Abstract
BH3 domains, classified initially as BCL2 homology domains, participate in both apoptosis and autophagy. Beclin-1 contains a BH3 domain, which is required for binding to antiapoptotic BCL2 homologs and BCL2-mediated inhibition of autophagy. BCL2-like 12 (BCL2L12) also harbors a BH3-like domain, which is 12 residues long and contains a LXXXAE/D motif. In a yeast two-hybrid system performed in the present study, BCL2L12 shared similar binding partnerships to antiapoptotic BCL2 homologs, such as Beclin-1. In addition, this BH3-like domain was involved in antiapoptosis and drug-induced autophagy in glioma cell lines. Mutations in S156 and hydrophobic L213 to alanine counteracted the antiapoptotic properties of BCL2L12 and downregulated the activation of microtubule associated protein 1 light chain 3B (LC3B), autophagy-related (ATG)12-ATG5 conjugates and Beclin-1, compared with a BCL2L12 wild-type group. Molecular dynamics simulations revealed that phosphorylation at Ser156 of BCL2L12 (within α-6 and α-7 helices) influenced the BH3-like domain conformation (α-9 helix), indicating that glycogen synthase kinase (GSK) 3β-mediated Ser156 phosphorylation modulated a BH3-like domain in BCL2L12. Altogether, the present findings indicated that BCL2L12 may participate in anti-apoptosis and autophagy via a BH3-like domain and GSK3β-mediated phosphorylation at Ser156. Furthermore, blockade of temozolomide (TMZ)-induced autophagy by 3-methyladenine (3-MA) resulted in enhanced activation of apoptotic markers, as well as tumor suppresor protein p53 (p53) expression in U87MG cells. The present results suggested that p53 and O6-methylguanine DNA methyltransferase activation, and BCL2, BCL-extra large, Beclin-1 and BCL2L12 expression may be used as a detection panel to determine which patients can benefit from TMZ and ABT-737 combination treatment.
Keywords: BCL2-like 12, Beclin-1, autophagy, BH3-like domain, anti-apoptosis, glioma
Introduction
BCL2-like 12 (BCL2L12), a new member of BCL2 family, is currently recognized as an apoptosis regulator (1). Its functional role appears contradictory in different types of cancer (2,3). In glioblastoma, BCL2L12 is highly expressed and associated with antiapoptotic properties. The BCL2L12-mediated antiapoptotic property is thought to be due to several mechanims: i) It attenuates caspase-dependent apoptosis through direct interaction with caspase-7 and indirect inactivation of caspase-3 activity via upregulating crystallin αB (CRYAB) expression. ii) BCL2L12 neutralizes TP53's fucntion at the transcriptional and translational levels as demonstrated by using co-immunoprecipitation (4,5). Downregulating BCL2L12 expression has been demonstrated to reduce glioblastoma growth in animal models (6). An RNAi-based therapy using spherical nucleic acid nanoparticle conjugates was reported to effectively knock down endogenous BCL2L12 mRNA and protein levels and sensitize glioma cells toward therapy-induced apoptosis by enhancing effector caspase and p53 activity (6). To investigate the antiapoptotic properties of BCL2L12, we previously demonstrated that it is as a substrate of glycogen synthase kinase (GSK) 3β, and that GSK3b-mediated phosphorylation at BCL2L12 Ser156 is crucial for anti-apoptosis (7). Conversely, treatment with LiCl, a GSK3β pharmaceutical inhibitor that diminishes GSK3b-mediated BCL2L12 phosphorylation, restored caspase activity and thus it might have great potential in treating glioblastoma multiforme (GBM) in addition to being a mood stabilizer in the clinic (7). Furthermore, a previous study using bioinformatics has demonstrated that in addition to harboring a BH2 domain, BCL2L12 contains a BH3-like domain (8). Our group further characterized the function of this BH3-like domain and its potential antiapoptotic properties. Exogenous expression of the BCL2L12 mutant that was directly site-mutated at the -4 or 0 hydrophobic residue demonstrated an elevated apoptotic marker expression when compared with the BCL2L12 wild-type group. In addition, it was demonstrated that this BH3-like domain may be associated with temozolomide (TMZ)-induced autophagy (9). Without this hydrophobic groove region, TMZ is unlikely to induce effective activation of autophagy markers.
In cell biology, cell proliferation, apoptosis and autophagy are the predominant activities that determine cell fate. The balance of these three cellular activities leads to different outcomes in cancer treatment. One of the main options for GBM treatment, TMZ, is usually co-administered with radiotherapy, generally with good response. More in-depth pretreatment and treatment evaluations are still needed for refining effective personalized medicine approaches. Traditionally, status of p53 and O6-methylguanine DNA methyltransferase (MGMT) is essential reference in order to determine the TMZ effect in GBM treatment (10,11). However, more advanced MGMT activity/methylation status, p53 expression, reactive oxygen species (ROS) production following chemotherapeutic intervention, measurement of apoptosis induction, and unwanted autophagy level (drug-induced pro-survival autophagy) are all critical indicators in modern cancer therapy. A previous study has revealed that glioma cells treated with high doses of TMZ may enhance ROS production and induce autophagy rather than apoptosis (12,13). This TMZ-induced autophagy is uncommon as a pre-death procedure but is linked to drug resistance and remains a significant challenge in GBM treatment. The canonical pathway of autophagy is governed by a subset of autophagy genes (14). One of these genes, Beclin-1 [also known as autophagy-related (Atg) 6, unlike the others, is known to regulate either apoptosis or autophagy based on its interactive partnerships; antiapoptotic when it interacts with BCL2 apoptosis regulator (BCL2) and BCL-extra large (BCL-XL) via the BH3 domain, or autophagy-inducing when it interacts with phosphatidylinositol 3-kinase Vps34 (Vps34) and phosphoinositide 3-kinase (PI3K) (15,16). As aforementioned, BCL2L12 harbors a BH3-like domain associated with TMZ-induced autophagy and antiapoptotic properties in GBM (9). In addition, BCL2L12 and Beclin-1 share a similar binding pattern to BCL2 and BCL-XL (15,17). Ectopic expression of BCL2L12 also enhanced Beclin-1 expression in glioma cell lines (9). Therefore, the present study hypothesized that the BCL2L12 BH3-like domain might functionally compensate for Beclin-1 in either regulating apoptosis or autophagy.
Materials and methods
Cloning
BCL2L12, BCL-XL, BCL2, BCL2 associated X (Bax), Beclin-1, induced myeloid leukemia cell differentiation protein MCL-1 (Mcl-1), and GSK3β were cloned into pACT2 and pAS2-1 vectors (Takara Bio Inc., Otsu, Japan) for yeast two-hybrid assay or into the pEGFP-C1 vector for overexpression. Polymerase chain reaction (PCR) technique was used to generate DNA fragments containing the desired gene sequences utilizing primers designed to contain the Xho I and Bam HI restriction enzyme recognition sites. For gene-specific PCR, 100 ng genomic DNA was used as template, and 2.5 µl 10X reaction buffer (Takara Bio Inc.), 4 µl 2.5 mM dNTPs (Takara Bio Inc.), 0.2 µl DNA polymerase (5 U/µl; Takara Bio Inc.), and 1 µl of 10 µM primers were added into a 25 µl reaction mix. The PCR was conducted on a GeneAmp 9,700 PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA), as follows: 95°C for 30 sec, followed by 30 sec annealing, and extension at 72°C for 35 cycles. The PCR products were separated on 2% agarose gel and visualized by staining with 0.5 µg/ml ethidium bromide. The PCR products with correct sizes were further purified using a PCR clean-up kit (GeneMark Technology Co., Ltd., Tainan, Taiwan) and subjected to restriction enzyme digestion. Plasmid DNA was further confirmed by DNA sequencing (Mission Biotech, Taipei, Taiwan).
Yeast two-hybrid assay
This assay is intended to screen an interaction between two proteins called 'bait' and 'prey'. In the present study, BCL2L12, Bax, Beclin-1, BCL2, BCL-XL, and Mcl-1, as well as several BH3 mutants of BCL2L12, were included in the assay in order to test their potential interactions. Standard techniques were used to perform the yeast two-hybrid screening (18-20), using the MATCHMAKER Two-Hybrid System 2 (Clontech Laboratories, Inc., Mountainview, CA, USA). YRG-2 yeast host cells (MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3 112 gal4-542 gal80-538 LYS2::UASGAL1-TATAGAL1-HIS3 URA3::UASGAL4 17mers(×3)-TATACYC1-lacZ) were purchased from Stratagene (Agilent Technologies, Inc., Santa Clara, CA, USA). The yeast cells were cotransfected with the pAS2-1 and pACT2 plasmids and then selected on G2 plates deficient in tryptophan and leucine, and on G3 plates further deficient in histidine. A positive interaction was determined by growth on G3 plates and by a visible blue color pattern in the subsequent colony filter lift assay (19). In the yeast two-hybrid system, YRG2 yeast cells were inoculated into 5 ml YPD medium and grown at 28°C, 240 rpm for 20 h. YPD medium (45 ml) was added to the original yeast culture to refresh their growth and further incubated for 4 h. To detect whether gene A (encoding 'bait' protein) interacts with gene B (encoding 'prey' protein), the pACT2 vector/gene A (1.2 µg) was premixed with pAS2-1 vector/gene B (1.2 µg), then further mixed with 10 µl boiled salmon sperm DNA and 800 µl of the yeast solution. The reaction was vortexed completely and incubated at 28°C, 7.7 × g for 30 min, and supplemented with 80 µl DMSO (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Then, the reaction was heat-shocked at 42°C for 15 min and placed on ice for 2-5 min. The reaction was further centrifuged at 25°C, 800 × g for 3 min, and the supernatant was discarded. The transformed yeast pellet, resuspended in sterilized water, was spread on nutritional deficient plates G2 and G3, respectively. The G2 or G3 plates were incubated at 28°C for colony growing. Afterwards, sterilized filter papers were used to cover the G2/G3 plates. The filter papers, with the yeast colonies attached and absorbed, were further dipped into liquid nitrogen for 40 sec to break any enclosed yeast cell walls. X-gal solution (1,600 µl) was added onto another filter and incubated at 28°C to detect whether the 'bait' protein would interact with 'prey' protein.
Site-directed mutagenesis
The specific primers for genes encoding the Bax L63A mutation and the BCL2L12 BH3-like domain mutations h1, h2, h3, charge, and h4, and the complementary sequences surrounding the mutation sites, were designed and synthesized as previously reported (19). Following PCR amplification, 1.5 µl Dpn I (10 U/µl) was added for digestion at 37°C for 10 min. Dpn I-treated DNA was transferred into 45 µl of prepared competent cells. Following incubation on ice for 30 min, heat shock was performed in a 42°C water bath for 30 sec. The reaction mixture was kept on ice for 2 min, and then 500 µl LB medium was added and incubated at 37°C, 5.37 × g, for 1 h. Following incubation, bacterial cultures were spread on selective plates containing antibiotics and incubated at 37°C for 16-18 h. Several colonies were picked for plasmid preparation, restriction enzyme digestion and sequencing to confirm the desired mutations.
Molecular dynamics simulations of the BCL2L12 phosphorylation at Ser156
The initial 3D model of BCL2L12 was obtained as previously described (9). Next, the Ser156 of BCL2L12 was phosphorylated with the AmberTools17 and amber parameter database (21). Then the model was inserted into the tip3p water box. The molecular dynamics (MD) simulations were performed in the canonical ensemble with a simulation temperature of 310 K, unless stated otherwise, by using the Verlet integrator with an integration time step of 0.002 ps and SHAKE constraints (22) of all covalent bonds involving hydrogen atoms. In the electrostatic interactions, Atom-based truncation was performed using the PME (23) method, and the switch van der Waals function was used with a 1.80 nm cutoff for atom-pair lists. The structure was minimized for 100,000 conjugate gradient steps and was then subjected to a 100 s isothermal, constant volume MD simulation. All the MD simulations were performed with Amber 16 (pmemd.cuda) program. The final structure was used in the BH3 domain comparisons (BCL2L12 and phosphorylation at Ser156 of BCL2L12).
Cell culture, transfection, and treatments
Human glioblastoma U87MG (cat. no. HTB-14), H4 (cat. no. HTB-148) and T98G (cat. no. CRL-1690) cell lines were purchased from American Type Culture Collection (Manassas, VA, USA). Cells were cultured using MEM (Gibco; Thermo Fisher Scientific Inc.) and DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and antibiotics (penicillin and streptomycin, 100 IU/l) at 37°C, 5% CO2 in air atmosphere. For transient transfections, cells were seeded into 100 mm diameter culture dishes (BD Biosciences, Franklin Lakes, NJ, USA) at a density of 2×106 cells per dish. DNA (1-2 µg/per well of 6 well cluster plate) was transfected into cells for 12-16 h using Lipofectamine 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.). To induce apoptosis, staurosporine (STS; 0.5 µM; Sigma-Aldrich; Merck KGaA) or TMZ (Sigma-Aldrich; Merck KGaA) (400 µM) was added to the culture for 24 and 48 h, respectively. STS treatment was the positive control for apoptosis induction. ABT-737 (Sigma-Aldrich; Merck KGaA) was applied alone or in combination with TMZ at a concentration of 50 µM.
Western blotting
RIPA Lysis buffer (Protech Inc., Taipei, Taiwan) was used to prepare the total lysates from cultured cells. For detecting cytochrome c efflux, the cytosolic fraction was prepared by a Mitochondria/Cytosol Fractionation kit (Merck Millipore, Billerica, MA, USA), according to the manufacturer's protocol. Protein concentration was determined using Protein Assay reagent (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Protein lysates (40 µg/lane) were separated on 12% SDS-PAGE and then transferred onto methanol-treated polyvinylidene fluoride membranes. The transferred membranes were blocked in TBST (TBS/0.1% Tween-20) with 5% non-fat dry milk and was agitated for 1 h, and then incubated with primary antibodies for 1 h. The primary antibodies targeting microtubule associated protein 1 light chain 3 β (cat. no. 2775; LC3B; recognizing both LC3B-I and II), Atg12-Atg5 conjugates (cat. no. 2010), Beclin-1 (cat. no. 3738), full length poly-ADP-ribose-polymerase (PARP; cat. no. 9542), cleaved PARP (cat. no. 9542), cleaved caspase-9/-3 (cat. nos. 9502 and 9662), cytochrome c (cat. no. 4272), BCL2 (cat. no. 2872), Bax (cat. no. 2774), BCL-XL (cat. no. 2762), Mcl-1 (cat. no. 4572), BCL2-like 11 (also known as Bim; cat. no. 2819), BCL2-related ovarian killer protein (Bok; cat. no. 4521) and BCL2-binding component 3 (also known as Puma; cat. no. 4976) were purchased from Cell Signaling Technology, Inc. (Beverly, MA, USA). The primary antobodies targeting green fluorescent protein (GFP, clone B-2; cat. no. sc-9996), p53 (clone DO-1; cat. no. sc-126) and β-actin (clone C4; cat. no. sc-47778) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Next, the membranes were incubated with secondary antibodies conjugated with horseradish peroxidase (HRP; Cell Signaling Technology, Inc.; anti-rabbit IgG, HRP-linked antibody cat. no. 7074 and anti-mouse IgG, HRP-linked antibody cat. no. 7076) for another 1 h. Both primary and secondary antibodies were diluted in 1% non-fat dry milk or 5% BSA in TBST. The protein signals were developed using enhanced chemiluminescence reagent (GE Healthcare, Chicago, IL, USA) and recorded using Fuji X-ray film Super RX (Fujifilm, Tokyo, Japan) for X-ray autoradiography.
Statistical analysis
The western blot analyses were quantified with ImageJ software (National Institutes of Health, Bethesda, MD, USA). Bar charts were generated using Sigma plot software version 12.3 (Systat Software Inc., Chicago, IL, USA). Data were expressed as the mean ± standard deviation. All data were analyzed using the SPSS for Windows 21.0 statistical software (IBM Corps., Armonk, NY, USA). Statistical significance between groups was examined with one-way analysis of variance for multiple comparisons followed by Bonferroni correction for adjusting the P-value of multiple tests. P<0.05 was considered to indicate a statistically significant difference.
Results
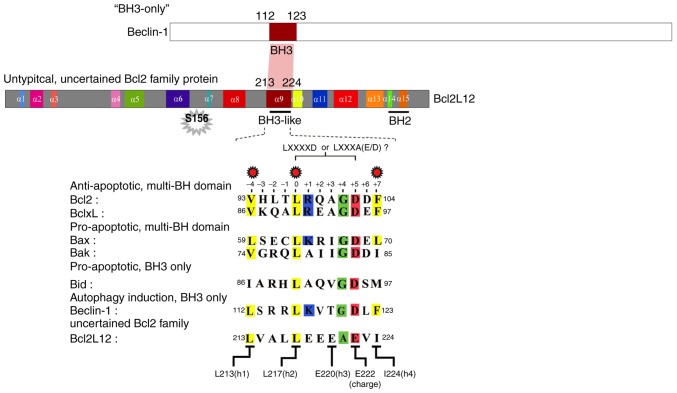
BCL2L12 contains a BH3-like domain on its α-9 helix, and this 12-residue motif is conserved among the BCL2 family proteins
The structural similarity of the BH3-like domain of BCL2L12 was compared to those of BCL2, BCL-XL, and Bax. As illustrated in Fig. 1, the α-9 helix of BCL2L12 is structurally similar to the α-2 helix of multiple BCL2 family proteins, including the antiapoptotic (BCL2 and BCL-XL) and proapoptotic (Bak and Bax) subgroups. To highlight the structural/functional similarity among these BH3 domains, five key amino acid residues were analyzed for their effects on the interaction between BCL2L12, BCL2 and BCL-XL. As reported previously, the L213 (-4), L217 (0) and I224 (+7) hydrophobic residues are crucial for the BCL2L12 interaction with BCL2 and BCL-XL in a yeast two-hybrid system (Fig. 1) (9). It was further determined that the BH3 domain most likely consists of a 12-residue-long core motif of LXXXAE/D in BCL2L12 instead of the canonical motif 'LXXXXD' in Bak or other BCL2 family proteins. Since BCL2L12 interacts with BCL2 and BCL-XL, which shares similar interacting partnerships with Bax and Beclin-1, it was hypothesized that the BCL2L12 BH3-like domain may be necessary for both autophagy and apoptosis regulation. Previously, it was reported that overexpressed BCL2L12 L213A and L217A mutants resulted in reactivation of apoptotic markers with or without STS treatment (9). Therefore, the present study investigated L213A as a representative BH3-like domain mutant in the subsequent cell-based assays.
Figure 1.
Domain structure and sequence of the BH3-like motifs in Beclin-1 and BCL2L12. The helix structure of BCL2L12 and the BH3 domain of both Beclin-1 and BCL2L12 are shown. A 12-residue region is diagramed to represent the BH3 domain defined in the present study. The LXXXXD (for other BCL2 family proteins) or LXXXAE/D (for BCL2L12) motif is drawn and critical residues comprising L213 (h1, position −4 corresponding to LXXXAE/D motif), L217 (h2, position 0) and I224 (h4, position +7) that affect interaction between BCL2L12 and BCL2/BCL-XL are highlighted in yellow. BCL2L12, BCL2-like 12; BCL2, BCL2 apoptosis regulator; BCL-XL, BCL-extra large.
BCL2L12 does not interact with Beclin-1, but shares a similar binding partnership with Beclin-1 to interact with BCL2 and BCL-XL, but not Mcl-1
The interactions between BCL2L12, Bax, Beclin-1 and anti-apoptotic BCL2 family proteins, BCL2, BCL-XL and Mcl-1, were investigated. GSK3β was used as positive control for its interaction with BCL2L12. Several BH3 mutants of BCL2L12 (I209A, L213A, L216A, L217A, E218A and E220A) (90-229, refers to the BCL2L12 truncated fragment spanning between the 90th amino acid residue to 229th residue. The plasmid harboring this BCL212 truncated fragment was further used as a parental vector to obtain mutants by a site-direct mutagenesis technique) as well as the binding-deficient mutant L63A of Bax (1-171), were tested in a yeast two-hybrid system, in order to determine whether they could abolish the interaction between BCL2L12, Bax, and their interacting proteins. As listed in Table I, both BCL2L12 (70-266) and Bax (1-171) could interact with BCL2 (1-211) and BCL-XL (1-209). Mutation on L213 of BCL2L12 (70-266) and L63A of Bax (1-171) resulted in a loss of binding to either BCL2 or BCL-XL. In addition, BCL2L12 (three different fragments comprising 70-266, 70-334 and 90-229) could also interact with Bax (1-171); however, BH3 mutants were unlikely to disrupt the binding between BCL2L12 (90-229) fragments and Bax (1-171). BCL2L12 (70-266) failed to interact with Beclin-1 (1-450). Mcl-1 could not interact with Beclin-1(1-450), Bax (1-171), BCL2L12 (70-266), and BCL2L12 (70-334). Altogether, based on the binding partnership in the present yeast two-hybrid system and the results from previous studies, BCL2L12 and Bax shared similar binding partnerships towards BCL2 and BCL-XL, probably through the BH3 domain; however, their interaction (BCL2L12-Bax) was independent of the BH3-like domain (Table I). BCL2L12 and Beclin-1 share positive binding to BCL2 and BCL-XL and non-binding to Mcl-1, even though they are unlikely to interact with each other.
Table I.
Yeast two-hybrid assay of BCL2L12 and Beclin-1 interaction with BCL2 family proteins and GSK3β.
| pGAL4-DBD vector(pAS2-1) | pGAL4-AD vector (pACT2)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| BCL-XL(1-209) | BCL2(1-211) | Beclin-1(1-450) | Bax(1-171) | BCL2L12(70-266) | BCL2L12(70-334) | BCL2L12(90-229) | BCL2L12(153-191) | BCL2L12(90-229), I209Aa | |
| BCL2L12(70-266) | B+ | B+ | – | B+ | ND | ND | ND | ND | ND |
| BCL2L12(70-266), L213A | – | – | ND | ND | ND | ND | ND | ND | ND |
| Bax(1-171) | B+ | B+ | ND | ND | B+ | B+ | B+ | ND | B+ |
| Bax(1-171), L63A | – | – | ND | ND | ND | ND | ND | ND | ND |
| Mcl-1(1-327) | ND | ND | – | – | – | – | ND | ND | ND |
| GSK3β | ND | ND | – | ND | B+ | B+ | B+ | B+ | ND |
BCL2L12 (90-229) L213A, L216A, L217A, E218A, E220A were all unable to affect the binding between BCL2L12 and Bax (data are not shown in the Table since their results are identical to that of the I209A mutant). BCL2L12, BCL2-like 12; BCL2, BCL2 apoptosis regulator; GSK3β, glycogen synthase kinase 3β; Bax, BCL2 associated X; Mcl-1, induced myeloid leukemia cell differentiation protein MCL-1; B+, interaction; –, no interaction; ND, not done, interaction is not tested in this assay. pGAL4-AD, pGAL4 transcription activation domain, pGAL4-DBD, factor's DNA-binding domain.
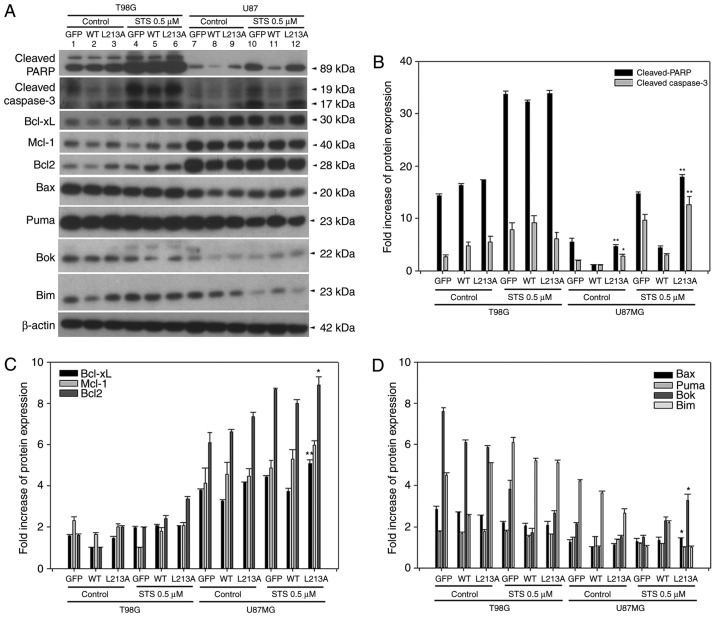
BH3-like domain L213A mutant restores apoptotic marker activities in U87MG, but not T98G cells
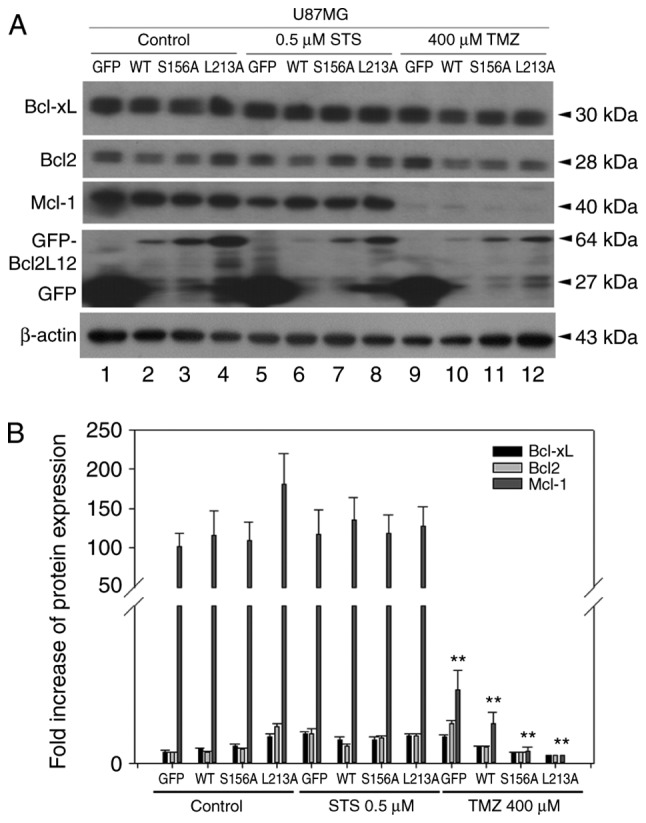
In two glioma cell lines overexpressing GFP, BCL2L12 wt and L213A mutant, cleaved-PARP and cleaved-caspase-3 were reactivated in the L213A overexpressing group compared with the BCL2L12 wild-type group (Fig. 2A and B). This phenomenon was evident in U87MG cells, but not in T98G cells (Fig. 2A and B). STS treatment further enhanced this effect (Fig. 2A and B). Anti-apoptotic BCL2 proteins, including BCL-XL, Mcl-1 and BCL2, were highly expressed in U87MG cells compared with T98G cells. Overexpression of the L213A mutant did not alter the expression levels of these antiapoptotic BCL2 proteins (Fig. 2C). By contrast, the proapoptotic and BH3-only BCL2 family proteins Bax, Puma, Bok, and Bim, appeared more abundantly expressed at the basal level and following STS treatment in T98G compared with U87MG cells (Fig. 2D). However, these proapoptotic and BH3-only BCL2 family protein expression levels are unlikely to be affected by BCL2L12 or BCL2L12 L213A mutant expression. Since high expression of BCL2, BCL-XL and Mcl-1 in cancer cells is reported to be associated with acquired resistance to chemotherapy, the present study investigated whether TMZ treatment caused any expression changes among these three proteins in glioma cells. As illustrated in Fig. 3, TMZ treatment significantly downregulated Mcl-1 expression, but not BCL-XL and BCL2 expression, and this effect was independent of the expression status of BCL2L12 and its mutant (S156A and L213A).
Figure 2.
BCL2L12 L213A is associated with reactivation of caspase-3 and the BCL2 family protein expression profiles are different in T98G and U87MG cell lines. (A) The protein expression levels of apoptotic markers and BCL2 family proteins were detected by western blotting. (B) Quantitative plot of cleaved PARP and cleaved caspase-3, (C) prosurvival proteins BCL-XL, Mcl-1, and BCL2, and (D) proapoptotic BCL2 family proteins expression in two glioma cell lines. All target protein expression levels were normalized to β-actin expression prior to conversion into fold increase measurements. *P<0.05 and **P<0.01 compared with WT group. BCL2L12, BCL2-like 12; BCL2, BCL2 apoptosis regulator; PARP, poly-ADP-ribose-polymerase; BCL-XL, BCL-extra large; Mcl-1, induced myeloid leukemia cell differentiation protein MCL-1; WT, wild-type; Bax, BCL2 associated X; Puma, BCL2-binding component 3; Bok, BCL2-related ovarian killer protein; Bim, BCL2-like 11.
Figure 3.
TMZ treatment downregulates Mcl-1, creating a desirable microenvironment for applying ABT-737. (A) The expression levels of prosurvival BCL2 family proteins were detected in different experimental groups of U87MG cells by western blotting. (B) Quantitative plot of BCL-XL, BCL2 and Mcl-1 expression in the different groups. **P<0.01 compared with control. TMZ, temozolomide; Mcl-1, induced myeloid leukemia cell differentiation protein MCL-1; BCL2, BCL2 apoptosis regulator; BCL-XL, BCL-extra large; BCL2L12, BCL2-like 12; STS, staurosporine; GFP, green fluorescent protein; WT, wild-type.
Combination treatment with TMZ and ABT-737 triggers enhanced apoptosis compared with each agent alone in U87MG cells
Because of the high levels of BCL2, BCL-XL and BCL2L12 expression in U87MG cells, it was postulated that BCL2L12 might interact with these antiapoptotic BCL2 family members and lead to enhanced outer membrane integrity of mitochondria. Therefore, BCL2L12 with BH3-like domain to trigger mitochondria-dependent apoptosis was preceded by the treatment of ABT-737, a mimetic agent of the BH3 domain of Bad, as well as a selective inhibitor of BCL2, BCL-XL, and BCL-w, alone or in combination with TMZ in U87MG cells. We previously reported that both TMZ and ABT-737 resulted in moderate apoptosis compared with an untreated GFP-alone group in U87MG cells, which express wild-type p53 (associated with the effects of TMZ) and high levels of BCL2 and BCL-XL. Bax was also expressed at an intermediate level with ABT-737 treatment in U87MG cells. However, induction of apoptosis by these two agents appears to be independent of BCL2L12 L217A expression, especially in combination use (9). When not treated with these two agents, the L217A group demonstrated an increased level of cleaved PARP like the L213A group compared with the untreated BCL2L12 wild-type and untreated GFP alone group in U87MG cells (9). Combination treatment with TMZ and ABT-737 resulted in higher levels of cleaved PARP and cleaved caspase-3 compared with each agent used alone (9). The induction level of apoptotic protein expression by these two agents whether used alone or in combination, the effect is increased compared with the effect contributed by the exogenously expressed L217A mutant (9). Thus, combination use of these two agents appears to be efficient in glioma cells with p53 wild-type, high BCL2 and BCL-XL expression, moderate Bax expression and null to low background MGMT activation. In addition, TMZ-induced downregulation of Mcl-1 creates a desirable microenvironment for the addition of ABT-737 in treating U87MG cells (Fig. 3), as mentioned in a previous study (9).
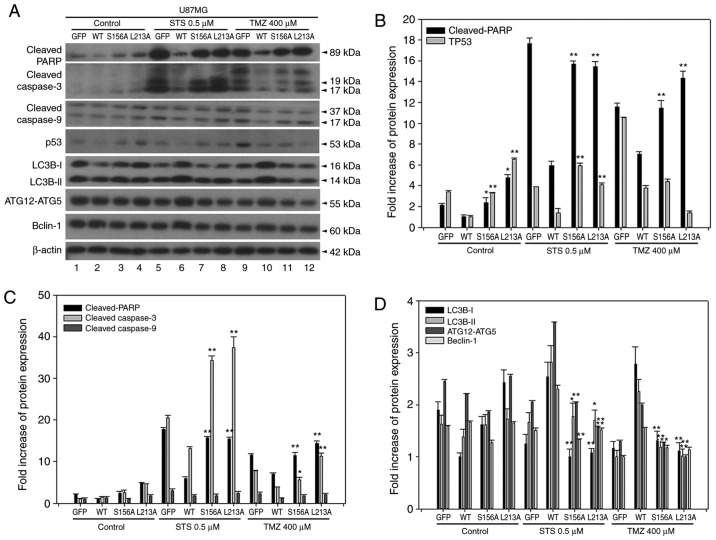
GSK3b-mediated phosphorylation on Ser156 and the BH3-like domain contributes to the antiapoptotic property of BCL2L12 and drug-induced autophagy in U87MG
Overexpressed BCL2L12 S156A and L213A increased the levels of cleaved PARP and cleaved caspase-3, but not cleaved caspase-9, compared with the BCL2L12 wild-type group (Fig. 4A and C). This effect was strengthened in the two drug treatment groups, where cleaved PARP and cleaved caspase-3 has dramatically altered expression levels. For cleaved caspase-9, reactivation of apoptotic markers was only observed in the TMZ-treated group (Fig. 4A and C). By contrast, p53 was reactivated in S156A and L213A-overexpressing cells in both the untreated and STS-treated group, but not in the TMZ-treated group (Fig. 4B). In the drug-treated group, overexpression of wild-type BCL2L12 resulted in an activation of autophagic markers, including LC3B-II, Atg12-Atg5 conjugates, and Beclin-1 in U87MG cells (Fig. 4A and D). However, following overexpression of the BCL2L12 S156A and L213A mutants, these autophagic markers were no longer activated and were observed at levels equal to the GFP group (Fig. 4A and D).
Figure 4.
BCL2L12 S156A and L213A mutants are involved in reactivation of apoptotic markers and drug-induced autophagy. (A) The expression profiles of p53 and of various apoptotic and autophagic markers were examined by western blotting. (B) Quantitative plot of relative cleaved PARP and p53 expression in U87MG cells with or without expression of BCL2L12 and its mutants (S156A or L213A). (C) Quantitative plot of relative expression of apoptotic markers cleaved PARP, cleaved caspase-3 and cleaved caspase-9 in U87MG cells with or without expression of BCL2L12 and its mutant proteins. (D) Quantitative plot of relative expression of autophagy-related proteins LC3B-I, II, ATG12-ATG5 conjugates and Beclin-1 in U87MG cells with or without expression of BCL2L12 and its mutant proteins. All target protein expression levels were normalized to β-actin expression prior to conversion into fold increase measurements. *P<0.05 and **P<0.01 compared with WT group. BCL2L12, BCL2-like 12; p53, tumor protein p53; PARP, poly-ADP-ribose-polymerase; LC3B, microtubule associated protein 1 light chain 3B; ATG, autophagy-related gene; STS, staurosporine; TMZ, temozolomide; GFP, green fluorescent protein; WT, wild-type.
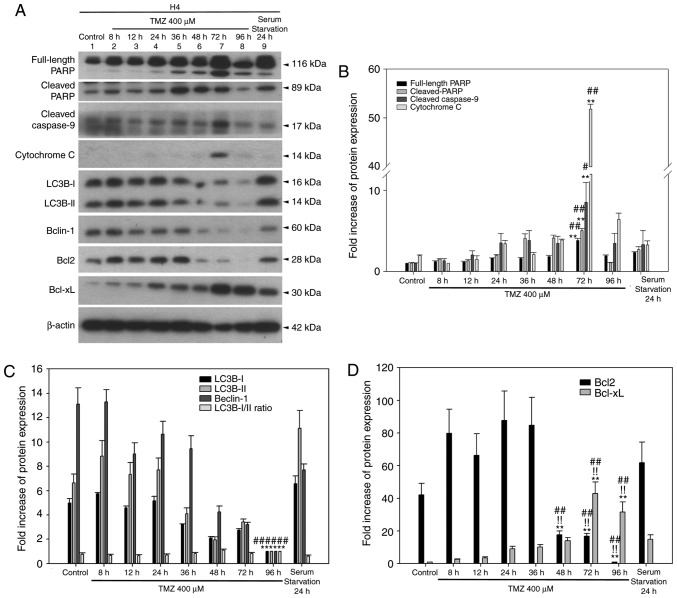
TMZ triggers an autophagy-apoptosis shift event in a time-dependent manner in H4 cells
A previous study revealed that a high concentration of TMZ may trigger autophagy, which is associated with ROS-mediated activation of the extracellular signal-regulated kinase (ERK) signaling pathway (12), as well as acquired resistance to chemotherapeutics. These experiments were repeated in the present study in three glioma cell lines in order to determine whether high-dose TMZ treatment induces an autophagy to apoptosis shift in a time-dependent manner. Similar to our previously published work in U87MG cells (9), the present data for U87MG cells demonstrated that cleaved PARP was activated at 24 h and gradually reached a peak at 48 h following treatment with 400 µM TMZ (9). Cleaved caspase-9 was detected even in the untreated group, and it also increased from 8 to 48 h. Regarding autophagy markers, LC3B-II was detected in the control group; following TMZ treatment its expression gradually increased from 8 to 36 h and then dropped at 72 h. Beclin-1 was also detected in the control group and progressively accumulated from 8 to 36 h. The expression levels of Beclin-1 at 36 to 72 h were not different between groups. The BCL-XL expression was affected by TMZ treatment, gradually increasing from 8 to 72 h. Altogether, the autophagy-apoptosis shift event in U87MG cells treated with TMZ appears to be at a high point at 36 h, and autophagy appears to be induced earlier than a subsequent apoptosis event. In T98G cells, cleaved PARP and caspase-9/-3 activation was observed in the untreated group from 8 to 72 h. However, in contrast to apoptotic markers, expression levels concerning LC3B-II, Beclin-1, and BCL-XL were unaltered by TMZ treatment (9). In H4 cells, activation of cleaved PARP and cleaved caspase-9 was detected in the untreated group and gradually increased until 36 h (cleaved PARP) or 72 h (cleaved caspase-9) following TMZ treatment (Fig. 5A and B). In addition, cytochrome c was more abundantly expressed at 72 h (Fig. 5A and B). Autophagy markers LC3B-II and Beclin-1 were consistently detected until 36 h and were decreased at time points beyond 36 h (Fig. 5C). The BCL2 expression pattern was highly similar to that of Beclin-1, and the BCL-XL expression pattern was gradually accumulative until 96 h with a peak at 72 h (Fig. 5D). Taken together, treating H4 cells with TMZ triggered an autophagy-apoptosis shift event similar to that observed in U87MG cells. Furthermore, TMZ treatment enhanced the protein expression of antiapoptotic BCL2 family members, such as BCL2 (before 36 h) and BCL-XL (after 48 h; Fig. 5D).
Figure 5.
Time-dependent TMZ-induced autophagy and apoptosis in H4 cells. (A) The expression profiles of apoptotic and autophagic markers, BCL2 and BCL-XL were determined by western blotting. (B) Quantitative plot of full-length PARP, cleaved-PARP, cleaved caspase-9 and cytochrome c expression in H4 cells. **P<0.01 compared with control and #P<0.05 and ##P<0.01 vs. the serum starvation group. (C) Quantitative plot of autophagic proteins LC3B-I, II, Beclin-1 expression and LC3B-I/II ratio. **P<0.01 compared with control. (D) Quantitative plot of BCL2 and BCL-XL expression. **P<0.01 indicate the significance of comparisons between the TMZ-treated 48, 72, 96 h group and control; !!P<0.01 vs. 8-36 h group; ##P<0.01 vs. the serum starvation. All target protein expression levels were normalized to β-actin expression prior to conversion into fold increase measurements. TMZ, temozolomide; BCL2, BCL2 apoptosis regulator; BCL-XL, BCL-extra large; PARP, poly-ADP-ribose-polymerase; LC3B, microtubule associated protein 1 light chain 3B.
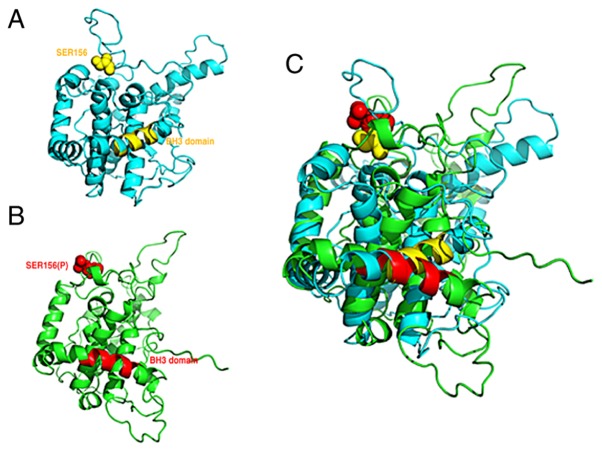
GSK3b-mediated phosphorylation on Ser156 influences the BH3-like domain and contributes to the antiapoptotic property of BCL2L12
As described above, both GSK3b-mediated phosphorylation on Ser156 and the BH3-like domain contribute to the antiapoptotic properties of BCL2L12 and drug-induced autophagy. Because of the prediction from the protein 3D structure of BCL2L12 containing 16 α-helixes that the BH3-like domain corresponded to the residues of 211–226 (α-9 helix) (9), it was hypothesized that phosphorylation at Ser156 may have a functional role on modulating the BH3-like domain. Molecular dynamics simulation data demonstrated that phosphorylation at Ser156 of BCL2L12 (within α-6 helix and α-7 helix) influences the BH3-like domain conformation (α-9 helix), suggesting that GSK3β-mediated Ser156 phosphorylation may function as an allosteric site to modulate the BH3-like domain of BCL2L12 (Fig. 6). The present results suggest that BCL2L12 participates in anti-apoptosis and autophagy via the BH3-like domain and that this BH3-like domain-mediated function is also further modulated by GSK3β-mediated phosphorylation at Ser156.
Figure 6.
Structural simulation of BCL2L12. (A) Ser156 site and BH3-like domain of BCL2L12 (yellow). (B) Phosphorylated Ser156 and BH3 domain of BCL2L12 (red). (C) Schematic of the structures from panels A and B superimposed. BCL2L12, BCL2-like 12; P, phosphorylated.
Blockage of TMZ-induced autophagy leads to a robust activation of apoptotic markers and p53 expression in U87MG cells
The null-MGMT expression of the U87MG glioma cell line (24) was further examined in the H4 cells (previously reported with a low MGMT mRNA expression) (25), because TMZ is better used in an MGMT-null background to determine its clinical relevance. Furthermore, the p53 background and endogenous BCL2L12 expression can influence treatment outcome in glioma, due to their reported physical interaction (2,26). In addition to retrieving the p53 expression status using the IARC p53 cell line database (http://p53.iarc.fr/CellLines.aspx), the endogenous MGMT and p53 expression in the three glioma cell lines used in the present study were investigated (Fig. 7). In contrast to T98G cells, U87MG and H4 cells were similar in MGMT and p53 expression, although the p53 expression in H4 cells was slightly higher compared with the U87MG cells (Fig. 7). The high expression of MGMT in T98G cells was consistent with a previous study (27). Therefore, the two major proteins that may confound the therapeutic effect of temozolomide (28) are controlled in H4 and U87MG cells. Similar results were reported in Figs. 4 and 5 for U87MG and H4 cell lines in the present study, therefore reported issues regarding cell line misidentification of U87MG ATCC when compared to U87MG Uppsala (29) are unlikely to affect the net results of the present study. More recently, a study also demonstrated that the U87MG ATCC cell line has several typical characteristics of glioblastoma through analyses of cell morphology and gene expression profile, even though its origin is unknown (30).
Figure 7.
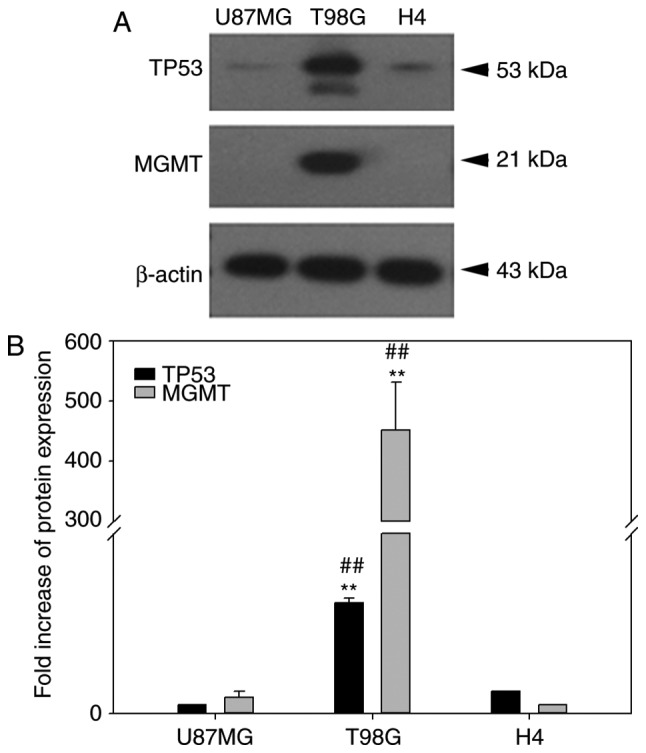
p53 and MGMT expression status of U87MG, T98G and H4 cell lines. (A) Representative plots from western blot results for p53 and MGMT protein expression levels in the three glioma cell lines. (B) Quantitative plot of p53 and MGMT expression levels following normalization to β-actin expression. TP53, tumor protein p53; MGMT, O6-methylguanine DNA methyltransferase. T98 G cells demonstrated an increased TP53 and MGMT expression compared with the two other cell lines. **P<0.001 vs. the U87MG, ##P<0.01 vs. the H4 cells.
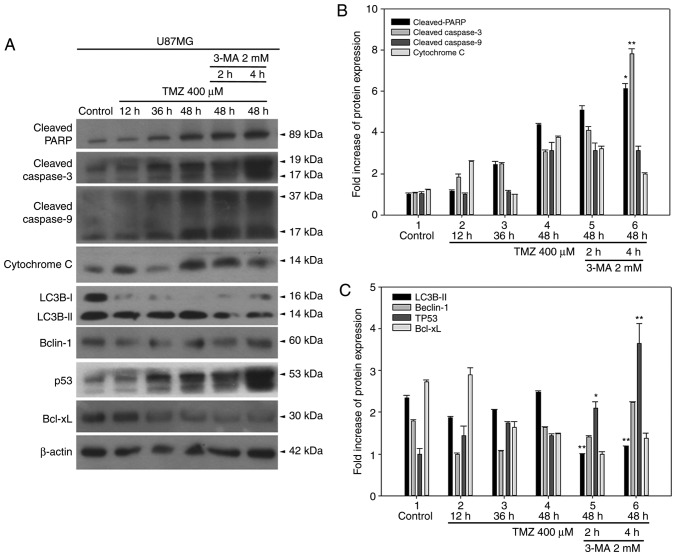
As illustrated in Fig. 8, apoptotic markers, including cleaved PARP, cleaved caspase-3, cleaved caspase-9, cytochrome c, and p53 were gradually activated from 12 to 48 h following TMZ treatment, wheraeas Beclin-1 and BCL-XL levels were unchanged. Treatment with 3-methyladenine (3-MA), an autophagy inhibitor, for 2 or 4 h at 2 mM concentration, downregulated LC3B-II, while the apoptotic markers p53, cleaved PARP and cleaved caspase-3 were activated (Fig. 8). This enhancement was more evident in the 4 h-treated group, indicating that inhibiting TMZ-induced autophagy may, in turn, activate apoptotic genes and may serve as a treatment option in GBM therapy.
Figure 8.
Inhibition of TMZ-induced autophagy by 3-MA leads to enhanced activation of apoptotic markers and p53 expression in U87MG cells. (A) The expression levels of apoptotic markers, autophagic markers, p53 and BCL-XL were detected by western blotting. (B) Quantitative plot of cleaved PARP and cleaved caspase-3 expression levels. (C) Quantitative plot of LC3B-II, Beclin-1, p53, and BCL-XL expression levels. *P<0.05 and **P<0.01 compared with TMZ alone. TMZ, temozolomide; 3-MA, 3-methyladenine; p53, tumor protein p53; BCL-XL, BCL-extra large; PARP, poly-ADP-ribose-polymerase; LC3B, microtubule associated protein 1 light chain 3B.
Discussion
BCL2L12 is a relatively new member of the BCL2 family and only shares a low amino acid similarity to other BCL2 family proteins. It is reported to interact with BCL2, BCL-XL and Bax (9), but not to interact with Mcl-1 in yeast two-hybrid assays, and interacts with GSK3β (7), p53 (27) and caspase-7 (26) in co-immunoprecipitation assays. BCL2L12 was reported to have two functional variants in 2008 (31), which increased to 9 variants by 2012 (8). More recently, using 3′RACE technique and new generation sequencing approaches, another 50 transcripts have been confirmed, but their functions have yet to be characterized (32). For the interaction between BCL2 proteins in regulating apoptosis, it is known that when BH3-only proteins, such as Bim, Bad or Noxa, are activated, their amphipathic α-helical BH3 domain inserts into the hydrophobic groove of pro-survival BCL2 homologs. This fundamental interaction initiates apoptosis, but cell death ensues only in cells that express Bax and Bak (33), the proapoptotic BCL2 members. When activated, Bax and Bak oligomerize on the mitochondrial outer membrane and result in loss of outer membrane integrity. The permeability alteration is accompanied by the release of apoptogenic proteins and cytochrome c efflux. These apoptogenic proteins promote activation of caspases, such as caspase-9, caspase-7 and caspase-3. The BH3-only BCL2 family proteins are effectors of canonical mitochondrial apoptosis. They discharge their proapoptotic functions through BH1-3 proapoptotic proteins, such as Bax and Bak, while their activity is suppressed by BH1-4 antiapoptotic BCL2 family members, such as BCL2, BCL-XL and Mcl-1. The precise mechanism by which BH3-only proteins mediate apoptosis remains unresolved, but three mutually non-exclusive models are proposed (34).
BCL2L12 contributes to apoptosis regulation, but has different roles in different types of cancer type. It has yet to be categorized in any distinct BCL2 protein subgroup, such as antiapoptotic, proapoptotic, sensitizer, activator and effector. In fact, as aforementioned, it shares only low amino acid similarity to the majority of BCL2 family proteins based on full-length amino acid sequence comparisons. However, its BH3-like domain does conserve essential residue compositions and retains a similar helix structure to BCL2, BCL-XL and Bax, as previously reported (9). BCL2L12 is relatively larger than many BCL2 family proteins, and so is Beclin-1; these two proteins are not as typical as their homologs. A BH3-only BCL2 protein like Beclin-1 is unlike Bid, Bim, and Bak; it is 450 amino acids in length and is suggested to not interact with Bax. Beclin-1 is a novel BH3-only BCL2 member and autophagy effector. Anti-apoptotic BCL2 homologs are known to downregulate autophagy through interactions with Beclin-1, an essential autophagy effector, and haploinsufficient tumor suppressor, through its BH3 domain, which is required for BCL2-mediated inhibition of autophagy (16). On the other hand, BCL2L12 is 334 amino acids in length, and only known to have a conserved BH2 domain before 2009 (1,8). Of note, the BCL2L12 BH3-like domain is located at the α-9 helix, while other BCL2 homolog BH3 domains are located at the α-2 helix. BCL2L12 harbors a BH3-like domain associated with TMZ-induced autophagy and antiapoptotic properties in glioma cells. Beclin-1 also contains a BH3 domain interacting with BCL2/BCL-XL in a context-specific dual role (autophagy and anti-autophagy). A dual role (apoptosis/autophagy) is also reported for another autophagy effector protein, autophagy and Beclin-1 regulator 1 (AMBRA1), which contains a BH3 domain at its C-terminus that is necessary for AMBRA1 binding with the antiapoptotic factor BCL2 for its anti-autophagy role. When the proapoptotic amplification loop is turned on, the caspase-cleaved form of AMBRA1 is bound to antiapoptotic BCL2 proteins, such as Mcl-1, BCL2 and BCL-XL, but not Bax and Bak (35). In addition to its BH3-like domain, it is noted that the N-terminus of BCL2Ll2, especially the 1-70 residues, is unable to interact with either BCL2 or BCL-XL (data not shown). A 3D simulation also reveals that BCL2L12 (1-173) does not overlap with BCL2, BCL-XL or Bax. In addition, BCL2L12 (90-229) interacts with Bax (1-171), and mutations on critical residues within the BH3-like domain are unable to disrupt the interaction between these two fragments in a yeast two-hybrid system (Table I). These findings indicate that the N-terminus of BCL2L12 may have uncharacterized functions. Whether BCL2L12 (70-153) serves as a Bax interacting region or has any regulating role in Bax's biofunctions needs further elucidation.
In the present study, overexpression of BCL2L12 Ser156A mutant resulted in reactivation of caspase activity and downregulation of autophagy markers. It was postulated that GSK3β-mediated phosphorylation at Ser156 of BCL2L12 may be related to BH3-like domain functionality since this site is in the middle of the α6 and α7 helix in proximity to the core motif LXXXAE/D of the BCL2L12 BH3-like domain at the α9 helix. Phosphorylation at a Serine site may cause conformational change and possibly more significant exposure of its BH3-like domain for accessing the hydrophobic groove of target proteins. We previously reported that GSK3β-mediated phosphorylation at Ser156 of BCL2L12 is associated with its antiapoptotic property. Addition of LiCl, a potent GSK3β inhibitor, resulted in reactivation of caspase-3, -7 and -9, which is identical to the process detected in U87MG cells over expressing BCL2L12 Ser156A (7). In the present study, the results demonstrated that following STS or TMZ treatments, overexpression of the BCL2L12 L156A mutant reversed autophagy, as the functional consequence of diminishing the BH3-like domain via overexpression of the BCL2L12 L213A mutant. Therefore, in U87MG cells, or in cells with wild-type p53, null or low MGMT activation, high BCL2, BCL-XL and Mcl-1 expression, drug-induced autophagy may be the initial step of anti-apoptosis events. Thus, it can be suggested that cytoprotective autophagy coordinated by both the BCL212 and Beclin-1 BH3 domain may counteract p53-dependent apoptotic or BCL2-mediated anti-autophagic signaling. Once the death signaling is irreversible, or the pro-survival autophagy is overwhelmed, cells may undergo apoptosis. When treated with TMZ or STS, a time-dependent autophagy-apoptosis shift event occurs, and this pro-survival autophagy is linked to acquired resistance to chemotherapeutics. Hence, inhibiting this pro-survival process, or inhibiting drug-induced autophagy events, may lead cancer cells to follow the fate of cell death (Fig. 9) (15,16,36).
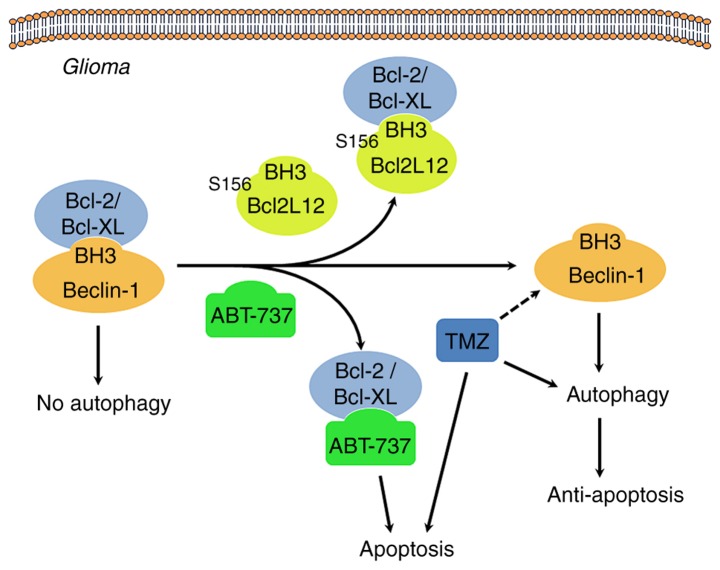
Figure 9.
Proposed mechanism of Ser156 phosphorylation as an allosteric site to modulate a BH3-like domain on BCL2L12 in glioma cells. When BCL2/BCL-XL interacts with the Beclin-1 BH3 domain, autophagy is inhibited. Overexpression of BCL2L12 may displace Beclin-1 in integrating with BCL2/BCL-XL via its BH3-like domain, leading to release of Beclin-1 and initiation of the autophagy process. In addition, since BCL2L12 occupies the hydrophobic groove of BCL2/BCL-XL, BH3 only BCL2 activator or sensitizer is unable to gain access, and the gross result of anti-apoptosis is observed. The BH3 domain mimetic agent, ABT-737, also binds to BCL2/BCL-XL, and hence competes and disrupts the interaction between BCL2/BCL-XL and BCL2L12, making tumor cells more vulnerable to apoptosis. Of note, GSK3β-mediated BCL2L12 S156 phosphorylation may affect BH3 domain function in glioma cells. BCL2L12, BCL2-like 12; BCL2, BCL2 apoptosis regulator; BCL-XL, BCL-extra large; GSK3β, glycogen synthase kinase 3β.
By contrast, in neuronal cells, sphingosine kinase 2 (SPK2) interacts with BCL2 via its BH3 domain, thereby dissociating it from Beclin-1 and activating autophagy. Tat-SPK2, a peptide designed from the BH3 domain of SPK2, activates autophagy and protects neural cells against oxygen-glucose deprivation injury. In primary murine cortical neurons and HT22 hippocampal neuronal cells, this cytoprotective autophagy appears beneficial to prolonging cell survival (36). Nevertheless, to further dissect the redundant role of BCL2L12 and Beclin-1 beyond cytoprotective autophagy, other experimental approaches are required. BCL2L12 may be tested in the future for potential interactions with well-known interacting partners of Beclin-1 in controlling autophagy, such as Vps34 PI3K (37) and UVRAG (38), in vitro and in vivo. Ectopically expressed wild-type BCL2L12 and BCL2L12 S156A mutant, but not BCL2L12A and BCL2L12 L213A mutants, restore the expression of autophagic markers in Beclin-1 shRNA knockdown glioma cell lines (unpublished data). This finding is more direct evidence that BCL2L12 may be another Beclin-1 that regulates autophagy under specific conditions.
Regarding GBM treatment, it may be necessary to modulate multiple molecular mechanisms in order to reverse its genetic antiapoptotic background and achieve better treatment outcomes than traditional chemotherapeutics or radiotherapy. Previous studies have pointed out that CRYAB and BCL2L12 are highly expressed in GBM (5). Thus, regardless of the expression of wild-type or mutant p53, p53 functions are compromised in GBM due to negative regulation by BCL2L12 of p53 at the transcriptional and translational levels. The first route of turning off p53 apoptotic events is directly binding to p53 to prevent its nuclear translocation leading to activation of downstream targets, such as Bax and Puma, an apoptosis modulator (2). BCL2L12 may also use three other routes to regulate apoptosis. First, it shuts down caspase-7 activation by directly binding and inactivating caspase-3 activity through upregulating its negative regulator CRYAB (5). Second, it interacts through its BH3-like domain with the hydrophobic groove region of BCL2 and BCL-XL, similar to the BH3 domain of Bax; occupancy of the hydrophobic groove of prosurvival BCL2 family proteins prevents access of the BH3-only apoptotic activator and sensitizer to this region to initiate apoptosis (9). Finally, its 90-229 region may be responsible for binding with Bax, scavenging active Bax to avoid Bax/Bak oligomer formation, thus leading to a gross antiapoptosis effect. Therefore, downregulation of BCL2L12 for treating GBM is a high priority goal (39). Previous studies also obtained good inhibitory results on tumor growth in animal models via small interfering (si)RNA directly against BCL2L12 (6) or SNA-miRNA-182 to downregulate BCL2L12 expression (40). Using TMZ is an option, but that involves p53 status and MGMT activation. TMZ-induced apoptosis depends on the expression status of p53 and MGMT, and TMZ-induced autophagy is linked to ERK signaling-associated ROS production (12). In addition, TMZ-induced autophagy occurs through mitochondrial damage and ER stress-dependent mechanisms to protect glioma cells from apoptotic damage (13). This drug-induced pro-survival autophagy is suggested to be an anti-apoptosis struggle with apoptosis stimuli from chemotherapeutic (TMZ) and STS (bacterial toxin, also protein kinase inhibitor) treatment (9).
ABT-737, a BH3 mimetic agent for treating cancer cells with a high BCL2, BCL-XL expression and Mcl-1 neutralization, may improve treatment outcome independent of p53 expression (41). We previously applied ABT-737 in combination with TMZ, since TMZ downregulates Mcl-1 expression in U87MG cells and thus creates a promising microenvironment for ABT-737. ABT-737 may compete with the BCL2L12 BH3-like domain for binding with BCL2 and BCL-XL to selectively inhibit their expression (9). Inhibition of interaction between BCL2 and Bax is helpful for Bax activation, forming Bax/Bak oligomerization to alter the outer membrane integrity of mitochondria and activate caspases. Although certain cell types lack Bax and Bak expression, ABT-737 may also induce dissociation of BCL2-Beclin-1 and autophagy (42). LiCl was also successfully applied to eliminate GSK3β-mediated phosphorylation at Ser156 of BCL2L12, resulting in reactivation of caspase-dependent apoptosis (7). In the present study, blocking TMZ-induced autophagy with 3-MA also revealed an enhancement of apoptosis induction. Using agents targeting mitochondria or ER, such as mitochondrial permeability transition pore inhibitor or 4-phenylbutyrate, an ER stress inhibitor, has also demonstrated proper apoptosis induction with TMZ (13).
By integrating the findings from our previous work and other studies, a combination of LiCl, siRNA of BCL2L12, ABT-737, autophagy inhibitor and TMZ are suggested to be applicable for GBM therapy. However, considering resistance to BCL2L12 targeting and ABT-737, as well as the low permeability of blood-brain barrier, potential application of these drugs in the clinic requires further investigation. To reach the goal of individualized medicine, knowing the p53 and MGMT background is essential for selecting which patients will respond to TMZ treatment. A quick screening of the expression levels of BCL2, BCL-XL, Mcl-1, Bax and Bak, in addition to p53 status and MGMT activation, is suggested to be essential in order to determine which patients will respond to the aforementioned combination therapeutic approaches.
Acknowledgments
The authors would like to thank Gary Mawyer, Managing Editor of Journal of the Wound, Ostomy and Continence Nurses Society, for language editing.
Abbreviations
- GBM
glioblastoma multiforme
- TMZ
temozolomide
- MGMT
O6-methylguanine DNA methyltransferase
- STS
staurosporine
- MD
molecular dynamics
- SPK2
sphingosine kinase 2
- 3-MA
3-methyladenine
Funding
The study was supported by a Ministry of Science and Technology (MOST) grant (no. 104-2320-B-037-030-MY3) and other funding sources from Kaohsiung Medical University, Kaohsiung Medical University Teaching Hospital, and National Sun Yat-sen University and Kaohsiung Medical University cooperative program (grant nos. KMU-DK105004, KMTTH-105-003 and NSYSUKMU-106-P004).
Availability of data and materials
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
The study was conceived and designed by JKL and YRH. BXH and CHC performed the yeast two-hybrid system experiments. Western blotting, Co-IP, and data analysis were performed by CWC, CHC, WSH, MCY, SJC and JKL. YTW performed protein conformational simulation. All other experiments were performed by CWC, BXH, and CHC. MCY and YRH wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Scorilas A, Kyriakopoulou L, Yousef GM, Ashworth LK, Kwamie A, Diamandis EP. Molecular cloning, physical mapping, and expression analysis of a novel gene, BCL2L12, encoding a proline-rich protein with a highly conserved BH2 domain of the Bcl-2 family. Genomics. 2001;72:217–221. doi: 10.1006/geno.2000.6455. [DOI] [PubMed] [Google Scholar]
- 2.Stegh AH, Brennan C, Mahoney JA, Forloney KL, Jenq HT, Luciano JP, Protopopov A, Chin L, Depinho RA. Glioma oncoprotein Bcl2L12 inhibits the p53 tumor suppressor. Genes Dev. 2010;24:2194–2204. doi: 10.1101/gad.1924710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzovaras A, Kladi-Skandali A, Michaelidou K, Zografos GC, Missitzis I, Ardavanis A, Scorilas A. BCL2L12: A promising molecular prognostic biomarker in breast cancer. Clin Biochem. 2014;47:257–262. doi: 10.1016/j.clinbiochem.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Stegh AH, Kim H, Bachoo RM, Forloney KL, Zhang J, Schulze H, Park K, Hannon GJ, Yuan J, Louis DN, et al. Bcl2L12 inhibits post-mitochondrial apoptosis signaling in glioblastoma. Genes Dev. 2007;21:98–111. doi: 10.1101/gad.1480007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stegh AH, Kesari S, Mahoney JE, Jenq HT, Forloney KL, Protopopov A, Louis DN, Chin L, DePinho RA. Bcl2L12-mediated inhibition of effector caspase-3 and caspase-7 via distinct mechanisms in glioblastoma. Proc Natl Acad Sci USA. 2008;105:10703–10708. doi: 10.1073/pnas.0712034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, Kouri FM, Merkel TJ, Luthi AJ, Patel PC, Cutler JI, et al. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci Transl Med. 2013;5:209ra152. doi: 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou CH, Chou AK, Lin CC, Chen WJ, Wei CC, Yang MC, Hsu CM, Lung FW, Loh JK, Howng SL, Hong YR. GSK3β regulates Bcl2L12 and Bcl2L12A anti-apoptosis signaling in glioblastoma and is inhibited by LiCl. Cell Cycle. 2012;11:532–542. doi: 10.4161/cc.11.3.19051. [DOI] [PubMed] [Google Scholar]
- 8.Kontos CK, Scorilas A. Molecular cloning of novel alternatively spliced variants of BCL2L12, a new member of the BCL2 gene family, and their expression analysis in cancer cells. Gene. 2012;505:153–166. doi: 10.1016/j.gene.2012.04.084. [DOI] [PubMed] [Google Scholar]
- 9.Yang MC, Loh JK, Li YY, Huang WS, Chou CH, Cheng JT, Wang YT, Lieu AS, Howng SL, Hong YR, Chou AK. Bcl2L12 with a BH3-like domain in regulating apoptosis and TMZ-induced autophagy: A prospective combination of ABT-737 and TMZ for treating glioma. Int J Oncol. 2015;46:1304–1316. doi: 10.3892/ijo.2015.2838. [DOI] [PubMed] [Google Scholar]
- 10.Blough MD, Beauchamp DC, Westgate MR, Kelly JJ, Cairncross JG. Effect of aberrant p53 function on temozolomide sensitivity of glioma cell lines and brain tumor initiating cells from glioblastoma. J Neurooncol. 2011;102:1–7. doi: 10.1007/s11060-010-0283-9. [DOI] [PubMed] [Google Scholar]
- 11.Srivastava A, Jain A, Jha P, Suri V, Sharma MC, Mallick S, Puri T, Gupta DK, Gupta A, Sarkar C. MGMT gene promoter methylation in pediatric glioblastomas. Childs Nerv Syst. 2010;26:1613–1618. doi: 10.1007/s00381-010-1214-y. [DOI] [PubMed] [Google Scholar]
- 12.Lin CJ, Lee CC, Shih YL, Lin TY, Wang SH, Lin YF, Shih CM. Resveratrol enhances the therapeutic effect of temozolomide against malignant glioma in vitro and in vivo by inhibiting autophagy. Free Radic Biol Med. 2012;52:377–391. doi: 10.1016/j.freeradbiomed.2011.10.487. [DOI] [PubMed] [Google Scholar]
- 13.Lin CJ, Lee CC, Shih YL, Lin CH, Wang SH, Chen TH, Shih CM. Inhibition of mitochondria- and endoplasmic reticulum stress-mediated autophagy augments temozolomide-induced apoptosis in glioma cells. PLoS One. 2012;7:e38706. doi: 10.1371/journal.pone.0038706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinha S, Levine B. The autophagy effector Beclin 1: A novel BH3-only protein. Oncogene. 2008;27(Suppl 1):S137–S148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SY, Song X, Zhang L, Bartlett DL, Lee YJ. Role of Bcl-xL/Beclin-1 in interplay between apoptosis and autophagy in oxaliplatin and bortezomib-induced cell death. Biochem Pharmacol. 2014;88:178–188. doi: 10.1016/j.bcp.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 19.Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: A method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu L. Yeast GAL4 two-hybrid system. A genetic system to identify proteins that interact with a target protein. Methods Mol Biol. 1997;63:173–196. doi: 10.1385/0-89603-481-X:173. [DOI] [PubMed] [Google Scholar]
- 21.Homeyer N, Horn AH, Lanig H, Sticht H. AMBER force-field parameters for phosphorylated amino acids in different protonation states: Phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J Mol Model. 2006;12:281–289. doi: 10.1007/s00894-005-0028-4. [DOI] [PubMed] [Google Scholar]
- 22.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 23.Darden T, York D, Pedersen L. Particle mesh Ewald: An N [center-dot] log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 24.Chahal M, Xu Y, Lesniak D, Graham K, Famulski K, Christensen JG, Aghi M, Jacques A, Murray D, Sabri S, Abdulkarim B. MGMT modulates glioblastoma angiogenesis and response to the tyrosine kinase inhibitor sunitinib. Neuro Oncol. 2010;12:822–833. doi: 10.1093/neuonc/noq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malley DS, Hamoudi RA, Kocialkowski S, Pearson DM, Collins VP, Ichimura K. A distinct region of the MGMT CpG island critical for transcriptional regulation is preferentially methylated in glioblastoma cells and xenografts. Acta Neuropathol. 2011;121:651–661. doi: 10.1007/s00401-011-0803-5. [DOI] [PubMed] [Google Scholar]
- 26.Stegh AH, DePinho RA. Beyond effector caspase inhibition: Bcl2L12 neutralizes p53 signaling in glioblastoma. Cell Cycle. 2011;10:33–38. doi: 10.4161/cc.10.1.14365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang JB, Svilar D, Trivedi RN, Wang XH, Goellne EM, Moore B, Hamilton RL, Banze LA, Brown AR, Sobol RW. N-methylpurine DNA glycosylase and DNA polymerase beta modulate BER inhibitor potentiation of glioma cells to temozolomide. Neuro Oncol. 2011;13:471–486. doi: 10.1093/neuonc/nor011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermisson M, Klumpp A, Wick W, Wischhusen J, Nagel G, Roos W, Kaina B, Weller M. O6-methylguanine DNA meth-yltransferase and p53 status predict temozolomide sensitivity in human malignant glioma cells. J Neurochem. 2006;96:766–776. doi: 10.1111/j.1471-4159.2005.03583.x. [DOI] [PubMed] [Google Scholar]
- 29.Allen M, Bjerke M, Edlund H, Nelander S, Westermark B. Origin of the U87MG glioma cell line: Good news and bad news. Sci Transl Med. 2016;8:354re3. doi: 10.1126/scitranslmed.aaf6853. [DOI] [PubMed] [Google Scholar]
- 30.Jia W, Jiang X, Liu W, Wang L, Zhu B, Zhu H, Liu X, Zhong M, Xie D, Huang W, et al. Effects of three-dimensional collagen scaffolds on the expression profiles and biological functions of glioma cells. Int J Oncol. 2018 Mar 20; doi: 10.3892/ijo.2018.4330. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong Y, Yang J, Wu W, Wang W, Kong X, Wang Y, Yun X, Zong H, Wei Y, Zhang S, Gu J. Knockdown of BCL2L12 leads to cisplatin resistance in MDA-MB-231 breast cancer cells. Biochim Biophys Acta. 2008;1782:649–657. doi: 10.1016/j.bbadis.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Adamopoulos PG, Kontos CK, Tsiakanikas P, Scorilas A. Identification of novel alternative splice variants of the BCL2L12 gene in human cancer cells using next-generation sequencing methodology. Cancer Lett. 2016;373:119–129. doi: 10.1016/j.canlet.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 33.Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, Korsmeyer SJ. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell. 2001;8:705–711. doi: 10.1016/S1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 34.Lomonosova E, Chinnadurai G. BH3-only proteins in apop-tosis and beyond: An overview. Oncogene. 2008;27(Suppl 1):S2–S19. doi: 10.1038/onc.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Rita A, Strappazzon F. AMBRA1, a Novel BH3-like protein: New insights into the AMBRA1-BCL2-family proteins relationship. Int Rev Cell Mol Biol. 2017;330:85–113. doi: 10.1016/bs.ircmb.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Song DD, Zhang TT, Chen JL, Xia YF, Qin ZH, Waeber C, Sheng R. Sphingosine kinase 2 activates autophagy and protects neurons against ischemic injury through interaction with Bcl-2 via its putative BH3 domain. Cell Death Dis. 2017;8:e2912. doi: 10.1038/cddis.2017.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 38.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stegh AH, Chin L, Louis DN, DePinho RA. What drives intense apoptosis resistance and propensity for necrosis in glioblastoma? A role for Bcl2L12 as a multifunctional cell death regulator. Cell Cycle. 2008;7:2833–2839. doi: 10.4161/cc.7.18.6759. [DOI] [PubMed] [Google Scholar]
- 40.Kouri FM, Ritner C, Stegh AH. miRNA-182 and the regulation of the glioblastoma phenotype-toward miRNA-based precision therapeutics. Cell Cycle. 2015;14:3794–3800. doi: 10.1080/15384101.2015.1093711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE, Willis SN, Scott CL, Day CL, Cory S, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pedro JM, Wei Y, Sica V, Maiuri MC, Zou Z, Kroemer G, Levine B. BAX and BAK1 are dispensable for ABT-737-induced dissociation of the BCL2-BECN1 complex and autophagy. Autophagy. 2015;11:452–459. doi: 10.1080/15548627.2015.1017191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.