 is bound by most bnAbs specific for the high-mannose patch. Also, the epitopes of PGT151 and 8ANC195 overlap in recognition of the glycan at position 637. The lysine-rich region at the apex is indicated by bright blue. Note that glycans in the structure might be truncated given that they were not completely resolved in the crystal structure.
The neutralizing antibodies described likely interfere with HIV entry by multiple mechanisms. Note that a positive neutralization signal simply indicates that fewer viral particles gain entry to target cells over the lifetime of the in vitro neutralization experiment. Importantly, it seems relatively clear that strong binding to native functional Env spikes is necessary and sufficient for neutralization (91, 92). High-affinity binding of non-neutralizing antibodies to functional Env spikes has not been described. As a corollary, binding of antibodies to functional spikes seems to be invariably associated with measurable neutralization. One mechanism for neutralization can thus be steric hindrance of the entry process via interference with productive virus attachment to target cells and/or with virus fusion. Such steric hindrance is perhaps easiest to visualize if entry involves the complex interaction of an array of Env spikes on the virus membrane with an array of CD4 and CCR5 receptor molecules on the target cell membrane. CD4bs-specific antibodies are expected to interfere with CD4 receptor binding and thereby prevent virus attachment and entry (93). The high-mannose patch antibody 2G12 interferes with CCR5 binding in particular (94). The HIV Env spike is intrinsically rather unstable, decaying and losing infectivity. A number of antibodies can accelerate this decay process, thereby reducing viral entry relative to the controls in the absence of antibody and yielding neutralization (95). The high-mannose patch bnAb PGT128 can neutralize HIV via this mechanism (96). Another high-mannose patch bnAb PGT121 can modulate CD4 binding allosterically, presenting another possible neutralization mechanism (97). Antibodies to the V2 apex (PG9) and the gp120-gp41 interface (PGT151) can cross-link protomers (85, 98, 99), leading to a further possible neutralization mechanism. Such cross-linking may freeze the trimer in its closed conformation and/or prevent structural rearrangement required for the induction of membrane fusion. MPER-specific monoclonal antibodies have very weak binding to the native trimer of tier 2 viruses and are believed to neutralize via recognition of a fusion intermediate that is formed following CD4 binding (100, 101). Binding of MPER-specific antibodies may lock the envelope protein in this intermediate conformation, thereby preventing the formation of the six-helix bundle and thus membrane fusion (101). In this discussion, it is worth noting that neutralization is observed irrespective of the location of the epitope recognized by antibody; antitag antibodies neutralize variant HIVs in which tags have been introduced at many different locations on the Env spike (102–104). Therefore, although a mechanism may be proposed for any given antibody as above, it may be difficult to unambiguously assert that antibodies of a given specificity neutralize by a specific mechanism.
As might be expected from their much greater potency in neutralization in vitro, the second generation of bnAbs has proven to be much more effective in vivo in monkey (105–109) and humanized mouse models (110–113). Sterilizing immunity has been achieved against SHIVs in the monkey model and against HIV in humanized mice at serum concentrations of a few micrograms per milliliter, in the range that might be achieved through vaccination. Again, serum concentrations of the order of 100 times in vitro neutralizing titer are typically associated with protection, although there is quite some variation (114). There are examples of antibodies that appear to “punch above their weight” by providing protection at relatively low serum neutralizing titers, most notably bnAb 10E8 (115). There is evidence, this time in humanized mouse models, of the importance of Fc receptor binding for antibody protection (110, 116).
The effects of the second generation of bnAbs on established HIV/SHIV infection are much more profound than the first. In humanized mice, rapid escape was again observed from single bnAbs, but cocktails of bnAbs were now able to control virus replication in many animals (110, 111). The results were even more dramatic in the SHIV/macaque model in which a single antibody, PGT121, was able to completely control virus in a number of animals and no neutralization escape was observed (107). Virus control was also observed in an independent study using the bnAbs 3BNC117 and 10-1074 (105). The bnAbs 3BNC117 and VRC01 have now been administered to humans in phase 1 studies and shown evidence of transient viral control (117, 118). There is great interest in pursuing bnAbs in human trials to establish proof-of principle for bnAb-based vaccines (Figure 1), to develop therapies, and to develop regimens of multiple agents to cure HIV infection.
HOW DO BROADLY NEUTRALIZING ANTIBODIES RECOGNIZE THEIR EPITOPES/HOW DO THEY DEFEAT THE VIRAL DEFENSE MECHANISMS?
To defeat HIV defense mechanisms, bnAbs must be able to recognize their epitopes on diverse isolates and be elicitable beginning with a naive human antibody repertoire. Clearly, the two are highly related, but we consider only the former in this section and the latter in a section below. Mature bnAbs face many obstacles in achieving high-affinity binding to Env spikes from multiple different isolates, primarily coping with glycans and amino acid variability close to or within the epitope. Thus glycans can be directly bound in the antibody combining site and contribute to the free energy of antibody binding to the spike. Alternatively, and this is observed frequently, antibodies can accommodate glycans, interacting with them, without this interaction necessarily contributing to the free energy of antibody binding. In fact, in many instances, loss of a given glycan shown to contact antibody in a crystal structure leads to enhanced neutralization of the corresponding virus. Coping with variability to some extent involves positioning antibody amino acid side chains to minimize contact with more variable residues. However, it is also true that the relatively conserved regions of protein surface recognized by bnAbs are often not extremely highly conserved, and the breadth of neutralization achieved to some degree reflects an ability to bind despite variation in that region. For example, the apex bnAbs bind to a lysine-rich region on the V2 loop that is not highly conserved in terms of amino acid sequence, but providing one lysine in particular is conserved and the lysine-rich character is preserved, then bnAb binding and neutralization are generally observed (119). Again, interaction with main chain atoms can also be a mechanism to cope with variability [e.g., PGT128 (96, 120)]. One of the consequences of dealing with glycans and variability is that bnAbs must frequently squeeze into rather narrow openings to access protein epitopes, and this likely explains the preponderance of long HCDR3s in bnAbs.
We now consider each bnAb antigenic region in turn, from the viral membrane to the apex of the trimer (Figures 5 and 6; Table 1). A major problem in the recognition of the gp41 MPER is the proximity of the viral membrane. The primary sequences of MPER bnAbs indicate they have rather long HCDR3s of notable hydrophobic character that may be involved in contacting the membrane and may be responsible for the tendency of this class of bnAbs to show varying degrees of polyreactivity (121, 123). Indeed, recent structures strongly support the notion that lipids are an integral part of the epitope recognized by MPER-specific antibody 4E10 (124). Given that these bnAbs also tend to recognize regions of continuous polypeptide sequence and at least parts of this sequence have been described in some cases in other host proteins, they can show some autoreactive behavior (125–129). The MPER antibodies must negotiate proximity to not only the membrane but also a number of complex glycans on gp41. The potential problems associated with these glycans are that they can be large relative to mannose glycans and they are more heterogeneous than mannose glycans (14). The latter property is probably responsible for the tendency of MPER antibodies to show incomplete neutralization (130, 131), i.e., neutralization of a given isolate by a given antibody that does not reach 100%.
Figure 6.
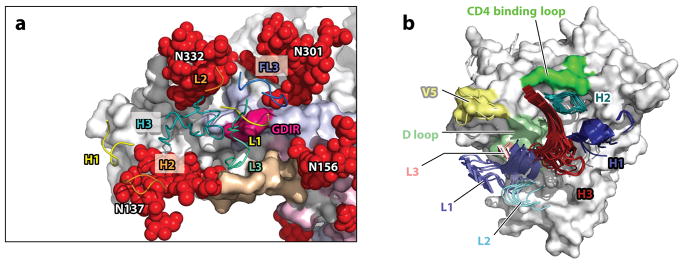
Recognition of Env by bnAbs. (a) Interaction of PGT122 with the high-mannose patch epitope of BG505 SOSIP [PDB 4NCO (22)]. Only CDR1 through 3 of the heavy and light antibody chains are shown, in yellow, orange, and teal, respectively. gp120 is shown in white, gp41 in gray. Glycans are depicted as red spheres, and gp120 hypervariable regions 1 through 2 are shown in tan, pink, and light blue, respectively. Close contact with multiple glycans is illustrated. The protein surface contacted is the GDIR amino acid sequence at the base of the V3 hypervariable loop. (b) Superimposition of the complementarity-determining regions of multiple VRC01-like antibodies bound to gp120 (PDB 4jpv, 4jpw, 4lsp, 4lsq, 4lsr, 4lss, 4lst, 4lsu, and 4lsv adapted from Reference 89). CDR L1 and H1 are indicated in light and dark blue, CDR L2 and H3 in light and dark teal, CDR L3 and H3 in light and dark red. The CD4 binding loop is in bright green, the D loop in pale green, and the V5 loop in yellow.
The gp120-gp41 interface is recognized by a number of bnAbs that interact with gp41 and gp120 to varying degrees (84–86, 98). The bnAb PGT151 is notable in that it reacts with two protomers each of gp120 and gp41 and is trimer and cleavage specific (85, 98). Its unique binding location leads to it stabilizing the Env trimer, a property that has been important in generating trimers from transfected cells for structural studies (A. Ward, personal communication). Again, proximity to gp41 leads to notable incomplete neutralization for the PGT151 family of bnAbs (85, 131).
The CD4bs is recognized by two classes of bnAbs, referred to as VRC01-like antibodies, after the prototype antibody VRC01, and loop binders, because their binding is much more dominated by the HCDR3 loop. Examples of the latter include CH103 (132) and b12 (133). The most potent CD4bs bnAbs belong to the VRC01-like class, and they have a number of common features that allow them to bind tightly to the CD4bs. First, they have a common VH from the VH1-2 family that, with suitable mutations, allows for binding to the CD4bs in a manner very similar to that of the terminal domain of CD4 (87–90). Second, they have VL domains that incorporate a number of critical features, most notably a very short LCDR3 of five amino acids and a number of mutations, including some in a framework region, that accommodate a glycan at N276 that is proximal to the CD4bs (79, 89). With these features, an antibody that is two Ig domains wide at the tip of the Fab arms is able to interact effectively with a site that, for infection, interacts with the CD4 molecule that is one immunoglobulin domain wide at its interacting extremity. Several other glycans are close to the CD4bs and may be obstacles to bnAb binding for any given isolate (glycan fence). However, incomplete neutralization does not seem to be associated with CD4bs bnAbs. Loop binders are not as restricted in gene usage or features as VRC01-like antibodies and are generally not as potent.
The high-mannose patch is found on the side of the outer domain of gp120 in the Env trimer and is so described because it has a preponderance of Man8/Man9 glycans in a dense region of the glycan shield. The region was first revealed by the bnAb 2G12 (55), which uses a unique domain-exchanged structure to recognize a cluster of mannose glycans centered on N332 (57, 134, 135). Later, a series of bnAbs were described that bind to overlapping sites on this region, again centered around N332 (75) but now penetrating through the glycan shield with long HCDR3 regions to access the protein surface, particularly the amino acid sequence GDIR around the base of the V3 loop (83, 96, 97, 136). These bnAbs have quite different angles of approach, and the region has been described as a “supersite of vulnerability” (136). Of note, the interaction by these bnAbs with glycans is complex: Breadth likely comes from the ability to tolerate glycan heterogeneity in some instances (83, 137) and even the ability to use alternate glycans under some circumstances, a phenomenon that has been described as glycan promiscuity (138). A consequence of promiscuity is that several high-mannose patch antibodies can recognize some isolates in the absence of the N332 glycan (138).
Apex bnAbs recognize a conserved lysine-rich region on V2 by penetrating between glycans at N160 of different protomers (74, 99). This mode of recognition requires very long HCDR3 regions (typically >25 amino acid residues) and acidic residues at the tip of the HCDR3 to bind to the basic lysine-rich region. The acidic residues are typically sulfated tyrosines or glutamates. The apex bnAbs generally bind along or close to the trimer threefold axis and are therefore generally dependent on trimer integrity, although some do show reactivity with certain gp120s.
DEVELOPMENT OF BROADLY NEUTRALIZING ANTIBODIES DURING NATURAL INFECTION
The journey from a germline B cell receptor (BCR), consisting of a heavy chain formed by VDJ recombination paired with a light chain formed by VJ recombination, to the corresponding mature HIV bnAb is generally a long one in evolutionary space. The first event might be anticipated as the activation of an appropriate BCR by Env. However, it was noted some time ago that inferred germline versions of bnAbs, sometimes referred to as unmutated common ancestors, often do not bind most Env molecules (139, 140). This has led to speculation about the triggering of bnAbs responses, with clear implications for vaccine design (141–143). One resolution of the apparent anomaly is that the appropriate BCR is triggered by an Env molecule but that this particular Env was not represented in the panel that was screened for reactivity with the germline version of the bnAb. Indeed, in the first study of coevolution of antibody and virus, it was shown that the founder virus Env does bind well to the unmutated common ancestor of the bnAb lineage (132). Alternatively, it is possible that triggering occurs via interaction with a different pathogen (144). Yet again, it should also be recognized that it is not absolutely clear whether there is a lower threshold of affinity (avidity) that can trigger a BCR in vivo, especially if the antigen is presented in a multivalent array (140). Another point of importance is that it appears that long HCDR3s are generated at the stage of VDJ recombination (145), and therefore, given the low frequency of long HCDR3s in naive B cells, the precursors to many bnAbs constitute a rare population in the naive repertoire. Once the appropriate B cell lineage has been activated, then extensive somatic hypermutation (SHM) generally occurs to generate the most potent bnAbs (132, 146, 147). Extensive SHM is frequently accompanied by insertions and deletions that are, in a number of instances, critical for broad neutralization, and breadth has been described to emerge after incorporation of the indel (148).
The first antibody-virus coevolution study focused on a CD4bs lineage of the loop-binding class (132). The lineage, CH103, was detectable as early as 14 weeks after infection, and the evolution of neutralization breadth was concomitant with extensive viral diversification near the CH103 binding site. A mature CH103 antibody isolated at 136 weeks neutralized just over 50% of a global isolate panel with an IC50 of around 5 μg/mL and a somewhat lower level of SHM than typically found among bnAbs (18%, nucleotide), although the antibody does have a 3–amino acid LCDR1 deletion. The theme of bnAbs with somewhat lower breadth and potency than the best bnAbs but lower SHM has also been described for lineage antibodies derived from next-generation sequencing studies of memory B cells from the PGT121 donor (149). For example, for a virus panel neutralized with a median IC50 of 0.03 μg/mL by PGT121 (18% nucleotide mutation in VH), inferred intermediate antibodies of 10% and 16% nt mutation in VH neutralized 40% of viruses with a median IC50 of 0.55 μg/mL and 72% of viruses with a median IC50 of 0.07 μg/mL, respectively. Again though, significant VL indels were present. Nevertheless, broad neutralization at lower levels of SHM is a welcome finding for vaccine design efforts. The second major antibody-virus coevolution study focused on a V2 apex bnAb lineage, CAP256-VRC26 (77). These antibodies had the characteristic features of V2 apex bnAbs, including a very long 35–amino acid anionic HCDR3. The lineage emerged prominently following superinfection of the donor, and again breadth developed concomitantly with extensive diversification of the infecting virus population. Breadth and potency were particularly notable against a panel of clade C isolates but much weaker against clade B isolates.
An interesting development with respect to the CH103 antibody evolution was the discovery of a second lineage, CH235, binding the D loop of the CD4bs (Figure 2), that selected for virus escape variants that resulted in enhanced bnAb lineage envelope binding as well as escape mutant neutralization (150). It was argued that the development of a helper B cell lineage in cooperation with the bnAb lineage might be critical for the development of bnAbs and a vaccine strategy might seek to mimic natural infection by inducing both types of lineage. A cooperative lineage has also been described for the evolution of high-mannose-patch bnAbs in natural infection (151).
BROADLY NEUTRALIZING ANTIBODIES AND VACCINE DESIGN
The structure of the native trimer, the interaction of bnAbs with the native trimer, and the evolution of bnAbs in natural infection likely hold the keys to successful HIV vaccine design aimed at inducing bnAbs. Naively, one might argue that a structure such as the SOSIP trimer that mimics the native trimer should induce bnAbs since it largely expresses bnAb epitopes mostly in the absence of non-neutralizing antibody epitopes. However, this ignores the presence of the relatively highly immunogenic, isolate-specific epitopes; and indeed SOSIP immunization generates autologous but not broadly neutralizing responses (152). Indeed, this mirrors natural infection in which autologous neutralizing antibody responses are elicited relatively early in infection. bnAb responses occur only after the encounter with multiple Env variants, as described above. Taking the lessons from natural infection, one vaccine strategy is to immunize with Envs from critical points along the pathway to bnAbs from an infected individual. Presumably the Envs would be in a configuration as close as possible to native trimers—i.e., some form of well-ordered trimers, although the use of monomeric gp120 has been considered. In this strategy in its purest form, an Env molecule identified from natural infection as binding to a germline precursor of a known bnAb would be used to initiate an appropriate response. One potential problem here is that the strategy relies on a suitable precursor corresponding to the known bnAb being available. A further potential problem is the observation that the opportunities for a given lineage to lose rather than acquire breadth appear to be many (81).
An alternative strategy to one based on mimicking natural infection is to critically examine the interaction of a class of bnAbs with Env at the molecular level and attempt to design a set of immunogens that will shepherd the antibody response between a number of key waypoints. For example, the first stage will be stimulating appropriate germline antibodies that may require the design of germline-triggering molecules such as modified well-ordered trimers or specifically designed molecules. An example of the latter is the eOD-GT8 molecule, which has been designed and selected to bind with moderate to high affinity to a large range of germline versions of VRC01-like CD4bs bnAbs (153). The final stage is likely to be immunization with a well-ordered trimer to mimic as closely as possible the conditions that must be met for antibody recognition of the trimer on virions. Intermediate stages are likely to be required, probably involving cocktails of Envs. We have referred to this type of strategy as reductionist. Clearly, there are likely to be many hybrids of reductionist and B cell lineage approaches.
For all approaches, evaluation of antibody responses in animal models is likely to be crucial. Conventional small animal models such as mice and rabbits are being used in immunization studies (e.g., 152), but they have considerable limitations given the many rare features associated with bnAbs. Thus mice, for example, do not appear to make antibodies with very long HCDR3 regions typically found in many bnAbs. Even monkeys have significantly different V gene repertoires compared to humans and may not respond to immunogens designed with a high degree of precision for humans, such as eOD-GT8. Therefore, mice expressing human antibodies, either as bnAb knock-in mice (125, 127–129, 153, 154) or as mice incorporating human antibody repertoires (155, 156), have become valuable tools. Early studies of various flavors of VRC01-like bnAb knock-in mice have provided promising leads. Knock-in mice with VRC01 germline VH and mouse wild-type VL domains immunized with eOD-GT8 showed activation of the VH1-2 germline, mutation of the VH in a desired direction, and strong selection of VL with a short 5–amino acid CDRL3 (153). No bnAbs were generated as expected. Knock-in mice with 3BNC60 (a VRC01-like antibody), germline VH, and mouse wild-type VL immunized with BG505 SOSIP trimer showed no activation of the germline as expected (157). However, immunization with SOSIP of 3BNC60 knock-in mice with mature VH and mouse wild-type VL did generate a level of bnAbs. Overall, the results support the notion of a germline-triggering molecule to initiate a VRC01-like response and a well-ordered trimer to finish it off. The challenge is to bridge the gap, i.e., to find immunogens that can take the response along the pathway between activation and completion. It should be emphasized that these experiments are done on mice with very restricted repertoires; much more rigorous immunogen evaluation will follow in mice with more complete, diverse human repertoires and ultimately in humans.
Finally, one concern with regard to inducing bnAbs is that, given findings of polyreactivity/ autoreactivity among some bnAbs, tolerance checkpoints may hinder the development of responses (126, 143, 144). Indeed, this may limit pathways to bnAbs in some instances, but one would presume that, in a polyclonal response, solutions avoiding overt polyreactivity/autoreactivity would appear and be selected.
CONCLUSIONS
The bnAb-based HIV vaccine field is entering one of its most exciting phases. The last few years have seen the discovery of many new bnAbs of exceptional potency that define new sites of vulnerability on the Env spike. Now that many bnAbs against most individual sites have been isolated, we have a much clearer view of what is required to recognize each site. Structures of the SOSIP trimer have revealed, for the first time, the conformation of the Env spike on the surface of the virus and the full extent of bnAb epitopes. With these structures, the role of glycans and variability in restricting or curtailing bnAb recognition of the Env spike is much better appreciated. This appreciation has enhanced awareness of the difficulties associated with eliciting bnAb responses through vaccination, but at the same time, we have been provided with data that can help to design effective immunogens and immunization strategies. Further, studies on bnAb-virus coevolution are providing new data that are also being incorporated into vaccine design and strategies. Advances in mechanistic understanding of the germinal center reaction and of Tfh cells hold promise for improved immunization protocols that lead more readily to the higher levels of SHM that appear to be associated with broad neutralization.
The many advances summarized have stimulated the design of many immunogens that are currently being evaluated. These can be loosely grouped into trimer-based and epitope-based immunogens. The SOSIP and other well-ordered trimers form excellent platforms for immunogen engineering. For example, SOSIP molecules can be engineered to bind to germline versions of bnAbs to attempt to initiate appropriate antibody responses or to eliminate induction of strain-specific responses. In the case of the latter, we note the proximity of the strain-specific epitopes to broadly neutralizing epitopes may make this extremely challenging without also curtailing bnAb responses. Epitope-based immunogens include, for example, eOD-GT8 for the CD4bs and peptide-glycan constructs for some of the glycan-dependent bnAb epitopes. Although we do not discuss it here, the presentation of immunogens in a multivalent format, e.g., on nanoparticles, and the choice of adjuvants are likely to be crucial factors in the generation of bnAb responses through immunization.
Finally, finding suitable animal models for evaluating immunogens is critical in developing bnAb-based vaccines given the likely necessity for considerable iteration to improve upon designs and, indeed, to discover optimal immunization strategies. Humanized mouse models are showing promise. Further, simply measuring serum ELISA or neutralizing titers alone following immunization is unlikely to be commensurate with the complexity of the problem being tackled. Rather, detailed analyses of responses, including isolating monoclonal antibodies and next-generation sequencing, are likely to be important contributors to the iterative design process.
Acknowledgments
Supported by the National Institute of Allergy and Infectious Diseases (D.R.B.), the Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (D.R.B. and L.H.), the International AIDS Vaccine Initiative (D.R.B.), the Bill and Melinda Gates Foundation (D.R.B.), and the Ragon Institute of MGH, MIT and Harvard (D.R.B.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol. 2013;31(1):705–42. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 2.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337(6091):183–86. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLOS Pathog. 2006;2(8):e83. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu P, Liu J, Bess J, Chertova E, Lifson JD, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441(7095):847–52. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 5.Brandenberg OF, Magnus C, Rusert P, Regoes RR, Trkola A. Different infectivity of HIV-1 strains is linked to number of envelope trimers required for entry. PLOS Pathog. 2015;11(1):e1004595. doi: 10.1371/journal.ppat.1004595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X, Kurteva S, Ren X, Lee S, Sodroski J. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J Virol. 2005;79(19):12132–47. doi: 10.1128/JVI.79.19.12132-12147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magnus C, Rusert P, Bonhoeffer S, Trkola A, Regoes RR. Estimating the stoichiometry of human immunodeficiency virus entry. J Virol. 2009;83(3):1523–31. doi: 10.1128/JVI.01764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klasse P-J. Modeling how many envelope glycoprotein trimers per virion participate in human immunodeficiency virus infectivity and its neutralization by antibody. Virology. 2007;369(2):245–62. doi: 10.1016/j.virol.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dintzis HM, Dintzis RZ, Vogelstein B. Molecular determinants of immunogenicity: the immunon model of immune response. PNAS. 1976;73(10):3671–75. doi: 10.1073/pnas.73.10.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachmann M, Rohrer U, Kundig T, Hengartner H, Zinkernagel R. The influence of antigen organization on B cell responsiveness. Science. 1993;262(5138):1448–51. doi: 10.1126/science.8248784. [DOI] [PubMed] [Google Scholar]
- 11.Decroly E, Vandenbranden M, Ruysschaert JM, Cogniaux J, Jacob GS, et al. The convertases furin and PC1 can both cleave the human immunodeficiency virus (HIV)-1 envelope glycoprotein gp160 into gp120 (HIV-1 SU) and gp41 (HIV-I TM) J Biol Chem. 1994;269(16):12240–47. [PubMed] [Google Scholar]
- 12.Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull. 2001;58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- 13.Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, et al. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. PNAS. 2010;107(31):13800–5. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek RA, et al. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLOS ONE. 2011;6(8):e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pritchard LK, Vasiljevic S, Ozorowski G, Seabright GE, Cupo A, et al. Structural constraints determine the glycosylation of HIV-1 envelope trimers. Cell Rep. 2015;11(10):1604–13. doi: 10.1016/j.celrep.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poignard P, Moulard M, Golez E, Vivona V, Franti M, et al. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J Virol. 2003;77(1):353–65. doi: 10.1128/JVI.77.1.353-365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore PL, Crooks ET, Porter L, Cayanan CS, Grise H, et al. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol. 2006;80(5):2515–28. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crooks ET, Tong T, Osawa K, Binley JM. Enzyme digests eliminate nonfunctional Env from HIV-1 particle surfaces, leaving native Env trimers intact and viral infectivity unaffected. J Virol. 2011;85(12):5825–39. doi: 10.1128/JVI.00154-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–59. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward AB, Wilson IA. Insights into the trimeric HIV-1 envelope glycoprotein structure. Trends Biochem Sci. 2015;40(2):101–7. doi: 10.1016/j.tibs.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartesaghi A, Merk A, Borgnia MJ, Milne JLS, Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol. 2013;20(12):1352–57. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Julien J-P, Cupo A, Sok D, Stanfield RL, Lyumkis D, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1477–83. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyumkis D, Julien J-P, de Val N, Cupo A, Potter CS, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1484–90. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514(7523):455–61. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Binley JM, Sanders RW, Clas B, Schuelke N, Master A, et al. A recombinant human immuno-deficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74(2):627–43. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76(17):8875–89. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanders RW, Derking R, Cupo A, Julien J-P, Yasmeen A, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLOS Pathog. 2013;9(9):e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, et al. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. PNAS. 2011;108(28):11440–45. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guttman M, Cupo A, Julien J-P, Sanders RW, Wilson IA, et al. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun. 2015;6:6144. doi: 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallo SA, Finnegan CM, Viard M, Raviv Y, Dimitrov A, et al. The HIV Env-mediated fusion reaction. Biochim Biophys Acta. 2003;1614(1):36–50. doi: 10.1016/s0005-2736(03)00161-5. [DOI] [PubMed] [Google Scholar]
- 31.Do Kwon Y, Pancera M, Acharya P, Georgiev IS, Crooks ET, et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol. 2015;22(7):522–31. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz JV, et al. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLOS Pathog. 2010;6(12):e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Lee JH, Ozorowski G, Ward AB. CryoEM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–48. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, et al. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J Virol. 2015;89(6):3380–95. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guenaga J, de Val N, Tran K, Feng Y, Satchwell K, et al. Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. PLOS Pathog. 2015;11(1):e1004570. doi: 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma SK, de Val N, Bale S, Guenaga J, Tran K, et al. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 2015;11(4):539–50. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei X, Decker JM, Wang S, Hui H, Kappes JC, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 37.Rusert P, Krarup A, Magnus C, Brandenberg OF, Weber J, et al. Interaction of the gp120 V1V2 loop with a neighboring gp120 unit shields the HIV envelope trimer against cross-neutralizing antibodies. 2011;208(7):1419–33. doi: 10.1084/jem.20110196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, et al. Vaccine-elicited tier 2 HIV-1 neutralizing antibodies bind to quaternary epitopes involving glycan-deficient patches proximal to the CD4 binding site. PLOS Pathog. 2015;11(5):e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doria-Rose NA, Georgiev I, O’Dell S, Chuang G-Y, Staupe RP, et al. A short segment of the HIV-1 gp120 V1/V2 region is a major determinant of resistance to V1/V2 neutralizing antibodies. J Virol. 2012;86(15):8319–23. doi: 10.1128/JVI.00696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455(7209):109–13. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyerson JR, Tran EEH, Kuybeda O, Chen W, Dimitrov DS, et al. Molecular structures of trimeric HIV-1 Env in complex with small antibody derivatives. PNAS. 2013;110(2):513–18. doi: 10.1073/pnas.1214810110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346(6210):759–63. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol. 2008;82(24):12449–63. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. PNAS. 2003;100(7):4144–49. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rong R, Bibollet-Ruche F, Mulenga J, Allen S, Blackwell JL, Derdeyn CA. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J Virol. 2007;81(3):1350–59. doi: 10.1128/JVI.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore PL, Williamson C, Morris L. Virological features associated with the development of broadly neutralizing antibodies to HIV-1. Trends Microbiol. 2015;23(4):204–11. doi: 10.1016/j.tim.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS. 2014;28(2):163–69. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simek MD, Rida W, Priddy FH, Pung P, Carrow E, et al. Human immunodeficiency virus type 1 elite neutralizers: Individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83(14):7337–48. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, et al. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39(4):758–69. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barbas CF, Bjorling E, Chiodi F, Dunlop N, Cababa D, et al. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. PNAS. 1992;89(19):9339–43. doi: 10.1073/pnas.89.19.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burton D, Pyati J, Koduri R, Sharp S, Thornton G, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266(5187):1024–27. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 52.Buchacher A, Predl R, Tauer C, Purtscher M, Gruber G, et al. Human monoclonal antibodies against gp41 and gp120 as potential agents for passive immunization. In: Brown F, Chanock R, Ginsberg HS, Lerner R, editors. Vaccines ‘92: Modern Approaches to New Vaccines Including Prevention of AIDS. Cold Spring Harbor, NY: Cold Spring Harb. Lab. Press; 1992. pp. 191–94. [Google Scholar]
- 53.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67(11):6642–47. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conley AJ, Kessler JA, Boots LJ, Tung JS, Arnold BA, et al. Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41-2F5, an anti-gp41 human monoclonal antibody. PNAS. 1994;91(8):3348–52. doi: 10.1073/pnas.91.8.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54a.Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75(22):10892–905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trkola A, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70(2):1100–8. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retrovir. 1994;10(4):359–69. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 57.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300(5628):2065–71. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 58.Huber M, Le KM, Doores KJ, Fulton Z, Stanfield RL, et al. Very few substitutions in a germ line antibody are required to initiate significant domain exchange. J Virol. 2010;84(20):10700–7. doi: 10.1128/JVI.01111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poignard P, Sabbe R, Picchio GR, Wang M, Gulizia RJ, et al. Neutralizing antibodies have limited effects on the control of established HIV-1 infection in vivo. Immunity. 1999;10(4):431–38. doi: 10.1016/s1074-7613(00)80043-6. [DOI] [PubMed] [Google Scholar]
- 60.Trkola A, Kuster H, Rusert P, Joos B, Fischer M, et al. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat Med. 2005;11(6):615–22. doi: 10.1038/nm1244. [DOI] [PubMed] [Google Scholar]
- 61.Baba TW, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, et al. Human neutralizing mono-clonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000;6(2):200–6. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 62.Shibata R, Igarashi T, Haigwood N, Buckler-White A, Ogert R, et al. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat Med. 1999;5(2):204–10. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 63.Hessell AJ, Rakasz EG, Tehrani DM, Huber M, Weisgrau KL, et al. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J Virol. 2010;84(3):1302–13. doi: 10.1128/JVI.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parren PWHI, Marx PA, Hessell AJ, Luckay A, Harouse J, et al. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J Virol. 2001;75(17):8340–47. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6(2):207–10. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 66.Hessell AJ, Hangartner L, Hunter M, Havenith CEG, Beurskens FJ, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449(7158):101–4. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 67.Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15(8):951–54. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nishimura Y, Igarashi T, Haigwood N, Sadjadpour R, Plishka RJ, et al. Determination of a statistically valid neutralization titer in plasma that confers protection against simian-human immunodeficiency virus challenge following passive transfer of high-titered neutralizing antibodies. J Virol. 2002;76(5):2123–30. doi: 10.1128/jvi.76.5.2123-2130.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gauduin M-C, Parren PWHI, Weir R, Barbas CF, Burton DR, Koup RA. Passive immunization with a human monoclonal antibody protects hu-PBL-SCID mice against challenge by primary isolates of HIV-1. Nat Med. 1997;3(12):1389–93. doi: 10.1038/nm1297-1389. [DOI] [PubMed] [Google Scholar]
- 70.Parren PWHI, Ditzel HJ, Gulizia RJ, Binley JM, Barbas CF, III, et al. Protection against HIV-1 infection in hu-PBL-SCID mice by passive immunization with a neutralizing human monoclonal antibody against the gp120 CD4-binding site. AIDS. 1995;9(6):F1–6. doi: 10.1097/00002030-199506000-00001. [DOI] [PubMed] [Google Scholar]
- 71.Poignard P, Moldt B, Maloveste K, Campos N, Olson WC, et al. Protection against high-dose highly pathogenic mucosal SIV challenge at very low serum neutralizing titers of the antibody-like molecule CD4-IgG2. PLOS ONE. 2012;7(7):e42209. doi: 10.1371/journal.pone.0042209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLOS Pathog. 2009;5(5):e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moldt B, Shibata-Koyama M, Rakasz EG, Schultz N, Kanda Y, et al. A nonfucosylated variant of the anti-HIV-1 monoclonal antibody b12 has enhanced FcγRIIIa-mediated antiviral activity in vitro but does not improve protection against mucosal SHIV challenge in macaques. J Virol. 2012;86(11):6189–96. doi: 10.1128/JVI.00491-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walker LM, Chan-Hui PY, Wagner D, Phung P, Goss JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–89. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477(7365):466–70. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Falkowska E, Ramos A, Feng Y, Moquin S, Walker LM, et al. PGV04, an HIV-1 gp120 CD4 binding site antibody, is broad and potent in neutralization but does not induce conformational changes characteristic of CD4. J Virol. 2012;86(8):4394–403. doi: 10.1128/JVI.06973-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509(7498):55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491(7424):406–12. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329(5993):856–61. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458(7238):636–40. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 81.Sok D, van Gils MJ, Pauthner M, Julien J-P, Saye-Francisco KL, et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. PNAS. 2014;111(49):17624–29. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Balla-Jhagjhoorsingh SS, Corti D, Heyndrickx L, Willems E, Vereecken K, et al. The N276 glycosylation site is required for HIV-1 neutralization by the CD4 binding site specific HJ16 monoclonal antibody. PLOS ONE. 2013;8(7):e68863. doi: 10.1371/journal.pone.0068863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. PNAS. 2012;109(47):E3268–77. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang J, Kang BH, Pancera M, Lee JH, Tong T, et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515(7525):138–42. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40(5):657–68. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Scharf L, Scheid JF, Lee JH, West AP, Chen C, et al. Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell Rep. 2014;7(3):785–95. doi: 10.1016/j.celrep.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333(6049):1633–37. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329(5993):811–17. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou T, Zhu J, Wu X, Moquin S, Zhang B, et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39(2):245–58. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou T, Lynch RM, Chen L, Acharya P, Wu X, et al. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell. 2015;161(6):1280–92. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roben P, Moore JP, Thali M, Sodroski J, Barbas CF, Burton DR. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68(8):4821–28. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sattentau QJ. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J Exp Med. 1995;182(1):185–96. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ugolini S, Mondor I, Parren PW, Burton DR, Tilley SA, et al. Inhibition of virus attachment to CD4+ target cells is a major mechanism of T cell line-adapted HIV-1 neutralization. J Exp Med. 1997;186(8):1287–98. doi: 10.1084/jem.186.8.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Platt EJ, Gomes MM, Kabat D. Kinetic mechanism for HIV-1 neutralization by antibody 2G12 entails reversible glycan binding that slows cell entry. PNAS. 2012;109(20):7829–34. doi: 10.1073/pnas.1109728109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ruprecht CR, Krarup A, Reynell L, Mann AM, Brandenberg OF, et al. MPER-specific antibodies induce gp120 shedding and irreversibly neutralize HIV-1. J Exp Med. 2011;208(3):439–54. doi: 10.1084/jem.20101907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang P-S, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334(6059):1097–103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Julien J-P, Sok D, Khayat R, Lee JH, Doores KJ, et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLOS Pathog. 2013;9(5):e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40(5):669–80. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McLellan JS, Pancera M, Carrico C, Gorman J, Julien J-P, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480(7377):336–43. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Frey G, Peng H, Rits-Volloch S, Morelli M, Cheng Y, Chen B. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. PNAS. 2008;105(10):3739–44. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chakrabarti BK, Walker LM, Guenaga JF, Ghobbeh A, Burton DR, Wyatt RT. Direct antibody access to the HIV-1 membrane-proximal external region positively correlates with neutralization sensitivity. J Virol. 2011;85(16):8217–26. doi: 10.1128/JVI.00756-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang X, Lipchina I, Cocklin S, Chaiken I, Sodroski J. Antibody binding is a dominant determinant of the efficiency of human immunodeficiency virus type 1 neutralization. J Virol. 2006;80(22):11404–8. doi: 10.1128/JVI.01102-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Laird ME, Desrosiers RC. Infectivity and neutralization of simian immunodeficiency virus with FLAG epitope insertion in gp120 variable loops. J Virol. 2007;81(20):10838–48. doi: 10.1128/JVI.00831-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pantophlet R, Wang M, Aguilar-Sino RO, Burton DR. The human immunodeficiency virus type 1 envelope spike of primary viruses can suppress antibody access to variable regions. 2009;83(4):1649–59. doi: 10.1128/JVI.02046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013;503(7475):277–80. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Moldt B, Rakasz EG, Schultz N, Chan-Hui P-Y, Swiderek K, et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. PNAS. 2012;109(46):18921–25. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503(7475):224–28. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Saunders KO, Wang L, Joyce MG, Yang Z-Y, Balazs AB, et al. Broadly neutralizing human immunodeficiency virus type 1 antibody gene transfer protects non-human primates from mucosal simian-human immunodeficiency virus infection. J Virol. 2015;89(16):8334–45. doi: 10.1128/JVI.00908-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shingai M, Donau OK, Plishka RJ, Buckler-White A, Mascola JR, et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med. 2014;211(10):2061–74. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Halper-Stromberg A, Lu C-L, Klein F, Horwitz JA, Bournazos S, et al. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell. 2014;158(5):989–99. doi: 10.1016/j.cell.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012;492(7427):118–22. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gruell H, Bournazos S, Ravetch JV, Ploss A, Nussenzweig MC, Pietzsch J. Antibody and antiretroviral preexposure prophylaxis prevent cervicovaginal HIV-1 infection in a transgenic mouse model. J Virol. 2013;87(15):8535–44. doi: 10.1128/JVI.00868-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pietzsch J, Gruell H, Bournazos S, Donovan BM, Klein F, et al. A mouse model for HIV-1 entry. PNAS. 2012;109(39):15859–64. doi: 10.1073/pnas.1213409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol. 2015;16(6):571–76. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pegu A, Boyington JC, Ko S-Y, Schmidt SD, McKee K, et al. Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor. Sci Transl Med. 2014;6(243):243ra88. doi: 10.1126/scitranslmed.3008992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bournazos S, Klein F, Pietzsch J, Seaman MS, Nussenzweig MC, Ravetch JV. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell. 2014;158(6):1243–53. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Caskey M, Klein F, Lorenzi JCC, Seaman MS, West AP, et al. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature. 2015;522(7557):487–91. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wadman M. HIV trial under scrutiny. Nature. 2013;17:279–80. doi: 10.1038/493279a. [DOI] [PubMed] [Google Scholar]
- 119.Andrabi R, Voss JE, Liang C-H, Briney B, McCoy LE, et al. Identification of common features in prototype broadly neutralizing antibodies to HIV envelope V2 apex to facilitate vaccine design. Immunity. 2015;43(5):959–73. doi: 10.1016/j.immuni.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Garces F, Sok D, Kong L, McBride R, Kim HJ, et al. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell. 2014;159(1):69–79. doi: 10.1016/j.cell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467(7315):591–95. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deleted in proof
- 123.Haynes BF, Moody MA, Verkoczy L, Kelsoe G, Alam SM. Antibody polyspecificity and neutralization of HIV-1: a hypothesis. Hum Antibodies. 2005;14(3–4):59–67. [PMC free article] [PubMed] [Google Scholar]
- 124.Irimia A, Sarkar A, Stanfield RL, Wilson IA. Crystallographic identification of lipid as an integral component of the epitope of HIV broadly neutralizing antibody 4E10. Immunity. 2016;44(1):21–31. doi: 10.1016/j.immuni.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Doyle-Cooper C, Hudson KE, Cooper AB, Ota T, Skog P, et al. Immune tolerance negatively regulates B cells in knock-in mice expressing broadly neutralizing HIV antibody 4E10. J Immunol. 2013;191(6):3186–91. doi: 10.4049/jimmunol.1301285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu M, Yang G, Wiehe K, Nicely NI, Vandergrift NA, et al. Polyreactivity and autoreactivity among HIV-1 antibodies. J Virol. 2015;89(1):784–98. doi: 10.1128/JVI.02378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Verkoczy L, Diaz M, Holl TM, Ouyang Y-B, Bouton-Verville H, et al. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. PNAS. 2010;107(1):181–86. doi: 10.1073/pnas.0912914107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Verkoczy L, Chen Y, Zhang J, Bouton-Verville H, Newman A, et al. Induction of HIV-1 broad neutralizing antibodies in 2F5 knock-in mice: Selection against membrane proximal external region-associated autoreactivity limits T-dependent responses. J Immunol. 2013;191(5):2538–50. doi: 10.4049/jimmunol.1300971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Verkoczy L, Chen Y, Bouton-Verville H, Zhang J, Diaz M, et al. Rescue of HIV-1 broad neutralizing antibody-expressing B cells in 2F5 VH × VL knockin mice reveals multiple tolerance controls. J Immunol. 2011;187(7):3785–97. doi: 10.4049/jimmunol.1101633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kim AS, Leaman DP, Zwick MB. Antibody to gp41 MPER alters functional properties of HIV-1 Env without complete neutralization. PLOS Pathog. 2014;10(7):e1004271. doi: 10.1371/journal.ppat.1004271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McCoy LE, Falkowska E, Doores KJ, Le K, Sok D, et al. Incomplete neutralization and deviation from sigmoidal neutralization curves for HIV broadly neutralizing monoclonal antibodies. PLOS Pathog. 2015;11(8):e1005110. doi: 10.1371/journal.ppat.1005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liao H-X, Lynch R, Gao F, Alam SM, Boyd SD, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496(7446):469–76. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445(7129):732–37. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, et al. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J Virol. 2002;76(14):7306–21. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Murin CD, Julien J-P, Sok D, Stanfield RL, Khayat R, et al. Structure of 2G12 Fab2 in complex with soluble and fully glycosylated HIV-1 Env by negative-stain single-particle electron microscopy. J Virol. 2014;88(17):10177–88. doi: 10.1128/JVI.01229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kong L, Lee JH, Doores KJ, Murin CD, Julien J-P, et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol. 2013;20(7):796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pritchard LK, Spencer DIR, Royle L, Bonomelli C, Seabright GE, et al. Glycan clustering stabilizes the mannose patch of HIV-1 and preserves vulnerability to broadly neutralizing antibodies. Nat Commun. 2015;6:7479. doi: 10.1038/ncomms8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sok D, Doores KJ, Briney B, Le KM, Saye-Francisco KL, et al. Promiscuous glycan site recognition by antibodies to the high-mannose patch of gp120 broadens neutralization of HIV. Sci Transl Med. 2014;6(236):236ra63. doi: 10.1126/scitranslmed.3008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xiao X, Chen W, Feng Y, Zhu Z, Prabakaran P, et al. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun. 2009;390(3):404–9. doi: 10.1016/j.bbrc.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ota T, Doyle-Cooper C, Cooper AB, Huber M, Falkowska E, et al. Anti-HIV B cell lines as candidate vaccine biosensors. J Immunol. 2012;189(10):4816–24. doi: 10.4049/jimmunol.1202165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kulp DW, Schief WR. Advances in structure-based vaccine design. Curr Opin Virol. 2013;3(3):322–31. doi: 10.1016/j.coviro.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stamatatos L. HIV vaccine design: the neutralizing antibody conundrum. Curr Opin Immunol. 2012;24(3):316–23. doi: 10.1016/j.coi.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 143.Haynes BF. New approaches to HIV vaccine development. Curr Opin Immunol. 2015;35:39–47. doi: 10.1016/j.coi.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Trama AM, Moody MA, Alam SM, Jaeger FH, Lockwood B, et al. HIV-1 envelope gp41 antibodies can originate from terminal ileum B cells that share cross-reactivity with commensal bacteria. Cell Host Microbe. 2014;16(2):215–26. doi: 10.1016/j.chom.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Briney BS, Willis JR, Crowe JE. Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes. PLOS ONE. 2012;7(5):e36750. doi: 10.1371/journal.pone.0036750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang Z, Schramm CA, Joyce MG, Do Kwon Y, Sheng Z, et al. Maturation and diversity of the VRC01-antibody lineage over 15 years of chronic HIV-1 infection. Cell. 2015;161(3):470–85. doi: 10.1016/j.cell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153(1):126–38. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kepler TB, Liao H-X, Alam SM, Bhaskarabhatla R, Zhang R, et al. Immunoglobulin gene insertions and deletions in the affinity maturation of HIV-1 broadly reactive neutralizing antibodies. Cell Host Microbe. 2014;16(3):304–13. doi: 10.1016/j.chom.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sok D, Laserson U, Laserson J, Liu Y, Vigneault F, et al. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLOS Pathog. 2013;9(11):e1003754. doi: 10.1371/journal.ppat.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gao F, Bonsignori M, Liao H-X, Kumar A, Xia S-M, et al. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell. 2014;158(3):481–91. doi: 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Haynes B. Ontogeny of broadly neutralizing antibodies during HIV infection. Presented at HIV Vaccines (X5) Keyst. Symp. Mol. Cell. Biol; Banff, Can. 2015. [Google Scholar]
- 152.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, et al. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349(6244):aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, et al. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349(6244):156–61. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ota T, Doyle-Cooper C, Cooper AB, Doores KJ, Aoki-Ota M, et al. B cells from knock-in mice expressing broadly neutralizing HIV antibody b12 carry an innocuous B cell receptor responsive to HIV vaccine candidates. J Immunol. 2013;191(6):3179–85. doi: 10.4049/jimmunol.1301283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pascal KE, Coleman CM, Mujica AO, Kamat V, Badithe A, et al. Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. PNAS. 2015;112(28):8738–43. doi: 10.1073/pnas.1510830112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee E-C, Liang Q, Ali H, Bayliss L, Beasley A, et al. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery. Nat Biotechnol. 2014;32(4):356–63. doi: 10.1038/nbt.2825. [DOI] [PubMed] [Google Scholar]
- 157.Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, et al. Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell. 2015;161(7):1505–15. doi: 10.1016/j.cell.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Klein F, Gaebler C, Mouquet H, Sather DN, Lehmann C, et al. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. J Exp Med. 2012;209(8):1469–79. doi: 10.1084/jem.20120423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rudicell RS, Kwon YD, Ko S-Y, Pegu A, Louder MK, et al. Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J Virol. 2014;88:12669–82. doi: 10.1128/JVI.02213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pantophlet R, Ollmann Saphire E, Poignard P, Parren PHI, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J Virol. 2003;77:642–58. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]