Abstract
The cytokine IFN-γ has well-established antibacterial properties against the bacterium Salmonella enterica in phagocytes, but less is known about the effects of IFN-γ on Salmonella-infected non-phagocytic cells, such as intestinal epithelial cells (IECs) and fibroblasts. Here we show that exposing human and murine IECs and fibroblasts to IFN-γ following infection with Salmonella triggers a novel form of cell death that is neither pyroptosis, nor any of the major known forms of programmed cell death. Cell death required IFN-γ-signaling via STAT1-IRF1-mediated induction of Guanylate Binding Proteins (GBPs) and the presence of live Salmonella in the cytosol. In vivo, ablating IFN-γ signaling selectively in murine IECs led to higher bacterial burden in colon contents, and increased inflammation in the intestine of infected mice. Together, these results demonstrate that IFN-γ signaling triggers release of Salmonella from the SCV into the cytosol of infected non-phagocytic cells, resulting in a form of non-pyroptotic cell death that prevents bacterial spread in the gut.
Keywords: Interferon-γ, Salmonella, GBPs, pyroptosis, necroptosis, apoptosis
Introduction
Salmonella enterica is a facultative intracellular bacterium that causes severe foodborne illness in humans worldwide (1, 2). Even though S. enterica infection is more common in underdeveloped countries, it is still very prevalent in the United States, infecting millions annually, usually through handling of raw or undercooked meats (3). Of the many serovars of S. enterica; two (Typhi and Typhimurium), are the most common causes of Salmonella sp. illnesses in humans (4, 5). S. Typhi infection is more commonly found in countries that lack sanitary food or drinking water, and infects via the fecal-oral route following ingestion the bacterium (6). If untreated, S. Typhi infection leads to severe fever and can be fatal (7). S. Typhimurium also infects humans but induces a self-limiting gastroenteritis that normally does not require treatment (8). S. Typhi does not significantly activate the host inflammasome machinery, and spreads systemically through the host, while S. Typhimurium, which induces a robust immune response, is rapidly cleared after limited spread in the intestine (9, 10). S. Typhi does not cause disease in mice, and therefore S. Typhimurium, which is pathogenic in mice, is used to model Salmonella infection in murine systems (11, 12).
Although the outcomes of infection are different between the two serovars, the initial stages of infection and immune mechanisms triggered are very similar. Upon ingestion, Salmonella first infects cells of the gut epithelium (13). Salmonella can invade these cells through use of an acquired pathogenicity island (SPI-1) that contains a Type III Secretion System (T3SS) (10, 14–16). SPI-1 has also recently been shown to prolong cell survival in non-phagocytic cells through Akt (17). After invasion, Salmonella induces formation of a cytosolic vacuole, called the Salmonella-containing vacuole (SCV), around the bacterium that allows for protection against host cytosolic anti-bacterial responses. For survival and replication in phagocytic cells, Salmonella utilizes a second pathogenicity island (SPI-2) that is required to survival in the low pH of these cell types. (18, 19).
The host macrophage has in place a mechanism of controlling Salmonella infection by preventing replication in the SCV. A family of IFN-inducible GTPases, called Guanylate Binding Proteins (GBPs), localize to the SCV after infection and lead to the formation of pores in the vacuole, releasing Salmonella into the cytosol of the infected macrophage (20, 21). LPS is sensed by the NLRC4 inflammasome machinery, or directly by caspase-11, triggering cleavage of caspase-1/11 and activation of Gasdermin D, leading to activation of pyroptosis, a pro-inflammatory form of cell death (22–26). In the absence of caspase-1, Salmonella can induce caspase-8 dependent cellular extrusion in intestinal epithelial cells (IECs) (27). Macrophages may also undergo necroptosis upon infection with Salmonella, dependent on Type I interferon (IFN) signaling (28).
Most studies of Salmonella-induced cell death have been conducted in macrophages and other phagocytes, and less is known about the role or mechanism of programmed cell death pathways in the control of Salmonella pathogenesis in non-phagocytic cells. As we and others have previously shown that interferons induce necroptosis in MEFs and other non-phagocytic cell types (29), and as Salmonella was reported to trigger IFN-induced necroptosis (28), we sought to test if Salmonella can induce cell death in non-phagocytic cells, and whether death was interferon-dependent necroptosis.
Here, we show that while Salmonella on its own does not induce cell death in non-phagocytic cells, exposure of infected cells to IFN-γ, but not to other cytokines tested, triggered robust cell death that could not be abrogated by preventing pyroptosis, apoptosis, necroptosis, nor any of the major forms of cell death or a combination of these. This pathway requires IRF-1-mediated induction of GBPs and subsequent SCV lysis and release of Salmonella into the cytosol of cells. In vivo, mice selectively deficient in IFN-γ signaling in IECs had higher levels of colonic bacteria and increased intestinal inflammation. Together, these results suggest that, as in phagocytic cells, non-phagocytic cells such as IECs and fibroblasts require IFN-γ signaling to undergo cell death and clear Salmonella infection from the colon. But, unlike in phagocytic cells, such cell death proceeds by a mechanism that is not reliant on any of the primary modes of programmed cellular demise. In addition to the ability of IFN-γ to control bacteria via macrophage dependent mechanisms, this study now identifies a non-pyroptotic form of IFN-γ dependent death in non-phagocytic cells.
Materials and Methods
Mice, Cells, and Reagents
Wild-type, ripk3−/− (30), tbk1−/− (31), gbpchr3−/− (32), and stat1−/− (33) MEFs were generated in-house from E14.5 embryos and used within five passages in experiments. In some studies, immortalized MEFs, generated by a 3T3 protocol (34), were used. Early passage irf1−/ −(Jovan Pavlovic), stinggt/gt, tnfr1−/− zbp1−/− (Jason Upton), ripk3−/−casp8−/−, fadd−/−mlkl−/− (Douglas Green) and trif −/− (Edward Mocarski) MEFs were obtained from the indicated laboratories. All other cell lines were obtained from the ATCC. Mice were housed in SPF facilities at the Fox Chase Cancer Center and experiments were conducted under protocols approved by the Committee on Use and Care of Animals at this institution. Reagents were obtained from the following sources: mIFN-γ (R&D systems), hIFN-γ (R&D systems), mIFN-β (PBL), mIL-1β (R&D systems), TNF-α (R&D systems), mIL-6 (R&D systems), IFN-α (R&D systems), JAK inhibitor I (Calbiochem), RIPK3 inhibitor GSK’843 (GSK), RIPK1 inhibitor GSK’963 (GSK), zVAD.fmk (Bachem), 3-MA (Sigma), Ferrostatin (Scott Dixon at Stanford University), LPS (Sigma), Streptomycin (Sigma) and YVAD (Enzo). Antibodies for immunoblotting: anti-IRF1 (1:1000, Santa Cruz), anti-GBP2 (1:1000, Santa Cruz), anti-β-actin (1:2000 Sigma). Antibodies for microscopy: anti-GBP2 [1:1000, gift of Jörn Coers (35)], anti-GFP (1:1000, ThermoFisher), fluorophore-conjugated secondary antibodies (1:500, Abcam and Jackson)
Generation of IEC-specific IFNGR2-deficient mice
IFNGR2flox/flox mice were generated using targeted ES cells obtained from the KOMP repository and injected into C57Bl6 Albino blastocysts by the FCCC Transgenic Facility. Chimeric mice were obtained and crossed to C57Bl6 Albino mice, and construct germline transmission was monitored by coat color and confirmed by PCR. Frt-site flanked beta-Gal and Neo cassettes were excised in vivo by crossing targeted germline transmitted mice with ACTA-FLP mice from Jackson Laboratories. The resultant heterozygous mice had Ifngr2 exon 3 flanked with loxP sites. To generate conditional knockout of IFNGR2 in the intestinal epithelium, IFNGR2 floxed mice were intercrossed with Villin-Cre mice (B6.Cg-Tg(Vil1-cre)997Gum/J; Jackson Labs) (36). Cre+ and Cre− littermate control mice were genotyped by standard PCR and used for subsequent experimentation. Detailed generation of mice will be reported elsewhere.
Bacterial strains
Salmonella Typhimurium strain SL1344 was used as the wild type in all experiments. Salmonella-GFP and Salmonella-RFP were obtained from Mary O’Riordan. ΔsifA, ΔAroC and ΔAroCsifA mutants were provided by David Holden. SPI-1 and 2 mutants have been described previously (37, 38).
Infection of cells
Salmonella was grown overnight shaking at 37°C in Luria Broth (LB) containing streptomycin. 1 mL of this culture was then grown in 100 mL LB without antibiotics for an additional 3 hours at 37°C, until an OD of 0.700 was reached. Cells were then pelleted (4000g for 10 min) and the pellet was resuspended in 50mL serum free DMEM. The OD600 of 1mL of this suspension was measured and used to determine MOI (1.00 OD600 = 1.00 × 109 cfu). Salmonella was added to cells in serum-free DMEM for 30 minutes. The medium was then removed and each well was washed 3× with serum free DMEM. Complete (10% fetal bovine serum) medium containing 50ug/mL gentamycin was then added to each well. After an additional 30 minutes, this medium was replaced with medium containing 5ug/mL gentamycin and IFN-γ for the remainder of the experiment. Cell viability was determined by Trypan Blue exclusion. To determine the proportion of infected cells by FACS, cells were infected with Salmonella-GFP and GFP positivity was measured using a Becton Dickinson FACScan scatter analyzer.
Immunofluorescence
An expression vector encoding LAMP1-GFP fusion protein (Addgene) was retrovirally transduced into immortalized wild-type MEFs, and populations stably expressing LAMP1-GFP were obtained by selection in hygromycin. These cells were plated on 4-well glass slides (Millipore). After infection with Salmonella and/or treatment with IFN-γ, the cells were fixed with 4% (w/v) paraformaldehyde, permeabilized in 0.2% (v/v) Triton-X, and blocked with 3% (w/v) BSA in PBS containing 0.1% Triton-X. Cells were then incubated overnight at 4°C with anti-GFP and/or anti-GBP2 antibodies. After 3× washes in PBS, samples were incubated with fluorophore conjugated secondary antibodies for 1 h at room temperature. Following an additional 3× washes in PBS, cells were mounted in Pro-long Gold antifade reagent (Invitrogen) and imaged by confocal microscopy on a Leica SP8 instrument.
Infection of mice
Eight- to ten-week-old sex-matched control (IFNGR2fl/fl) and IFNGR2-ΔIEC (IFNGR2ΔIEC/ΔIEC) mice were treated with 20 mg streptomycin by oral gavage 24 hours prior to infection. Mice were inoculated intragastrically with either 0.1 ml of sterile LB (mock infection) or an equal volume of LB containing S. Typhimurium (109 CFU/mouse) grown in LB broth at 37°C overnight with shaking. Mice were sacrificed 48 hours after infection. To determine the colony forming units of S. Typhimurium, tissue samples of liver, spleen, colon contents and cecum were collected, weighed and homogenized in 5 ml of sterile PBS. Bacteria were enumerated by plating 10-fold serial dilutions of tissue homogenates on LB agar plates supplemented with streptomycin (100mg/ml in water). The cecum and segments of the colon were fixed in 10% formalin and embedded in paraffin for histopathological analysis.
Histopathological analyses
Segments of the ileum, cecum, and colon were collected and fixed in 10% phosphate-buffered formalin (or 4% formaldehyde; 37% saturated formaldehyde = 100% formalin) 24–48 hrs, dehydrated and embedded in paraffin. Paraffin blocks were cut into 5-µm sections, mounted on microscope slides and stained with hematoxylin and eosin. Histopathological evaluation was performed in a blinded manner, using the following histopathoogical scoring scheme (39), (i) Submucosal edema. Submucosal edema was scored as follows: 0 = no pathological changes; 1 = mild edema (the submucosa is <0.20 mm wide and accounts for <50% of the diameter of the entire intestinal wall (tunica muscularis to epithelium); 2 = moderate edema; the submucosa is 0.21 to 0.45 mm wide and accounts for 50 to 80% of the diameter of the entire intestinal wall; and 3 = profound edema (the submucosa is >0.46 mm wide and accounts for >80% of the diameter of the entire intestinal wall). The submucosa widths were determined by quantitative microscopy and represent the averages of 30 evenly spaced radial measurements of the distance between the tunica muscularis and the lamina mucosalis mucosae. (ii) PMN infiltration into the lamina propria. Polymorphonuclear granulocytes (PMN) in the lamina propria were enumerated in 8 high-power fields (×400 magnification), and the average number of PMN/high-power field was calculated. The scores were defined as follows: 0 = <5 PMN/high-power field; 1 = 5 to 20 PMN/high-power field; 2 = 21 to 60/high-power field; 3 = 61 to 100/high-power field; and 4 = >100/high-power field. Transmigration of PMN into the intestinal lumen was consistently observed when the number of PMN was >60 PMN/high-power field. (iii) Epithelial integrity. Epithelial integrity was scored as follows: 0 = no pathological changes detectable in 10 high-power fields (×400 magnification); 1 = epithelial desquamation; 2 = erosion of the epithelial surface (gaps of 1 to 10 epithelial cells/lesion); and 3 = epithelial ulceration (gaps of >10 epithelial cells/lesion; at this stage, there is generally granulation tissue below the epithelium).
Statistics
Statistical significance was determined by use of Student’s t-test. Significance of in vivo data was determined by two-sided Wilcoxon Rank Sum test. P values of 0.05 or lower were considered significant. Graphs were generated using GraphPad 6.0 Prism software.
Results
IFN-γ sensitizes non-phagocytic cells to Salmonella-triggered cell death
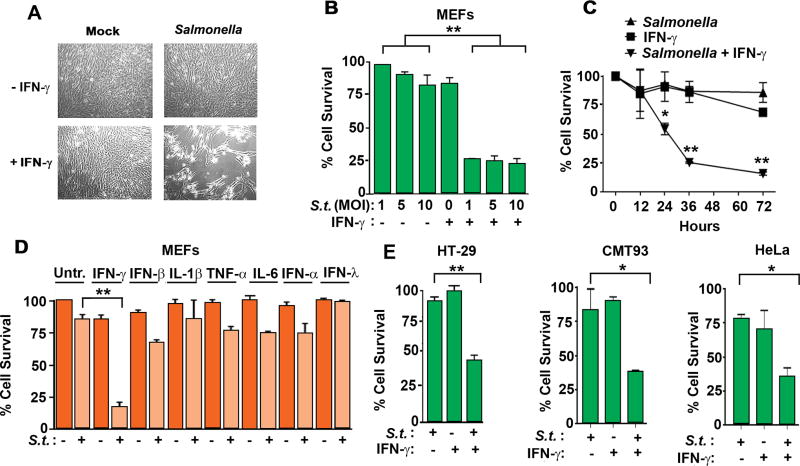
While examining the effects of Salmonella infection on murine embryonic fibroblasts (MEFs), we made the unexpected observation that, while Salmonella on its own did not cause much cell death, subsequent exposure of infected cells to IFN-γ (1 hour post infection in this and later experiments) triggered rampant cell death (Fig. 1A). To further evaluate this phenomenon, we infected MEFs with a broad dose range of Salmonella; from this analysis, we found that, while cells that were not treated with IFN-γ remained resistant to Salmonella mediated cell death up to MOIs of 25, an MOI of 1 robustly killed MEFs when IFN-γ was added to cells after infection (Fig. 1B). Cell death was first observed ~12 hours after infection and most cells were dead by 36 hours (Fig. 1C). Remarkably, this effect was unique to IFN-γ, as neither type I/III IFNs (IFN-α4, IFN-β, IFN-λ3) nor the pro-inflammatory cytokines IL-1β, TNF-α, or IL-6, sensitized MEFs to Salmonella-induced cell death (Fig. 1D). Although Salmonella undergoes its full replication cycle in fibroblasts, it is not a cell type commonly encountered during the course of infection. We therefore also tested more physiologically relevant cell types such as HT-29 human epithelial cells, CMT93 murine colorectal cells, and HeLa human cervical carcinoma cells (Fig. 1E) and found that these cells also succumbed to the combination of Salmonella and IFN-γ, but not significantly to either stimulus, when these were deployed singly. Together, these data demonstrate that, while Salmonella does not trigger much cell death on its own in non-phagocytic cells, the addition of IFN-γ after Salmonella infection leads to robust cell death over a 36 hour timeframe.
Figure 1. IFN-γ sensitizes non-phagocytic cells to Salmonella-triggered cell death.
(A) Photomicrographs of wild-type MEFs either mock-infected (left) or infected with Salmonella (MOI 10, right), and subsequently treated with IFN-γ (10ng/ml, bottom panels). Cells were exposed to IFN-γ 1h after infection and photomicrographs were taken 48h after infection. (B) Cell viability of MEFs infected with Salmonella (MOIs 1, 5 and 10) and subsequently exposed to IFN-γ (10ng/ml). (C) Kinetics of cell death induced by Salmonella (MOI 10) in the presence or absence of IFN-γ (10ng/ml). (D) Viability of wild-type MEFs infected with Salmonella (MOI 10) and treated with IFN-γ, IFN-β, IL-1β, TNF-α, IL-6, IFN-α (α4) and IFN-λ (λ3). All cytokines were used at 10 ng/ml (E) Viability of HT-29 cells (MOI 10; left), CMT93 cells (MOI 50; center) and HeLa cells (MOI 10; right) infected with Salmonella in the presence or absence of human IFN-γ (10ng/ml). Viability data shown in this figure are representative of at least three independent experiments. Error bars represent mean +− SD. *p<0.05, **p<0.005
IFN-γ promotes a novel form of cell death in Salmonella-infected non-phagocytic cells
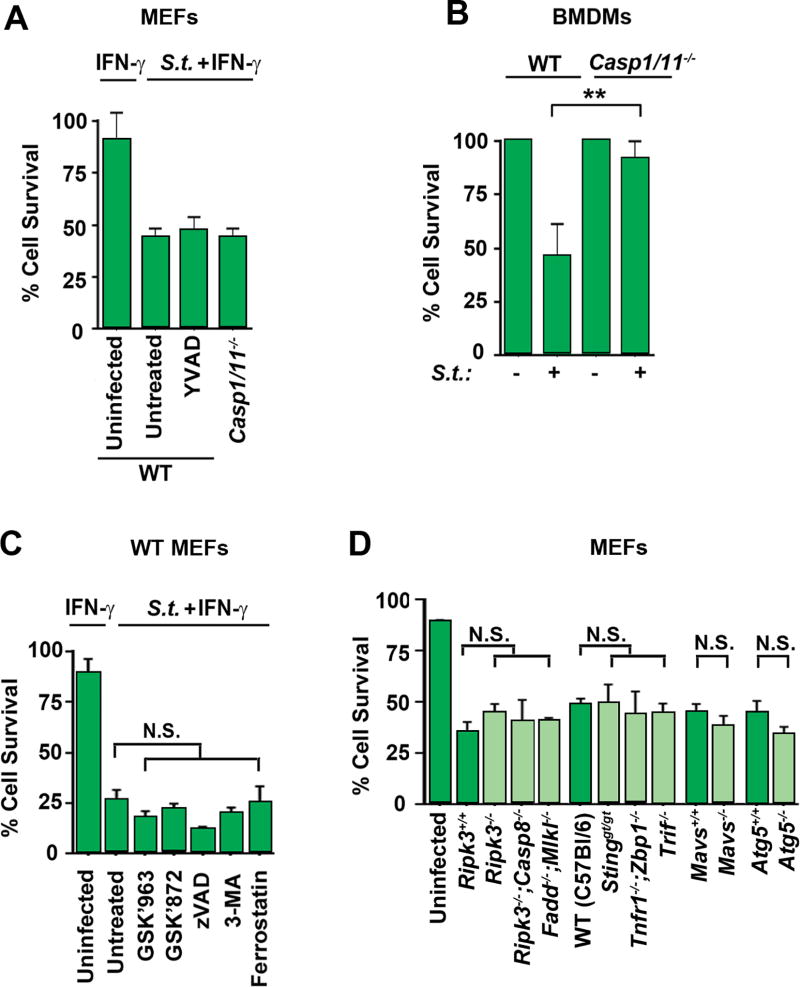
To determine if such cell death was pyroptosis, we infected MEFs from pyroptosis-deficient (caspase-1/11 null) mice with Salmonella before exposing them to IFN-γ. Surprisingly, these MEFs were as susceptible to Salmonella-triggered cell death as WT MEFs (Fig. 2A), even though macrophages from these mice were resistant to death after 90 minutes following Salmonella infection (Fig. 2B). Moreover, treatment of cells with the caspase-1 inhibitor YVAD.fmk, at doses shown to block pyroptosis in macrophages (40), did not block cell death seen in MEFs (Fig. 2A), indicating that Salmonella-induced death in MEFs was neither canonical nor non-canonical pyroptosis. We next tested MEFs either treated with small-molecule inhibitors of other known cell death pathways, or lacking essential components of major pathways of programmed cell death and/or innate signaling, for protection against the combination of Salmonella + IFN-γ. We found that treating wild-type MEFs with inhibitors of necroptosis (RIPK1 inhibitor GSK’963 and RIPK3 inhibitor GSK’843), apoptosis (pan-caspase inhibitor zVAD.fmk), autophagy (PI3K inhibitor 3-MA) or ferroptosis (erastin inhibitor ferrostatin) could not rescue cells from Salmonella + IFN-γ mediated cell death (Fig. 2C). Similarly, MEFs lacking essential components of necroptosis (Ripk3−/−), or both necroptosis and death-receptor-mediated apoptosis (Ripk3−/−;Casp8−/−, Fadd−/−;Mlkl−/−) also succumbed to Salmonella with kinetics and magnitude not significantly different from wild type MEFs, indicating that cell death induced by Salmonella + IFN-γ was neither necroptosis, apoptosis, autophagy, nor ferroptosis (Fig. 2D). In agreement, neither necroptosis (as measured by examining phospho-MLKL and phospho-RIPK3 by immunoblotting), apoptosis (cleaved caspase-3) or pyroptosis (cleaved caspase-1) was activated to any detectable extent by Salmonella + IFN-γ in MEFs (Fig. S1). Moreover, MEFs lacking functional STING (Goldenticket; Stinggt/gt), DAI (zbp1−/−), TNF (tnfr1−/−;zbp1−/−), TRIF (trif −/−) or RLR (mavs−/−) signaling succumbed to Salmonella + IFN-γ, indicating that none of these signaling pathways were essential for cell death induced by Salmonella in cells exposed to IFN-γ (Fig. 2D). Together, these findings demonstrate that Salmonella-induced death after exposure to IFN-γ requires GBP induced SCV lysis and is not singly mediated by any of the known major innate pathways of programmed cell death.
Figure 2. IFN-γ promotes a novel form of cell death in Salmonella-infected non-phagocytic cells.
(A) Wild-type or caspase1−/−caspase11−/− double knockout MEFs were infected with Salmonella (MOI 10) and exposed to IFN-γ (10ng/ml) with or without the presence of caspase-1 inhibitor YVAD (10uM), and cell viability was determined 48 h.p.i. (B) Wild-type and caspase1/11 double knockout bone-marrow derived macrophages (BMDMs) were infected with Salmonella (MOI 25) and cell viability was determined after 90 minutes. (C) Wild-type MEFs were infected with Salmonella (MOI 10) and exposed to IFN-γ (10ng/ml) in the presence of RIPK1 inhibitor GSK’963 (5uM), RIPK3 inhibitor GSK’843 (5uM), pan-caspase inhibitor zVAD (50uM), PI3K inhibitor 3-MA (5mM), erstatin inhibitor Ferrostatin (2.5uM) and cell viability was determined 48 h.p.i. (D) ripk3−/−, ripk3−/−casp8−/− double knockout, fadd−/−mlkl−/− double knockout, sting goldenticket mutant, tnfr1−/−zbp1−/− double knockout trif−/−, mavs−/− or atg5−/− knockout MEFs, along with wild-type controls, were infected with Salmonella in the presence of IFN-γ (10ng/ml) and cell viability was determined 48 h.p.i. Viability data shown in this figure are representative of at least three independent experiments. Error bars represent mean +− SD. **p<0.005
IFN-γ sensitizes Salmonella-infected non-phagocytic cells to death via Jak/STAT1-mediated induction of IRF-1 and GBPs
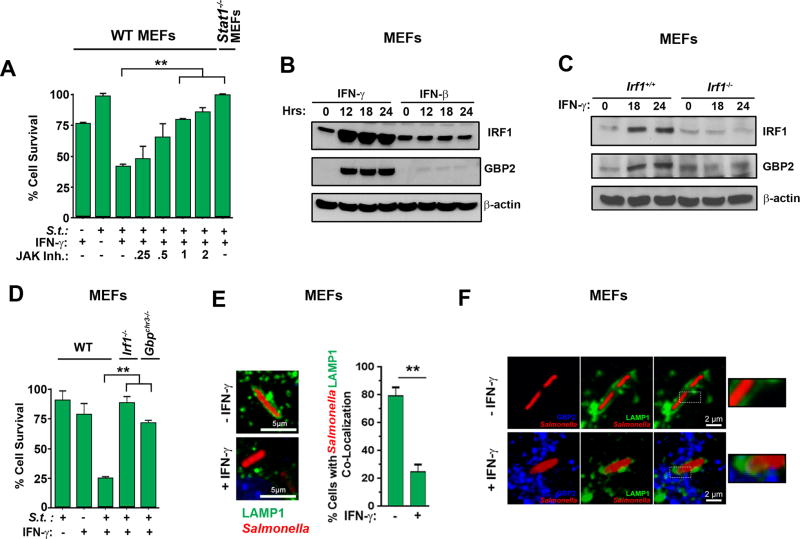
IFN-γ typically mediates its effects via a Jak1/2-STAT1-mediated transcriptional program that activates the expression of hundreds of genes, called interferon-stimulated genes, or ISGs (41). To test if IFN-γ required Jak/STAT signaling to sensitize non-phagocytic cells to Salmonella-triggered cell death, we infected MEFs with Salmonella, following which we treated them with IFN-γ in the presence of a potent inhibitor of JAK1/2 kinase activity (JAK Inhibitor I). Co-treatment with this inhibitor, in a dose dependent manner, efficiently protected cells from Salmonella-triggered death (Fig. 3A). Similarly, MEFs lacking STAT1 were completely protected against cell death induced by Salmonella and IFN-γ (Fig. 3A). Previous studies have shown that IFN-γ sensitizes macrophages to pyroptosis by induction of the ISGs encoding IRF1 and GBPs (4, 20, 21). In a two-step Jak/STAT-mediated process, IFN-γ first induces the rapid production of the transcription factor IRF1, which then drives induction of the genes encoding GBPs. These GBPs traffic to the SCV and promote its rupture, releasing Salmonella into the cytosol. The inflammasome machinery senses cytosolic Salmonella, resulting in pyroptosis (20, 21, 23, 42). To examine if IFN-γ promoted death of Salmonella-infected cells also involved IRF1 and GBPs, we determined expression levels of these proteins in MEFs. Neither IRF1 nor a representative GBP (GBP2) were expressed at significant levels in unstimulated MEFs, but both IRF1 and GBP2 were induced to high levels within 12 hours by IFN-γ, but not by IFN-β in uninfected cells (Fig. 3B). Indeed, IRF1 was induced in as few as 30 minutes after treatment with IFN-γ (data not shown). GBP2 was not induced to any significant extent in IRF1-deficient MEFs, demonstrating that IRF1 was required for production of GBP2 by IFN-γ (Fig. 3C) Importantly, MEFs lacking irf1 or harboring a deletion in the genetic locus on murine chromosome 3 encoding GBPs 1,2,3,5,7,2ps (gbpChr3−/−) were largely resistant to Salmonella-triggered death in the presence of IFN-γ (Fig. 3D). Reconstituting gbpchr3−/− MEFs by stable retroviral reintroduction of GBP2 restored susceptibility to Salmonella + IFN-γ induced cell death (Fig. S2). Thus, as in phagocytic cells, IFN-signaling via a Jak/STAT1-IRF-1 axis activates GBPs that lead to eventual death of the infected fibroblast. To determine if IFN-γ-induced GBPs promote lysis of the SCV and release of Salmonella into the cytosol of cells, we stably expressed the SCV marker LAMP1-GFP (43) in MEFs, infected these cells with Salmonella-RFP, exposed them to IFN-γ (10ng/ml), and examined integrity of their SCVs. We observed that ~80% of infected cells not treated with IFN-γ displayed intact SCVs, as measured by uniform encapsulation of Salmonella-RFP by LAMP1-GFP (Fig. 3E). On the other hand, only ~25% of infected cells exposed to IFN-γ contained intact SCVs; the rest of these cells showed cytosolic distribution of Salmonella-RFP that did not localize with GFP signal (Fig. 3E). Endogenous GBP2 was undetectable in untreated cells, but was readily observed in a punctate cytosolic pattern in cells exposed to IFN-γ. In these cells, a subset of GBP2 co-localized with LAMP1 and Salmonella (Fig. 3F). Collectively, these findings demonstrate that IFN-γ induces GBPs to disrupt the SCV and release Salmonella into the host cell cytosol.
Figure 3. IFN-γ sensitizes Salmonella-infected non-phagocytic cells to death via Jak/STAT1-mediated induction of IRF1 and GBPs.
(A) Viability of wild-type MEFs infected with Salmonella (MOI 10) and treated with IFN-γ (10 ng/ml) in the presence of increasing amounts of JAK inhibitor I. Viability of stat1−/− MEFs infected with Salmonella (MOI 10) and treated with IFN-γ (10 ng/ml) is also shown (rightmost bar). (B) Wild-type MEFs treated with IFN-γ or IFN-β (10ng/ml each) for the indicated times were examined for IRF1 and GBP2 by immunoblotting. β-actin was used as a loading control. (C) Irf1+/+ and irf1−/− MEFs were treated with IFN-γ (10ng/ml) for the indicated times and examined for IRF1 and GBP2 by immunoblotting. (D) Wild-type, irf1−/− and gbpchr3−/− MEFs were infected with Salmonella (MOI 10) in the presence or absence of subsequent IFN-γ treatment (10ng/ml) and cell viability was determined 48 h.p.i. (E) Wild-type MEFs stably expressing LAMP1-GFP were infected with Salmonella-RFP in the presence or absence of IFN-γ (10ng/ml), and localization of LAMP1-GFP and Salmonella-RFP was determined by confocal microscopy. Representative images of co-localization (left) and quantification of cells showing co-localized Salmonella with LAMP1-GFP, indicative of intact SCVs (right) are shown. (F) Wild-type MEFs stably expressing LAMP1-GFP were infected with Salmonella-RFP and subsequently treated with IFN-γ (10ng/ml). Localization of Salmonella (red), LAMP1 (green) and GBP2 (blue) was determined by confocal microscopy. Enlarged images of boxed sections are shown to the right. Note that LAMP1-GFP encapsulates Salmonella in the absence of IFN-γ. Upon IFN-γ treatment, LAMP1-GFP encapsulation is lost and GBP2-LAMP1-Salmonella co-localization becomes evident. Viability data shown in this figure are representative of at least three independent experiments. Error bars represent mean +− SD. **p<0.005.
Induction of cell death requires live Salmonella in the cytosol
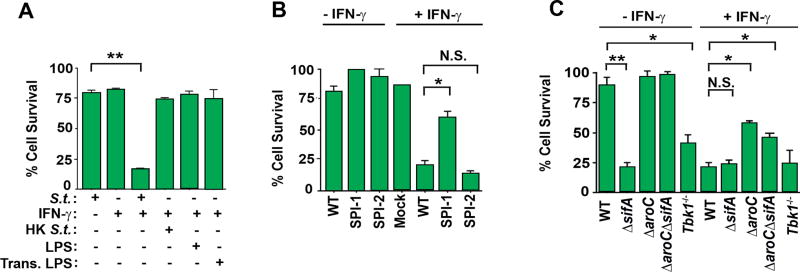
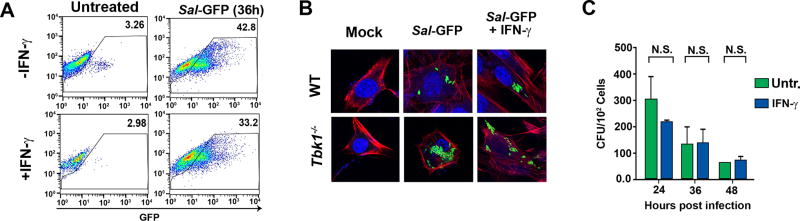
To identify Salmonella determinants required for induction of cell death, we first tested the requirement for live Salmonella in this process. We found that, while live Salmonella + IFN-γ induced robust cell death in wild-type MEFs, treating these cells with heat killed Salmonella (HK S.t.) or exposing MEFs to the gram-negative bacterial cell wall component LPS did not trigger cell death in the presence of IFN-γ (Fig. 4A). Transfecting LPS into the cytosol of cells, capable of activating the pyroptotic machinery in macrophages (23), also did not kill cells in the presence of IFN-γ, indicating that live Salmonella was necessary for cell death (Fig. 4A). A mutant of Salmonella lacking the first of its T3SS (invA mutant, called ΔSPI-1 hereafter) was largely unable to kill cells, even in the presence of IFN-γ, while a mutant lacking the second T3SS (ssaR mutant, called ΔSPI-2 hereafter) was able to robustly kill cells following exposure to IFN-γ, similar to wild-type Salmonella (Fig. 4B). As Salmonella SPI-1, but not SPI-2, is required for the invasion of MEFs (44), these findings demonstrate that Salmonella must invade cells in order to induce IFN-γ mediated cell death. Furthermore, as ΔSPI-2 Salmonella behaves in this experiment as the wild-type bacterium does, induction of cell death in non-phagocytic cells was not dependent on a SPI-2 effector protein(s) (Fig. 4B). A mutant of Salmonella that can directly enter the cytosol of cells without forming the SCV (ΔsifA) (45), killed MEFs without the need for IFN-γ, suggesting that the primary role of IFN-γ was to lyse the SCV and release Salmonella into the cytosol. Notably, a Salmonella mutant that can invade MEFs and enter the cytosol, but cannot replicate once in the cell (ΔsifA;ΔaroC) was defective in its capacity to induce cell death, demonstrating that, while live Salmonella in the cytosol was required for death, active replication was also necessary. In line with the argument that cell death necessitated live Salmonella in the cytosol of infected cells, infecting MEFs lacking TBK1, a host kinase required for the formation of the SCV (46), with wild-type Salmonella triggered cell death without the need for IFN-γ (Fig. 4C).
Figure 4. Induction of cell death requires live Salmonella in the cytosol.
(A) Wild-type MEFs were infected with Salmonella (MOI 10), heat-killed Salmonella (MOI 10), treated with LPS (4 ng/ml), or transfected with LPS (4 ng/ml) in the presence or absence of IFN-γ (10ng/ml) and cell viability was determined 48 h.p.i. (B) Wild-type MEFs infected with either wild-type Salmonella (MOI 10), Salmonella lacking its first pathogenicity island (ΔSPI-1), or its second pathogenicity island (ΔSPI-2) were exposed to IFN-γ (10ng/ml) and cell viability was determined 48 h.p.i. (C) Wild-type MEFs were infected with either wild-type Salmonella (MOI 10), Salmonella lacking sifA (ΔsifA) (MOI 10), aroC (ΔaroC) or both sifA and aroC (ΔsifAΔaroC) (MOI 10), exposed to IFN-γ (10ng/ml), and cell viability was determined 48 h.p.i. In parallel, MEFs lacking tbk1 were infected with Salmonella (MOI 10) and exposed to IFN-γ (10ng/ml); cell viability was determined 48 h.p.i. Viability data shown in this figure are representative of at least three independent experiments. Error bars represent mean +− SD. *p<0.05, **p<0.005
IFN-γ does not significantly increase Salmonella replication
In other settings, the host cytosol has been shown to be permissive to Salmonella hyper-replication (47). To test if IFN-γ sensitized cells to death by licensing increased Salmonella infectivity or replication, we first examined if IFN-γ altered the proportion of infected MEFs, compared to untreated cells. IFN-γ did not result in a significant increase in the proportion of infected cells (Fig. 5A). IFN-γ also did not significantly alter the number of intracellular Salmonella, when examined by immunofluorescence (Fig. 5B, top row). In contrast, loss of TBK1 resulted in Salmonella hyper-replication, as previously reported (46, 48) Overall bacterial replication rates were largely indistinguishable between MEFs infected with Salmonella alone, or subject to the combination of Salmonella + IFN-γ, as demonstrated by direct measurement of intracellular bacterial numbers (Fig. 5C). These findings demonstrate that Salmonella + IFN-γ mediated cell death requires live Salmonella in the cytosol of infected cells, but that IFN-γ does not much alter overall replication rates of Salmonella; rather, it appears to increase the proportion of live Salmonella in the cytosol.
Figure 5. IFN-γ does not significantly increase Salmonella replication.
(A) FACS analysis of WT MEFs infected with Salmonella-GFP (MOI 25) for 36 hr. The y-axis shows side scatter. Numbers within each FACS panel show percent GFP-positive cells. Mock-infected cells showed negligible (<2%) GFP positivity. (B) Immunofluorescence staining of wild type or tbk1−/− MEFs infected with Salmonella-GFP (MOI 25) with or without the treatment of IFN-γ (10ng/ml) 36 h.p.i. Phalloidin staining is shown in red, DAPI staining is blue and Salmonella-GFP is green. (C) Colony counts per cell of wild-type MEFs infected with Salmonella (MOI 10) with and without IFN-γ. Colony count data shown in this figure are representative of at least three independent experiments. Error bars represent mean +− SD.
IFN-γ signaling in IECs is required to control Salmonella Replication in vivo
To examine the role of IFN-γ in controlling Salmonella specifically in non-phagocytic cells, we generated mice selectively deficient in IFN-γ signaling in cells of the intestinal epithelium. We achieved this by first producing IFNGR2fl/fl mice, which we then crossed into an IEC-specific Villin-Cre deletor strain to selectively ablate IFNGR2 expression (and IFN-γ signaling) in IECs (Fig. 6A). Histological analyses of cecum samples of Salmonella-infected (1 × 109 cfu by oral gavage) control (IFNGR2fl/fl) and IFNGR2-ΔIEC (IFNGR2ΔIEC/ΔIEC) mice after 48 hours showed that IFNGR2-ΔIEC mice had higher levels of polymorphonuclear granulocytes (PMN) infiltration and erosion of the intestinal lining (black arrows) compared to controls (blue arrows) (Fig. 6B). IFNGR2-ΔIEC mice infected with Salmonella had significantly higher overall intestinal damage, as measured by scoring epithelial integrity, PMN infiltration into the lamina propria, and edema, compared to control mice (Fig. 6C,D).
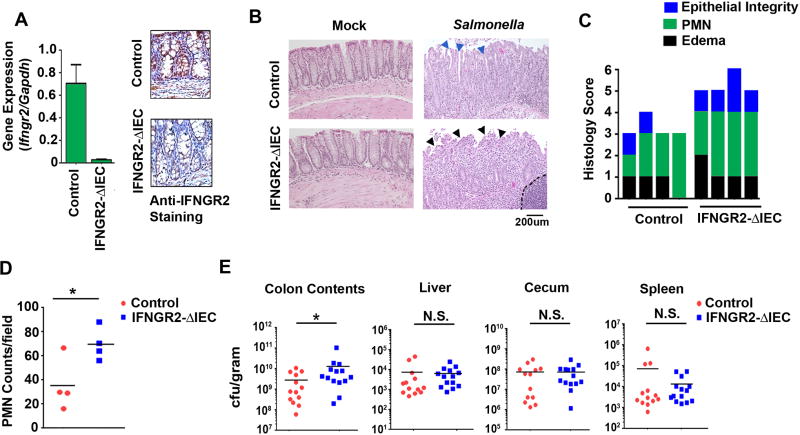
Figure 6. Ablating IFN-γ signaling in IECs increases Salmonella spread and pathology in vivo.
(A) RT-PCR for expression of IFNGR2 gene expression in colons of IFNGR2fl/fl and IFNGR2ΔIEC/ΔIEC mice, and histological staining of colons from IFNGR2 (shown in brown) in IFNGR2fl/fl and IFNGR2ΔIEC/ΔIEC mice. (B) Representative 200× H&E stained sections of intestines from uninfected (Mock) or Salmonella infected (1 × 109 cfu) control and IFNGR2-ΔIEC mice 48 h.p.i. Blue arrows indicates intact intestinal lining in control mice and black arrows depicts erosion of the intestinal lining in IFNGR2-ΔIEC mice. (C) Histological scoring of uninfected (Mock), and Salmonella (1 × 109 cfu) infected control and IFNGR2-ΔIEC mice after 48 hours. Histological scoring consists of submucosal edema (black), polymorphonuclear granulocyte infiltration into the lamina propria/high-powered field (PMN, green) and epithelial integrity (blue) scores of H&E stained colonic sections (D) PMN/high power field counts in control and IFNGR2-ΔIEC mice. (E) Colony counts (cfu/gram) of the colon contents, liver, cecum and spleen from control and IFNGR2-ΔIEC mice infected with Salmonella (1 × 109 cfu) 48 h.p.i. Error bars represent mean +− SD. *p<0.05.
Importantly, Salmonella replicated to higher levels in IFNGR2-ΔIEC mice, as evidenced by significantly higher bacterial loads in the colon contents (Fig. 6E). However, deleting IFN-γ signaling in IECs did not notably affect bacterial dissemination beyond the colon, as similar bacterial counts were observed in the cecum, liver and spleen in IFNGR2-ΔIEC mice versus controls (Fig. 6E). Together, these results demonstrate that IFN-γ signaling in IECs limits Salmonella spread and alleviates tissue damage in the infected colon.
Discussion
Here we show that IFN-γ sensitizes non-phagocytic cells, including IECs and fibroblasts, to death upon infection with Salmonella, and that such cell death may help to control bacterial colonization in the gut in vivo. Mechanistically, we show that IFN-γ, via a Jak-STAT1 axis, induces IRF1 and up-regulates GBPs to promote disruption of the SCV and release of Salmonella into the cytosol, where the bacterium triggers cell death. Distinct from macrophages, GBP-mediated release of Salmonella into the cytosol of non-phagocytic cells activates a form of cell death that is neither caspase-1/11 driven pyroptosis previously shown in macrophages, nor any of the major known forms of programmed cell death.
Our data suggest that, as in macrophages, the dominant role of IFN-γ in facilitating the death of Salmonella-infected cells is to induce expression of GBPs, which leads to the lysis of the SCV, releasing Salmonella into the cytosol, where the bacterium triggers cell death. A mutant of Salmonella (ΔsifA) that can directly enter the cytosol of cells without forming an SCV kills cells without the need for IFN-γ; similarly, MEFs lacking TBK1, a protein required for stabilization of the SCV, also succumb to Salmonella-mediated cell death (46), supporting the idea that IFN-γ-driven disruption of the SCV underlies this cytokine’s ability to promote Salmonella-induced cell death. In agreement with the findings of Broz and colleagues (20), the cell death we observe requires live Salmonella, as heat-killed Salmonella, LPS or cytosolic LPS does not lead to cell death. Notably, exposure to IFN-γ does not lead to an appreciable increase in bacterial replication, as evidenced by FACS, microscopy, and measurement of intracellular bacterial numbers. As suggested by Holden and colleagues (45), the cytosol of the wild type fibroblast does not appear to be permissive to Salmonella hyper-replication.
Although the mechanism of cell death requires the release of Salmonella into the cytosol of cells, how cytosolic Salmonella induces cell death, and whether such cell death is programmed, remains unknown. In other settings, NLRP3/caspase-11 inflammasomes detect cytosolic Salmonella for activation of pyroptosis (20, 23), and RLR systems sense Salmonella nucleic acid for production of IFN (49); as-yet unknown host innate pathway(s), similar to these may sense Salmonella to activate cell death in non-phagocytes exposed to IFN-γ. Alternatively, a bacterial metabolite or other product may simply toxify the host cell cytosol, resulting in unprogrammed death; these possibilities remain to be examined.
In vivo, our data suggest that IFN-mediated destruction of Salmonella-infected IECs is required for local control of Salmonella infection. Mice lacking IFN-γ signaling selectively in the intestinal epithelium have significantly higher bacterial burden in their colonic contents and manifest increased inflammation and epithelial damage in their colons. Previous reports have demonstrated that caspase-11 (caspase-4 in humans) induces epithelial cell extrusion in IECs after Salmonella infection, which aids in the clearance of the bacterium (50, 51). A more recent study demonstrates that caspase-8 can also induce IEC extrusion in the absence of caspase-1 (27). These processes happen quickly after infection, are caspase-driven and possess hallmarks of pyroptosis (27, 50, 51), whereas the mechanism of cell death reported in the current study appears to be distinct from these reports, as cell death occurs over a more-delayed time course of 36–48 hours, and cells lacking caspase-1/11 and caspase-8, or cells in which caspase-dependent cell death pathways have been pharmacologically inhibited, still undergo cell death upon exposure to Salmonella + IFN-γ.
In conclusion, our results demonstrate that IFN-γ promotes death of Salmonella-infected epithelial cells and clearance of Salmonella in vivo. We propose a model in which Salmonella entry into intestine activates a robust inflammatory response that recruits IFN-γ-producing immune cells, such as NK cells and helper T cells to the sites of infection. IFN-γ secreted by these cells in the vicinity of infected intestinal epithelial cells leads to the induction of a STAT1-IRF1 signaling cascade that induces expression of GBPs, which then lyse the SCV, releasing Salmonella into the cytosol of epithelial cells, and triggering cell death. Such cell death likely exposes Salmonella to an innate host defense pathway(s) that destroys the infected cell, limiting Salmonella spread and consequent tissue damage in the colons of infected mice.
Supplementary Material
Acknowledgments
We gratefully acknowledge Jörn Coers, Chris Dillon, Doug Green, William Kaiser, Edward Mocarski, Jovan Pavlovic, Glenn Rall, and Jason Upton for cells and antibodies. We also thank Mary O’Riordan, David Holden and Denise Monack for recombinant and mutant strains of Salmonella. We are grateful to the Fox Chase Laboratory Animal Facility for assistance in production of IFNGR2fl/fl mice, and help with in vivo experiments.
Funding: This work was supported by NIH R00 DK088589 and Pew Scholar in Biomedical Sciences Award (to S.G.), and NIH grants CA168621, CA190542 and AI113469 to S.B. Additional funds were provided by NIH Cancer Center Support Grant P30CA006927. S.B. is a consultant for Ascend Biopharmaceuticals.
References
- 1.Pham OH, McSorley SJ. Protective host immune responses to Salmonella infection. Future Microbiol. 2015;10:101–110. doi: 10.2217/fmb.14.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scanu T, Spaapen RM, Bakker JM, Pratap CB, Wu LE, Hofland I, Broeks A, Shukla VK, Kumar M, Janssen H, Song JY, Neefjes-Borst EA, te Riele H, Holden DW, Nath G, Neefjes J. Salmonella Manipulation of Host Signaling Pathways Provokes Cellular Transformation Associated with Gallbladder Carcinoma. Cell Host Microbe. 2015;17:763–774. doi: 10.1016/j.chom.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Schwille-Kiuntke J, Unverdorben A, Weimer K, Schlarb AA, Gulewitsch MD, Ellert U, Enck P. Bacterial infections in childhood: A risk factor for gastrointestinal and other diseases? United European Gastroenterol J. 2015;3:31–38. doi: 10.1177/2050640614558346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunn JS, Marshall JM, Baker S, Dongol S, Charles RC, Ryan ET. Salmonella chronic carriage: epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol. 2014;22:648–655. doi: 10.1016/j.tim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bula-Rudas FJ, Rathore MH, Maraqa NF. Salmonella Infections in Childhood. Adv Pediatr. 2015;62:29–58. doi: 10.1016/j.yapd.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Dougan G, Baker S. Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever. Annu Rev Microbiol. 2014;68:317–336. doi: 10.1146/annurev-micro-091313-103739. [DOI] [PubMed] [Google Scholar]
- 7.Parry CM, Hien TT, Dougan G, White N, Farrar JJ. Typhoid Fever. New England Journal of Medicine. 2002;347:1770–1782. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- 8.Grassl GA, Finlay BB. Pathogenesis of enteric Salmonella infections. Curr Opin Gastroenterol. 2008;24:22–26. doi: 10.1097/MOG.0b013e3282f21388. [DOI] [PubMed] [Google Scholar]
- 9.Wilson RP, Raffatellu M, Chessa D, Winter SE, Tukel C, Baumler AJ. The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cell Microbiol. 2008;10:876–890. doi: 10.1111/j.1462-5822.2007.01090.x. [DOI] [PubMed] [Google Scholar]
- 10.Bruno VM, Hannemann S, Lara-Tejero M, Flavell RA, Kleinstein SH, Galan JE. Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS Pathog. 2009;5:e1000538. doi: 10.1371/journal.ppat.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song J, Willinger T, Rongvaux A, Eynon EE, Stevens S, Manz MG, Flavell RA, Galan JE. A mouse model for the human pathogen Salmonella typhi. Cell Host Microbe. 2010;8:369–376. doi: 10.1016/j.chom.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Brien AD, Rosenstreich DL, Taylor BA. Control of natural resistance to Salmonella typhimurium and Leishmania donovani in mice by closely linked but distinct genetic loci. Nature. 1980;287:440–442. doi: 10.1038/287440a0. [DOI] [PubMed] [Google Scholar]
- 13.de Jong HK, Parry CM, van der Poll T, Wiersinga WJ. Host-pathogen interaction in invasive Salmonellosis. PLoS Pathog. 2012;8:e1002933. doi: 10.1371/journal.ppat.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee CA, Jones BD, Falkow S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci U S A. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lara-Tejero M, Galan JE. Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect Immun. 2009;77:2635–2642. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen-Wester I, Hensel M. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 2001;3:549–559. doi: 10.1016/s1286-4579(01)01411-3. [DOI] [PubMed] [Google Scholar]
- 17.Finn CE, Chong A, Cooper KG, Starr T, Steele-Mortimer O. A second wave of Salmonella T3SS1 activity prolongs the lifespan of infected epithelial cells. PLoS Pathog. 2017;13:e1006354. doi: 10.1371/journal.ppat.1006354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srikanth CV, Mercado-Lubo R, Hallstrom K, McCormick BA. Salmonella effector proteins and host-cell responses. Cell Mol Life Sci. 2011;68:3687–3697. doi: 10.1007/s00018-011-0841-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McEwan DG, Richter B, Claudi B, Wigge C, Wild P, Farhan H, McGourty K, Coxon FP, Franz-Wachtel M, Perdu B, Akutsu M, Habermann A, Kirchof A, Helfrich MH, Odgren PR, Van Hul W, Frangakis AS, Rajalingam K, Macek B, Holden DW, Bumann D, Dikic I. PLEKHM1 regulates Salmonella-containing vacuole biogenesis and infection. Cell Host Microbe. 2015;17:58–71. doi: 10.1016/j.chom.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 20.Meunier E, Dick MS, Dreier RF, Schurmann N, Kenzelmann Broz D, Warming S, Roose-Girma M, Bumann D, Kayagaki N, Takeda K, Yamamoto M, Broz P. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–370. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- 21.Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 22.Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, Monack DM. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med. 2010;207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 24.Kayagaki N, Stowe IB, Lee BL, O'Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, Dixit VM. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 25.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 26.Ruhl S, Broz P. Caspase-11 activates a canonical NLRP3 inflammasome by promoting K(+) efflux. Eur J Immunol. 2015;45:2927–2936. doi: 10.1002/eji.201545772. [DOI] [PubMed] [Google Scholar]
- 27.Rauch I, Deets KA, Ji DX, von Moltke J, Tenthorey JL, Lee AY, Philip NH, Ayres JS, Brodsky IE, Gronert K, Vance RE. NAIP-NLRC4 Inflammasomes Coordinate Intestinal Epithelial Cell Expulsion with Eicosanoid and IL-18 Release via Activation of Caspase-1 and-8. Immunity. 2017;46:649–659. doi: 10.1016/j.immuni.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson N, McComb S, Mulligan R, Dudani R, Krishnan L, Sad S. Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat Immunol. 2012;13:954–962. doi: 10.1038/ni.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thapa RJ, Nogusa S, Chen P, Maki JL, Lerro A, Andrake M, Rall GF, Degterev A, Balachandran S. Interferon-induced RIP1/RIP3-mediated necrosis requires PKR and is licensed by FADD and caspases. Proc Natl Acad Sci U S A. 2013;110:E3109–3118. doi: 10.1073/pnas.1301218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newton K, Sun X, Dixit VM. Kinase RIP3 is dispensable for normal NF-kappa Bs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Mol Cell Biol. 2004;24:1464–1469. doi: 10.1128/MCB.24.4.1464-1469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, Akira S. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto M, Okuyama M, Ma JS, Kimura T, Kamiyama N, Saiga H, Ohshima J, Sasai M, Kayama H, Okamoto T, Huang DC, Soldati-Favre D, Horie K, Takeda J, Takeda K. A cluster of interferon-gamma-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity. 2012;37:302–313. doi: 10.1016/j.immuni.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 33.Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, Carver-Moore K, DuBois RN, Clark R, Aguet M, Schreiber RD. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 34.Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol. 2005;Chapter 28(Unit 28):21. doi: 10.1002/0471142727.mb2801s70. [DOI] [PubMed] [Google Scholar]
- 35.Haldar AK, Saka HA, Piro AS, Dunn JD, Henry SC, Taylor GA, Frickel EM, Valdivia RH, Coers J. IRG and GBP host resistance factors target aberrant, "non-self" vacuoles characterized by the missing of "self" IRGM proteins. PLoS Pathog. 2013;9:e1003414. doi: 10.1371/journal.ppat.1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 37.Brumell JH, Rosenberger CM, Gotto GT, Marcus SL, Finlay BB. SifA permits survival and replication of Salmonella typhimurium in murine macrophages. Cell Microbiol. 2001;3:75–84. doi: 10.1046/j.1462-5822.2001.00087.x. [DOI] [PubMed] [Google Scholar]
- 38.Galan JE, Curtiss R., 3rd Distribution of the invA, -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect Immun. 1991;59:2901–2908. doi: 10.1128/iai.59.9.2901-2908.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smalley C, Bechelli J, Rockx-Brouwer D, Saito T, Azar SR, Ismail N, Walker DH, Fang R. Rickettsia australis Activates Inflammasome in Human and Murine Macrophages. PLoS One. 2016;11:e0157231. doi: 10.1371/journal.pone.0157231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 42.Man SM, Karki R, Sasai M, Place DE, Kesavardhana S, Temirov J, Frase S, Zhu Q, Malireddi RK, Kuriakose T, Peters JL, Neale G, Brown SA, Yamamoto M, Kanneganti TD. IRGB10 Liberates Bacterial Ligands for Sensing by the AIM2 and Caspase-11-NLRP3 Inflammasomes. Cell. 2016;167:382–396. e317. doi: 10.1016/j.cell.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajashekar R, Liebl D, Chikkaballi D, Liss V, Hensel M. Live cell imaging reveals novel functions of Salmonella enterica SPI2-T3SS effector proteins in remodeling of the host cell endosomal system. PLoS One. 2014;9:e115423. doi: 10.1371/journal.pone.0115423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steele-Mortimer O, Brumell JH, Knodler LA, Meresse S, Lopez A, Finlay BB. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol. 2002;4:43–54. doi: 10.1046/j.1462-5822.2002.00170.x. [DOI] [PubMed] [Google Scholar]
- 45.Beuzon CR, Salcedo SP, Holden DW. Growth and killing of a Salmonella enterica serovar Typhimurium sifA mutant strain in the cytosol of different host cell lines. Microbiology. 2002;148:2705–2715. doi: 10.1099/00221287-148-9-2705. [DOI] [PubMed] [Google Scholar]
- 46.Radtke AL, Delbridge LM, Balachandran S, Barber GN, O'Riordan MX. TBK1 protects vacuolar integrity during intracellular bacterial infection. PLoS Pathog. 2007;3:e29. doi: 10.1371/journal.ppat.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, Steele-Mortimer O. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc Natl Acad Sci U S A. 2010;107:17733–17738. doi: 10.1073/pnas.1006098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmolke M, Patel JR, de Castro E, Sanchez-Aparicio MT, Uccellini MB, Miller JC, Manicassamy B, Satoh T, Kawai T, Akira S, Merad M, Garcia-Sastre A. RIG-I detects mRNA of intracellular Salmonella enterica serovar Typhimurium during bacterial infection. MBio. 2014;5:e01006–01014. doi: 10.1128/mBio.01006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knodler LA, Crowley SM, Sham HP, Yang H, Wrande M, Ma C, Ernst RK, Steele-Mortimer O, Celli J, Vallance BA. Noncanonical inflammasome activation of caspase-4/caspase-11 mediates epithelial defenses against enteric bacterial pathogens. Cell Host Microbe. 2014;16:249–256. doi: 10.1016/j.chom.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sellin ME, Muller AA, Felmy B, Dolowschiak T, Diard M, Tardivel A, Maslowski KM, Hardt WD. Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe. 2014;16:237–248. doi: 10.1016/j.chom.2014.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.