Abstract
Background/Aims
Probiotics are expected to modify the composition of gut microbiota. We aimed to investigate the changes in the composition and diversity of gut microbiota by the administration of probiotics in healthy individuals.
Methods
Twelve healthy volunteers with age range of 30–42 years provided baseline fecal samples. Subsequently, they took commercially available probiotic capsules (a mixture for Bifidobacterium, Lactobacillus, and Enterococcus) for 4 weeks. Fecal samples were collected at 4 weeks of administration and 2 weeks after the stop of administration. Fecal microbiota was analyzed via 16S ribosomal RNA gene sequencing.
Results
The mean Shannon index was not significantly altered by the 4-week administration of probiotics (4.365 vs 4.556, P > 0.05). The proportion of Bacteroidetes, Actinobacteria, Firmicutes, and Proteobacteria was not significantly changed by the 4-week administration of probiotics. At the genus level, the proportions of Lactobacillus (2.138% vs 2.773%, P = 0.028) and Enterococcus (0.022% vs 2.758%, P = 0.004) significantly increased 4 weeks after the administration of probiotics, but reduced 2 weeks after the stop of administration (2.773% vs 3.292%, P = 0.064 and 2.758% vs 0.001%, P = 0.001).
Conclusions
The diversity of fecal microbiota is not significantly affected by 4 weeks of probiotics administration. The proportion of fecal microbiota at the genus level is significantly altered by the administration of probiotics. However, this effect does not seem to last long, probably because of homeostasis or dietary influence.
Keywords: Composition, Diversity, Fecal microbiota, Probiotics
Introduction
A lot of microorganisms live in the lower intestine. These microbes are known to be critical for host physiology and constitute the ecosystem within the intestinal tract. The intestinal microbiota comprises hundreds of bacterial species and approximately 1014 organisms/g of feces.1–4 The intestinal microbiota affects the health of the host through various mechanisms. The microorganisms are considered to play a role in digesting food, blocking pathogens, synthesizing nutrients, detoxifying potential carcinogens, and maintaining mucosal and systemic immunity.5–7
The intestinal microbiota has a close relationship with the mucosal immune function of the gastrointestinal (GI) tract.8 In healthy individuals, intestinal microbes ferment undigested food to supply nutrients and energy to the host and maintain immune homeostasis. The composition of the intestinal microbiota changes after events such as diarrhea or antibiotic administration.9,10 Dysbiosis of the intestinal microbiota alters the host immune response and causes symptoms or diseases. Studies have revealed that the composition or diversity of the intestinal microbiota in patients with irritable bowel syndrome (IBS) and inflammatory bowel disease (IBD) is different from that of healthy individuals.11–14
Probiotics are live microorganisms that are believed to influence gut microbiota. Probiotics are used for maintaining health or treating diseases such as IBS and IBD. Data regarding the effect of probiotics are still limited. However, probiotics are presumed to be able to modify the composition of the intestinal microbiota in disease-related disequilibrium. Some studies have also suggested that probiotics have a preventive or therapeutic role in the treatment of T-helper 17 cells-related diseases such as IBD and immune-related diseases.8,15,16 Probiotics are popular health supplements in the general population. However, to our knowledge, the influence of probiotics on the diversity or composition of gut microbiota has not been fully evaluated. The duration of the effects of probiotics administration is still unclear.
Therefore, we investigated the changes in the composition and diversity of gut microbiota by the administration of probiotics in healthy individuals.
Materials and Methods
Study Participants
Twelve healthy volunteers who did not have organic diseases, systemic diseases, or chronically recurrent abdominal symptoms participated in the present study. The exclusion criteria included past history of medical diseases, use of medication (including probiotics) in the last 6 months, and recurrent abdominal symptoms in the last 3 months. During the study period, the routine lifestyles of the participants were checked daily, and activities that could affect gut microbiota, including the change of eating habits and alcohol consumption, were not allowed. The study protocol was approved by the institutional review board of Ajou University Hospital (AJIRB-BMR-SMP-16-211). All participants provided written informed consent.
Study Design
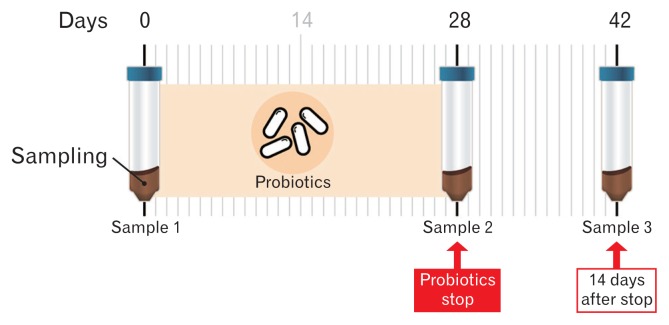
Twelve healthy volunteers took 2 capsules of a commercially available probiotic (Duolac; Cell Biotech Co, Ltd, Gimpo, Korea) daily for 4 weeks. Duolac is a probiotic mixture containing 3 strains of Bifidobacterium (B. brevis, B. lactis, and B. longum), 3 strains of Lactobacillus (L. acidophilus, L. plantarum, and L. rhamnosus), and 1 strain of Enterococcus (E. faecium). Each capsule contains 5 billion bacteria (approximately 700 million bacteria for each strain). Used probiotic mixture is double-coated with peptides and polysaccharides. This protective technology can protect the ingested bacteria from unfavorable conditions such as low pH, protease-rich conditions, and bile acid exposure. The polypeptide acts in a pH-dependent way, and coated cells remain mostly intact at low pH conditions (pH 4.0 in the stomach). The cells start getting released from the coats at pH 6.0 and are totally released in the intestine (pH 7.0). Therefore, the ingested coated bacteria can successfully reach the intestine alive and perform their functions. The subjects took the provided probiotics 30 minutes after a meal with water, twice a day (1 capsule: 500 mg) for 4 weeks. Participants provided baseline fecal samples (Sample 1) at the start of the experiment. Subsequently, they took the probiotic capsules for 4 weeks. Fecal samples were collected at 4 weeks (Sample 2) after the start of probiotics administration. Additional fecal samples were collected at 2 weeks (Sample 3) after the stop of probiotics administration (Fig. 1).
Figure 1.
The protocol of the study. Twelve healthy volunteers participated in the study and provided fecal samples 3 times.
The sampling process to all the enrolled subjects for adequate fecal sampling without contamination. The process was as follows: subjects were educated to try urinating before attaching the paper towel, that we provided, to the toilet. This was followed by washing hands with a cleansing agent. After the adhesive tape was removed from the paper, it was to be stuck in a shape such that it hung in the toilet. The stool sample was to be inserted using the spoon provided in the tube. Finally, the paper was removed (the paper melts in water). Fecal samples were collected in a Falcon tube and stored at −20°C, and then transferred to −80°C within 24 hours. For measurement of fecal calprotectin levels, additional fecal samples were collected in a stool box, stored at 1°C, and then transferred to the analysis center.
DNA Extraction and Sequencing
The extraction method for bacterial DNA was performed using a PowerMax Soil DNA Isolation Kit (Mo Bio, San Diego, CA, USA). Each sequenced sample was prepared according to the Illumina 16S Metagenomic Sequencing Library protocols to amplify the V3 and V4 region (519F-806R). The DNA quality was measured by PicoGreen and Nanodrop (Thermo Fisher Scientific, Inc, Pittsburgh, PA, USA). Input genomic DNA (10 ng) was polymerase chain reaction (PCR) amplified. The barcoded fusion primers 519F (5′-CCTACGGGNGGCWGCAG-3′) and 816R (5′-GACTACHVGGGTATCTAATCC-3′) were used for amplification. The final purified product was then quantified using quantitative PCR (qPCR) according to the qPCR Quantification Protocol Guide (KAPA Library Quantification kits for Illumina Sequencing platforms), and the quality was checked using the Lab-Chip GX HT DNA High Sensitivity Kit (Perkin Elmer, Hopkinton, MA, USA). Next, paired-end (2 × 300 bp) sequencing was performed on the MiSeq platform (Illumina, San Diego, CA, USA).
Operational Taxonomic Unit Analysis
After sequencing of the MiSeq raw data of 36 samples, a FASTQ file was created using bcl2fastq (v1.8.4), and the PhiX sequence was removed with Burrows-Wheeler Aligner (BWA). The paired-end data separated by each sample were extracted using fast length adjustment of short reads (FLASH 1.2.11), and only high-quality sequences with 120–160 bp overlaps and a final length of 440–460 bp were selected. Using a cluster database at high identity with tolerance operational taxonomic unit (OUT), the obtained sequence was clustered with sequences with > 97% similarity after eliminating low-quality, ambiguous, and chimeric sequences, which were regarded as sequencing errors.17,18
A representative sequence from each OTU was used for basic local alignment search tool (BLASTN v2.4.0). Nucleotide analysis in a reference database (National Center for Biotechnology Information 16S, Maryland, USA) and taxonomic assignment were performed based on sequence similarity. When the query coverage of the best hit that matched the database was < 85% and the identity of the matched area was < 85%, the taxonomy was not defined.
Diversity Analysis
A comparative analysis of various microbial communities was performed using quantitative insights into microbial ecology (QI-IME v1.8) with OTU abundance and taxonomy information. The Shannon Index and Inversed Simpson Index were determined to confirm species diversity and the uniformity of microbial communities in samples.19
Statistical Methods
The Wilcoxon-signed rank test (R software program v3.1.2), a non-parametric test, was performed to compare the proportion of fecal microbiota at the phylum, genus, and species.20 The differences in the relative abundance of microbes before, during, and after probiotic use were visualized using heat maps (log application). P < 0.05 was considered to be statistically significant.
Results
Baseline Characteristics
The baseline characteristics of the 12 healthy volunteers (8 males and 4 females) are described in Table. The median age was 32 years with range of 30–42 years. The body mass index was 23.01 ± 2.94 kg/m2. A rarefaction model was used to evaluate the species richness of the sampling data. The results revealed that appropriate sampling had been performed in all samples.
Table.
Characteristics of the 12 Study Participants
| Parameters | |
|---|---|
| Sex (n [%]) | |
| Male | 8 (66.7) |
| Female | 4 (33.3) |
| Age (yr) | |
| Median | 32 |
| Range | 30–42 |
| BMI (kg/m2) | |
| Mean ± SD | 23.01 ± 2.94 |
| Smoking history, n (%) | |
| None | 10 (83.3) |
| Past | 0 (0.0) |
| Current | 2 (16.7) |
| Alcohol history (n [%]) | |
| None | 2 (16.7) |
| Social | 10 (83.3) |
| More than twice a week | 0 (0.0) |
BMI, body mass index.
The Diversity of Fecal Microbiota
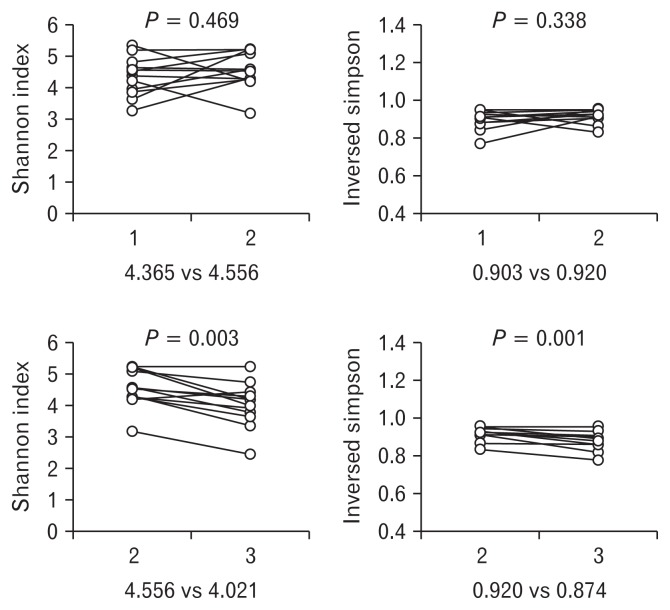
The Shannon index and inverse Simpson index were not significantly changed by the administration of probiotics, compared with the baseline fecal samples (4.365 [Sample 1] vs 4.556 [Sample 2], P = 0.469 and 0.903 [Sample 1] vs 0.920 [Sample 2], P = 0.338, respectively). Both indexes significantly decreased 2 weeks after the stop of administration (4.556 [Sample 2] vs 4.201 [Sample 3], P = 0.003 and 0.920 [Sample 2] vs 0.874 [Sample 3], P = 0.001) (Fig. 2).
Figure 2.
Shannon index and inversed Simpson index representing overall taxa diversity. The Both indexes were not significantly changed by the administration of probiotics, but significantly decreased 2 weeks after the stop of administration.
The Composition of Fecal Microbiota at the Phylum Level
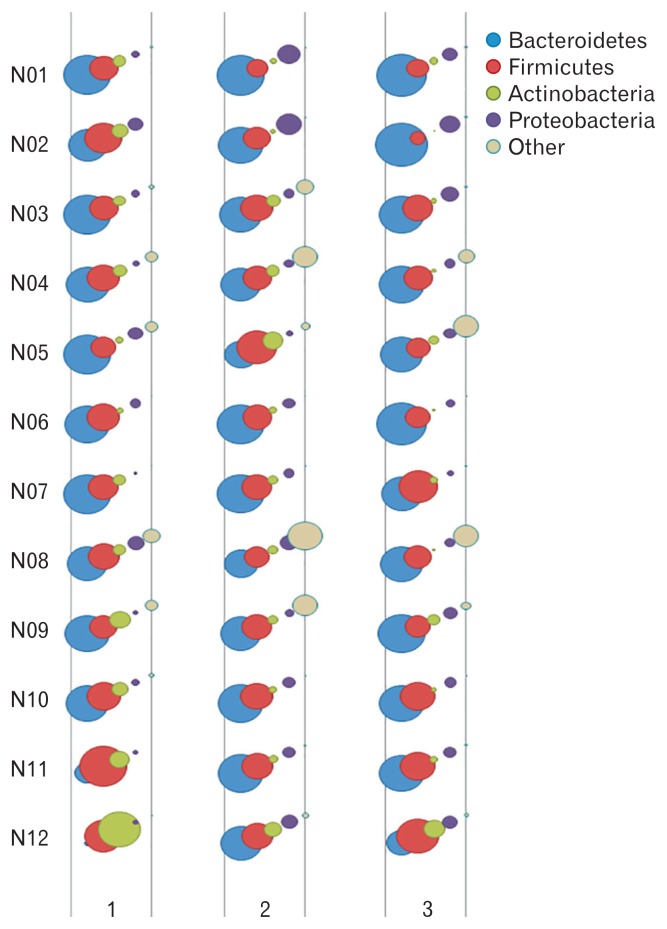
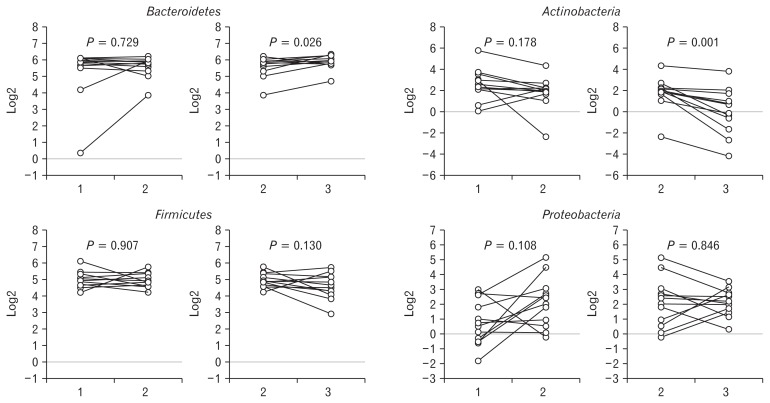
Figure 3 illustrated a graphical bubble chart showing the change in the proportion of fecal microbiota at the phylum level in all subjects. The top 4 phyla were Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria. The composition of the 4 phyla were not significantly changed by the administration of probiotics, compared with the baseline fecal samples (Bacteroidetes: 51.8% vs 51.9%, P = 0.729; Firmicutes: 33.1% vs 31.7%, P = 0.907; Actinobacteria: 10.1% vs 5.1%, P = 0.178; Proteobacteria: 2.7% vs 8.1%, P = 0.108). The proportion of Bacteroidetes significantly increased (51.9% vs 61.1%, P = 0.026) and that of Actinobacteria significantly decreased (5.1% vs 2.4%, P = 0.001) 2 weeks after the stop of administration (Fig. 4).
Figure 3.
A graphical bubble chart showing the change in the proportion of fecal microbiota at the phylum level in all subjects. The top 4 phyla were Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria.
Figure 4.
The composition of fecal microbiota at the phylum level. The composition of Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria was not significantly changed by the administration of probiotics. However, the proportion of Bacteroidetes significantly increased and that of Actinobacteria significantly decreased 2 weeks after the stop of administration.
The Composition of Fecal Microbiota at the Genus Level
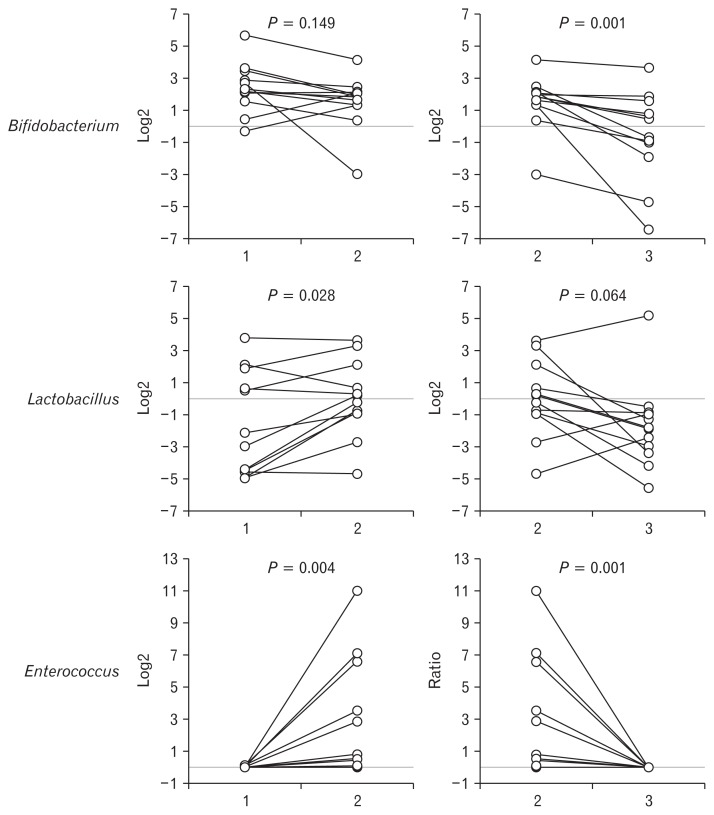
The probiotics used in the present study were composed of Bifidobacterium, Lactobacillus, and Enterococcus. The proportion of Bifidobacterium did not significantly differ between Samples 1 and 2 (9.4% vs 4.4%, P = 0.149). The proportions of Lactobacillus (2.1% vs 2.8%, P = 0.028) and Enterococcus (0.0% vs 2.7%, P = 0.004) significantly increased in Sample 2, compared with Sample 1. The proportions of Bifidobacterium (4.4% vs 2.1%, P = 0.001), Lactobacillus (2.7% vs 3.3%, P = 0.064), and Enterococcus (2.7% vs 0.0%, P = 0.001) significantly decreased or tended to decrease in Sample 3, compared with Sample 2 (Fig. 5).
Figure 5.
The composition of fecal microbiota at the genus level. The proportions of Lactobacillus and Enterococcus significantly increased in Sample 2, compared with Sample 1. The proportions of Bifidobacterium, Lactobacillus, and Enterococcus significantly decreased or tended to decrease in Sample 3, compared with Sample 2.
Fecal Calprotectin Levels
There was no significant difference in the level of fecal calprotectin between Samples 1 and 2 (18.9 ± 16.9 μg/g vs 17.8 ± 22.0 μg/g of feces, P = 0.893), and between Samples 2 and 3 (17.8 ± 22.0 μg/g vs 15.3 ± 8.7 μg/g of feces, P = 0.917).
Discussion
This study was conducted to investigate the change in the composition and diversity of fecal microbiota before, during, and after the administration of probiotics in healthy individuals. Nowadays, probiotics are commonly used for improving the general health, preventing or treating diseases. It is generally believed that probiotics affect the composition and diversity of gut microbiota. However, scientific evidence showing the effect of commercially available probiotics on the intestinal microflora is lacking. In the present study, changes in the diversity and composition of fecal microbiota associated with probiotic administration were serially analyzed in healthy individuals. The main observations of the current study were: (1) the overall diversity of fecal microbiota was not significantly altered by the 4-week administration of probiotics, but significantly decreased after the stop of administration, (2) the composition of fecal microbiota was not significantly changed by the 4-week administration of probiotics at the phylum level, but the proportions of Bacteroidetes and Actinobacteria significantly increased and decreased, respectively, 2 weeks after the stop of administration, (3) the proportions of Lactobacillus and Enterococcus were significantly increased by the 4-week administration of probiotics, but the proportions of Bifidobacterium, Lactobacillus, and Enterococcus decreased 2 weeks after the stop of administration, and (4) fecal calprotectin levels were not significantly affected by the administration of probiotics.
Several factors influence the composition of intestinal microbiota, such as diet, host genetics, diseases, and medications. Indigestible components in the food, for example fiber, provide the substrates for intestinal microbiota.21 Actually, studies have shown that the human intestinal microbiota is affected by diet, lifestyle, and environment.22 Therefore, the appropriate control of those factors is necessary for the study of intestinal microbiota. In the present study, we minimized the influence of diet, host genetics, diseasess, medications, and environmental factors on fecal microbiota. All participants were ethnically homogeneous Koreans and healthy without diseases. Their age, working place, job, routine activities, and lifestyles were similar. Furthermore, medications, alcohol drinking, smoking, eating habits, and working time were checked and controlled during the study period.
Whether or not fecal samples can reflect the entire microbial community in the intestine has been the subject of debate. Lyra et al23 reported that fecal samples do not reflect the bacterial counts in the intestinal mucosa. However, biopsy for investigating the microbiota of the intestinal mucosa requires adequate bowel preparation for colonoscopy, which may alter the microbiome composition. In addition, biopsy is an invasive method that can damage the intestinal mucosa. Brush sampling has been devised to overcome these problems of biopsy. It covers a larger surface area, and is less traumatic to the colonic epithelium.24 However, bowel preparation is usually necessary for brush sampling. Accordingly, fecal samples are considered to be the easiest method to analyze intestinal microbiota, and have been used in many studies.
Probiotics have been used to control symptoms in various GI diseases or to prevent some diseases. Studies have been reported the effect of probiotics on several conditions such as infectious diarrhea, IBS, and IBD.25–27 Furthermore, healthy individuals take probiotics for improving general health or preventing diseases. However, guidelines regarding the strains, duration, and dosage of probiotics for those kinds of purposes are not clear. In the present study, we demonstrated that the proportion of a part of strains administered by probiotics in fecal microbiota significantly increased. When considering that the proportion of these strains significantly decreased 2 weeks after the stop of administration, this effect does not seem to last long.
The probiotics used in the present study are composed of 3 strains of Bifidobacterium (B. brevis, B. lactis, and B. longum), 3 strains of Lactobacillus (L. acidophilus, L. plantarum, and L. rhamnosus), and one strain of Enterococcus (E. faecium). Each capsule contains approximately 700 million bacteria for each strain. However, regular intake of probiotics for 1 month in healthy individuals does not affect the overall diversity of fecal microbiota. It may be attributed to the relatively larger size and number of overall intestinal microbiota, compared with probiotics administered. When considering that the diversity indexes significantly decreased 2 weeks after the stop of administration, the administration of probiotics might have the effect of preventing the decrease of diversity.
In the present study, the proportion of Bacteroidetes significantly increased, whereas that of Actinobacteria significantly decreased 2 weeks after the stop of administration. The Firmicutes or Bacteroidetes phyla are the main microbes reside in the GI tract. Bacteroidetes have colonized in the colon, and account for about 50% of the 16S ribosomal RNA sequences detected in the mucosal tissues from healthy individuals.4 The tendency for the increase in the proportion of Bacteroidetes members in Sample 3, that were not present in the administered probiotics, might be associated with returning to the original proportion or homeostasis. Conversely, Actinobacteria species were included in the administered probiotics (B. brevis, B. lactis, and B. longum). The reason for the decrease in the proportion of Actinobacteria 2 weeks after stopping the probiotic intake is unclear. However, it might also be associated with homeostasis, whereby the composition of intestinal microbiota is actively regulated to remain nearly constant. Actinobacteria occupied a relatively small portion of fecal microbiota, and its proportion decreased after stopping the probiotic intake, probably due to homeostasis or the original influence of the usual environmental factors, including diet.
The proportion of Bifidobacterium did not significantly differ between Samples 1 and 2 (P = 0.149). However, between Samples 2 and 3, the proportion of Bifidobacterium significantly decreased (4.407% vs 2.164%, P = 0.001). This reduction might also be related to homeostasis or the influence of the usual environmental factors, including diet. Before performing our study, we wondered whether the proportion of genera was maintained when the probiotics were discontinued. We found that there was no change in the diversity 2 weeks after stopping probiotic intake, but a change in the proportion of microbiota was observed. The reduction in the proportion of Bifidobacterium can happen after discontinuation of probiotics, because probiotics are not administered any more. This suggests that the effect of probiotics administration does not last long.
Firmicutes was included in probiotics administered (L. acidophilus, L. plantarum, L. rhamnosus, and E. faecium). At the phyla level, the proportion of Firmicutes did not change significantly, probably because it occupied a large proportion of fecal microbiota. In the current study, changes in the microbiota composition at the genus level were more apparent than those at the phylum level. Our results provide evidence that probiotics strains can affect the composition of intestinal microbiota at the genus level. An imbalance of intestinal microbiota has been observed in various diseases. The administration of probiotics may be helpful in patients who suffer from conditions associated with imbalance of intestinal microbiota.
We additionally measured fecal calprotectin levels in each participant. Fecal calprotectin is a calcium-containing protein released into the intestinal lumen that is excreted in the feces during acute and chronic inflammation. Fecal calprotectin levels have been reported to reflect inflammation in the intestine.28,29 In the present study, we did not find significant changes in fecal calprotectin levels before and after probiotics administration. This may be attributable to the fact that this study was conducted in healthy individuals without symptoms or intestinal inflammation. This might be explained by another factor that fecal calprotectin levels may be determined by various factors including gut microbiota. This negative result is not likely to indicate the clinical useless of the probiotics. Further studies are required to evaluate the effect of probiotics on intestinal inflammation in patients with elevated fecal calprotectin levels.
This study has several limitations. The microbiota in healthy individuals is influenced by a wide variety of factors including diet and age.30 The participants were not admitted for the study, and a standardized diet was not provided. However, we tried to control various conditions to minimize these confounding factors. The routine lifestyles of the participants were checked daily, and activities that could affect gut microbiota, including the change of eating habits and alcohol consumption. The age range of the participants was limited. The environment was also limited. All subjects spent more than 12 hours a day in the same working space. In addition, at least 2 meals a day were the same, and drinking of alcohol was strictly restricted. We used the same dosage that the company recommended, and did not compare the effect of different dosages of probiotics. Further studies with a larger sample size or using different strains are warranted. Because this study was performed in healthy individuals, the baseline sample (Sample 1) was regarded as the control, and a comparative study was conducted. However, we did not compare the effect of probiotics with a placebo.
In conclusion, the diversity of fecal microbiota is not significantly affected by 4 weeks of probiotics administration. The proportion of fecal microbiota at the genus level is significantly altered by the administration of probiotics. However, this effect does not seem to last long, probably because of homeostasis or dietary influence.
Footnotes
Financial support: None.
Conflicts of interest: None.
Author contributions: Choong-Kyun Noh, Bong Sun Kim, Gana Hong, Jae Youn Cheong, and Kwang Jae Lee contributed to study design; Choong-Kyun Noh, Bong Sun Kim, Gana Hong, and Kwang Jae Lee contributed to data collection; Choong-Kyun Noh and Kwang Jae Lee wrote the manuscript and evaluated the eligibility of subjects; and Kwang Jae Lee provided critical revision.
References
- 1.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flores R, Shi J, Gail MH, Gajer P, Ravel J, Goedert JJ. Assessment of the human faecal microbiota: II. Reproducibility and associations of 16S rRNA pyrosequences. Eur J Clin Invest. 2012;42:855–863. doi: 10.1111/j.1365-2362.2012.02659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han DS. Current status and prospects of intestinal microbiome studies. Intest Res. 2014;12:178–183. doi: 10.5217/ir.2014.12.3.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Relman DA. Microbial genomics and infectious diseases. N Engl J Med. 2011;365:347–357. doi: 10.1056/NEJMra1003071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host Microbe. 2011;10:324–335. doi: 10.1016/j.chom.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanabe S. The effect of probiotics and gut microbiota on Th17 cells. Int Rev Immunol. 2013;32:511–525. doi: 10.3109/08830185.2013.839665. [DOI] [PubMed] [Google Scholar]
- 9.Stecher B, Robbiani R, Walker AW, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156(Pt 11):3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 11.Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 12.Willing B, Halfvarson J, Dicksved J, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 13.Chassard C, Dapoigny M, Scott KP, et al. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment Pharmacol Ther. 2012;35:828–838. doi: 10.1111/j.1365-2036.2012.05007.x. [DOI] [PubMed] [Google Scholar]
- 14.Rajilić-Stojanović M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 15.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarra M, Pallone F, Macdonald TT, Monteleone G. IL-23/IL-17 axis in IBD. Inflamm Bowel Dis. 2010;16:1808–1813. doi: 10.1002/ibd.21248. [DOI] [PubMed] [Google Scholar]
- 17.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, Fu L, Niu B, Wu S, Wooley J. Ultrafast clustering algorithms for metagenomic sequence analysis. Brief Bioinform. 2012;13:656–668. doi: 10.1093/bib/bbs035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauer DF. Constructing confidence sets using rank statistics. J Am Stat Assoc. 1972;67:687–690. doi: 10.1080/01621459.1972.10481279. [DOI] [Google Scholar]
- 21.Graf D, Di Cagno R, Fåk F, et al. Contribution of diet to the composition of the human gut microbiota. Microb Ecol Health Dis. 2015;26:26164. doi: 10.3402/mehd.v26.26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnorr SL, Candela M, Rampelli S, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyra A, Forssten S, Rolny P, et al. Comparison of bacterial quantities in left and right colon biopsies and faeces. World J Gastroenterol. 2012;18:4404–4411. doi: 10.3748/wjg.v18.i32.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huse SM, Young VB, Morrison HG, et al. Comparison of brush and biopsy sampling methods of the ileal pouch for assessment of mucosa-associated microbiota of human subjects. Microbiome. 2014;2:5. doi: 10.1186/2049-2618-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu R, Chen K, Zheng YY, et al. Meta-analysis of the efficacy of probiotics in Helicobacter pylori eradication therapy. World J Gastroenterol. 2014;20:18013–18021. doi: 10.3748/wjg.v20.i47.18013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ki Cha B, Mun Jung S, Hwan Choi C, et al. The effect of a multispecies probiotic mixture on the symptoms and fecal microbiota in diarrhea-dominant irritable bowel syndrome: a randomized, double-blind, placebo-controlled trial. J Clin Gastroenterol. 2012;46:220–227. doi: 10.1097/MCG.0b013e31823712b1. [DOI] [PubMed] [Google Scholar]
- 27.Ojima M, Motooka D, Shimizu K, et al. Metagenomic analysis reveals dynamic changes of whole gut microbiota in the acute phase of intensive care unit patients. Dig Dis Sci. 2016;61:1628–1634. doi: 10.1007/s10620-015-4011-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalla R, Kennedy NA, Ventham NT, et al. Serum calprotectin: a novel diagnostic and prognostic marker in inflammatory bowel diseases. Am J Gastroenterol. 2016;111:1796–1805. doi: 10.1038/ajg.2016.342. [DOI] [PubMed] [Google Scholar]
- 29.Lee J. [Fecal calprotectin in inflammatory bowel disease]. Korean J Gastroenterol. 2016;67:233–237. doi: 10.4166/kjg.2016.67.5.233. [Korean] [DOI] [PubMed] [Google Scholar]
- 30.Claesson, Marcus, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]