SUMMARY
A standard approach in the identification of transcriptional enhancers is the use of transgenic animals carrying DNA elements joined to reporter genes inserted randomly in the genome. We examined elements near Tbx5, a gene required for forelimb development in humans and other vertebrates. Previous transgenic studies reported a mammalian Tbx5 fore-limb enhancer located in intron 2 containing a putative retinoic acid response element and a zebrafish tbx5a forelimb (pectoral fin) enhancer located downstream that is conserved from fish to mammals. We used CRISPR/Cas9 gene editing to knockout the endogenous elements and unexpectedly found that deletion of the intron 2 and downstream elements, either singly or together in double knockouts, resulted in no effect on forelimb development. Our findings show that reporter transgenes may not identify endogenous enhancers and that in vivo genetic loss-of-function studies are required, such as CRISPR/Cas9, which is similar in effort to production of animals carrying reporter transgenes.
In Brief
Forelimb development requires Tbx5. Using CRISPR/Cas9 gene editing to create homozygous deletions, Cunningham et al. show that two Tbx5 forelimb enhancers identified with reporter transgenes are not required for Tbx5 activation or forelimb development. These observations demonstrate that knockout studies are required to identify endogenous enhancers necessary for biological processes.
INTRODUCTION
An accurate understanding of gene regulation requires identification of DNA control elements that function as transcriptional enhancers in vivo (Smith and Shilatifard, 2014). A common technique in enhancer identification is the production of transgenic reporter animals that carry putative DNA control elements joined to reporter genes that are randomly inserted in the genome (Andrey and Mundlos, 2017; Catarino and Stark, 2018). Here, we examined putative enhancers for Tbx5, a gene mutated in some humans suffering from forelimb and heart developmental defects (Bruneau et al., 2001).
During forelimb development, mouse Tbx5 and its zebrafish homolog tbx5a are both expressed in the early forelimb field, where they are required for initiation of forelimb outgrowth, i.e., arms in mouse and pectoral fins in zebrafish; genetic loss-of-function results in complete absence of forelimbs or pectoral fins (Ahn et al., 2002; Agarwal et al., 2003; Rallis et al., 2003). Transgenic mouse studies identified a DNA element lying within intron 2 of mouse Tbx5 that conferred transgene expression specifically in the forelimb field (Minguillon et al., 2012). A minimal element needed for transgene expression was reported to contain six HOX-binding sites in a 361-bp region, with four additional HOX-binding sites located close upstream (Figure 1), supporting the hypothesis that Hox4 and Hox5 genes known to be expressed in the forelimb field may function to activate Tbx5 in this location (Minguillon et al., 2012; Nishimoto et al., 2014). Further studies on the minimal 361-bp DNA element reported that it also contains a Tcf/Lef (Wnt/β-catenin) binding site and a retinoic acid response element (RARE) (Nishimoto et al., 2015). The report of a Tcf/Lef (Wnt/β-catenin) binding site was supported by β-catenin conditional knockout studies showing partial loss of forelimb Tbx5 expression (Nishimoto et al., 2015). However, the suggestion that a RARE may be required for forelimb Tbx5 activation (Nishimoto et al., 2015) is in contradiction to mouse and zebrafish RA genetic loss-of-function studies showing that RA is not directly required to activate Tbx5 in the forelimb field but that RA is required to repress Fgf8 in the forelimb field in order to permit forelimb Tbx5 activation by an unknown activator (Cunningham et al., 2013).
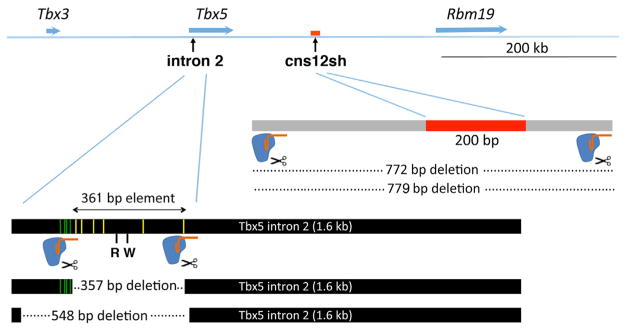
Figure 1. Strategy for Knockout of Putative Mouse Forelimb Tbx5 Enhancer.
CRISPR/Cas9 gene editing was used to delete the Tbx5 intron 2 DNA element (Minguillon et al., 2012) and the downstream cns12sh element (Adachi et al., 2016) using two sgRNAs flanking each element. Yellow and green bars within the intron 2 element designate reported HOX-binding sites within and upstream of the minimal 361-bp sequence (Minguillon et al., 2012; Nishimoto et al., 2014). Additional reported control elements include a RARE (R) and a Tcf/Lef Wnt/β-catenin control element (W) (Nishimoto et al., 2015). See also Figures S1 and S2.
Whereas the intron 2 Tbx5 enhancer is conserved only in mammals (Minguillon et al., 2012), transgenic reporter studies in zebrafish described a tbx5a forelimb (pectoral fin) enhancer (cns12sh) located downstream that is conserved from fish to mammals (Adachi et al., 2016). Transgenic zebrafish carrying either the zebrafish cns12sh sequence or the conserved mouse cns12sh sequence were able to drive gene expression in the zebrafish pectoral fin field, demonstrating conservation of cns12sh activity in the forelimb (Adachi et al., 2016). The Tbx5 cns12sh enhancer was not analyzed to identify any potential DNA-binding transcription factors, and its sequence is not similar to the Tbx5 intron 2 enhancer.
Further studies were performed here to determine whether either of these elements is required for activation of the endogenous Tbx5 or tbx5a genes in the forelimb fields of mouse or zebrafish, respectively.
RESULTS
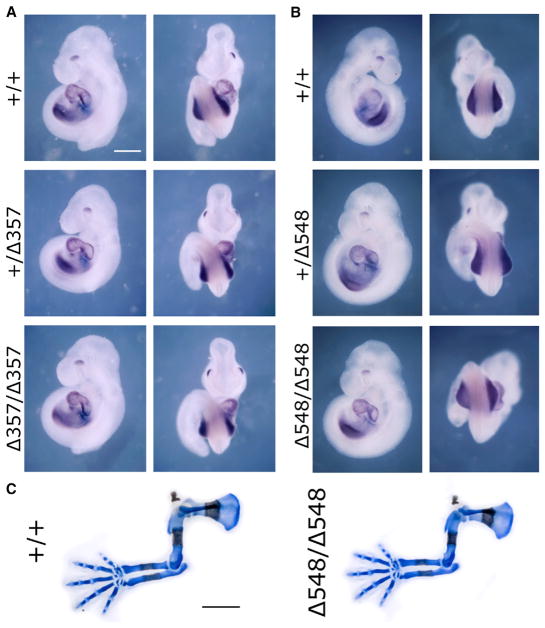
In order to further examine the potential functions of the intron 2 and cns12sh Tbx5 forelimb enhancers, including a putative RARE, we first performed CRISPR/Cas9 gene editing to delete the endogenous elements in mouse (Figure 1). With regard to the Tbx5 forelimb enhancer reported to reside inside intron 2, we obtained one deletion mutant that removed 357 bp, including most of the conserved 361-bp element, and another deletion mutant that removed 548 bp, including the entire conserved 361-bp element plus the four additional upstream HOX-binding regions (Figure S1). Mouse lines homozygous for each deletion produced offspring that were healthy and reproductive. For each homozygous deletion, E9.5 embryos exhibited normal expression of Tbx5 in the forelimb field and normally sized forelimb buds (357-bp deletion, n = 12; 548-bp deletion, n = 10) (Figures 2A and 2B). Examination of embryos at embryonic day 14.5 (E14.5) showed that both homozygous deletion mutants exhibited normal forelimb skeleton formation (357-bp deletion, n = 7; 548-bp deletion, n = 10) (Figure 2C). Thus, loss of the Tbx5 intron 2 DNA element had no effect on forelimb Tbx5 expression, forelimb initiation, or fore-limb development. Our results suggest that the HOX-binding sites, RARE, and Tcf/Lef Wnt/β-catenin elements that reside in the intron 2 element are not required for forelimb Tbx5 activation.
Figure 2. Analysis of Mice Carrying Deletions of the Tbx5 Intron 2 DNA Element.
(A and B) Expression of Tbx5 in E9.5 wild-type (+/+) embryos compared to embryos carrying either a 357-bp deletion (Δ357) (A) or a 548-bp deletion (Δ548) (B) that eliminates the putative Tbx5 fore-limb enhancer within intron 2; scale bar: 0.3 mm (same for all panels).
(C) Skeletal staining of E14.5 deletion mutants compared to wild-type; scale bar: 1 mm (same for all panels). See Figure 3 for quantification.
As our studies show that the mouse Tbx5 intron 2 DNA element is not required for forelimb development, another DNA element must be required. The Tbx5 intron 2 DNA element is conserved only in mammals (Minguillon et al., 2012); however, other studies performed in zebrafish discovered a ~200-bp DNA element called cns12sh located ~30 kb downstream of tbx5a that confers forelimb-specific expression using zebrafish transgenic enhancer analysis; sequence alignment showed cns12sh is conserved from jawed fishes to mammals, including humans (Adachi et al., 2016). No evidence was presented for putative transcriptional regulatory proteins that may bind the cns12sh element. In mouse, the cns12sh element is located ~100 kb downstream of Tbx5 in a 200-bp region (Figure 1). Using CRISPR/Cas9 gene editing, we generated a mouse line carrying a homozygous 779-bp deletion, including the conserved cns12sh element (Figures 1 and S2). Mice carrying the 779-bp deletion were used to generate homozygous deletion embryos (cns12 KO), which exhibited normal forelimb expression of Tbx5 and normal forelimb initiation at E9.5 (n = 10), plus normal forelimb skeleton formation at E14.5 (n = 4), similar to the intron 2 element homozygous deletion (in2 KO) (Figures 3A and 3B). Thus, like the intron 2 element, the cns12sh element is not required for mouse forelimb development.
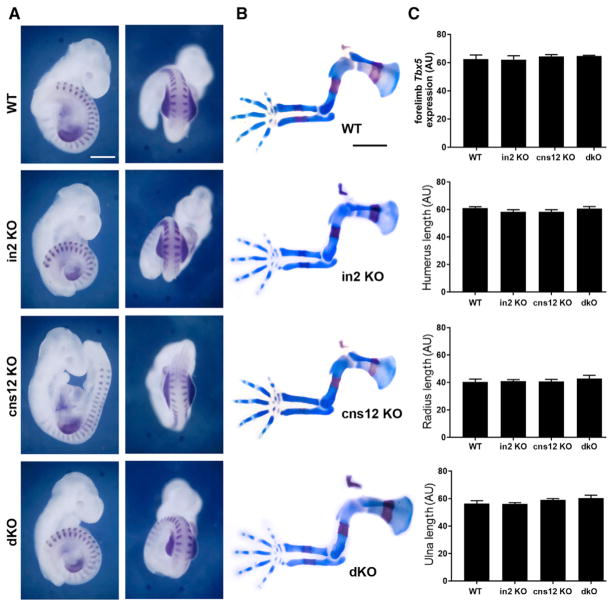
Figure 3. Analysis of Mice Carrying Deletions of the Tbx5 cns12sh Element or Both Intron 2 and cns12sh Elements.
(A) Expression of both Tbx5 and Uncx (somites) in E9.5 embryos carrying homozygous deletions of putative Tbx5 forelimb enhancers; cns12 KO, cns12 element 779-bp deletion; dKO, double KO containing intron 2 element 548-bp deletion and cns12 element 772-bp deletion; in2 KO, intron 2 element 548-bp deletion; WT, wild-type; scale bar: 0.3 mm (same for all panels).
(B) Skeletal staining of E14.5 homozygous deletion mutants compared to WT; scale bar: 1 mm (same for all panels).
(C) Comparison of forelimb Tbx5 expression in stage-matched E9.5 embryos (24 or 25 somites detected by Uncx expression) and forelimb skeletal element lengths (humerus, radius, and ulna) at E14.5 across the indicated genotypes; data are expressed as mean ± SD. For all comparisons, p > 0.05 (not significant difference) using one-way ANOVA non-parametric test (n = 3 biological replicates for each genotype, stage-matched).
In order to determine whether the Tbx5 intron 2 and downstream cns12sh elements may function redundantly, we generated double mutants. As these elements are located relatively close together on the same chromosome, we used an iterative approach. Fertilized oocytes derived from animals carrying the 548-bp deletion within intron 2 were used for a second round of CRISPR/Cas9 gene editing for the cns12sh element, which resulted in generation of a 772-bp deletion, including cns12sh, very similar to the location of the 779-bp deletion in the single mutant (Figures 1 and S2). Surprisingly, embryos carrying homozygous deletions of both the intron 2 and cns12sh elements (double knockout [dKO]) exhibited normal forelimb expression of Tbx5 and normal forelimb initiation at E9.5 (n = 7), as well as normal forelimb skeleton formation at E14.5 (n = 5) (Figures 3A and 3B). A comparison of forelimb Tbx5 expression and skeletal element lengths in the single and double knockouts shows no significant difference with wild-type (Figure 3C). These results demonstrate that the intron 2 and cns12sh elements do not exhibit a redundant function with each other that is required for activation of Tbx5 expression in the mouse forelimb.
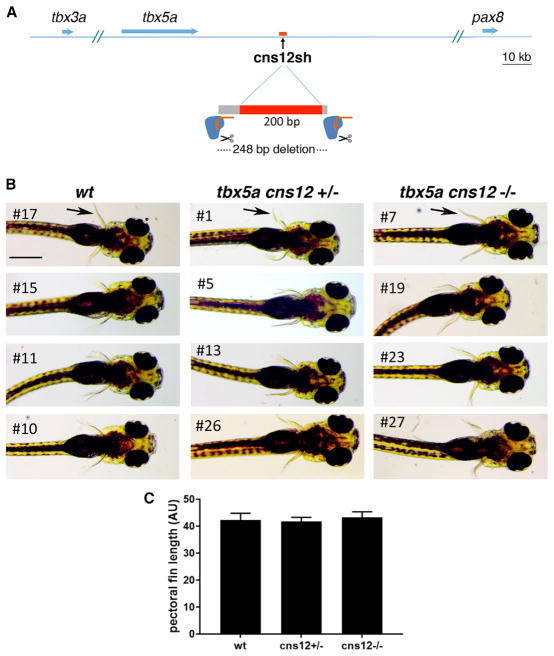
As the cns12sh element was originally reported to be a fore-limb tbx5a enhancer in transgenic zebrafish (Adachi et al., 2016), we wondered whether this element may have a function in initiation of zebrafish pectoral fin outgrowth that was lost during later evolution of mammals. We generated a line of zebrafish carrying a 248-bp deletion, including the conserved cns12sh element (Figures 4A and S3). From matings of heterozygous deletion mutant parents, we obtained zebrafish offspring homozygous for the 248-bp cns12sh deletion that all exhibited normal pectoral fins; (first mating offspring: wild-type [WT] n = 4, +/− n = 18, −/− n = 6; second mating offspring: WT n = 7, +/− n = 11, −/− n = 6) (Figure 4B). A comparison of pectoral fin length shows no significant difference between the cns12sh knockout and wild-type (Figure 4C). These findings demonstrate that the cns12sh element is not required for initiation of zebrafish pectoral fin outgrowth.
Figure 4. Deletion of Zebrafish tbx5a cns12sh DNA Element.
(A) CRISPR/Cas9 gene editing strategy to delete the zebrafishtbx5a cns12shelement (Adachi et al., 2016). (B) Zebrafish 5-day-old embryos showing pectoral fins (arrows) in wild-type as well as heterozygous and homozygous cns12sh deletion mutants; scale bar: 1 mm (same for all panels).
(C) Comparison of pectoral fin lengths; data are expressed as mean ± SD. For all comparisons, p > 0.05 (not significant difference) using one-way ANOVA non-parametric test (n = 4 biological replicates for each genotype, stage-matched).
See also Figure S3.
DISCUSSION
For more than 30 years, numerous transgenic animal studies have been used to provide in vivo evidence for DNA elements that function as transcriptional enhancers. Identification of presumed enhancers in transgenic animals is considered highly superior to analysis of enhancers in cell lines; however, this technique involves ligation of a DNA element to a reporter gene and random insertion in the genome far from its normal location. With the CRISPR/Cas9 single and double knockout studies presented here, we show that one cannot rely only on reporter transgenes to identify enhancers that are biologically necessary. Our findings suggest that DNA regulatory elements that are closely linked to a basal promoter and randomly inserted into the genome may exhibit enhancer functions that do not exist in their normal location, often far from the nearest promoter. Thus, although reporter transgenes can identify potential enhancers, identification of a true enhancer that is necessary for gene regulation requires the observation of a defect after knocking out the endogenous DNA element in its normal location or knocking it out along with one or more additional DNA elements in the case of enhancer redundancy.
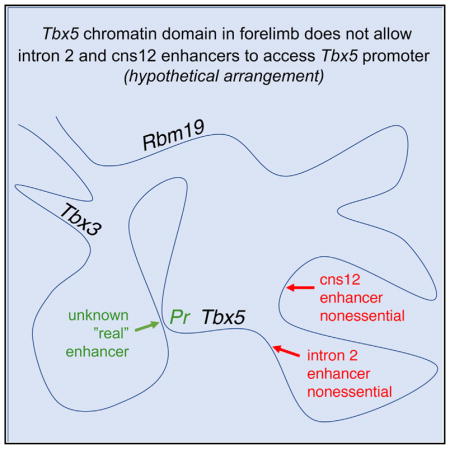
Here, we were unable to build upon four previously published transgene studies on two presumed Tbx5 enhancers independently identified in two vertebrate animals (Minguillon et al., 2012; Nishimoto et al., 2014, 2015; Adachi et al., 2016). Instead, our findings indicate that at least one other unknown DNA element must be required for activation of forelimb Tbx5 in both mouse and zebrafish. In addition, our mouse double knockout studies show that the intron 2 and cns12sh elements do not share redundant or additive functions for forelimb development with each other, i.e., not even a partial effect on Tbx5 expression that is enough to effect forelimb development in the double knockout; in contrast, the Tbx5 knockout results in complete loss of forelimbs (Ahn et al., 2002; Agarwal et al., 2003; Rallis et al., 2003). It may be possible that one or both of these DNA elements share a redundant function with an unknown third DNA element. However, recent enhancer knockout studies by others demonstrated that, when two or more endogenous DNA elements exhibit redundancy for gene activation in vivo, they typically function additively instead of completely redundantly (Will et al., 2017; Catarino and Stark, 2018; Dickel et al., 2018; Osterwalder et al., 2018). Thus, as we see no effect on Tbx5 expression when both of the Tbx5 DNA elements are deleted, it is possible that neither of these DNA elements are required for activation of Tbx5 or initial forelimb outgrowth, i.e., they may not share a redundant function with an unknown third DNA element that is yet to be discovered. We suggest that, even though the intron 2 and cns12sh elements can activate transcription when placed near basal promoters in ectopic genomic locations, in their normal genomic locations they may not be properly positioned within their topologically associated domain (TAD) to control forelimb Tbx5 activation. Thus, the Tbx5 DNA elements we examined may be vestigial enhancers or pseudoenhancers, i.e., enhancers that once had a required function in forelimb activation that has been lost during evolution. Further knockout studies on another potential forelimb Tbx5 enhancer are needed to resolve this issue. Additional enhancer candidates may be obtained by analyzing the Tbx5 genomic landscape during forelimb initiation using chromatin conformation capture and other chromatin profiling techniques. One such study that included an analysis of H3K27ac marks near mouse Tbx5 estimated it may be regulated by at least four enhancers in developing limbs at E11.5 (Osterwalder et al., 2018); we observed that the intron 2 enhancer is included within a very large region marked by H3K27ac, whereas the cns12sh enhancer is not marked by H3K27ac; however, analysis at E11.5 may be too late to make conclusions on potential enhancers, as Tbx5 is activated at E8.5 in mouse (Agarwal et al., 2003).
Our observations indicate that enhancer reporter transgene findings need to be validated by enhancer knockout studies before concluding that factors binding the enhancer are required for gene regulation. Importantly, as the RARE reported to exist in the mouse Tbx5 intron 2 enhancer (Nishimoto et al., 2015) was deleted in our studies without any effect on forelimb Tbx5 expression, the RARE enhancer data can no longer be used to support a role for RA receptors and RA in activation of Tbx5 in the forelimb. Our observation of no effect on forelimb Tbx5 expression following loss of the RARE is consistent with previous studies demonstrating that RA is not directly required for Tbx5 activation in zebrafish or mouse but instead that RA is required for repression of Fgf8 to permit activation of Tbx5 by some other mechanism in both zebrafish and mouse (Cunningham et al., 2013). Our knockout studies also demonstrate that the Hox binding and Tcf/Lef (Wnt/β-catenin) binding sites previously reported in the Tbx5 intron 2 DNA element (Minguillon et al., 2012; Nishimoto et al., 2014, 2015) are not required for Tbx5 forelimb activation.
Several RAREs have been previously studied in vivo using reporter transgenes, but only a few have been subjected to genetic loss of function to determine whether they are required for regulation of a nearby gene (Cunningham and Duester, 2015). Traditional knockout studies using embryonic stem cells identified RAREs that are required to activate Cdx1 (Houle et al., 2003) and Hoxa1 (Dupe et al., 1997). Recent CRISPR/Cas9 gene editing was used to identify a silencer RARE that is required for RA repression of Fgf8 (Kumar et al., 2016). In the future, we recommend that in vivo knockout analyses be performed before concluding that a DNA element is required as a RARE.
Our double knockout studies provide a compelling case for moving to reliance on genetic disruption of endogenous noncoding DNA elements to provide a definitive answer on whether a DNA element is required for gene regulation. Reporter transgenes can identify candidate enhancers, but verification of function by knockout is necessary to move the field forward in a positive direction. A CRISPR/Cas9 knockout approach requires about the same time, effort, and cost compared to the traditional strategy of generating mice or zebrafish carrying transgenic reporters, and CRISPR facilitates the ability to generate double knockouts in the case of redundancy. Validation of enhancers or silencers with in vivo knockout studies will allow the field of epigenetics to focus on changes observed in DNA elements that are necessary for gene regulation in vivo.
EXPERIMENTAL PROCEDURES
Animal Studies
All mouse and zebrafish studies conformed to the regulatory standards adopted by the Institutional Animal Care and Use Committee at the SBP Medical Discovery Institute, which approved this study under Animal Welfare Assurance Number A3053-01. Age and gender were not relevant for this study, as analysis was performed on embryos.
Generation of Mutant Mice
CRISPR/Cas9 gene editing was performed as previously described (Kumar et al., 2016), using two single-guide RNA (sgRNAs) flanking either the mouse Tbx5 intron 2 DNA element or the downstream cns12sh DNA element in order to delete these elements (see Figures S1 and S2). Fertilized mouse oocytes injected with Cas9mRNA and sgRNAs targetingeither the Tbx5intron 2 DNA element (Minguillon et al., 2012) or the Tbx5 cns12 downstream element (Adachi et al., 2016) were allowed to proceed to birth to generate mouse lines. In order to generate double mutants carrying deletions of both the Tbx5 intron 2 element and the cns12 downstream element, a mouse line homozygous for the intron 2 DNA element deletion was used to prepare fertilized oocytes that were then injected with sgRNAs for the cns12 downstream element deletion. Mutant F0 adult mice were identified by DNA sequencing of PCR products overlapping the mutation from analysis of tail DNA. Further matings to wild-type mice generated F1 heterozygous mutants, and F1 matings generated F2 homozygous deletion mutants. Embryos at stages E9.5 and E14.5 were generated from timed matings of mice carrying heterozygous or homozygous deletion alleles; embryos were genotyped by PCR analysis of yolk sac DNA.
Generation of Mutant Zebrafish
CRISPR/Cas9 gene editing was performed in zebrafish as previously described (Gagnon et al., 2014; Irion et al., 2014), using EnGen Cas9 NLS protein from S. pyogenes (New England Biolabs) and two sgRNAs flanking the tbx5a cns12sh conserved DNA element (see Figure S3). Adult F0 fish were genotyped using PCR analysis and DNA sequencing of tail clips, and further matings of positive F0 fish generated F1 heterozygotes and F2 homozygotes. Embryos (5 days old) were imaged and genotyped by PCR analysis of genomic DNA isolated from whole embryos.
PCR Primers for Genotyping of Mice and Zebrafish
Genotyping of mice carrying a deletion of the Tbx5 intron 2 enhancer:
Tbx5-in2-Fwd 5′-TTCAGCTTGGAGTGAAGGGT-3′
Tbx5-in2-Rev 5′-TCAGTGATGGGTCTTAGCGG-3′
Genotyping of mice carrying a deletion of the Tbx5 cns12sh enhancer:
Tbx5-cns12sh-Fwd 5′-GTCCTCGGCACTACATACGA-3′
Tbx5-cns12sh-WT-Rev 5′-CCCGGGGTCATCTGTTTTCA-3′ (to detect wild-type)
Tbx5-cns12sh-mut-Rev 5′-TGTAGCCTTGGTTGGTCTGA-3′ (to detect mutant).
Genotyping of zebrafish carrying a deletion of the Tbx5 intron 2 enhancer:
ztbx5a-cns12sh-Fwd 5′-GCTCAGACAAATTCCAAGCGT-3′
ztbx5a-cns12sh-Rev 5′-TGTGAACAAAGTTTTCGGTTGA-3′
Detection of mRNA and Limb Skeletal Elements
Detection of Tbx5 and Uncx mRNAs was performed by whole-mount in situ hybridization as previously described (Sirbu and Duester, 2006). ImageJ software (https://imagej.net/welcome; Schneider et al., 2012) was used to quantitate forelimb Tbx5 mRNA in photographs of E9.5 mouse embryos that were stage matched (embryos with 24 or 25 somites detected by Uncx expression), stained the same day for an equal length of time, and photographed at the same magnification; briefly, the ImageJ area selection tool was used to select regions of equal size in the forelimb domains of each embryo and then average pixel density was measured. Alcian blue and Alizarin red staining of cartilage and bone was performed as previously described (Rigueur and Lyons, 2014) to enable quantification of forelimb skeletal element length in E14.5 mouse embryos; skeletal element lengths were measured in photographs of forelimbs taken at the same magnification for all genotypes.
Statistical Analysis
Statistical analysis was performed using one-way ANOVA (non-parametric test) to compare across all genotypes with data presented as mean ± SD and with p > 0.05 indicating non-significance.
Supplementary Material
Highlights.
Putative Tbx5 forelimb enhancers were deleted in mouse and zebrafish using CRISPR/Cas9
Double knockouts of the two enhancers show both are nonessential and nonredundant
A retinoic acid response element in one enhancer is nonessential for forelimb growth
Knockout studies are needed to infer necessity of proposed transcriptional enhancers
Acknowledgments
We thank the Animal Resources Core Facility at SBP Medical Discovery Institute for conducting timed matings to generate mouse embryos and Judy Wade for help generating fertilized mouse oocytes from Tbx5 intron 2 (Δ548) mutants to use for iterative CRISPR/Cas9 editing to also introduce the cns12sh mutation to produce double mutants. This work was funded by NIH grant R01 AR067731 (to G.D.).
Footnotes
Supplemental Information includes three figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.05.052.
AUTHOR CONTRIBUTIONS
T.J.C. and G.D. designed the study, analyzed the data, and wrote the paper. T.J.C., J.J.L., and M.B. performed all the experiments. J.J.L. and P.D.S.D. provided zebrafish for the studies. All authors discussed the results and commented on the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Adachi N, Robinson M, Goolsbee A, Shubin NH. Regulatory evolution of Tbx5 and the origin of paired appendages. Proc Natl Acad Sci USA. 2016;113:10115–10120. doi: 10.1073/pnas.1609997113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P, Wylie JN, Galceran J, Arkhitko O, Li C, Deng C, Grosschedl R, Bruneau BG. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development. 2003;130:623–633. doi: 10.1242/dev.00191. [DOI] [PubMed] [Google Scholar]
- Ahn DG, Kourakis MJ, Rohde LA, Silver LM, Ho RK. T-box gene tbx5 is essential for formation of the pectoral limb bud. Nature. 2002;417:754–758. doi: 10.1038/nature00814. [DOI] [PubMed] [Google Scholar]
- Andrey G, Mundlos S. The three-dimensional genome: regulating gene expression during pluripotency and development. Development. 2017;144:3646–3658. doi: 10.1242/dev.148304. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, Seidman JG. A murine model of Holt-Oram syndrome defines roles of the t-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Catarino RR, Stark A. Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes Dev. 2018;32:202–223. doi: 10.1101/gad.310367.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham TJ, Duester G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat Rev Mol Cell Biol. 2015;16:110–123. doi: 10.1038/nrm3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham TJ, Zhao X, Sandell LL, Evans SM, Trainor PA, Duester G. Antagonism between retinoic acid and fibroblast growth factor signaling during limb development. Cell Rep. 2013;3:1503–1511. doi: 10.1016/j.celrep.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickel DE, Ypsilanti AR, Pla R, Zhu Y, Barozzi I, Mannion BJ, Khin YS, Fukuda-Yuzawa Y, Plajzer-Frick I, Pickle CS, et al. Ultra-conserved Enhancers Are Required for Normal Development. Cell. 2018;172:491–499. e15. doi: 10.1016/j.cell.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupé V, Davenne M, Brocard J, Dollé P, Mark M, Dierich A, Chambon P, Rijli FM. In vivo functional analysis of the Hoxa-1 3′ retinoic acid response element (3′RARE) Development. 1997;124:399–410. doi: 10.1242/dev.124.2.399. [DOI] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle M, Sylvestre JR, Lohnes D. Retinoic acid regulates a subset of Cdx1 function in vivo. Development. 2003;130:6555–6567. doi: 10.1242/dev.00889. [DOI] [PubMed] [Google Scholar]
- Irion U, Krauss J, Nüsslein-Volhard C. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development. 2014;141:4827–4830. doi: 10.1242/dev.115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Cunningham TJ, Duester G. Nuclear receptor core-pressors Ncor1 and Ncor2 (Smrt) are required for retinoic acid-dependent repression of Fgf8 during somitogenesis. Dev Biol. 2016;418:204–215. doi: 10.1016/j.ydbio.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minguillon C, Nishimoto S, Wood S, Vendrell E, Gibson-Brown JJ, Logan MP. Hox genes regulate the onset of Tbx5 expression in the forelimb. Development. 2012;139:3180–3188. doi: 10.1242/dev.084814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto S, Minguillon C, Wood S, Logan MP. A combination of activation and repression by a colinear Hox code controls forelimb-restricted expression of Tbx5 and reveals Hox protein specificity. PLoS Genet. 2014;10:e1004245. doi: 10.1371/journal.pgen.1004245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto S, Wilde SM, Wood S, Logan MP. RA Acts in a Coherent Feed-Forward Mechanism with Tbx5 to Control Limb Bud Induction and Initiation. Cell Rep. 2015;12:879–891. doi: 10.1016/j.celrep.2015.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M, Barozzi I, Tissières V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu Y, Plajzer-Frick I, Pickle CS, et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 2018;554:239–243. doi: 10.1038/nature25461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallis C, Bruneau BG, Del Buono J, Seidman CE, Seidman JG, Nissim S, Tabin CJ, Logan MPO. Tbx5 is required for forelimb bud formation and continued outgrowth. Development. 2003;130:2741–2751. doi: 10.1242/dev.00473. [DOI] [PubMed] [Google Scholar]
- Rigueur D, Lyons KM. Whole-mount skeletal staining. Methods Mol Biol. 2014;1130:113–121. doi: 10.1007/978-1-62703-989-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu IO, Duester G. Retinoic-acid signalling in node ectoderm and posterior neural plate directs left-right patterning of somitic mesoderm. Nat Cell Biol. 2006;8:271–277. doi: 10.1038/ncb1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. Enhancer biology and enhanceropathies. Nat Struct Mol Biol. 2014;21:210–219. doi: 10.1038/nsmb.2784. [DOI] [PubMed] [Google Scholar]
- Will AJ, Cova G, Osterwalder M, Chan WL, Wittler L, Brieske N, Heinrich V, de Villartay JP, Vingron M, Klopocki E, et al. Composition and dosage of a multipartite enhancer cluster control developmental expression of Ihh (Indian hedgehog) Nat Genet. 2017;49:1539–1545. doi: 10.1038/ng.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.