Abstract
Myocardial ischemic injury results from severe impairment of the coronary blood supply and may lead to metabolic and ultrastructural changes, thereby causing irreversible damage. MicroRNA (miR)-20b-5p has been demonstrated to be involved in malignancies of the breast, colorectum, stomach, blood and oropharynx. The present study aimed to investigate the effects of miR-20b-5p on ventricular remodeling following myocardial ischemia-reperfusion (IR) injury in rats by targeting small mothers against decapentaplegic homolog 7 (Smad7) via the transforming growth factor-β (TGF-β)/Smad signaling pathway. A total of 70 adult male Sprague-Dawley rats were divided into seven groups: Sham group, IR group, negative control group, miR-20b-5p mimics group, miR-20b-5p inhibitors group, small interfering RNA (siRNA)-Smad7 group, and miR-20b-5p inhibitors + siRNA-Smad7 group. Dual luciferase reporter gene assays were used to verify the association between miR-20b-5p and Smad7. Myocardial infarction size, myocardial collagen volume fraction and perivascular collagen area were detected separately using triphenyltetrazolium chloride and Masson's staining. The rate of positive expression of Smad7 was detected using immunohistochemistry, and the expression levels of miR-20b-5p, TGF-β1, Smad3 and Smad7 were detected using reverse transcription-quantitative polymerase chain reaction and western blot analyses. The findings revealed that miR-20b-5p inhibited Smad7. Compared with the sham group, the other six groups had increased myocardial infarction size, myocardial collagen, and expression of miR-20b-5p, TGF-β1 and Smad3, and decreased expression of Smad7. Compared with the IR group, the miR-20b-5p mimics group and the siRNA-Smad7 group had increased myocardial infarction size and myocardial collagen, increased expression of TGF-β1 and Smad3, and decreased expression of Smad7. The expression of miR-20b-5p was markedly increased in the miR-20b-5p mimics group, but did not differ significantly from that in the siRNA-Smad7 group. The results demonstrated that miR-20b-5p promoted ventricular remodeling following myocardial IR injury in rats by inhibiting the expression of Smad7 through activating the TGF-β/Smad signaling pathway.
Keywords: microRNA-20b-5p, small mothers against decapentaplegic homolog 7, small mothers against decapentaplegic homolog 3, transforming growth factor-β1/small mothers against decapentaplegic, myocardial ischemia-reperfusion injury, ventricular remodeling, myocardial collagen, myocardial infarction
Introduction
Myocardial ischemia-reperfusion (IR) injury typically occurs when patients are experiencing an ST segment elevation myocardial infarction (MI) (1). Thrombolytic therapy or primary percutaneous coronary intervention may be the optimal method to reduce acute myocardial ischemic injury and limit the size of the MI (2). However, further cardiomyocyte death may arise in the process of myocardial reperfusion, which is termed myocardial IR injury (3). The four recognized forms of myocardial IR include reversible forms (reperfusion-induced arrhythmias and myocardial stunning) and irreversible forms (microvascular obstruction and lethal myocardial reperfusion injury) (4,5). Several critical factors have been identified by experimental investigations as potential mediators of the detrimental effects of myocardial IR, including oxidative stress, inflammation, intracellular Ca2+ overload, rapid restoration of physiological pH at the time of reperfusion, mitochondrial permeability transition pore, and late myocardial reperfusion injury (6-9). In addition to improvements in earlier reperfusion and advancements in percutaneous coronary intervention technology, there are also emerging therapeutic strategies, including ischemic preconditioning or postconditioning, remote ischemic preconditioning, therapeutic hyperoxemia and hypothermia, and pharmacologic agents for preventing myocardial IR injury (10). Previously, Fan and Yang suggested that microRNAs (miRs) may be potential therapeutic targets in IR injury by altering key signaling elements (11).
miRs, a class of endogenous RNAs of ~22 nucleotides in length, can inhibit the translation of target mRNAs by pairing to sites in the 3′ untranslated region (3′-UTR) (12,13). He et al detected miR expression in a myocardial IR model in Sprague-Dawley (SD) rats following reperfusion and found that miRs had an effect on myocardial IR (14). It has been shown that miR-20b is involved in cardiac remodeling and antagomiR-20b reduces IR-induced vascular endothelial growth factor (15). Circulating levels of miR-20b have also been found to be useful as a novel diagnostic indicator in hypertension-induced heart failure (16). miR-20b-5p has been reported to be dysregulated in blood malignancies (17). It is known that miR expression is modulated by small mother against decapentaplegic (Smad)s through transcriptional and post-transcriptional mechanisms, and miRs appear to have an important effect on the physiological activity of transforming growth factor (TGF)-β signaling (18). Smad7 belongs to the third type of Smad, the inhibitory Smads, with Smad6, however, Smad7 is more potent than Smad6 at inhibiting TGF-β signaling (19). A previous study demonstrated that Smad7 has important functions in maintaining cellular homeostasis in terms of anti-inflammatory and antifibrotic activity under physiological conditions (20). Therefore, the present study aimed to investigate whether miR-20b-5p promotes ventricular remodeling following myocardial IR injury in rats via inhibiting the expression of Smad7 through activating the TGF-β/Smad signaling pathway.
Materials and methods
Ethics statement
The present study was approved by the Laboratory Animal Ethics Committee of The First Affiliated Hospital of Harbin Medical University (Harbin, China), and was performed in accordance with the principles of animal protection, welfare and ethics and the National Laboratory Animal Welfare Ethics regulations.
Dual luciferase reporter gene assay
A search was performed at www.microRNA.org to analyze and predict the target genes of miR-20b-5p, and the fragment containing the binding sites was obtained. In strict accordance with the instructions of the TIANamp Genomic DNA kit (Tiangen Biotech Co., Ltd., Beijing, China), DNA was extracted from 293T cells (Wuhan Cell Bank, Wuhan, China). Smad7-3′-UTR-wild-type (wt) and Smad7-3′-UTR-mutant (mut) without the miR-20b-5p binding site were designed. The recombinant luciferase reporter vector was constructed to transfect the 293T cells with the miR-20b-5p mimics and miR-20b-5p inhibitors, and luciferase activity was measured with a dual luciferase reporter assay system (Promega Corporation, Madison, WI, USA). Following transfection for 48 h, the growth medium was removed from the cultured cells, which were washed twice with PBS. Passive lysis buffer (100 µl) was added to each well containing the cells, and the plates were gently rocked at room temperature for 15 min to collect cell lysates. Subsequently, the program was set to pre-read the sample for 2 sec and to read its value for 10 sec following each injection of 100 µl LARIIStop&Glo® reagent (Promega Corporation). The prepared LARIIStop&Glo® reagent was added into luminous tubes or wells containing lysate (20 µl of each sample). Signals in the luminous tubes or wells were detected with a Modulus™ single tube multimode reader (TumerBioSystems, Sunnyvale, CA, USA). Firefly luciferase (LUC) activity and Renilla luciferase (RL) activity were detected. RL activity reflected the transfection efficiency of each well, and the ratio of LUC activity to RL activity (LUC/RL) was calculated as the relative luciferase expression.
Model generation
A total of 70 healthy male SD rats (250-300 g) of similar age (7-8 weeks) were purchased from the Experimental Animal Center of the Military Medical Science Academy of the PLA (Beijing, China). The rats were fed normal feed and had free access to water at 20-25°C, and a 12-h light/dark cycle at 22°C, and there was a 72-h adaptation period prior to the experiment. A total of 60 SD rats were randomly selected to generate a model of IR. Prior to surgery, the rats were anesthetized with 10% pentobarbital sodium (90 mg/kg, cat. no. wS20060401, Shanghai Westang Biotechnology Co., Ltd., Shanghai, China). Electrocardiographic (ECG) monitoring electrodes (BeneHearth R3; Mindray, Wuhan, China) were connected to the rat limbs, and the animals were ventilated with an animal respirator (R407; RWD Biotech Co., Ltd., Shenzhen, China). The gas source was room air, the ventilator frequency was 60 breaths/min, and the tidal volume was 13-15 ml/kg. The heart was exposed via a left thoracotomy through the fourth intercostal space, and the left anterior descending (LAD) coronary artery was ligated with a 6/0 atraumatic suture. ST-segment elevation by ECG monitoring indicated that the vessel was successfully occluded. Following occlusion for 60 min, the coronary artery was recanalized by suture removal, and ST-segment resolution indicated that the rat model of IR injury was successfully established.
Animal grouping
The 10 rats that did not undergo IR injury were used as the sham group. The rats in the sham group were ligated with a 6/0 atraumatic suture in the LAD coronary artery following thoracotomy without blocking the vessels. The modeled rats were randomly divided into six groups (n=10 in each group) as follows: i) IR group: Left coronary artery was ligated and blocked with a 6/0 atraumatic suture; 60 min later, the vessels were opened; ii) negative control (NC) group: Intramyocardial injection of the negative control sequence (average of five injections, 2 µg per injection to a total of 10 µg) was performed 24 h prior to occlusion of blood vessels, and the remaining procedures were performed at the same time as in the IR group; iii) miR-20b-5p mimics group: Intramyocardial injection of miR-20b-5p mimics (average of five injections, 2 µg per injection to a total of 10 µg) was performed 24 h prior to occlusion of blood vessels, and the remaining procedures were conducted at the same time as in the IR group; iv) miR-20b-5p inhibitors group: Intramyocardial injection of miR-20b-5p inhibitors (average of five injections, 2 µg per injection to a total of 10 µg) was performed 24 h prior to occlusion of blood vessels, and the remaining procedures were performed at the same time as in the IR group; v) siRNA-Smad7 (si-Smad7) group: Intramyocardial injection of si-Smad7 (average of five injections, 2 µg per injection to a total of 10 µg) was performed 24 h prior to occlusion of blood vessels, and the remaining procedures were performed at the same time as in the IR group; vi) miR-20b-5p inhibitors + si-Smad7 group: Intramyocardial injection of miR-20b-5p inhibitors and si-Smad7 (average of five injections, 2 µg per injection to a total of 10 µg) was performed 24 h prior to occlusion of blood vessels, and the remaining procedures were performed at the same time as in the IR group. All injection sequences were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The mortality rate of the rats was ~5.9% during the surgical procedure.
Transthoracic echocardiography
At 28 days post-surgery, the mortality rate of the rats was ~10%. In accordance with a random number table, five rats in each group were randomly selected to be anaesthetized with an intraperitoneal injection of 10% pentobarbital sodium (90 mg/kg, cat. no. wS20060401, Shanghai Westang Biotechnology Co., Ltd.) and confirmed with transthoracic echocardiography. Acuson Sequoia 512 color Doppler ultrasonography was performed by professionals; ultrasonography was performed with a 6C2-S probe at a frequency of 8.5 mHz, and the scanning speed was adjusted to 100 mm/sec. The detection methods were as follows: Rats were anesthetized with 10% pentobarbital sodium (90 mg/kg,) and fixed on the testing platform, following which the papillary muscle level M curve of the left ventricular long axis and left ventricular short axis were measured. Left ventricular diastolic diameter (LVEDD; mm), left ventricular systolic diameter (LVESD; mm), left ventricular end-diastolic volume (LVEDV) and left ventricular end-systolic volume (LVESV; µl) were measured continuously in three cardiac cycles, and the mean value was calculated. According to Simpson's method, the left ventricular ejection fraction (LVEEF) was calculated using the following formula: (LVEDV-LVESV)/LVEDV ×100%, and the left ventricular fractional shortening (LVEFS) was measured using the following formula: (LVEDD-LVESD)/LVEDD ×100% (21). One end of the catheter was connected to a pressure sensor, and the other end of the catheter was inserted into the left ventricle through the right common carotid artery to measure the left ventricular systolic pressure (LVESP; mmHg) and the left ventricular end-diastolic pressure (LVEDP; mmHg). In addition, rat cardiac function was evaluated by analyzing the association between the above indicators and cardiac function.
Triphenyltetrazolium chloride (TTC) staining
Following confirmation with transthoracic ECG, the heart was isolated for gross examination. The necrotized myocardium was stained with TTC and visualized under a transmission electron microscope (TEM). The left ventricular wall was separated into several sections at a thickness of 5-mm from the apex to the bottom of the heart, perpendicular to the long axis of the left ventricle; these sections were subjected to TTC staining, TEM, hematoxylin and eosin (HE) staining, Masson's staining, immunohistochemistry and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis. The apical tissue of the left ventricular wall was randomly selected from three rats in each group. Ultrathin sections (50-60 nm) were cut using a vibratome along the long axis of left ventricle. The myocardium of the left ventricular wall was stained with TTC, and the ultrastructure was observed. The sections were stained in 1% TTC phosphate-buffered solution (CAS no. 2530-85-0; Guidechem Shanghai, China). The infarcted myocardium was off-white, and the surviving myocardium was a normal color. The infarcted myocardium and the surviving myocardium were separated. Images of the sections were captured using a camera (LEICA digital camera 480; Leica Microsystems GmbH, Wetzlar, Germany), and myocardial infarct size was measured using ImageJ 1.26 image analysis software (National Institutes of Health, Bethesda, MD, USA). Myocardial infarct size (mm2) was calculated as myocardial infarction area (mm2)/left ventricular total area (mm2) ×100% (22,23).
TEM
Samples were prepared using a conventional TEM sample preparation technique. The myocardial tissues situated 5 mm above the apex of the left ventricular wall were randomly selected from three rats in each group and were cut into small sections of ~1 mm3. These sections were fixed in 4% polyformaldehyde solution (CAS no. 30525-89-4; Nanjing Guochen Chemicals Co., Ltd., Nanjing, China) for 2 h at 4°C, washed in 0.1 mol/l PBS, and fixed in 1% osmic acid at room temperature (CAS: 20816-12-0, Shanghai Fusheng Industrial Co., Ltd., Shanghai, China) for 2 h, followed by standard procedures of dehydration in an ethanol gradient (30% for 5 min, 50% for 5 min, 70% for 10 min, 80% for 15 min, 95% for 15 min, and 100% for 15 min for twice) at room temperature. The dehydrated myocardial sections were permeabilized, embedded in epoxy resin (cat. no. NPES-301, Yueyang Liqiang Chemical Co., Ltd., Yueyang, China) and cut into ultra-thin sections (50-60 nm) with a microtome (Leica EM UC7; Leica Microsystems GmbH). Following staining with uranyl acetate (CAS no. 6159-44-0; Hubei Chushengwei Chemistry Co., Ltd., Hubei, China) and citrate (CAS no. 512-26-5; Shanghai Jinjinle Industrial Co., Ltd., Shanghai, China), the sections were observed with TEM (JSM-840A SEM; JEOL, Ltd., Tokyo, Japan), and the ultrastructure was recorded by image capture.
HE staining
Myocardial tissue sited 10 mm above the apex of the left ventricular wall was randomly selected from three rats in each group, fixed in 4% formaldehyde (volume percentage) for 6 h at 4°C, embedded in paraffin, and sectioned at 3-µm thickness. Following heating at 60°C overnight, the sections were successively dewaxed for 20 min in Xylene I (CAS no. 14936-97-1; Eykits Research Biological Technology Co., Ltd., Shanghai, China) and Xylene II (CAS no. 523-67-1; Yuduo Biological Technology Co., Ltd., Shanghai, China), washed in distilled water for 5 min, and dehydrated in a 100, 95, 80 and 70% ethanol series. The sections were stained in hematoxylin (CAS no. 474-07-7; Qingdao Jieshikang Biotechnology Co., Ltd., Qingdao, China) at room temperature for 10 min, washed in tap water at room temperature for 15 min, counterstained in eosin (cat. no. RY0648, Qingdao Jieshikang Biotechnology Co., Ltd.) at room temperature for 30 sec and washed in double distilled water to remove the red color. The sections were then dehydrated in alcohol, cleared in xylene, and sealed with neutral balsam. HE staining and histopathologic examination were used to observe the color of the myocardial tissues of the rats, the distribution range and staining intensities. Using the morphological image analysis system (JD801; Jieda Technology Development Co., Ltd., Nanjing, China), different groups were selected and analyzed at ×200 magnification. Pathological changes, including necrosis and edema, were observed in the HE-stained sections. The images were randomly collected, and the experiment was repeated three times.
Masson's staining
Myocardial tissue sited 15 mm above the apex of the left ventricular wall was randomly selected from three rats in each group and fixed in 4% paraformaldehyde at 4°C for 24 h. Following routine steps of desiccation, clearing, embedding and slicing (3-µm), the specimens were stained with Picric acid-Sirius red at room temperature for 30 min and inhibited by hematoxylin at room temperature for 2 min (CAS no. PT003; Shanghai Bogoo Biotechnology Co., Ltd., Shanghai, China). Images of the myocardial sections were obtained under a polarized light microscope (XPT-480; Shanghai Zhongheng, Co., Ltd., Shanghai, China) and were analyzed with Image-Pro 6 software (Media Cybernetics Inc., Bethesda, MD, USA). Five high-magnification fields were randomly selected in each section, and the myocardial collagen volume fraction (CVF) was quantitatively analyzed. Five transverse arterioles were randomly selected in each specimen, and the perivascular collagen area (PVCA) was measured. The CVF was calculated using the following formula: CVF (%)=collagen area/entire area ×100%, wherein the collagen area did not include the area surrounding the vessels. The PVCA (%) was measured as the collagen area surrounding the vessel/total vessel wall area ×100% (24).
Immunohistochemistry
Myocardial tissue sited 20 mm above the apex of the left ventricular wall was randomly selected from three rats in each group and fixed in 4% paraformaldehyde at 4°C for 24 h. Following paraffin embedding, a microtome was used to cut 3-µm serial sections. According to the conventional method of immunohistochemical staining, following the elimination of endogenous peroxidase activity in 0.3% H2O2− methanol solution at room temperature for 10 min, a 1:100 dilution of rabbit anti-rat Smad7 antibody (cat. no. BA1399; 1:100; Wuhan Boster Biological Technology Co., Ltd., Wuhan, China) was added to the cardiac tissue for 16 h at 4°C. The sections were then incubated with rabbit anti-rat IgG (cat. no. BA1058; 1:100; Wuhan Boster Biological Technology Co., Ltd.) antibody labeled with horseradish peroxidase for 2 h at room temperature. At 5 min post-DAB staining (cat. no. AR1000; Wuhan Boster Biological Technology Co., Ltd.), the sections were observed and imaged were captured under light microscopy. The digital images were processed by Image-Pro plus 6.0 software (Media Cybernetics, Inc.). Five visual fields were randomly selected in each slice under ×400 magnification, and the percentage of positive cells in each field was then calculated. PBS, as a control for the primary antibody, was used as the negative control, and normal tissues were used as the positive control; yellow or brown staining in the cytoplasm or cell membrane indicated a positive cell. Four high-power fields (magnification, ×400) in each slice were selected, the number of positive cells of 200 cells in each field was calculated to obtain the percentage of the positive cells: Positive myocardial cells/total myocardial cells. If the percentage was >10%, the sample was considered positive (+); otherwise, it was considered negative (−) (25). The experiment was repeated three times, and the mean value was calculated.
RT-qPCR analysis
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to extract total RNA from the myocardial tissues in each group. Ultrapure water treated with diethylpyrocarbonate (Sangon Biotech Co., Ltd., Shanghai, China) was used to dissolve RNA, and the absorbance at 260 and 280 nm was detected by ND-1000 ultraviolet-visible spectrophotometry (NanoDrop; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). Subsequently, the quality of total RNA was determined, and the RNA concentration was adjusted. According to the kit (Fermentas; Thermo Fisher Scientific, Inc.), reverse transcription of the extracted RNA was completed by the two-step method. The reaction conditions were as follows: 70°C for 10 min, ice bath for 2 min, 42°C for 60 min, 70°C for 10 min, and temporary hold at −80°C. RT-qPCR was performed using the TaqMan probe method, the reaction system was used in accordance with the kit instructions (Fermentas; Thermo Fisher Scientific, Inc.), and the primer sequences are shown in Table I. The reaction system contains 2.0 µl cDNA template, 1.0 µl each of primer and probe mix, 10.0 µl TaqMan® Fast Advanced Master Mix (2X), and nuclease-free water was added up to 20 µl. The reaction conditions were as follows: Pre-denaturation at 95°C for 30 sec; 40 cycles of denaturation at 95°C for 10 sec; annealing at 60°C for 20 sec and extension at 70°C for 10 sec. The RT-qPCR system (Bio-Rad iQ5; Bio-Rad; Laboratories, Inc., Hercules, CA, USA) was used for detection, U6 was used as the internal reference for miR-20b-5p (26), and β-actin was used as the internal reference for other target genes. The value was calculated by the relative quantitative method, and the relative expression of each target gene was determined with the 2−∆∆Cq method (27) Each experiment was repeated three times.
Table I.
Reverse transcription-quantitative polymerase chain reaction primer sequences.
| Gene | Primer sequence |
|---|---|
| miRNA-20b-5p | F: 5′-CCTAGTAGTGCCAAAGTGCT-3′ |
| R: 5′-CCAGGAGTACTAGAAGTGATCA-3′ | |
| U6 | F: 5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
| R: 5′-CGCTTCACGAATTTGCGTGTCAT-3′ | |
| Smad3 | F: 5′-CATTACCATCCCCAGGTCAC-3′ |
| R: 5′-AGGCTCTACTGTGTCCAA-3′ | |
| Smad7 | F: 5′-ACTCTTGTTGTCCGAATTGA-3′ |
| R: 5′-ACTCTTGTTGTCCGAATTGA-3′ | |
| TGF-β1 | F: 5′-TATAGCAACAATTCCTGGCGTTAC-3′ |
| R: 5′-TGTATTCCGTCTCCTTGGTCA-3′ | |
| β-actin | F: 5′-CACCCGCGAGTACAACCTTC-3′ |
| R: 5′-CCCATACCCACCATCACACC-3′ |
F, forward; R, reverse; miR-20b-5p, microRNA-20b-5p; Smad3, small mothers against decapentaplegic homolog 3; Smad7, small mothers against decapentaplegic homolog 7; TGF-β1, transforming growth factor-β1.
Western blot analysis
Myocardial tissue sited 25 mm above the apex of the left ventricular wall was randomly selected from rats of each group, and 1X SDS lysis buffer was added to the samples (cat. no. P0013G; Beyotime Institute of Biotechnology, Shanghai, China), which were homogenized at 1,006 × g at room temperature to achieve complete cleavage. The samples were then placed on ice for 30 min at 4°C and centrifuged at 16,099 × g at 4°C for 4 min. The supernatant was separated and stored at −80°C. The proteins were extracted using RIPA lysis buffer (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), and the protein concentration was measured according to the instructions of the BCA kit (cat. no. AR0146, Wuhan Boshide Company, Wuhan, China); and, the concentration of each sample was adjusted to 3 µg/µl. Sample buffer (30 µg per well) was added to the extracted protein, and the resulting mixture was boiled for 10 min at 95°C and separated by 10% polyacrylamide gel electrophoresis. The proteins were transferred onto a polyvinylidende difluoride membrane via a semi-dry transfer method (cat. no. P2438, Sigma-Aldrich; Merck KGaA). The membranes were blocked with 5% bovine serum albumin (BSA; Fermentas; Thermo Fisher Scientific, Inc.) at room temperature for 1 h, following which the blocking solution was discarded, and the membrane was placed into a plastic container. Subsequently, 5% BSA with the appropriate concentration of rat anti-TGF-β1 primary antibody (cat. no. ab2792, Abcam, Cambridge, MA, USA) was added at a dilution of 1:1,000, rabbit anti-Smad3 primary antibody (cat. no. ab40854, Abcam) was added at a dilution of 1:1,000, rabbit anti-Smad7 primary antibody (cat. no. ab216428, Abcam) was added at a dilution of 1:1,000, and rat anti-β-actin primary antibody (cat. no. ab189146, Abcam) was added at a dilution of 1:500. Subsequently, with the transfer side up, the samples were placed in the refrigerator at 4°C overnight. The following day, the samples were rinsed with 0.05% TBST (three times, 10 min each), and the goat anti-rat IgG secondary antibody labeled with horseradish peroxidase was added at a dilution of 1:2,000 (cat. no. ab6789, Abcam) for incubation at 4°C for 4-6 h and then washed with TBS-T (three times, 15 min each). The chemiluminescence reagents A and B (Yanhui Biotechnology Co., Ltd., Shanghai, China) were mixed at a 1:1 ratio, and the mixture was evenly added to the nitrocellulose membrane. Relative light density analysis was performed on all the western blot bands using Quantity One 4.6 software (Bio-Rad Laboratories, Inc.).
Statistical analysis
All the data were analyzed utilizing the Statistical Package for the Social Sciences version 21.0 (SPSS; IBM Corps., Armonk, NY, USA). All experiments were repeated three times. Continuous data are presented as the mean ± standard deviation. Multiple groups were compared using one-way analysis of variance with the least significant difference test. Pairwise comparisons among multiple groups were performed with the least significant difference t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
miRNA-20b-5p targets Smad
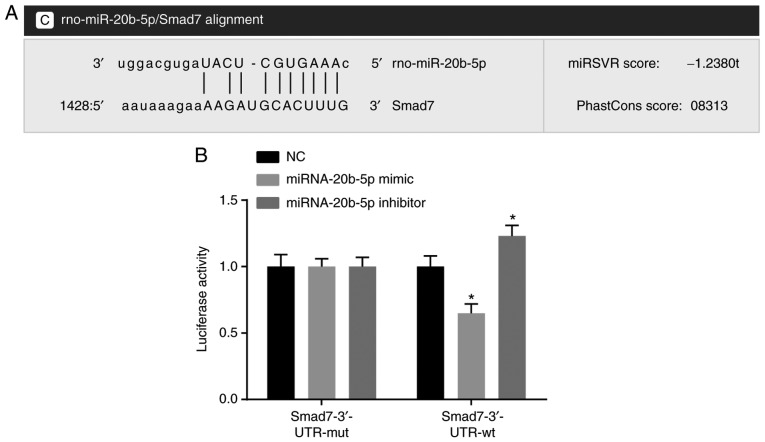
The results of the statistical analysis showed that there was a specific binding region between the 3′UTR 1458-1464 sequence and the miR-20b-5p sequence of Smad7, and Smad7 was found to be a potential target gene of miR-20b-5p (Fig. 1A). Luciferase reporter gene assays were used to verify this gene as a target of miR-20b-5p (Fig. 1B), and the results indicated that miR-20b-5p mimics had no significant effect on the luciferase activity of the Smad7-3′-UTR-mut group (P>0.05), but significantly decreased the luciferase activity in the Smad7-3′-UTR-wt group (P<0.05). miR-20b-5p was found to specifically bind to the Smad7-3′-UTR and downregulated expression of Smad7 was observed. The effect of the miR-20b-5p inhibitor was evaluated by dual luciferase reporter gene assays. The results (Fig. 1B) showed that luciferase activity was significantly increased in the Smad7-3′-UTR-wt group, compared with that in the NC group (P<0.05), suggesting that miR-20b-5p inhibitor upregulated the expression of Smad7. Taken together, miR-20b-5p was found to specifically bind Smad7 and negatively regulate the expression of Smad7.
Figure 1.
Target association verification between miR-20b-5p and Smad7. (A) Predicted binding site of miR-20b-5p in the Smad7 3′UTR. (B) Detection of luciferase activity by dual luciferase reporter gene assays. *P<0.05, compared with the NC group. miR-20b-5p, microRNA-20b-5p; Smad7, small mothers against decapentaplegic homolog 7; mut, mutant; wt, wild-type; NC, negative control.
Animal model generation
In establishing the rat model of myocardial IR injury, the data indicated no significant change in the ST segment of the ECG in the sham group at the preoperative, intraoperative or postoperative stages. The ST segment of the ECG of rats of the model group was significantly higher following ligation than pre-ligation, and decreased following reperfusion, which indicated that opening the chest without blocking the blood vessels had no significant effect on the cardiac electrophysiology of the rats. The changes in the ST segment were significant following ligation of the anterior descending coronary artery. Following reperfusion, cardiac electrophysiology was restored, which indicated that the rat model of myocardial IR injury was successfully established. All the experiments were performed in surviving rats without malignant arrhythmia. The survival rates of the rats in each group following 28 days of modeling are shown in Table II.
Table II.
Survival rates of rats.
| Group | Pre-modeling (n) | 28 days post-modeling (n) | Survival rate (%) |
|---|---|---|---|
| Sham | 10 | 10 | 100 |
| IR | 10 | 9 | 90 |
| NC | 10 | 9 | 90 |
| miR-20b-5p mimics | 10 | 10 | 100 |
| miR-20b-5p inhibitors | 10 | 8 | 80 |
| si-Smad7 | 10 | 9 | 90 |
| miR-20b-5p inhibitors + si-Smad7 | 10 | 10 | 100 |
IR, ischemia-reperfusion; NC, negative control; miR-20b-5p, microRNA-20b-5p; Smad7, small mothers against decapentaplegic homolog 7; si-, small interfering RNA.
Increased expression of miR-20b-5p and decreased expression of Smad7 are unfavorable for cardiac function in IR injury model rats
Compared with the sham group, the other six groups had significantly increased LVEDV, LVEDD, LVESV and LVESD (all P<0.05). Compared with the IR group, the NC group and the miR-20b-5p inhibitors + si-Smad7 group showed no significant differences in LVEDV, LVEDD, LVESV and LVESD (all P>0.05); the miR-20b-5p inhibitors group exhibited a significant decrease in LVEDV, LVEDD, LVESV and LVESD (all P<0.05); and the miR-20b-5p mimics group and the si-Smad7 group exhibited a significant increase in LVEDV, LVEDD, LVESV and LVESD (all P<0.05). No significant difference in these values were found between the miR-20b-5p mimics group and the si-Smad7 group (all P>0.05). These results are shown in Table III.
Table III.
Results of LVEDV, LVEDD, LVESV and LVESD by echocardiography (n=5).
| Group | LVEDV (µl) | LVEDD (mm) | LVESV (µl) | LVESD (mm) |
|---|---|---|---|---|
| Sham | 226.22±48.92 | 5.68±0.54 | 81.21±15.12 | 3.25±0.33 |
| IR | 738.17±92.36a | 11.37±2.10a | 266.40±11.22a | 7.83±1.00a |
| NC | 739.10±89.25a | 11.40±2.00a | 267.40±11.12a | 7.84±1.10a |
| miR-20b-5p mimics | 939.25±111.32a,b,c | 31.40±2.00a,b,c | 366.40±11.22a,b,c | 10.83±1.00a,b,c |
| miR-20b-5p inhibitors | 414.79±89.25a,b,c | 8.47±2.02a,b,c | 116.45±45.04a,b,c | 5.62±2.23a,b,c |
| si-Smad7 | 938.25±120.02a,b,c | 31.30±2.02a,b,c | 356.40±11.22a,b,c | 9.83±1.00a,b,c |
| miR-20b-5p inhibitors + si-Smad7 | 733.17±110.23a | 11.39±2.00a | 270.40±11.22a | 7.90±1.00a |
P<0.05, compared with the sham group;
P<0.05, compared with the IR group;
P<0.05, compared with the miR-20b-5p inhibitors + si-Smad7 group. LVEDD, left ventricular diastolic diameter; LVESD, left ventricular systolic diameter; LVEDV, left ventricular end-diastolic volume; LVESV, left ventricular end-systolic volume; IR, ischemia-reperfusion; NC, negative control; miR-20b-5p, microRNA-20b-5p; Smad7, small mothers against decapentaplegic homolog 7; si-, small interfering RNA.
Compared with the sham group, the LVEEF and LVEFS were significantly decreased in the other six groups (all P<0.05). Compared with the IR group, no significant differences in LVEEF or LVEFS were found between the NC group and the miR-20b-5p inhibitors + si-Smad7 group (P>0.05). The miR-20b-5p inhibitors group had significantly increased LVEEF and LVEFS (P<0.05), whereas the miR-20b-5p mimics group and the si-Smad7 group had significantly decrease in LVEEF and LVEFS (P<0.05). There were no differences in these values between the miR-20b-5p mimics group and the si-Smad7 group (P>0.05). These results are presented in Table IV.
Table IV.
Echocardiograph results of LVEEF and LVEFS (n=5).
| Group | LVEEF (%) | LVEFS (%) |
|---|---|---|
| Sham | 79.41±4.78 | 49.65±6.19 |
| IR | 57.41±3.28a | 29.21±5.12a |
| NC | 56.51±3.22a | 28.54±4.56a |
| miR-20b-5p mimics | 40.23±2.12a,b,c | 19.12±3.34a,b,c |
| miR-20b-5p inhibitors | 68.23±2.34a,b,c | 37.34±4.01a,b,c |
| si-Smad7 | 39.67±2.45a,b,c | 18.17±3.96a,b,c |
| miR-20b-5p inhibitors + si-Smad7 | 57.67±1.45a | 28.03±3.96a |
LVEEF = (LVEDV-LVESV)/LVEDV ×100%; LVEFS = (LVEDD-LVESD)/LVEDD ×100%.
P<0.05, compared with the sham group;
P<0.05, compared with the IR group;
P<0.05, compared with the miR-20b-5p inhibitors + si-Smad7 group. LVEEF, left ventricular ejection fraction; LVEFS, left ventricular fractional shortening; IR, ischemia-reperfusion; NC, negative control; miR-20b-5p, microRNA-20b-5p; Smad7, small mothers against decapentaplegic homolog 7; si-, small interfering RNA.
Compared with the sham group, the other six groups exhibited a decrease in LVESP and an increase in LVEDP (all P<0.05). Compared with the IR group, no significant differences in LVESP or LVEDP were found between the NC group and the miR-20b-5p inhibitors + si-Smad7 group (P>0.05). The miR-20b-5p inhibitors group exhibited an increase in LVESP and a decrease in LVEDP (P<0.05), and the miR-20b-5p mimics group and the si-Smad7 group exhibited a decrease in LVESP and an increase in LVEDP (P<0.05). There were no differences in these values between the miR-20b-5p mimics group and the si-Smad7 group (P>0.05), as shown in Table V. Taken together, the overexpression of miR-20b-5p and downregulated expression of Smad7 were critical in the cardiac function of rats with IR injury.
Table V.
Echocardiograph results of LVESP and LVEDP (n=5).
| Group | LVESP (mmHg) | LVEDP (mmHg) |
|---|---|---|
| Sham | 128.11±4.02 | 3.51± 0.87 |
| IR | 92.42±8.22a | 8.92±1.32a |
| NC | 91.21±8.50a | 9.05±1.25a |
| miR-20b-5p mimics | 76.58±3.22a,b,c | 13.19±1.48a,b,c |
| miR-20b-5p inhibitors | 111.45±7.23a,b,c | 6.16±0.92a,b,c |
| si-Smad7 | 74.42±3.33a,b,c | 14.02±1.45a,b,c |
| miR-20b-5p inhibitors + si-Smad7 | 90.52±8.02a | 8.92±1.13a |
P<0.05, compared with the sham group;
P<0.05, compared with the IR group;
P<0.05, compared with the miR-20b-5p inhibitors + si-Smad7 group. LVESP, left ventricular systolic pressure; LVEDP, left ventricular end-diastolic pressure; IR, ischemia-reperfusion; NC, negative control; miR-20b-5p, microRNA-20b-5p; Smad7, small mothers against decapentaplegic homolog 7; si-, small interfering RNA.
Increased expression of miR-20b-5p and decreased expression of Smad7 enlarge myocardial infarct size in rats
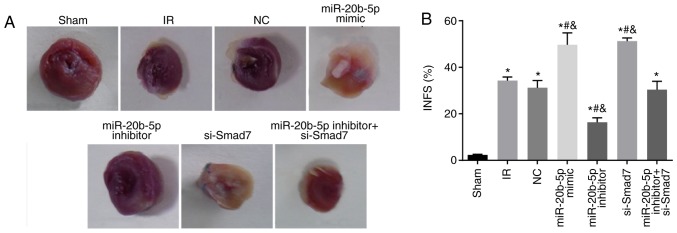
Compared with the sham group, the other sex groups exhibited a significant increase in myocardial infarction area (all P<0.05). Compared with the IR group, no significant differences in myocardial infarction area were found in the miR-20b-5p inhibitors + si-Smad7 group or the NC group (P>0.05), whereas miR-20b-5p inhibitors group exhibited a significant decrease in myocardial infarction area (P<0.05), and the miR-20b-5p mimics group and the si-Smad7 group exhibited significant increases in myocardial infarction area (P<0.05). There was no significant difference in this value between the miR-20b-5p mimics group and the si-Smad7 group (P>0.05), as shown in Fig. 2A and B. The overexpression of miR-20b-5p and downregulation of Smad7 increased myocardial infarct size in the rats.
Figure 2.
Myocardial infarct size in rats in each group. (A) Triphenyltetrazolium chloride staining results of rats in each group. (B) Comparison of myocardial infarct size in rats in each group. *P<0.05, compared with the sham group; #P<0.05, compared with the IR group; &P<0.05, compared with the miR-20b-5p inhibitors + si-Smad7 group. miR-20b-5p, microRNA-20b-5p; Smad7, small mothers against decapentaplegic homolog 7; si-, small interfering RNA; NC, negative control; IR, ischemia-reperfusion; INFS, infarct size.
Increased expression of miR-20b-5p and decreased expression of Smad7 inhibit the growth of cardiomyocytes in rats with IR injury
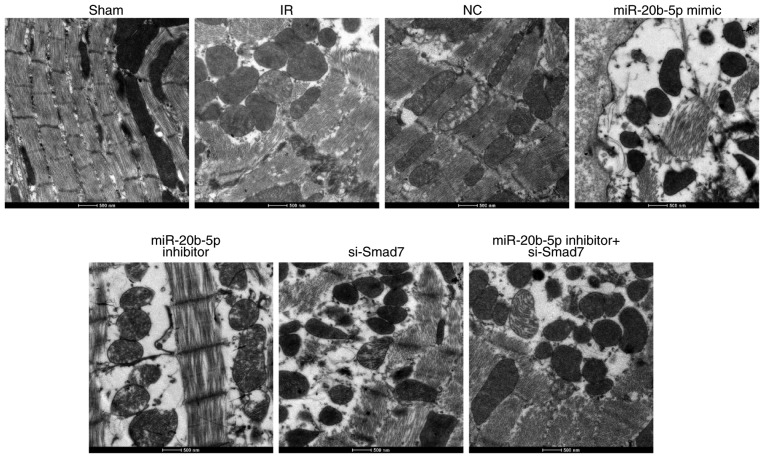
The ultrastructural examination of cardiomyocytes in each group showed the orderly arrangement of myofilaments, normal shape and structure of mitochondria, and orderly arrangement and clear structure of cristae in rat cardiomyocytes in the sham group. In the IR group, the myofilaments of rat cardiomyocytes were broken and dissolved, and the majority of these were loosely arranged and displaced. The electron density of mitochondria was increased, and regions of the cristae membranes were blurred and dissolved. The myocardial ultrastructure in the NC group and the miR-20b-5p inhibitors + si-Smad7 group was similar to that in the IR group. Compared with the IR group, the miR-20b-5p inhibitors group showed marginal recovery with orderly arrangement of myofilaments and distinguished mitochondria; in the miR-20b-5p mimics group and the si-Smad7 group, the myofilament rupture and dissolution were more severe, and the majority of the filaments were loosely arranged and displaced. The electron density of mitochondria was higher, and the majority of the cristae of mitochondria were indistinct, with evidence of dissolution (Fig. 3). The low expression of miR-20b-5p and high expression of Smad7 promoted the growth of cardiomyocytes in the rats with IR injury.
Figure 3.
Ultrastructure of cardiomyocytes in rats in each group (n=3). Images at magnification, ×3,000. miR-20b-5p, microRNA-20b-5p; Smad7, small mothers against decapentaplegic homolog 7; si-, small interfering RNA; NC, negative control; IR, ischemia-reperfusion.
Increased expression of miR-20b-5p and decreased expression of Smad7 accelerate myocardial necrosis in rats with IR injury
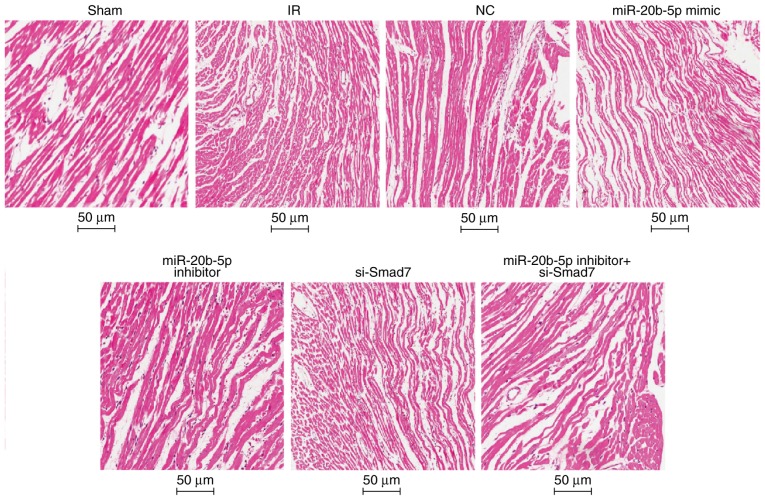
Under the light microscope, the myocardial fibers were orderly and tightly arranged with no edema or hemorrhage in the sham group. In the IR group, the myocardial fibers were disordered, the cells were swollen, interstitial edema was obvious, and myocardial cells showed marked necrosis. The results of the HE staining were similar in the NC group, the miR-20b-5p inhibitors + si-Smad7 group and the IR group. Compared with the IR group, the myocardial fiber arrangement was more orderly and myocardial cell necrosis was significantly reduced in the miR-20b-5p inhibitors group. In the miR-20b-5p mimics group and the si-Smad7 group, the myocardial fibers were disordered, and the necrotic area was significantly increased (Fig. 4). The high expression of miR-20b-5p and low expression of Smad7 enhanced myocardial necrosis in rats with IR injury.
Figure 4.
Hematoxylin and eosin staining of the myocardium in rats in each group (n=3). Images at magnification, ×200. miR-20b-5p, microRNA-20b-5p; Smad7, small mothers against decapentaplegic homolog 7; si-, small interfering RNA; NC, negative control; IR, ischemia-reperfusion.
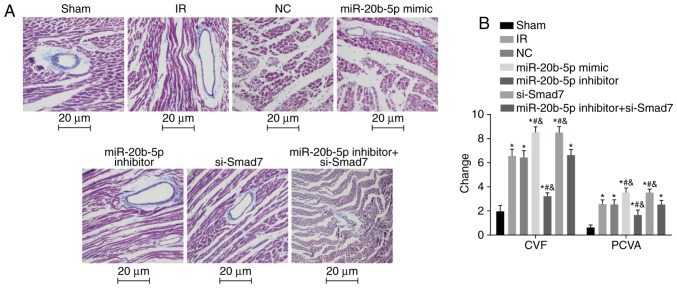
Increased expression of miR-20b-5p and decreased expression of Smad7 result in increased CVF and PVCA
The results of the Masson's staining of myocardial tissue in each group are shown in Fig. 5A and B. Compared with the sham group, the other six groups exhibited significantly increased CVF and PVCA (P<0.05). Compared with the IR group, there were no significant differences in CVF or PVCA in the NC group or the miR-20b-5p inhibitors + si-Smad7 group (P>0.05), the miR-20b-5p inhibitors group exhibited decreased CVF and PVCA (P<0.05), and the miR-20b-5p mimics group and the si-Smad7 group exhibited increased CVF and PVCA (P<0.05). The CVF and PVCA values did not differ significantly between the miR-20b-5p mimics group and the si-Smad7 group (P>0.05). The overexpression of miR-20b-5p and downregulation of Smad7 increased myocardial collagen in the rats with IR injury.
Figure 5.
Masson's staining of myocardial tissue from rats in each group. (A) Masson's staining of myocardial tissue from rats in each group (magnification, ×400). (B) Changes in CVF and PVCA in rats in each group are shown in a bar graph. *P<0.05, compared with the sham group; #P<0.05, compared with the IR group; &P<0.05, compared with the miR-20b-5p inhibitors + si-Smad7 group. CVF, myocardial collagen volume fraction; PVCA, perivascular collagen area; miR-20b-5p, microRNA-20b-5p; Smad7, small mothers against decapentaplegic homolog 7; si-, small interfering RNA; NC, negative control; IR, ischemia-reperfusion.
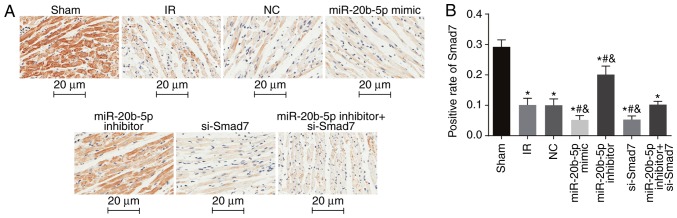
Smad7-positive rate is low in myocardial tissue of rats with IR injury
The positive expression of Smad7 in the myocardial cytosol and membrane were indicated by brown staining (Fig. 6A). Compared with the sham group, the other six groups had a significantly decreased Smad7-positive rate (P<0.05). Compared with the IR group, there was no significant difference in the Smad7-positive rate in the NC group or the miR-20b-5p inhibitors + si-Smad7 group (P>0.05), whereas the Smad7-positive rate was significantly increased in the miR-20b-5p inhibitors group, and decreased in the miR-20b-5p mimics group and the si-Smad7 group (P<0.05). There was no significant difference between the miR-20b-5p mimics group and the si-Smad7 group (P>0.05), as shown in Fig. 6B. The overexpression of Smad7 had a protective effect in the rats with IR injury.
Figure 6.
Immunohistochemical results of rats in each group (n=3). (A) Immunohistochemical results (magnification, ×400). (B) Comparison of the Smad7-positive rate among all rat groups. miR-20b-5p, microRNA-20b-5p; Smad7, small mothers against decapentaplegic homolog 7; si-, small interfering RNA; NC, negative control; IR; ischemia-reperfusion; *P<0.05, compared with the sham group; #P<0.05, compared with the IR group; &P<0.05, compared with the miR-20b-5p inhibitors + si-Smad7 group.
Expression of miR-20b-5p and the mRNA expression of TGF-β1 and Smad3 are upregulated, and the mRNA expression of Smad7 is decreased in the myocardium of rats with IR injury
Compared with the sham group, the other six groups exhibited significantly increased expression of miR-20b-5p and mRNA expression of TGF-β1 and Smad3, whereas the mRNA expression of Smad7 was significantly decreased (all P<0.05; Fig. 7). Compared with the IR group, there was no significant difference in these markers in the NC group (P>0.05); the miR-20b-5p inhibitors + si-Smad7 group showed no significant difference in the mRNA expression of TGF-β1, Smad7 or Smad3 (P>0.05), however, a significant decrease in the expression of miR-20b-5p was observed (P<0.05); the miR-20b-5p inhibitors group exhibited a decrease in the expression of miR-20b-5p and the mRNA expression of TGF-β1 and Smad3, and an increase in the mRNA expression of Smad7 (all P<0.05); the miR-20b-5p mimics group exhibited a marked increase in expression of miR-20b-5p and the mRNA expression of TGF-β1 and Smad3, and a significant decrease in the mRNA expression of Smad7 (all P<0.05); the si-Smad7 group showed no significant difference in the expression of miR-20b-5p (P>0.05), but there were significant increases in the mRNA expression of TGF-β1 and Smad3, and a significant decrease in the mRNA expression of Smad7 (all P<0.05). These results are shown in Fig. 7. Activation of the TGF-β1/Smad signaling pathway promoted ventricular remodeling in the rats with IR injury.
Figure 7.
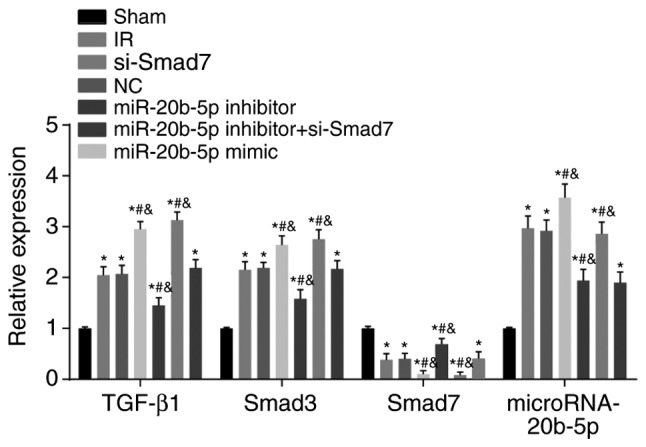
Expression of miR-20b-5p and mRNA expression of TGF-β1/Smad signaling pathway-related proteins in myocardial tissue from rats in each group. *P<0.05, compared with the sham group; #P<0.05, compared with the IR group; &P<0.05, compared with the miR-20b-5p inhibitors + si-Smad7 group. miR-20b-5p, microRNA-20b-5p; Smad3, small mothers against decapentaplegic homolog 3; Smad7, small mothers against decapentaplegic homolog 7; si-, small interfering RNA; TGF-β1, transforming growth factor-β1; NC, negative control; IR, ischemia-reperfusion.
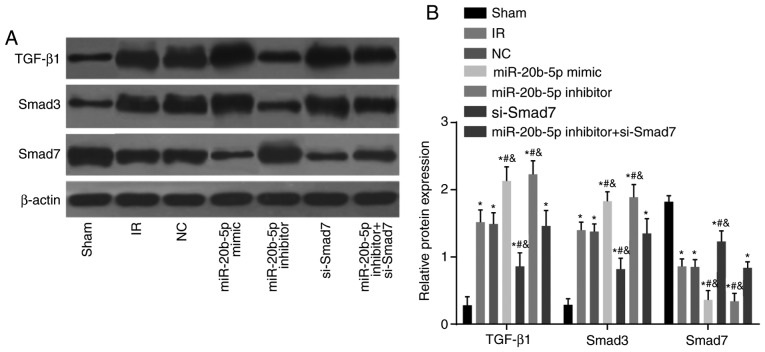
Protein expression levels of TGF-β1 and Smad3 are upregulated and the protein expression of Smad7 is decreased in the myocardium of rats with IR injury
Compared with the sham group, the other six groups showed a significant increase in the protein expression of TGF-β1 and Smad3, whereas the protein expression of Smad7 decreased (all P<0.05; Fig. 8A and B). Compared with the IR group, no significant difference in the protein expression of TGF-β1 and Smad3 were observed in the NC group or the miR-20b-5p inhibitors + si-Smad7 group (P>0.05). The miR-20b-5p inhibitors group exhibited a marked decrease in the expression of TGF-β1 and Smad3 and a significant increase in the expression of Smad7 (P<0.05), and the miR-20b-5p mimics group and the si-Smad7 group exhibited a marked increase in the expression of TGF-β1 and Smad3 and a decrease in the expression of Smad7 (P<0.05). No significant differences (P>0.05) in these values were found between the miR-20b-5p mimics group and the si-Smad7 group (Fig. 8). The low expression of miR-20b-5p and high expression of Smad7 inhibited the TGF-β1/Smad signaling pathway.
Figure 8.
Expression of TGF-β1/Smad signaling pathway components in myocardial tissue from rats in each group. (A) Banding diagram of the expression of TGF-β1/Smad signaling pathway components in myocardial tissue from rats in each group, (B) Histogram of the expression of TGF-β1/Smad signaling pathway components in myocardial tissue from rats in each group. *P<0.05, compared with the sham group; #P<0.05, compared with the IR group; &P<0.05, compared with the miR-20b-5p inhibitors + si-Smad7 group. miR-20b-5p, microRNA-20b-5p; Smad3, small mothers against decapentaplegic homolog 3; Smad7, small mothers against decapentaplegic homolog 7; si-, small interfering RNA; TGF-β1, transforming growth factor-β1; NC, negative control; IR, ischemia-reperfusion.
Acknowledgments
Not applicable.
Discussion
With the development of thrombolysis, percutaneous transluminal coronary angioplasty and coronary artery bypass grafting, myocardial IR injury has become the main obstacle in obtaining the optimal therapeutic effect from ischemic heart disease during reperfusion treatment (28). Following myocardial IR therapy, the heart is confronted with further injury through ventricular remodeling (29,30). Therefore, it is important to introduce effective myocardial protection measures to reduce ventricular remodeling following myocardial IR therapy (31). Following myocardial IR therapy in rats, the present study found that myocardial ischemia was involved with the process of cardiac remodeling following treatment, and the infarct size and myocardial collagen content were increased. The results showed that miR-20b-5p inhibited the expression of Smad7, activated the TGF-β signaling pathway, and promoted ventricular remodeling following myocardial IR injury in the rats.
Following myocardial ischemia, the sudden recovery of myocardial blood flow aggravates ischemic myocardial injury, which clinically manifests as arrhythmia, myocardial stunning, heart dysfunction and other symptoms, and is termed myocardial IR injury (10). Increasing the concentration of glycolytic substrates in the reperfusion blood can reduce the damage to ischemic myocardial cells and promote the recovery of myocardial function, whereas inhibiting the glycolytic process can increase the overload of myocardial intracellular calcium and delay the recovery of myocardial function (32). Galang et al (33) reported that, when IR occurred in the isolated rat heart, cardiomyocyte apoptosis was reduced by erythrocuprein and catalase. The present study found that, compared with that in the sham group, the area of myocardial infarction following reperfusion in the other six groups was significantly higher. Following the downregulation of miR-20b-5p, the infarct size decreased significantly. These results showed that myocardial IR injury led to the apoptosis of myocardial cells, and that miR-20b-5p was significant in promoting apoptosis in myocardial IR injury.
Ventricular remodeling is the internal cause of cardiac insufficiency and heart failure (29). Numerous basic and clinical studies have shown that ventricular remodeling can be organized or reversed (34). Ventricular remodeling is usually associated with fibrosis (35). A number of studies have indicated that the canonical TGF-β1/Smad3 pathway is critically involved in the pathogenesis of fibrosis in several tissues (36). Long-term ventricular remodeling eventually manifests as increased heart rate, LVEDV and LVRDV, and decreased systolic volume and LVEEF (37). The ventricular remodeling following myocardial infarction has been verified in a number of studies, however, ventricular remodeling following myocardial IR is rare; therefore, the present study established a rat model of myocardial IR injury model to observe the dynamic process of ventricular remodeling following myocardial IR injury. The results of transthoracic echocardiography showed that, following reperfusion, LVEEF and LVEFS decreased significantly. These results suggested that the heart underwent a process of ventricular remodeling following myocardial IR injury in rats, resulting in a decline in cardiac function.
miRs, by combining with target mRNAs, regulate target mRNA translation and are involved in the regulation of genes that are widely involved in cell differentiation, proliferation and apoptosis (38,39). The expression of miRs has strict temporal and tissue specificity, and the cardiovascular system has its specific miR expression profile (40). It has been revealed that miRs expressed in the cardiovascular system are involved in the development of the cardiovascular system and the course of disease (41). Mukhopadhyay et al (15) suggested that increased miR-20b (anti-angiogenic) was linked with cardiac remodeling. Smads are considered to be mediators and regulators of TGF-β signaling (42). Smad7 has been suggested as an important inhibitory protein in the TGF-β signaling pathway (18). In the present study, RT-qPCR and western blot analyses were used to detect the expression of miR-20b-5p, TGF-β1, Smad3 and Smad7, and the results showed that the expression of miR-20b-5p, TGF-β1 and Smad3 were significantly increased, whereas that of Smad7 was significantly decreased following myocardial IR. Smad7 was found to be essential for cardiac development at the embryonic stage and for cardiac function in adults (43). In addition, Smad7 can reduce angiotensin II-induced hypertensive cardiac remodeling, and it may be a therapeutic target for hypertensive cardiovascular diseases (44). The promoter region of Smad7 is stimulated by the ectopic expression of Smad3, and active TGF-β and activin receptors, indicating that the transcription of Smad7 is regulated by these two receptors (45). Smad3 activates myostatin, thereby inducing the expression of Smad7, whereas Smad7 mediates the myostatin signal transduction pathway through a negative feedback mechanism (46). These results suggested that miR-20b-5p upregulated the expression of Smad7 in the process of ventricular remodeling following myocardial IR injury.
In conclusion, the present in vivo model of myocardial IR injury in rats verified the process of ventricular remodeling following myocardial IR, indicating that miR-20b-5p promoted ventricular remodeling following myocardial IR injury in rats via inhibiting the expression of Smad7 through activating the TGF-β/Smad signaling pathway. However, due to limited time and funds, the present study failed to observe longer term myocardial remodeling following myocardial IR injury. Verification of these results is required in future investigations.
Funding
No funding was received.
Availability of data and materials
The datasets generated/analysed during the current study are available.
Authors' contributions
ZGL and HY contributed in the conception of the work, conducting the study, preparing the manuscript. RSX collated the data, designed and developed the database, and CLG been involved in reviewing the results and discussions. YT analyzed and interpreted the data, and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Laboratory Animal Ethics Committee of The First Affiliated Hospital of Harbin Medical University, and was performed in accordance with the principles of animal protection, welfare and ethics and the National Laboratory Animal Welfare Ethics regulations.
Consent for publication
Consent for publication was obtained from the participants.
Competing interests
The authors declare that they have no competing interests.
References
- 1.De Roeck L, Vandamme S, Everaert BR, Hoymans V, Haine S, Vandendriessche T, Bosmans J, Ronsyn MW, Miljoen H, Van Berendoncks A, et al. Adiponectin and ischemia-reperfusion injury in ST segment elevation myocardial infarction. Eur Heart J Acute Cardiovasc Care. 2016;5:71–76. doi: 10.1177/2048872615570770. [DOI] [PubMed] [Google Scholar]
- 2.Selmer R, Halvorsen S, Myhre KI, Wisløff TF, Kristiansen IS. Cost-effectiveness of primary percutaneous coronary intervention versus thrombolytic therapy for acute myocardial infarction. Scand Cardiovasc J. 2005;39:276–285. doi: 10.1080/14017430510035988. [DOI] [PubMed] [Google Scholar]
- 3.Ong SB, Samangouei P, Kalkhoran SB, Hausenloy DJ. The mitochondrial permeability transition pore and its role in myocardial ischemia reperfusion injury. J Mol Cell Cardiol. 2015;78:23–34. doi: 10.1016/j.yjmcc.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Fröhlich GM, Meier P, White SK, Yellon DM, Hausenloy DJ. Myocardial reperfusion injury: Looking beyond primary PCI. Eur Heart J. 2013;34:1714–1722. doi: 10.1093/eurheartj/eht090. [DOI] [PubMed] [Google Scholar]
- 5.Wick G, Grundtman C, Mayerl C, Wimpissinger TF, Feichtinger J, Zelger B, Sgonc R, Wolfram D. The immunology of fibrosis. Annu Rev Immunol. 2013;31:107–135. doi: 10.1146/annurev-immunol-032712-095937. [DOI] [PubMed] [Google Scholar]
- 6.Neri M, Fineschi V, Di Paolo M, Pomara C, Riezzo I, Turillazzi E, Cerretani D. Cardiac oxidative stress and inflammatory cytokines response after myocardial infarction. Curr Vasc Pharmacol. 2015;13:26–36. doi: 10.2174/15701611113119990003. [DOI] [PubMed] [Google Scholar]
- 7.Zhang M, Xu YJ, Saini HK, Turan B, Liu PP, Dhalla NS. TNF-alpha as a potential mediator of cardiac dysfunction due to intracellular Ca2+-overload. Biochem Biophys Res Commun. 2005;327:57–63. doi: 10.1016/j.bbrc.2004.11.131. [DOI] [PubMed] [Google Scholar]
- 8.Hausenloy DJ, Yellon DM. The mitochondrial permeability transition pore: Its fundamental role in mediating cell death during ischaemia and reperfusion. J Mol Cell Cardiol. 2003;35:339–341. doi: 10.1016/S0022-2828(03)00043-9. [DOI] [PubMed] [Google Scholar]
- 9.hao ZQ, Nakamura M, Wang NP, Velez DA, Hewan-Lowe KO, Guyton RA, Vinten-Johansen J. Dynamic progression of contractile and endothelial dysfunction and infarct extension in the late phase of reperfusion. J Surg Res. 2000;94:133–144. doi: 10.1006/jsre.2000.6029. [DOI] [PubMed] [Google Scholar]
- 10.Hausenloy DJ, Yellon DM. Myocardial ischemia-reperfusion injury: A neglected therapeutic target. J Clin Invest. 2013;123:92–100. doi: 10.1172/JCI62874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan ZX, Yang J. The role of microRNAs in regulating myocardial ischemia reperfusion injury. Saudi Med J. 2015;36:787–793. doi: 10.15537/smj.2015.7.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki HI, Miyazono K. Emerging complexity of microRNA generation cascades. J Biochem. 2011;149:15–25. doi: 10.1093/jb/mvq113. [DOI] [PubMed] [Google Scholar]
- 13.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 14.He B, Xiao J, Ren AJ, Zhang YF, Zhang H, Chen M, Xie B, Gao XG, Wang YW. Role of miR-1 and miR-133a in myocardial ischemic postconditioning. J Biomed Sci. 2011;18:22. doi: 10.1186/1423-0127-18-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukhopadhyay P, Das S, Ahsan MK, Otani H, Das DK. Modulation of microRNA 20b with resveratrol and longevinex is linked with their potent anti-angiogenic action in the ischaemic myocardium and synergestic effects of resveratrol and γ-tocotrienol. J Cell Mol Med. 2012;16:2504–2517. doi: 10.1111/j.1582-4934.2011.01480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickinson BA, Semus HM, Montgomery RL, Stack C, Latimer PA, Lewton SM, Lynch JM, Hullinger TG, Seto AG, van Rooij E. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur J Heart Fail. 2013;15:650–659. doi: 10.1093/eurjhf/hft018. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Chen D, Jin L, Liu J, Su Z, Li Y, Gui Y, Lai Y. MicroRNA-20b-5p functions as a tumor suppressor in renal cell carcinoma by regulating cellular proliferation, migration and apoptosis. Mol Med Rep. 2016;13:1895–1901. doi: 10.3892/mmr.2015.4692. [DOI] [PubMed] [Google Scholar]
- 18.Blahna MT, Hata A. Smad-mediated regulation of microRNA biosynthesis. FEBS Lett. 2012;586:1906–1912. doi: 10.1016/j.febslet.2012.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanyu A, Ishidou Y, Ebisawa T, Shimanuki T, Imamura T, Miyazono K. The N domain of Smad7 is essential for specific inhibition of transforming growth factor-beta signaling. J Cell Biol. 2001;155:1017–1027. doi: 10.1083/jcb.200106023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zorzi F, Angelucci E, Sedda S, Pallone F, Monteleone G. Smad7 antisense oligonucleotide-based therapy for inflammatory bowel diseases. Dig Liver Dis. 2013;45:552–555. doi: 10.1016/j.dld.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Cozzolino D, Sasso FC, Salvatore T, Torella M, Gentile S, Torella R, Giugliano D. Acute effects of beta-endorphin on cardiovascular function in patients with mild to moderate chronic heart failure. Am Heart J. 2004;148:E13. doi: 10.1016/j.ahj.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 22.Takagawa J, Zhang Y, Wong ML, Sievers RE, Kapasi NK, Wang Y, Yeghiazarians Y, Lee RJ, Grossman W, Springer ML. Myocardial infarct size measurement in the mouse chronic infarction model: Comparison of area- and length-based approaches. J Appl Physiol. 2007;102:2104–2111. doi: 10.1152/japplphysiol.00033.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Timmers L, Lim SK, Arslan F, Armstrong JS, Hoefer IE, Doevendans PA, Piek JJ, El Oakley RM, Choo A, Lee CN, et al. Reduction of myocardial infarct size by human mesenchymal stem cell conditioned medium. Stem Cell Res. 2007;1:129–137. doi: 10.1016/j.scr.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Li XW, Wang XM, Li S, Yang JR. Effects of chrysin (5,7-dihydroxyf lavone) on vascular remodeling in hypoxia-induced pulmonary hypertension in rats. Chin Med. 2015;10:4. doi: 10.1186/s13020-015-0032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown RS, Wahl RL. Overexpression of Glut-1 glucose transporter in human breast cancer. An immunohistochemical study. Cancer. 1993;72:2979–2985. doi: 10.1002/1097-0142(19931115)72:10<2979::AID-CNCR2820721020>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 26.Li R, Yan G, Li Q, Sun H, Hu Y, Sun J, Xu B. MicroRNA-P145 protects cardiomyocytes against hydrogen peroxide (H2O2)-induced apoptosis through targeting the mitochondria apoptotic pathway. PLoS One. 2012;7:e44907. doi: 10.1371/journal.pone.0044907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Liu H, Shang J, Chu F, Li A, Wu B, Xie X, Liu W, Yang H, Tong T. Protective effects of Shen-Yuan-Dan, a traditional Chinese medicine, against myocardial ischemia/reperfusion injury in vivo and in vitro. Evid Based Complement Alternat Med. 2013;2013:956397. doi: 10.1155/2013/956397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohn JN, Ferrari R, Sharpe N. Cardiac remodeling-concepts and clinical implications: A consensus paper from an international forum on cardiac remodeling. Behalf of an international forum on cardiac remodeling. J Am Coll Cardiol. 2000;35:569–582. doi: 10.1016/S0735-1097(99)00630-0. [DOI] [PubMed] [Google Scholar]
- 30.Ito Y, Ito K, Shiroto T, Tsuburaya R, Yi GJ, Takeda M, Fukumoto Y, Yasuda S, Shimokawa H. Cardiac shock wave therapy ameliorates left ventricular remodeling after myocardial ischemia-reperfusion injury in pigs in vivo. Coron Artery Dis. 2010;21:304–311. doi: 10.1097/MCA.0b013e32833aec62. [DOI] [PubMed] [Google Scholar]
- 31.Hagiwara S, Iwasaka H, Shingu C, Matumoto S, Hasegawa A, Noguchi T. Heat shock protein 47 (HSP47) antisense oligonucleotides reduce cardiac remodeling and improve cardiac function in a rat model of myocardial infarction. Thorac Cardiovasc Surg. 2011;59:386–392. doi: 10.1055/s-0030-1250658. [DOI] [PubMed] [Google Scholar]
- 32.Apstein CS. Increased glycolytic substrate protection improves ischemic cardiac dysfunction and reduces injury. Am Heart J. 2000;139:S107–S114. doi: 10.1067/mhj.2000.103920. [DOI] [PubMed] [Google Scholar]
- 33.Galang N, Sasaki H, Maulik N. Apoptotic cell death during ischemia/reperfusion and its attenuation by antioxidant therapy. Toxicology. 2000;148:111–118. doi: 10.1016/S0300-483X(00)00201-8. [DOI] [PubMed] [Google Scholar]
- 34.Fedak PW, Verma S, Weisel RD, Skrtic M, Li RK. Cardiac remodeling and failure: From molecules to man (Part III) Cardiovasc Pathol. 2005;14:109–119. doi: 10.1016/j.carpath.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 35.Donekal S, Venkatesh BA, Liu YC, Liu CY, Yoneyama K, Wu CO, Nacif M, Gomes AS, Hundley WG, Bluemke DA, Lima JA. Interstitial fibrosis, left ventricular remodeling, and myocardial mechanical behavior in a population-based multiethnic cohort: The multi-ethnic study of Atherosclerosis (MESA) study. Circ Cardiovasc Imaging. 2014;7:292–302. doi: 10.1161/CIRCIMAGING.113.001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu M, Chen J, Huang Y, Ke J, Li L, Huang D, Wu W. Triptolide alleviates isoprenaline-induced cardiac remodeling in rats via TGF-β1/Smad3 and p38 MAPK signaling pathway. Pharmazie. 2015;70:244–250. [PubMed] [Google Scholar]
- 37.Kasama S, Toyama T, Hatori T, Sumino H, Kumakura H, Takayama Y, Ichikawa S, Suzuki T, Kurabayashi M. Evaluation of cardiac sympathetic nerve activity and left ventricular remodelling in patients with dilated cardiomyopathy on the treatment containing carvedilol. Eur Heart J. 2007;28:989–995. doi: 10.1093/eurheartj/ehm048. [DOI] [PubMed] [Google Scholar]
- 38.Fazi F, Nervi C. MicroRNA: Basic mechanisms and transcriptional regulatory networks for cell fate determination. Cardiovasc Res. 2008;79:553–561. doi: 10.1093/cvr/cvn151. [DOI] [PubMed] [Google Scholar]
- 39.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 40.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 42.Kretzschmar M, Massague J. SMADs: Mediators and regulators of TGF-beta signaling. Curr Opin Genet Dev. 1998;8:103–111. doi: 10.1016/S0959-437X(98)80069-5. [DOI] [PubMed] [Google Scholar]
- 43.Chen Q, Chen H, Zheng D, Kuang C, Fang H, Zou B, Zhu W, Bu G, Jin T, Wang Z, et al. Smad7 is required for the development and function of the heart. J Biol Chem. 2009;284:292–300. doi: 10.1074/jbc.M807233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei LH, Huang XR, Zhang Y, Li YQ, Chen HY, Yan BP, Yu CM, Lan HY. Smad7 inhibits angiotensin II-induced hypertensive cardiac remodelling. Cardiovasc Res. 2013;99:665–673. doi: 10.1093/cvr/cvt151. [DOI] [PubMed] [Google Scholar]
- 45.Nagarajan RP, Zhang J, Li W, Chen Y. Regulation of Smad7 promoter by direct association with Smad3 and Smad4. J Biol Chem. 1999;274:33412–33418. doi: 10.1074/jbc.274.47.33412. [DOI] [PubMed] [Google Scholar]
- 46.Zhu X, Topouzis S, Liang LF, Stotish RL. Myostatin signaling through Smad2, Smad3 and Smad4 is regulated by the inhibitory Smad7 by a negative feedback mechanism. Cytokine. 2004;26:262–272. doi: 10.1016/j.cyto.2004.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated/analysed during the current study are available.