Abstract
Roquin-1, a RING finger E3 ubiquitin ligase, functions as a modulator of inflammation; however, nothing is known about how Rc3h1 expression is regulated. Here, we describe an opposing relationship between Roquin-1 and the IL-17 proinflammatory cytokine by demonstrating that enforced expression of Rc3h1 restricts Il17a expression, and that exposure of T cells to IL-10, a cytokine with immunosuppressive activity, increases Rc3h1 expression. Luciferase reporter assays conducted using eight transcription factor plasmids (STAT1, STAT3, STAT5, GATA2, c-Rel, IKZF1, IKZF2, and IKZF3) demonstrated that STAT1, STAT3, GATA2, and c-Rel increased Rc3h1 promoter activity, whereas IKZF2 decreased activity. Gene expression of those five transcription factors increased in T cells exposed to IL-10. Transcription factor-specific siRNAs suppressed the IL-10 effect on Rc3h1 transcription. These findings identify a role for IL-10 in regulating Rc3h1 transcription, and they have implications for understanding how Roquin-1 controls the immune response.
Keywords: cytokine, IL-10, gene expression, luciferase, promoter, siRNA, transcription factor
1. Introduction
Roquin-1 is a RING finger E3 ubiquitin ligase characterized by a CCCH zinc finger that enables binding to target mRNAs to facilitate RNA degradation. Roquin-1 localizes to Processing bodies (P bodies) and stress granules, two areas involved in RNA processing and turnover (Athanasopoulos et al., 2010; Kulkarni et al., 2010; Vinuesa et al., 2005). Moreover, Rck and Edc4, notable proteins localized to P bodies and involved in mRNA decapping, were identified as interacting partners of Roquin-1 (Glasmacher et al., 2010). A constitutive decay element present in many mRNAs that contributes to mRNA recognition by Roquin-1 has been identified (Leppek et al., 2013). These studies confirm a role for Roquin-1 as an RNA processing protein with critical targets important for immune function and inflammation.
In a screen of N-ethyl-N-nitrosourea-treated mice, a mutation in Rc3h1 resulted in an M199R substitution in the ROQ domain of the Roquin-1 protein (Vinuesa et al., 2005). Roquinsan/san mice (also referred to as sanroque mice), which bear the Rc3h1 mutation, develop extensive and severe immunological perturbations that manifest as splenomegaly, systemic lupus erythematosus-like disease, necrotizing hepatitis, glomerulonephritis, autoimmune thrombocytopenia, anemia, small intestinal inflammation, exaggerated numbers of germinal centers and CD4+ follicular helper T (Tfh) cells, and increased expression of the inducible costimulator (ICOS) and OX40 T cell activation molecules (Glasmacher et al., 2010; Heissmeyer and Vogel, 2013; Lin and Mak, 2007; Linterman et al., 2009; Pratama et al., 2013; Schaefer et al., 2013; Schaefer et al., 2011; Vinuesa et al., 2005; Vogel et al., 2013; Yu et al., 2007). Mice in which Rc3h1 and its paralog, Rc3h2, were disrupted developed a sanroque-like phenotype (Bertossi et al., 2011; Pratama et al., 2013; Vogel et al., 2013). Small intestinal inflammation occurred in mice in which Rc3h1 expression was ablated by insertion of a gene trap (Schaefer et al., 2013). In IL-10−/− mice, levels of Rc3h1 mRNA were decreased and Icos and Il17 mRNAs were increased in colonic intraepithelial lymphocytes (cIELs) (Schaefer et al., 2010; Schaefer et al., 2011), and treatment of cIELs with IL-10 increased Rc3h1expression and decreased Icos and Il17 expression (Schaefer et al., 2010; Schaefer et al., 2011). Those findings demonstrate a role for Roquin-1 in modulating the inflammatory response and controlling autoimmunity, and they indicate that IL-10, a cytokine with immunosuppressive activity, influences Rc3h1 expression.
Although many features of the effector response whereby Roquin-1 controls autoimmunity have been characterized, nothing is yet known about the regulatory elements that are responsible for Rc3h1 expression or how IL-10 contributes to that process. In the present study we identified five transcription factors that target the Rc3h1 promoter, and demonstrated that IL-10 enhances the expression of those transcription factors. These findings provide new insights into how levels of the Roquin-1 protein are maintained, and they open ways for developing strategies to sustain Roquin-1 expression in the face of autoimmunity and other chronic inflammatory responses.
2. Materials and methods
2.1 Mice, cell culture, staining, and flow cytometry
Female C57BL/6 mice, 6-8 weeks of age, were purchased from Harlan Laboratories, Indianapolis, IN. Mice were used in accord with the University of Texas Health Science Center institutional animal welfare guidelines. Murine EL4 T cells were maintained in 10% FBS, 1 U/ml penicillin-streptomycin, RPMI 1640 medium in a 37°C incubator with 5% CO2. For stimulation experiments, cells were cultured for 0-48 hr at a concentration of 1×106 cells/ml and stimulated with 50 ng/ml murine rIL-10 (eBioscience, San Diego, CA) or with mouse recombinant transforming growth factorβ (rTGFβ) (Shenandoah Biotechnology, Warwick, PA). Cells were collected and processed for qRT-PCR.
Mesenteric lymph node (MLN) T cells were isolated using a T cell enrichment column (R&D Systems, Minneapolis, MN), and stimulated using a Th17 induction protocol consisting of culture in the presence of 0.05 μg/ml hamster anti-CD3ε antibody onto 1 μg/ml mouse plate-bound anti-hamster antibody (eBioscience) in the presence of 5 ng/ml rTGFβ (Shenandoah Biotechnology), 10 ng/ml rIL-23 (eBioscience), and 20 ng/ml rIL-6 (Shenandoah Biotechnology) for 24 and 48 hr.
Cells were stained using a protocol similar to that previously described by our laboratory (Montufar-Solis et al., 2007). Antibodies used were: purified anti-hamster IgG, purified anti-mouse CD3ε, FITC-control IgG, monensin intracellular protein block (BD Biosciences, San Jose, CA), FITC anti-mouse CD4, Alexafluor 660 anti-mouse IL-17A, PE anti-mouse RORγt, Alexafluor 647 IgG, PE IgG, and anti-mouse CD16/32 (eBioscience). 1 μl/ml monensin was added for 5 hr to cell cultures to block intracellular protein transport. Cells were collected and reacted with anti-CD16/32 for 15 min followed by anti-CD4 mAb for 30 min at 4°C. Cells were washed in PBS and suspended in 250 μl cytofix/cytoperm (BD Bioscience) for 20 min at room temperature, washed in 1× permwash (BD Biosciences), and reacted with anti-IL-17, anti-RORγt, or control mAbs for 30 min at 4°C. Cells were washed 1× with permwash, suspended in 2% formaldehyde, and analyzed on FACScan flow cytometer using Cell-Quest software (BD Biosciences).
2.2 Luciferase reporter assay
293T and 293FT cells were grown overnight in 24-well plates with serum-supplemented DMEM. Initial experiments used 293T cells; however, 293FT cells were subsequently used. No difference was observed in experimental results depending upon the cell type used. The next day, cells were transfected using Fugene HD (Promega, Madison, WI) with 200 ng of a Rc3h1 promoter luciferase reporter plasmid cloned into the pGLuc-Basic plasmid (New England Biolabs, Ipswich, MA) by our laboratory, along with 100 ng of a plasmid encoding for either constitutively-active STAT1 (eGFP STAT1 S727E, Addgene, Cambridge, MA), constitutively-active STAT3 (Stat3-C Flag pRc/CMV, Addgene), constitutively-active STAT5 (CA-STAT5-GFP-RV (Yang et al., 2011), a gift of Dr. Jinfan Zhu), c-Rel (c-Rel cFlag pcDNA3, Addgene), GATA2 (pFlag-GATA2, Addgene), IKZF1, IKZF2, and IKZF3 transcription factors (cloned into the pEF6/V5-His plasmid in our laboratory), and 100 ng of a β-galactosidase plasmid (pSV-β-galactosidase). 48 hr post-transfection, cell supernatants and cell pellets were collected. A TD 20/20 Luminometer was used to measure luciferase activity in 20 μl of supernatant using the BioLux Gaussia Luciferase Assay Kit (New England Biolabs). Cell pellets were lysed and β-galactosidase activity was measured in 20 μl of the cell lysate. Relative luciferase light units were calculated from the luciferase reading divided by β-galactosidase activity. Data are fold induction of luciferase activity of transcription factor-transfected cells relative to non-transcription factor transfected cells.
The Rc3h1 promoter constructs were generated by PCR cloning. Briefly, the Rc3h1 promoter sequence was TOPO cloned from BALB/c genomic DNA into the pCR8 TOPO vector. Traditional restriction digest cloning, using HindIII and BamHI restriction sites included in the cloning primers, was used to transfer the promoter sequence into the pGLuc-Basic plasmid and create the final reporter plasmids. Primers used were −2.1 kb Rc3h1: CCAAGCTTAAACTGAAGGAGAATAGTACAGAACC; −1.3 kb Rc3h1: CCCAAGCTTGCAGAGGTTCAGAACTTCCAG; −0.2 kb Rc3h1: CGGTGACTCAGAGCTTTCCT; and Rc3h1 reverse: CCCAAGCTTAGGGGCCATCTTGTTGGT.
2.3 Quantitative realtime PCR (qRT-PCR)
Total RNA was isolated using the mRNeasy Minikit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Life Technologies, Grand Island, NY) from 1 μg of total RNA. The Power SYBR Green PCR Master Mix (Life Technologies) was used to measure Rc3h1, Il17, and transcription factor (Stat1, Stat3, c-Rel, Gata2, and Ikzf2) transcript levels. Samples were analyzed using the StepOnePlus real-time thermal cycler (Life Technologies) and software. Relative gene expression was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001) and normalized to Gapdh. Rc3h1, Il17, transcription factor, and Gapdh specific primers were designed and purchased from Integrated DNA Technologies (Coralville, IA). Primers are as follows: Rc3h1, 5′-GGCTGCTCGATCTTTAGGTG-3′ and 5′-TGTTCTCTCCTCAGAGCTTCG-3′; Il17, 5′-CTCCAGAAGGCCCTCAGACTAC-3′ and 5′-GGGTCTTCATTGCGGTGG-3′; Stat1, 5′-ACAACATGCTGGTGACAGAGCC-3′ and 5′-TGAAAACTGCCAACTCAACACCTC-3′; Stat3, 5′-ATCCTAAGCACAAAGCCCCC-3′ and 5′-ATAGCCCATGATGATTTCAGCAA-3′; c-Rel, 5′- ACCTCAATGTGGTGAGGCTG-3′ and 5′- TGGGGCACGGTTGTCATAAA-3′; Gata2, 5′-AGACGACAACCACCACCTTA-3′ and 5′-TCCTTCTTCATGGTCAGTGG-3′; Ikzf2, 5′- CAGCGAGGTGGCCGACAACAG-3′ and 5′- GGCCGCTCACCAGTGTGACTCC -3′; Gapdh, 5′-AGAACATCATCCCTGCATCC-3′ and 5′-AGCCGTATTCATTGTCATACC-3′.
2.4 small inhibitory RNA (siRNA) treatment
50 pmols of transcription factor-specific (Santa Cruz Biotechnology, Santa Cruz, CA) or control siRNA oligonucleotides (Integrated DNA Technologies, Coralville, IA) were transfected into EL4 cells using Lipofectamine RNAiMAX (Life Technologies). Four hr post-transfection, 50 ng/ml of rIL-10 were added to the culture medium. Cells were harvested 48 hr later, RNA was isolated, and qRT-PCR was conducted to determine Rc3h1 expression.
2.5 Western blot
EL4 cells were transfected using an Amaxa Nucleofector with a C-009 program and Roquin-1 pEF6 or LacZ pEF6 plasmids prepared in our laboratory. Cells were collected 72 hr later and processed for Roquin-1 western blotting and gene expression. Cells were lysed and homogenized in RIPA buffer (50mM Tris-HCl pH 7.4, 150mM NaCl, 1% NP40, 1% SDS, 0.5% Na-deoxycholate, 1mM PMSF, 50mM NaF, 1 μg/ml each of: aprotinin, leupeptin, pepstatin). 40 μgs of lysate were electrophoresed through 8% SDS-polyacrylamide gel. Following overnight transfer of the separated proteins to Immun-Blot PVDF membrane (Bio-Rad, Hercules, CA), the membranes were blocked for 2 hr in 5% milk TBS containing 0.1% Tween-20 (TBS-T0.1). The membrane was probed with a Roquin-1 antibody [Bethyl Laboratories, Montgomery, TX; A300-514A, (1:560)] overnight at 4°C with rocking. Following 3 rounds of washing with TBS-T0.1, the membrane was incubated with HRP-conjugated Goat anti-Rabbit antibody [Vector Laboratories, Burlingame, CA; (1:1500)]. Enhanced chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate, Thermo Fisher Scientific, Rockford, IL) was used to detect the protein bands. Ponceau S stain was used to identify proteins in samples.
The Roquin-1 expression plasmid was created using the pEF6/V5-His TOPO kit (Invitrogen Grand Island, NY) according to the manufacturer’s instructions. The following primers were used to PCR clone Rc3h1from BALB/c cDNA: CCGGAATTCCACCATGCCTGTACAAGCTCCACAAT and CCGGAATTCCTAGGGAGCAGAATTGGAAACAA. The LacZ pEF6 plasmid is a positive control included with the kit.
2.6 Chromatin immunoprecipitation (ChIP) assay
Plasmids (1000 ng) encoding either the STAT1 or GATA2 transcription factors were transfected into 293FT cells grown in supplemented DMEM medium using Fugene HD (Promega). 48 hr post-transfection, chromatin was crosslinked by incubating the 293FT cells for 10 minutes with 1% formaldehyde. After quenching with glycine (125 mM final concentration), the cells were collected and pelleted. The Pierce Agarose ChIP Kit (Thermo Fisher Scientific) was used according to manufacturer’s instructions to perform the ChIP. Micrococcal Nuclease digestion followed by sonication was used in order to obtain chromatin fragments of 400 to 600 kb in size. Chromatin was precleared with Protein A/G Plus agarose for 1 hr before being incubated at 4°C overnight with either GATA-2 antibody or normal rabbit IgG (Santa Cruz Biotechnology). Antibody complexes were captured using Protein A/G Plus agarose and washed four times with kit buffers. Chromatin was eluted and the crosslinking was reversed by incubating the samples in elution buffer plus proteinase K at 65°C overnight. DNA was column purified and analyzed by SYBR green (Life Technologies) real-time PCR. ChIP primer sequences used were: RC3H1 GATA −1.8 kb, 5′- GCTCACTACAACCTCCAC CT -3′ and 5′- CATTTTAGGAGGCTGAGGGG -3′; RC3H1 GATA -2.2 kb, 5′- CCAAAGTGCTGGGATTACAA -3′ and 5′- TCCCCAAACCCAAATTCA T -3′; RC3H1 GATA −1.0 kb, 5′- TGCCAAGAGTTTAGGAAGGTG -3′ and 5′- AGGAGAGAGAGAGTCCCCAAA -3′; and RC3H1 GATA −0.4 kb, 5′- GTAGTGTGGGGTTCGATGGA -3′ and 5′- GCAAAGCTCTGAGTCACCG -3′. Primers used for the mouse ChIP qPCR were: EpiTect ChIP qPCR Primer Assay For Rc3h1, NM_001024952.1, −2000 bp and −1000 bp (Qiagen, Valencia, CA).
2.7 Statistical analysis
Calculations of statistical significance of replicate data sets were done using a one-tailed t-test with equal variance.
3. Results
3.1 Roquin-1 modulates expression of the IL-17 proinflammatory cytokine
The opposing relationship between Roquin-1 and IL-17 was demonstrated using MLN T cells cultured under Th17-inducing conditions as described in the Materials and Methods. Generation of Th17 cells was confirmed by flow cytometry. 48 hr post-stimulation, 4.8% of CD4+ MLN T cells co-expressed IL-17 and the retinoic acid related orphan receptor γt (RORγt) transcription factor for Il17a (Fig 1A). In contrast, cells stimulated through CD3 without Th17 induction did not express IL-17 or RORγt (Fig. 1B). Following 24 and 48 hr of stimulation, Il17a transcription was significantly increased, and Rc3h1 transcription was significantly decreased, compared to control cultures (plate-bound anti-hamster antibody with hamster IgG) (Fig. 1C-D). In previous studies, the mouse EL4 T cell line has been used for studies into the role of Roquin-1 in the regulation of cytokine synthesis (Kim et al., 2012; Schaefer et al., 2011). We demonstrate that exposure of EL4 cells to IL-10 results in increased Rc3h1 transcription at 24 and 48 hr (Fig. 1E). That EL4 cells were responsive to IL-10 was confirmed by an increase in SOCS3 gene expression following exposure to IL-10 (Fig. 1F); SOCS3 is known to operate within the IL-10 signaling pathway. To determine if enforced Rc3h1 expression would modulate Il17a expression, IL-17-producing EL4 cells were transfected with a Roquin-1 pEF6 expression plasmid; control cells were transfected with a LacZ pEF6 expression plasmid. Increased Roquin-1 protein expression in transfected cells was confirmed by western blotting (Fig. 1G). Relative protein levels were comparable based on Ponceau S staining (Fig. 1G). Roquin-1 pEF6 transfection resulted in increased Rc3h1 transcription and lower Il17a transcript levels (Fig. 1H-I). These findings collectively demonstrate the ability of Roquin-1 to suppress IL-17 synthesis, and the capacity of IL-10 to increase Rc3h1 expression.
Fig. 1.
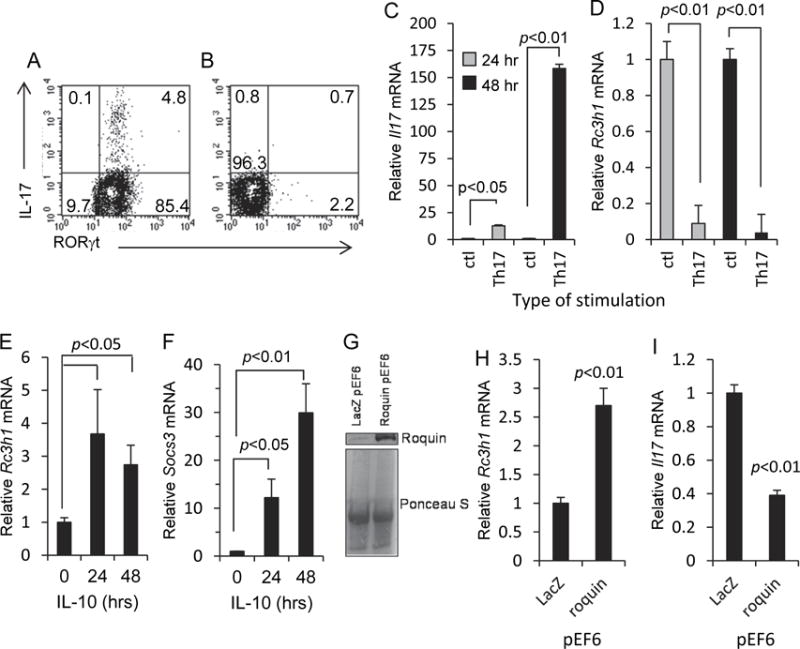
Opposing relationship between Roquin-1 and IL-17. (A) MLN T cells 48 hr after stimulation using the Th17 induction regimen described in the Materials and Methods. (B) MLN T cells 48 hr after CD3 stimulation in the absence of Th17 induction. Cells were gated onto the CD4+ cell population. Representative histograms from three experiments. (C, D) Increased Il17a and decreased Rc3h1 transcription in MLN T cells following 24 and 48 hr of Th17 induction. Control cultures consisted of plate-bound anti-hamster antibody with normal hamster IgG. (E) Increased Rc3h1 transcription in EL4 cells following exposure to IL-10. (F) Exposure of EL4 cells to IL-10 resulted in increased Socs3 transcription. (G) Western blot showing increased levels of Roquin-1 protein expression in EL4 cells transfected with the Roquin-1 pEF6 expression plasmid. Ponceau S staining was done to compare sample protein levels. (H, I) Roquin-1 pEF6 transfection of EL4 cells resulted in increased Rc3h1 transcription and decreased Il17a transcription. Data are mean values ± SEM of at least three experiments.
3.2 Identification of Rc3h1 transcription factors
To identify transcriptional elements involved in Rc3h1 expression, we made −2.1 kb, −1.3 kb, and −0.2 kb Rc3h1 promoter constructs, and cloned them into luciferase reporter plasmids. In addition to the STAT canonical IL-10 transcription factors, we sought to understand the contribution of other transcription factors to the regulation of Roquin-1. Using the TFSEARCH web-based program with a threshold of 0.8, we identified additional potential transcription factor binding elements (Heinemeyer et al., 1998). We chose to focus on the STAT, IKZF, c-Rel, and GATA2 binding elements as each of these are involved in the regulation of immune signaling. IL-10 classically signals through the JAK- STAT pathway, thus making STAT1 and STAT3 possible targets. The IKZF and GATA family of transcription factors are involved in hematopoietic cell differentiation with IKZF2 and IKZF1 in particular affecting IL-10 and IL-17 production (Akimova et al., 2011; Himmel et al., 2013; Suzuki et al., 2013; Wong et al., 2013; Yap et al., 2005). c-Rel is classically involved in T and B cell signaling. These binding elements are depicted in Fig. 2A. This identified STAT, GATA, IKZF (Ikaros family members), and c-Rel as potential transcription factors binding to various portions of the promoter (Fig. 2A and Tables 1 and 2).
Fig. 2.
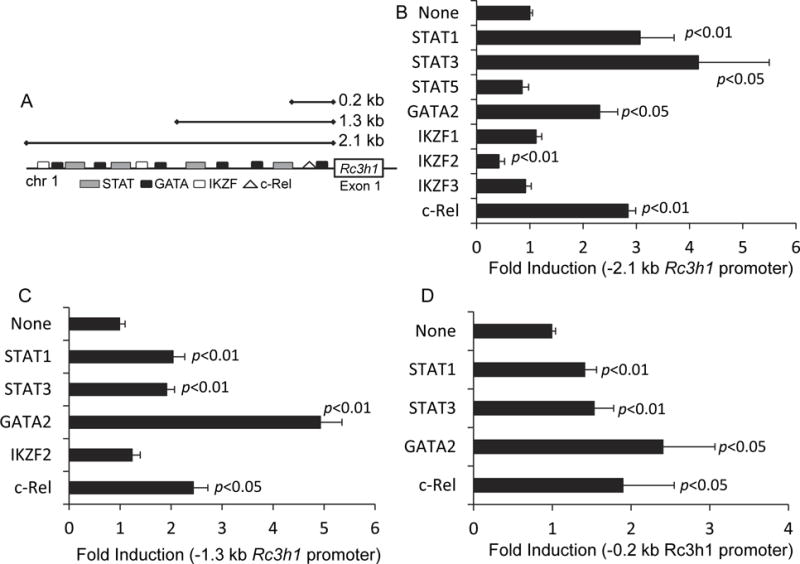
Transcriptional regulation of Rc3h1. (A) A web-based search for transcriptional factors that bind to the Rc3h1 promoter identified binding sites for STATs, GATA, IKZF and c-Rel. Luciferase reporter assays were used to evaluate the activational effects of eight transcription factors on the Rc3h1 promoter regions. Using a (B) -2.1 kb Rc3h1 promoter region, STAT1, STAT3, GATA2, and c-Rel resulted in a significant increase in luciferase activity; IKZF2 resulted in a significant decrease. STAT1, STAT3, GATA2, and c-Rel also resulted in increased luciferase activity using the (C) -1.3 kb and (D) -0.2 kb Rc3h1 promoter regions. Values are fold induction of luciferase activity of transcription factor-transfected cells relative to non-transcription factor transfected cells. Data are mean values ± SEM of 3-4 independent experiments.
Table 1.
Predicted transcription factor binding elements on mouse Rc3h1 promoter
| Transcription factor | Position (bp) | Sequence |
|---|---|---|
| c-Rel | −134 | TGACTCAGAGCTTTCCTCCC |
| IKZF | −2036 | AGGGTATGTTCCCAACTGTT |
| IKZF | −1509 | TTTAATCCCAGCACTTGGAA |
| IKZF | −1206 | TCTGTTCCGGGAATAAAACA |
| IKZF | −598 | GGGGTGGGATGCAAAACAGA |
| GATA2 | −2137 | CCAAGAGATAACGTTTAGAT |
| GATA2 | −2019 | CAGCAGATGACTAAACTGAA |
| GATA2 | −1787 | AAATTTCAGATAAGTGGGGA |
| GATA2 | −1609 | GGAAAATGTGTGATATTTCA |
| GATA2 | −904 | TTCTAGGATAGCCAAGGGTA |
| GATA2 | −548 | AACGCCATCATACACACACA |
| GATA2 | −38 | GCCTTCATCTCCGCCCCAGT |
| STAT | −1852 | TGAATTATTTCTGGAATTTT |
| STAT | −1625 | TCCAAGTTCAGGAAAATGTG |
| STAT | −1206 | TCTGTTCCGGGAATAAAACA |
| STAT | −307 | AGCTTACGGGAACCGCCTGG |
Table 2.
Predicted transcription factor binding elements on human RC3H1 promoter
| Transcription factor | Position (bp) | Sequence | ChIP Primers |
|---|---|---|---|
| c-Rel | −360 | ACGAAAGAAATTCCCCC | |
| IKZF | −2192 | AAAGTGCTGGGATTACAAGC | |
| IKZF | −1755 | CTAAAATGTTGGGATTACAG | |
| IKZF | −530 | GGCTGAACTCCCAGAGGGGT | |
| GATA2 | −2157 | CGGCCTTATCCCCTGTATCT | −2.2 kb |
| GATA2 | −1774 | TTTAGTAGAGATGGGGTTTC | −1.8 kb |
| GATA2 | −1673 | CTGCATCTATCTGTCCTTTA | |
| GATA2 | −1037 | CACTATCTGTGATAGCGATTC | −1.0 kb |
| GATA2 | −908 | AAGGTGATAGCGCCGAGT | −1.0 kb |
| GATA2 | −339 | CTCGCCATCTGGCCACT | −0.4 kb |
| STAT | −1992 | ATTTGGGTTTGGGGAAGCGC | |
| STAT | −1601 | TCATACTGCTTAAGGGAACC | |
| STAT | −733 | CCTCCCATTCCCCAAAACCA | |
| STAT | −357 | AAATTCCCCCAATTTACTCT |
Using the −2.1 kb promoter construct in the luciferase reporter assay, constitutively-active STAT1 and constitutively-active STAT3, as well as GATA2 and c-Rel increased promoter activity (Fig. 2B). Those findings also held true for the −1.3 kb and the −0.2 kb Rc3h1 promoter regions (Fig. 2C-D), indicating that there were multiple binding sites for those transcription factors throughout the Rc3h1 promoter. IKZF2 lowered the Rc3h1 promoter activity for the −2.1 kb Rc3h1 promoter (Fig. 2B) but had no effect on the −1.3 kb (Fig. 2C) or the −0.2 kb (not shown) promoter regions, indicating that binding of IKZF2 occurred between the −1.3 kb and −2.1 kb regions. IKZF1, IKZF3, and constitutively-active STAT5 had no effect on Rc3h1 promoter activation (Fig. 2B). The presence of STAT1 and STAT3 promoter activity in the −0.2 kb promoter region (Fig. 2D), despite the lack of a predicted binding site (Fig. 2A), likely reflects additional STAT sites that were below the 0.8 threshold used in the TFSEARCH program.
Because GATA2 had greater luciferase activity for the −1.3 kb Rc3h1 promoter region (Fig. 2C) compared to the −2.1 kb and −0.2 kb regions (Fig. 2B and D), we conducted ChIP assays to further dissect GATA2 binding to the Rc3h1 promoter. Amplification of anti-GATA2 immunoprecipitated DNA was done using primer sets for −2.2, −1.8, −1.0, and −0.4 kb Rc3h1 promoter regions. Products were produced for all regions from GATA2 ChIP experiments (Fig. 3A-D). Control experiments consisting of mock-transfected cells and constitutively-active STAT1-transfected cells resulted in no appreciable fold enrichment. These findings confirm that GATA2 binds at multiple sites throughout the Rc3h1 promoter, as predicted in Fig. 2A.
Fig. 3.
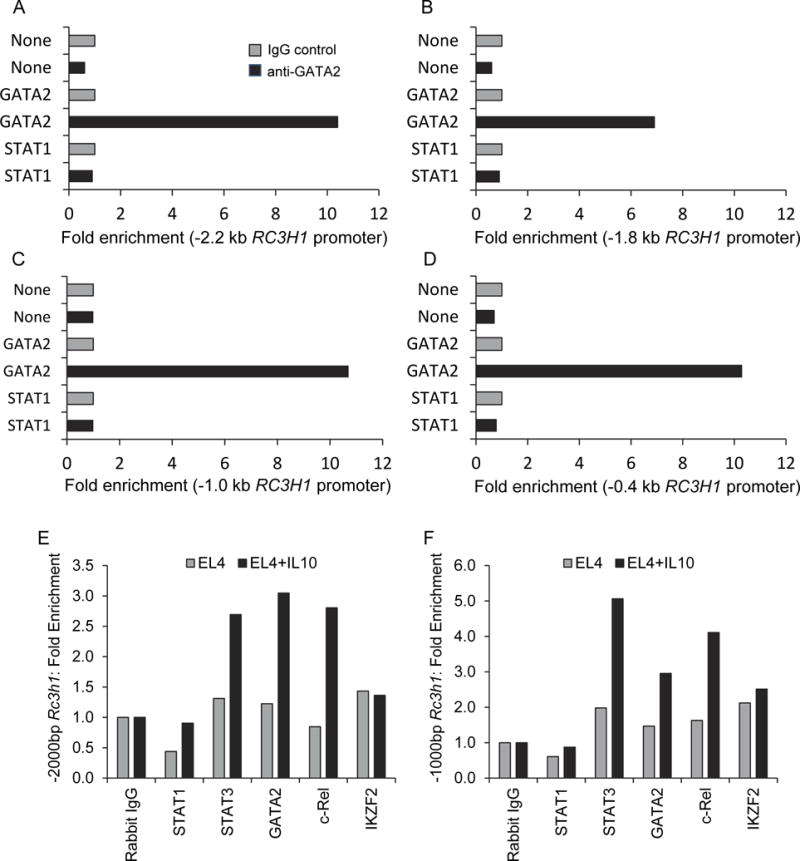
ChIP assays showing GATA2 interactions with: (A) -2.2 kb, (B) -1.8 kb, (C) -1.0 kb, and (D) -0.4 kb RC3H1 promoter regions. Data in each panel represent one ChIP experiment; the average values of all four data entries for the GATA2 ChIP assays (anti-GATA2 antibody) was significantly greater (p<0.001) than that of ChIP assay results done with None (mock) transfections, or STAT1 transcription factor transfections. (E, F) ChIP assays showing the effect of IL-10 on transcription factor interactions with the Rc3h1 promoter.
To determine if IL-10 induced increased binding of the relevant transcription factors to the Rc3h1 promoter region, ChIP analysis was performed on EL4 cells treated with or without IL-10 for 48 hr. STAT3, GATA2, and c-Rel had increased occupation of the −2000 bp and −1000 bp region of the Rc3h1 promoter following IL-10 treatment (Fig. 3E-F). This suggested that STAT3, GATA2, and c-Rel were actively recruited to the Rc3h1 promoter in response to IL-10.
3.3 IL-10 increases Rc3h1 transcription factor expression
Based on the finding that exposure of T cells to IL-10 led to increased Rc3h1 expression (Fig. 1E) (Schaefer et al., 2011), and to further understand the mechanisms by which Rc3h1 is regulated, we examined gene expression of Stat1, Stat3, Gata2, c-Rel, and Ikzf2 in EL4 cells at 0, 24, and 48 hr after exposure to rIL-10, or to rTGFβ as a control cytokine. At 48 hr, there was a statistically-significant increase in gene expression of Stat1 and Gata2 (p<0.01), and Stat3 and c-Rel (p<0.05). Ikzf2 expression increased to almost a statistically-significant level (p=0.06) (Fig. 4A).
Fig. 4.
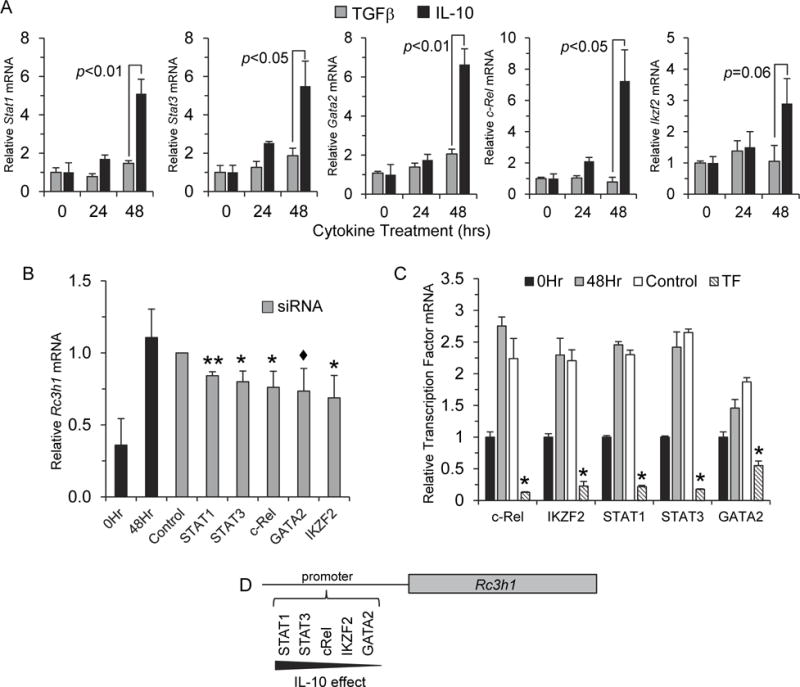
IL-10 increased Rc3h1 transcription factor expression. (A) EL4 cells were cultured in the presence of mouse rIL-10, or rTGFβ as a control cytokine. Gene expression was measured at 0, 24, and 48 hr. Compared to the rTGFβ control cytokine, exposure of cells to IL-10 resulted in a significant increase in gene expression of STAT1, STAT3, GATA2, and c-Rel, as well as a non-statistical increase in IKZF2 48 hr post-IL-10 treatment. Data are mean values ± SEM of 3 experiments. (B) EL4 cells were transfected with siRNAs to either STAT1, STAT3, GATA2, c-Rel, IKZF2, a scramble control siRNA, or no siRNA for 4 hr followed by exposure to rIL-10 as described in the Materials and Methods. Cells were harvested and Rc3h1 expression was measured by qRT-PCR. All transcription factor-specific siRNAs decreased Rc3h1 expression relative to the control siRNA-transfected cells. For STAT1 and STAT3, this occurred in a statistically-significant manner. (C) Efficiency of knockdown of transcription factor expression. Data are mean values ± SEM of 3 experiments. ** p<0.01, * p<0.05, ◆ p<0.1 and >0.05. (D) A model whereby IL-10 exerts an effect on the transcriptional regulation of Rc3h1.
To determine if IL-10 affected Rc3h1 expression through STAT1, STAT3, GATA2, c-Rel, and/or IKZF2, EL4 cells were transfected with transcription factor-specific siRNAs and cultured for 48 hr in the presence of IL-10. siRNAs to STAT1, STAT3, c-Rel, and IKZF2 significantly decreased (p<0.05) Rc3h1 expression compared to control siRNA-transfected cells (Fig. 4B). Suppression also occurred using GATA2 siRNA, though the level of suppression did not reach statistical significance (p=0.08) (Fig. 4B). The mild effect of GATA2 siRNA on Rc3h1 expression could be due to inadequate silencing of GATA2. To determine the efficiency of silencing by siRNA treatment, expression of each transcription factor was assayed by qPCR in relation to the matching siRNA. Efficient knockdown of transcription factor gene expression was observed for all five transcription factors (Fig. 4C). However, the efficiency of the GATA2-specific siRNA to repress GATA2 was less in comparison to the other transcription factor-specific siRNAs. These findings collectively suggest that IL-10 increases Rc3h1 expression by enhancing the expression of Rc3h1 transcription factors.
4. Discussion
The findings reported here have direct implications for understanding how Rc3h1 is regulated. A model of this is shown in Fig. 3C, which indicates a spectrum whereby IL-10 influences the capacity of transcription factors to act on the Rc3h1 promoter, with IL-10 having a strong effect on STAT1, STAT3, c-Rel, and IKZF2, and a modest effect of IL-10 on GATA2. The role of IL-10 in this pathway is of particular interest because of the wide range of cells capable of producing it, including monocytes and T cells as well as non-hematopoietic cells such as intestinal epithelial cells. Collectively, this would result in a two-fold effect on the suppression of autoimmunity. First, IL-10 would limit the expression of proinflammatory cytokines such as interferon-γ, tumor necrosis factor-α, and IL-17. Second, IL-10 would have a potentiating effect on Roquin-1 protein expression. Higher levels of Roquin-1 protein would suppress IL-17 production and inhibit the expression of T cell activation molecules such as ICOS and OX40. Additionally, it points to a mechanism whereby IL-10-treatment of cIELs from IL-10−/− mice, an animal model of inflammatory bowel disease, down-regulates the inflammatory response.
These findings also bear on the process of Tfh cell regulation, a cell population that contributes significantly to the autoimmune process in sanroque mice. Although IL-21 is a signature cytokine of Tfh cells, IL-10 also has been reported to be produced by Tfh cells (King et al., 2008; Luthje et al., 2012; Vogelzang et al., 2008; Zhang et al., 2013). Further, it has been reported that T cells with a deficiency in IL-10 signaling via deletion of the IL-10Rβ subunit were able to more readily differentiate into Tfh cells (Cai et al., 2012). Hence, Tfh cell self-regulation by IL-10 which feedbacks and activates transcription factors that drive Rc3h1 expression would limit Tfh cell proliferation. IL-10-induced increases in Roquin-1 protein levels would curtail ICOS translation, the net effect of which would be interference in a key signaling pathway used in Tfh cell development, thereby keeping Tfh cells in check and preventing autoantibody production. In the absence of a strong Roquin-1 response, unrestricted growth of Tfh cells and the development of autoantibody-mediated immunopathology would ensue.
In summary, we have identified regulatory elements used in Rc3h1 expression. These findings are expected to open the way for additional studies into the mechanisms by which Roquin-1 controls the inflammatory response.
Acknowledgments
We wish to thank Niyati Nakra for assistance with some of the luciferase assays. This work was supported by Career Development Award No. 3627 from the Crohn’s and Colitis Foundation of America to JSS, and NIH grants DK035566 and AI100159 to JRK.
Abbreviations
- ChIP
chromatin immunoprecipitation
- cIELs
colonic intraepithelial lymphocytes
- ICOS
inducible costimulator
- MLN
mesenteric lymph node
- RORγt
retinoic acid related orphan receptor γt
- siRNA
small inhibitory RNA
- Tfh
follicular helper T cell
- TGFβ
transforming growth factorβ
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Akimova T, Beier UH, Wang L, Levine MH, Hancock WW. Helios expression is a marker of T cell activation and proliferation. PloS one. 2011;6:e24226. doi: 10.1371/journal.pone.0024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasopoulos V, Barker A, Yu D, Tan AH, Srivastava M, Contreras N, Wang J, Lam KP, Brown SH, Goodnow CC, Dixon NE, Leedman PJ, Saint R, Vinuesa CG. The ROQUIN family of proteins localizes to stress granules via the ROQ domain and binds target mRNAs. The FEBS journal. 2010;277:2109–2127. doi: 10.1111/j.1742-4658.2010.07628.x. [DOI] [PubMed] [Google Scholar]
- Bertossi A, Aichinger M, Sansonetti P, Lech M, Neff F, Pal M, Wunderlich FT, Anders HJ, Klein L, Schmidt-Supprian M. Loss of Roquin induces early death and immune deregulation but not autoimmunity. J Exp Med. 2011;208:1749–1756. doi: 10.1084/jem.20110578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G, Nie X, Zhang W, Wu B, Lin J, Wang H, Jiang C, Shen Q. A regulatory role for IL-10 receptor signaling in development and B cell help of T follicular helper cells in mice. J Immunol. 2012;189:1294–1302. doi: 10.4049/jimmunol.1102948. [DOI] [PubMed] [Google Scholar]
- Glasmacher E, Hoefig KP, Vogel KU, Rath N, Du L, Wolf C, Kremmer E, Wang X, Heissmeyer V. Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat Immunol. 2010;11:725–733. doi: 10.1038/ni.1902. [DOI] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic acids research. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heissmeyer V, Vogel KU. Molecular control of Tfh-cell differentiation by Roquin family proteins. Immunol Rev. 2013;253:273–289. doi: 10.1111/imr.12056. [DOI] [PubMed] [Google Scholar]
- Himmel ME, MacDonald KG, Garcia RV, Steiner TS, Levings MK. Helios+ and Helios- cells coexist within the natural FOXP3+ T regulatory cell subset in humans. J Immunol. 2013;190:2001–2008. doi: 10.4049/jimmunol.1201379. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Ji YR, Kim MO, Yu DH, Shin MJ, Yuh HS, Bae KB, Park S, Yi JK, Kim NR, Park SJ, Yoon du H, Lee WH, Lee S, Ryoo ZY. The role of Roquin overexpression in the modulation of signaling during in vitro and ex vivo T-cell activation. Biochem Biophys Res Commun. 2012;417:280–286. doi: 10.1016/j.bbrc.2011.11.101. [DOI] [PubMed] [Google Scholar]
- King C, Tangye SG, Mackay CR. T follicular helper (TFH) cells in normal and dysregulated immune responses. Annual review of immunology. 2008;26:741–766. doi: 10.1146/annurev.immunol.26.021607.090344. [DOI] [PubMed] [Google Scholar]
- Kulkarni M, Ozgur S, Stoecklin G. On track with P-bodies. Biochemical Society transactions. 2010;38:242–251. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]
- Leppek K, Schott J, Reitter S, Poetz F, Hammond MC, Stoecklin G. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell. 2013;153:869–881. doi: 10.1016/j.cell.2013.04.016. [DOI] [PubMed] [Google Scholar]
- Lin AE, Mak TW. The role of E3 ligases in autoimmunity and the regulation of autoreactive T cells. Current opinion in immunology. 2007;19:665–673. doi: 10.1016/j.coi.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Linterman MA, Rigby RJ, Wong RK, Yu D, Brink R, Cannons JL, Schwartzberg PL, Cook MC, Walters GD, Vinuesa CG. Follicular helper T cells are required for systemic autoimmunity. J Exp Med. 2009;206:561–576. doi: 10.1084/jem.20081886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−DDCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luthje K, Kallies A, Shimohakamada Y, Belz GT, Light A, Tarlinton DM, Nutt SL. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat Immunol. 2012;13:491–498. doi: 10.1038/ni.2261. [DOI] [PubMed] [Google Scholar]
- Montufar-Solis D, Wang HC, Klein JR. Stimulatory and costimulatory effects of IL-18 directed to different small intestinal CD43 T cell subsets. J Leukoc Biol. 2007;82:1166–1173. doi: 10.1189/jlb.0207108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratama A, Ramiscal RR, Silva DG, Das SK, Athanasopoulos V, Fitch J, Botelho NK, Chang PP, Hu X, Hogan JJ, Mana P, Bernal D, Korner H, Yu D, Goodnow CC, Cook MC, Vinuesa CG. Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity. 2013;38:669–680. doi: 10.1016/j.immuni.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Schaefer JS, Montufar-Solis D, Nakra N, Vigneswaran N, Klein JR. Small intestine inflammation in roquin-mutant and roquin-deficient mice. PloS one. 2013;8:e56436. doi: 10.1371/journal.pone.0056436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer JS, Montufar-Solis D, Vigneswaran N, Klein JR. ICOS promotes IL-17 synthesis in colonic intraepithelial lymphocytes in IL-10−/− mice. J Leukoc Biol. 2010;87:301–308. doi: 10.1189/jlb.0409238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer JS, Montufar-Solis D, Vigneswaran N, Klein JR. Selective upregulation of microRNA expression in peripheral blood leukocytes in IL-10−/− mice precedes expression in the colon. J Immunol. 2011;187:5834–5841. doi: 10.4049/jimmunol.1100922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kobayashi-Osaki M, Tsutsumi S, Pan X, Ohmori S, Takai J, Moriguchi T, Ohneda O, Ohneda K, Shimizu R, Kanki Y, Kodama T, Aburatani H, Yamamoto M. GATA factor switching from GATA2 to GATA1 contributes to erythroid differentiation. Genes to cells : devoted to molecular & cellular mechanisms. 2013;18:921–933. doi: 10.1111/gtc.12086. [DOI] [PubMed] [Google Scholar]
- Vinuesa CG, Cook MC, Angelucci C, Athanasopoulos V, Rui L, Hill KM, Yu D, Domaschenz H, Whittle B, Lambe T, Roberts IS, Copley RR, Bell JI, Cornall RJ, Goodnow CC. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435:452–458. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- Vogel KU, Edelmann SL, Jeltsch KM, Bertossi A, Heger K, Heinz GA, Zoller J, Warth SC, Hoefig KP, Lohs C, Neff F, Kremmer E, Schick J, Repsilber D, Geerlof A, Blum H, Wurst W, Heikenwalder M, Schmidt-Supprian M, Heissmeyer V. Roquin paralogs 1 and 2 redundantly repress the Icos and Ox40 costimulator mRNAs and control follicular helper T cell differentiation. Immunity. 2013;38:655–668. doi: 10.1016/j.immuni.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Vogelzang A, McGuire HM, Yu D, Sprent J, Mackay CR, King C. A fundamental role for interleukin-21 in the generation of T follicular helper cells. Immunity. 2008;29:127–137. doi: 10.1016/j.immuni.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Wong LY, Hatfield JK, Brown MA. Ikaros sets the potential for Th17 lineage gene expression through effects on chromatin state in early T cell development. J Biol Chem. 2013;288:35170–35179. doi: 10.1074/jbc.M113.481440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G, Kanno Y, O’Shea JJ, Laurence A. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12:247–254. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap WH, Yeoh E, Tay A, Brenner S, Venkatesh B. STAT4 is a target of the hematopoietic zinc-finger transcription factor Ikaros in T cells. FEBS letters. 2005;579:4470–4478. doi: 10.1016/j.febslet.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Yu D, Tan AH, Hu X, Athanasopoulos V, Simpson N, Silva DG, Hutloff A, Giles KM, Leedman PJ, Lam KP, Goodnow CC, Vinuesa CG. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450:299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ing S, Fraser A, Chen M, Khan O, Zakem J, Davis W, Quinet R. Follicular helper T cells: new insights into mechanisms of autoimmune diseases. The Ochsner journal. 2013;13:131–139. [PMC free article] [PubMed] [Google Scholar]


