Abstract
Williams syndrome (WS) is a neurodevelopmental disorder involving hemideletion of as many as 26–28 genes, resulting in a constellation of unique physical, cognitive and behavior phenotypes. The haploinsufficiency effect of each gene has been studied and correlated with phenotype(s) using several models including WS subjects, animal models, and peripheral cell lines. However, links for most of the genes to WS phenotypes remains unclear. Among those genes, general transcription factor 2I (GTF2I) is of particular interest as its haploinsufficiency is possibly associated with hypersociability in WS. Here, we describe studies of atypical WS cases as well as mouse models focusing on GTF2I that support a role for this protein in the neurocognitive and behavioral profiles of WS individuals. We also review collective studies on diverse molecular functions of GTF2I that may provide mechanistic explanation for phenotypes recently reported in our relevant cellular model, namely WS induced pluripotent stem cell (iPSC)-derived neurons. Finally, in light of the progress in gene-manipulating approaches, we suggest their uses in revealing the neural functions of GTF2I in the context of WS.
Keywords: GTF2I, Williams syndrome, hypersociability, TRPC3
1.1 Williams syndrome
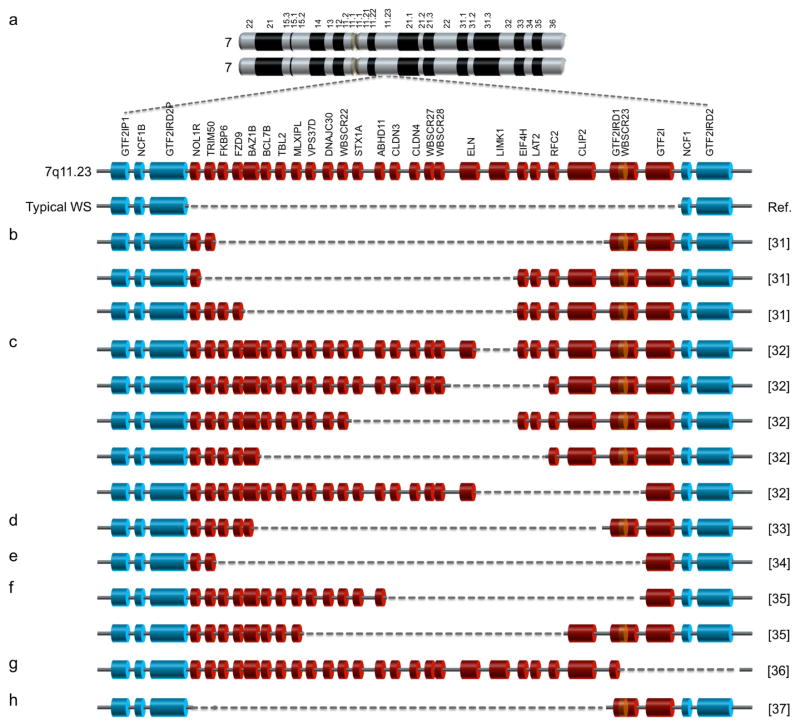
Williams syndrome is a rare multigenic neurodevelopmental disorder with the prevalence of 1 in 7,500 [1]. It is caused by hemizygous deletion of 26–28 genes on chromosome band 7q11.23 [2] (Fig. 1a) as a result of unequal non-allelic homologous recombination between low-copy repeats flanking the region of deletion [3, 4] during meiosis [2]. Due to haploinsufficiency of all these genes, classical (or typical) WS individuals, accounting for 98% of diagnosed cases [5], exhibit clinical symptoms including craniofacial (elfin) features, cardiovascular abnormalities especially supravalvular aorta stenosis (SVAS), hypercalcemia, and overall brain volume reduction [6–9]. In terms of cognitive profiles, typical WS subjects exhibit mild to moderate mental retardation, overuse of expressive language, and visuospatial deficits [10, 11]. Strikingly, they share a very unique behavioral profile, i.e. over-friendliness or hypersociability [12, 13], which is opposite the characteristics of autism spectrum disorders (ASD). Rarer atypical WS cases, especially the ones whose hemizygous deletion spans less than 26–28 genes, exhibit a partial spectrum of typical WS phenotypes [14–16]. These atypical cases provide vital clues for genotype–phenotype correlation studies, enabling the dissection of the role of each gene in WS etiology. Haploinsufficiency of ELN, for example, results in decreased elasticity of blood vessels and thereby leading to SVAS [14], the major cause of death in WS individuals [17]. In the past few decades, several attempts have been made to establish the links between other hemideleted genes and WS neurocognitive and behavioral phenotypes.
Figure 1. WS genetics and atypical WS cases in genotype–phenotype correlation studies.
a, WS is caused by hemizygous deletion of multiple genes on chromosome band 7q11.23. A typical (classical) WS deletion encompasses every gene in red (NOL1R to GTF2I). b–h, Individuals with atypical partial WS deletions (b–f, h) [31–35, 37] and a subject with an extended distal WS deletion (g) [36] have been reported in genotype–phenotype correlation studies, suggesting a link between GTF2I and multiple WS phenotypes (see text for detail).
In addition to microdeletion, non-allelic homologous recombination during meiosis could result in duplication of this region, causing a disorder called 7q11.23 microduplication syndrome (7dup) [18]. 7dup individuals harbor three alleles of each of these 26–28 genes as opposed to one allele in WS patients. Interestingly, 7dup individuals are characterized by ASD-like phenotypes including developmental delay, language impairment, and poor social skills [18, 19], which are the opposite of WS characteristics, suggesting a gene-dosage effect. Mutations in MECP2, FMR1, SHANK3, CDKL5, CACNA1C and TRPC6, have been independently reported contributing to syndromic forms of ASD, i.e. Rett syndrome [20], fragile X syndrome [21], Phelan-McDermid syndrome [22], CDKL5-related disorder [23], Timothy syndrome [24], and TRPC6-related disorder [25], respectively. At first, proteins encoded by these genes may seem to act in different pathways, but their mutations ultimately result in common ASD phenotypes. These phenotypes are also shared by individuals with non-syndromic ASD, which make up the majority of ASD cases, yet the genetic bases for these phenotypes remain unknown. With relevant established models, studies of rare disorders with well-defined genetic mutations like WS and 7dup, which differ only in gene copy number, offer powerful insight into the functions of those genes in 7q11.23 as well as common mechanisms underlining neurocognitive profiles and social behaviors in ASD.
The use of neurons generated from patient-derived iPSCs has recently become a routine in neurological disease modeling [26–28] as genetic mutations in patients’ somatic cells can be captured and passed on to neurons, which can then be manipulated and investigated for altered neuronal characteristics. We are the first group to report WS phenotypes in WS iPSC-derived neural progenitor cells (NPCs), specifically an increase in apoptosis as a result of frizzled9 haploinsufficiency, and in WS iPSC-derived neurons, including changes in morphology comparable to those observed in postmortem neurons as well as increases in calcium transient frequency when compared to NPCs and neurons derived from typical developing (TD) subjects’ iPSCs, respectively [29]. In order to specifically look further into the role of each gene in the WS context and form appropriate hypotheses, information must be adequately gathered from previous studies using several different systems including atypical WS individuals as well as mouse and other cellular models. Among those 26–28 WS genes, GTF2I is of our interest as several genotype-phenotype correlation studies in WS individuals with partial deletions suggest its importance in regulating neurocognitive profiles and social behavior. Findings from those studies are, here, reviewed.
1.2 Atypical WS subjects and mouse models: Clues from genotype–phenotype correlation studies of GTF2I
GTF2I is one of the 26–28 hemizygously deleted genes in WS, being located at the telomeric end of the deleted region (Fig. 1a). It encodes general transcription factor 2I, which was first described in 1991 as a transcription initiation factor that binds to pyrimidine-rich initiator (Inr) elements at the transcription start site [30]. However, its role in WS etiology was implicated only when atypical WS cases with both alleles of GTF2I retained in the genome were reported for the first time in the early 2000s. Hirota et al. described three atypical WS subjects whose deletions were smaller than those with typical WS and did not include GTF2I and GTF2IRD1 (Fig. 1b) [31]. Compared to typical WS subjects, these atypical WS individuals still have SVAS but do not exhibit elfin facies and visuospatial deficits [31]. Morris et al. reported five families, with members affected by SVAS, each having different deletions in 7q11.23 including ELN and some WS genes (Fig. 1c) [32]. In the genomes of all individuals analyzed, only FKBP6 and GTF2I had two preserved alleles. Besides variation in WS cognitive profiles, none of the individuals had mental retardation [32]. However, social behaviors were not assessed in these two studies [31, 32]. Ferrero et al. reported atypical WS subject whose deletion was similar to one of those three patients described by Hirota et al. except that FKBP6, FZD9 and partial BAZB1 were retained in his genome (Fig. 1d) [33]. The patient exhibited partial craniofacial features, SVAS and particular cognitive deficits but no hypercalcemia and hypersociability [33]. Dai et al. described a very unique atypical WS subject whose deletions spanned almost all typical WS genes except GTF2I (Fig. 1e) [34]. The individual showed typical WS physical and developmental phenotypes as expected, but did not exhibit overly social behavior [34]. Antonell et al. reported another two families with different partial WS deletions preserving GTF2I (Fig. 1f) [35]. Despite variable cardiovascular abnormalities, members of both families exhibited normal visuospatial ability and, surprisingly, hypersociability [35]. Edelmann et al. described an individual with hemideletion of GTF2IRD1 (partial), GTF2I and another 14 genes outside the typical WS deletion region towards the telomeric end of chromosome 7 (Fig. 1g) [36]. While the subject met the criteria for ASD, including abnormalities in comprehension of simple language and conversation, she also exhibited WS cognitive and behavioral profiles, including visuospatial deficits, hypersociability, non-social anxiety, and language delay [36]. No heart defects, hypercalcemia, or typical craniofacial features were observed due to preservation of most typical WS genes [36]. Delgado et al. described another atypical case with deletion almost opposite to the individual in previous study, i.e. only partial GTF2IRD1 and GTF2I were not hemizygously deleted (Fig. 1h). While the individual showed cardiovascular abnormalities and craniofacial features, her neuropsychological phenotypes, including hypersociability, visuospatial deficits and developmental delay, were, unfortunately, not assessed in this study [37].
Increasing studies of variation in single-nucleotide polymorphisms (SNPs) in GTF2I coupled with brain response using functional magnetic resonance imaging in different populations also suggest the gene’s association with particular traits linked to certain brain regions. Jabbi et al. found that a minor allele of particular GTF2I SNP is associated with dorsolateral prefrontal cortex activity responding to aversive stimuli processing in the TD population [38]. Moreover, Malenfant et al. reported the association of another two GTF2I SNPs and their haplotype with social skill impairment and repetitive behaviors in ASD population [39]. Recently, Swartz et al. showed that one of these 2 alleles is also associated with reduction in amygdala activity relating to threat in female TD participants [40].
In addition to human studies, mouse models have also provided vital clues regarding the role of the murine homolog Gtf2i through genetic manipulation and whole animal characterization. Typical WS mice, with hemizygous deletion of their human gene counterparts in conserved syntenic region in reverse orientation on mouse chromosome band 5G2, were successfully generated [41]. This was achieved by breeding an atypical WS mouse with a proximal deletion (Gtf2i to Limk1) to another atypical WS mouse with a distal deletion (Limk1 to Fkbp6) [41]. The typical WS progeny mice recapitulate phenotypes observed in typical WS individuals while atypical WS mice exhibit partial traits, allowing mapping of those features to a set of genes in either the distal or proximal deletion region. Using these mice, Li et al. found that the proximal genes were associated with elevated sociability and hypersensitivity to sounds, but not with cognitive defect and brain volume reduction [41]. Single-gene deficient mice also offer insight into the haploinsufficiency effect of specific gene of interest. Sakurai et al. reported that homozygous deletion of Gtf2i is embryonic lethal due to abnormal neural tube closure during embryonic development, while mice with heterozygous deletion of Gtf2i appeared to be physically normal accompanied by normal learning and memory and no increase in anxiety [42]. However, these animals exhibited hypersociability as indicated by significant increase in time exploring unfamiliar mouse and decreased habituation [42]. Lucena et al. generated mouse model expressing truncated form of GTF2I, i.e. only first 140 amino acids were deleted [43]. Instead of embryonic lethality, homozygous mutant mice showed compromised viability [43]. Interestingly, homozygous and, to a lesser extent, heterozygous mutants exhibited craniofacial phenotypes. Additional phenotypes reported for heterozygous mice include hypersensitivity to sound and high anxiety [43]. Another study by Mervis et al., in which mice with varying copy numbers of Gtf2i were generated, demonstrated that a significant increase in separation anxiety was observed in mouse pups with more than two copies of Gtf2i relative to pups with one or two copies, both of which surprisingly exhibited comparable levels of separation anxiety [44]. Borralleras et al. characterized different mutant mice (intragenic deletion of Gtf2i, proximal and complete deletions of WS critical region) [45]. All mutants exhibited poor motor coordination, anxiety and hypersociability [45]. In addition, as GTF2I is phosphorylated by SRC tyrosine kinase and, consequently, translocated to nucleus [46], mutation in Src disrupting its kinase activity also negatively affects GTF2I function. Mice with homozygous mutation of Src showed WS phenotypes including visuospatial deficits, craniofacial features and hypersociability [47].
These genotype–phenotype correlation studies have suggested association of GTF2I in the manifestation of many WS phenotypes such as social behavior, anxiety, visuospatial deficits, hypersensitivity to sound, mental retardation as well as craniofacial features. Nevertheless, in order to establish unambiguous links between this gene and any of these potential phenotypes, further investigations of more rare atypical WS individuals, SNPs in different populations, and transgenic mice with a deletion or duplication of GTF2I are needed.
1.3 Function of GTF2I: Collective studies in different cellular models
GTF2I together with the GTF2I repeat domain-containing protein 1 (GTF2IRD1) and 2 (GTF2IRD2) genes are members of the transcription factor 2I family [48]. GTF2IRD1, located upstream of GTF2I, is also hemizygously deleted in typical WS individuals, unlike GTF2IRD2 which is located downstream of GTF2I (Fig. 1a). GTF2I contains a leucine zipper at the N terminus for homodimerization, a nuclear localization signal, a basic region (DNA binding domain), and six repeated regions (I-repeats) with a helix-loop-helix motif in each repeat for protein-protein interactions [49]. Through alternative splicing, there are at least four human GTF2I isoforms (α, β, γ, and Δ) [50], with the γ isoform being highly expressed in neuronal cells [51]. These isoforms can interact with one another to form homomers and heteromers [50] in various combinations that may enable binding to different target promoters and, therefore, drive expression of different genes [50]. As suggested by its distinct domains for both DNA-protein and protein-protein interaction, GTF2I is multifunctional transcription factor that binds to both Inr core promoter element for basal transcription and to upstream elements for signal-induced transcription [52, 53]. c-fos promoter is described as first transcriptional target of GTF2I [54]. In mouse fibroblasts, it is demonstrated that, at resting state, the Δ isoform is found in the cytoplasm while the β isoform resides in the nucleus, controlling basal transcription of c-fos. Upon signaling-induced phosphorylation at tyrosine residues, the Δ isoform is translocated to the nucleus, recruits other activators and activates c-fos gene, while the β isoform is transported to the cytoplasm [55]. GTF2I has been shown to be involved in several cellular processes, including endoplasmic reticulum stress response, cell cycle, apoptosis or even virus transcription, etc. through its interaction with multiple proteins. Examples of binding partners of GTF2I and its corresponding transcriptional targets and cellular processes that have been reported are demonstrated in Table 1. Microarray analysis in mouse embryonic fibroblasts overexpressing GTF2I followed by qRT-PCR and chromatin immunoprecipitation of selected candidates confirmed that primary downstream targets of GTF2I were Bax, Shrm, Cfl-1, Ezh2, Epc1, Ccnd3 and Hdac1 [56]. Another microarray analysis comparing between mouse cortex of heterozygous mutants expressing truncated GTF2I, generated by Lucena et al. [43], and that of wild-type animals as well as between XS0353, embryonic stem cell line with heterozygous mutation for Gtf2i, and AB2.2, wild-type embryonic stem cell line, indicated that downstream target of GTF2I are genes in phosphatidylinositol 3-kinase signaling pathways, e.g. Pik3r1, Cab39l, and Eif4b, involved in dendritic spine formation during development and synaptic plasticity, as well as genes in transforming growth factor beta signaling pathways including Shc1 and Snw1 [57]. In terms of particular WS phenotypes, direct targets of GTF2I, namely Cfdp1, Sec23a and Nsd1, genes strongly associated with craniofacial development, have been identified [58].
Table 1.
Roles of GTF2I in transcription and signal transduction through interaction with different binding partners
| Binding partners | Inducing Signal | Action | Cell type | GTF2I target promoter /sequence | Processes | Ref. |
|---|---|---|---|---|---|---|
| BTK | BCR | Phosphorylates GTF2I | B cells | c-fos | B-cell development | [72, 73] |
| ITK | TCR | Phosphorylates GTF2I | T cells | c-fos | T-cell activation and function | [74] |
| CTCF | - | Form complex with GTF2I | WEHI-231 | metabolic genes | Epigenetic processes | [75] |
| SMAD2 | TGFβ/activ in signaling | Form complex with GTF2I | P19 | Gsc | Development and patterning | [76] |
| ERK | Serum stimulation | Phosphorylates GTF2I | Mouse fibroblasts | c-fos | Cell growth | [77] |
| JAK2 | Serum stimulation | Phosphorylates GTF2I | Mouse fibroblasts | c-fos | Cell growth and proliferation | [78] |
| c-Src | ER stress | Phosphorylates GTF2I | Mouse fibroblasts | Grp78 | Cell survival and proliferation | [79, 80] |
| c-Src | α2M | Phosphorylates GTF2I | 1-LN | Grp78 | Cell survival and proliferation | [81] |
| SRF, | Serum | Phosphorylates | HeLa cells | c-fos | Cell growth | [54] |
| Phox1 | stimulation | GTF2I | ||||
| USF | Ras/MAPK | Phosphorylates GTF2I | T cells | HIV-1 LTR | Viral transcription | [82] |
| PLC-γ | RTK | Binds to phosphorylated GTF2I | PC12 | N/A | Inhibition of agonist- induced calcium entry | [59] |
Btk, Bruton’s tyrosine kinase; BCR, B-cell receptor; ITK, inducible tyrosine kinase; TCR, T-cell receptor; CTCF, epigenetic regulatory protein; P19, mouse embryonic carcinoma cells; Gsc, Goosecoid; ERK, extracellular signal-regulated kinase; ER, endoplasmic reticulum; PC12, mouse neural cells; 1-LN, human prostate cancer cells; α2M, alpha(2)-macroglobulin; SRF, serum response factor; USF, upstream stimulatory factor; MAPK, mitogen-activated protein kinase; RTK, receptor tyrosine kinase; N/A, not applicable
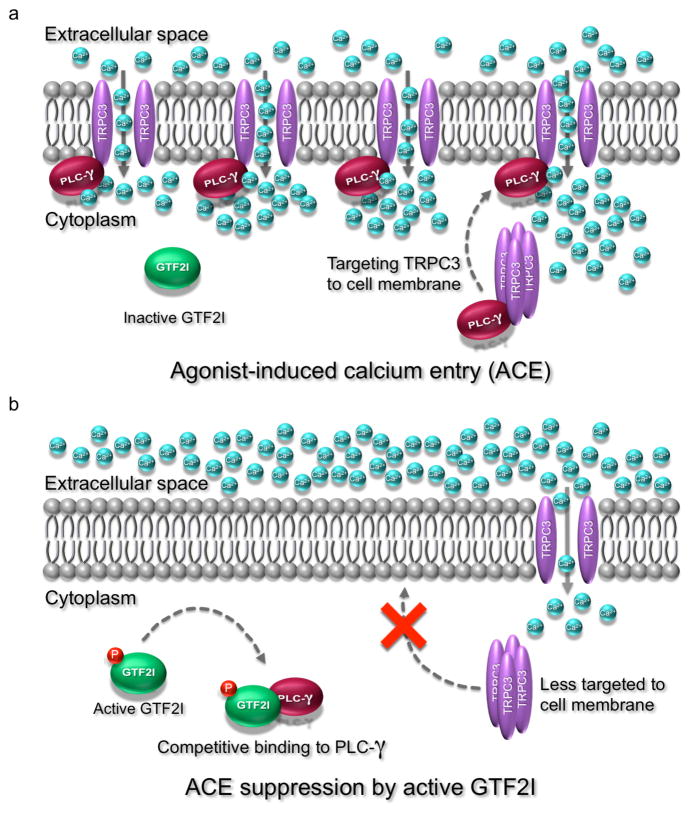
One of the cytosolic functions of GTF2I relatable to WS etiology is its negative regulation of agonist-induced calcium entry (ACE), which may be associated with hypercalcemia described in WS individuals. Using rat neural cell lines, Caraveo et al. reported that, upon binding of agonists (uridine triphosphate and bradykinin) to G protein-coupled receptors on cell surface, phospholipase C-gamma (PLC-γ) becomes activated and its split pleckstrin homology (PH) domain binds to the PH-like domain of a calcium channel protein, namely transient receptor potential cation channel subfamily C member 3 (TRPC3), leading to an increase in surface expression of TRPC3, and, therefore, Ca2+ influx (Fig. 2a) [59]. As GTF2I also contains a PH like domain, activation by phosphorylation enables it to compete against TRPC3 for binding to PLC-γ at the split PH domain as well as to Src homology 2 (SH2) domain of PLC-γ, leading to decreased surface accumulation of TRPC3, and, hence, reduced Ca2+ influx (Fig. 2b) [59].
Figure 2. The potential role of GTF2I in agonist-induced calcium entry suppression in neurons.
a, When GTF2I is inactive, activated PLC-γ binds to TRPC3, stimulating recruitment of more TRPC3 to the cell membrane. This results in an increase in calcium influx via agonist-induced calcium entry (ACE). b, When its tyrosine is phosphorylated, GTF2I becomes active and compete with TRPC3 for binding to activated PLC-γ, leading to a decrease in TRPC3 surface accumulation and, therefore, a reduction in ACE. GTF2I, general transcription factor 2I; TRPC3, transient receptor potential cation channel subfamily C member 3; PLC-γ, phospholipase C gamma; P, phosphorylation; Ca2+, calcium ions.
There are intriguing hints that these molecular data may have physiological relevanc. TRPC3 has been functionally implicated in synaptic transmission in murine cerebellar Purkinje neurons [60]. On the human side, TRPC3 dysregulation has been described in WS patient, where hypercalcemia has been associated with TRPC3 overexpression of TRPC3 in intestines and kidney tissues [61]. Taken together, one can speculate that haploinsufficiency of GTF2I, as seen in WS individuals causes an increase in surface expression of TRPC3, and, consequently, an increase in ACE in human Purkinje neurons. This hypothesis needs further investigation.
1.4 Uncovering the function of GTF2I in brain cells: the future path to understanding WS
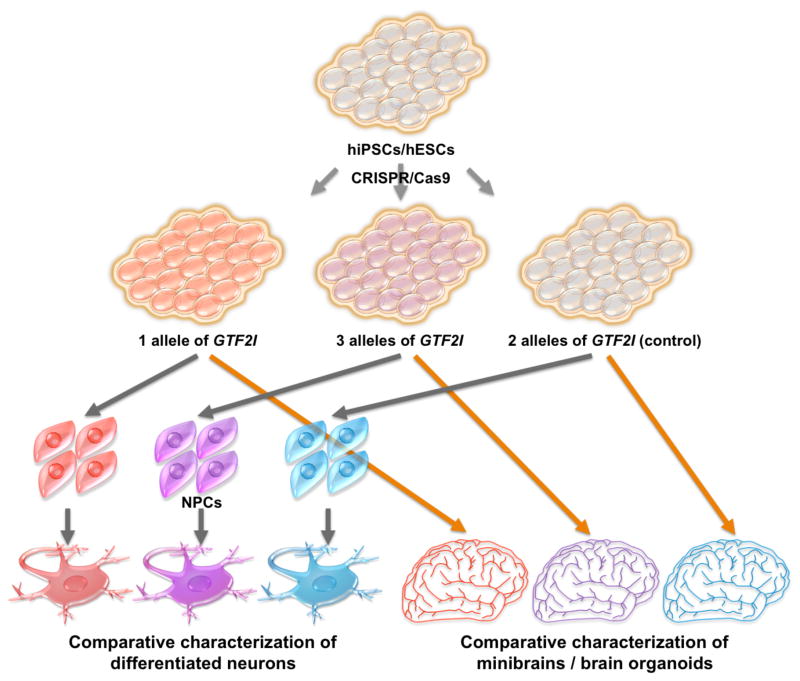
Although multiple functions of GTF2I have been reported in several cellular models, the lack of human neurons and astrocytes has been a major obstacle hindering investigation of its cell lineage-specific roles, leaving mechanisms underlying WS etiology unclear. Recently, however, Adamo et al. investigated the GTF2I complexes in WS- and 7dup-derived iPSCs by mass spectrometry analysis and identified BEND4, a transcription factor highly expressed in brain and involved in neural processes, as one of direct targets of GTF2I [62]. As pluripotent stem cell technology, including iPSCs and embryonic stem cells (ESCs), together with established differentiation protocols has allowed researchers to generate not only 2D culture of desired brain cells, including neurons [63, 64], glia [65] and oligodendrocytes [66], but also 3D culture, i.e. mini brains or brain organoids [67, 68], both transcriptional targets and direct target proteins of GTF2I in, for example, cortical neurons, derived from iPSCs/ESCs can be identified. It also offers opportunities to determine whether candidate proteins, e.g. BEND4, described in non-neural cells actually interact with GTF2I in any brain cells. According to expression pattern of GTF2I in mouse, it is ubiquitously expressed in developing brain and becomes exclusively restricted to neurons with high levels of expression in cerebellar Purkinje neurons and hippocampal interneurons [69]. As iPSC-derived neurons in culture are considered temporally comparable to the ones in developing brain, we should be able to determine whether previously reported increase in calcium oscillation frequency observed in WS iPSC-derived cortical neurons [29] is caused by the increase in surface accumulation of TRPC3 as a result of reduction in GTF2I expression. Similar experiment should also be performed in human iPSC-derived Purkinje neurons as both GTF2I and TRPC3 expression was reported in mouse Purkinje neurons [69, 70]. Furthermore, single-gene deficient cellular model could be now established using genome-editing techniques such as CRISPR/Cas9 [71], facilitating the functional studies of specific gene of interest. By combining these 2 technologies, researchers could generate and characterize specific subtypes of neurons harboring one or three working alleles of GTF2I (Fig. 3). These studies would identify true targets of GTF2I in neural cells, reveal altered molecular mechanisms underlying WS etiology, and ultimately help establish the link between gene, brain, and behavior observed in WS individuals.
Figure 3. Pluripotent stem cell and CRISPR/Cas9 approaches in revealing the role of GTF2I in brain cells.
In order to study the neural effects of GTF2I copy number, CRISPR/Cas9 genome-editing technique can be used to generate hiPSCs/hESCs with one (as observed in WS) and three (as in 7dup) GTF2I alleles. Generated cells can be differentiated into NPCs and neurons, for example, or minibrains (brain organoids). Comparative characterization of these cells can reveal cellular and molecular differences in neurons as a result of haploinsufficiency or duplication of GTF2I. hiPSC, human induced pluripotent stem cells; hESCs, human embryonic stem cells; NPCs, neural progenitor cells.
Highlights.
Copy number variation of genes on 7q11.23 results in converse phenotypes
Gene-phenotype correlation studies in atypical WS suggest role of GTF2I in WS
GTF2I is a candidate for neurocognitive and behavioral profiles of WS
GTF2I plays roles in both transcription and signal transduction
GTF2I hemideletion is possibly responsible for calcium alteration in WS neurons
Acknowledgments
We thank Dr. Samaporn Teeravechyan for critical reading and language editing of our manuscript. This work was supported by grants from the California Institute for Regenerative Medicine (CIRM) TR4-06747, the National Institutes of Health through the P01 NICHD033113, 156MH109587, U19MH107367 and a NARSAD Independent Investigator Grant to A.R.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stromme P, Bjornstad PG, Ramstad K. Prevalence estimation of Williams syndrome. Journal of child neurology. 2002;17(4):269–71. doi: 10.1177/088307380201700406. [DOI] [PubMed] [Google Scholar]
- 2.Urban Z, Helms C, Fekete G, Csiszar K, Bonnet D, Munnich A, Donis-Keller H, Boyd CD. 7q11.23 deletions in Williams syndrome arise as a consequence of unequal meiotic crossover. American journal of human genetics. 1996;59(4):958–62. [PMC free article] [PubMed] [Google Scholar]
- 3.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends in genetics : TIG. 2002;18(2):74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 4.Bayes M, Magano LF, Rivera N, Flores R, Perez Jurado LA. Mutational mechanisms of Williams-Beuren syndrome deletions. American journal of human genetics. 2003;73(1):131–51. doi: 10.1086/376565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris CA, Mervis CB. Williams syndrome and related disorders. Annual review of genomics and human genetics. 2000;1:461–84. doi: 10.1146/annurev.genom.1.1.461. [DOI] [PubMed] [Google Scholar]
- 6.Williams JC, Barratt-Boyes BG, Lowe JB. Supravalvular aortic stenosis. Circulation. 1961;24:1311–8. doi: 10.1161/01.cir.24.6.1311. [DOI] [PubMed] [Google Scholar]
- 7.Beuren AJ, Apitz J, Harmjanz D. Supravalvular aortic stenosis in association with mental retardation and a certain facial appearance. Circulation. 1962;26:1235–40. doi: 10.1161/01.cir.26.6.1235. [DOI] [PubMed] [Google Scholar]
- 8.Burn J. Williams syndrome. Journal of medical genetics. 1986;23(5):389–95. doi: 10.1136/jmg.23.5.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reiss AL, Eliez S, Schmitt JE, Straus E, Lai Z, Jones W, Bellugi U. IV. Neuroanatomy of Williams syndrome: a high-resolution MRI study. Journal of cognitive neuroscience. 2000;12(Suppl 1):65–73. doi: 10.1162/089892900561986. [DOI] [PubMed] [Google Scholar]
- 10.Farran EK, Jarrold C. Visuospatial cognition in Williams syndrome: reviewing and accounting for the strengths and weaknesses in performance. Developmental neuropsychology. 2003;23(1–2):173–200. doi: 10.1080/87565641.2003.9651891. [DOI] [PubMed] [Google Scholar]
- 11.Bellugi U, Lichtenberger L, Jones W, Lai Z, St George M. I. The neurocognitive profile of Williams Syndrome: a complex pattern of strengths and weaknesses. Journal of cognitive neuroscience. 2000;12(Suppl 1):7–29. doi: 10.1162/089892900561959. [DOI] [PubMed] [Google Scholar]
- 12.Doyle TF, Bellugi U, Korenberg JR, Graham J. "Everybody in the world is my friend" hypersociability in young children with Williams syndrome. American journal of medical genetics. Part A. 2004;124A(3):263–73. doi: 10.1002/ajmg.a.20416. [DOI] [PubMed] [Google Scholar]
- 13.Jones W, Bellugi U, Lai Z, Chiles M, Reilly J, Lincoln A, Adolphs R. II. Hypersociability in Williams Syndrome. Journal of cognitive neuroscience. 2000;12(Suppl 1):30–46. doi: 10.1162/089892900561968. [DOI] [PubMed] [Google Scholar]
- 14.Curran ME, Atkinson DL, Ewart AK, Morris CA, Leppert MF, Keating MT. The elastin gene is disrupted by a translocation associated with supravalvular aortic stenosis. Cell. 1993;73(1):159–68. doi: 10.1016/0092-8674(93)90168-p. [DOI] [PubMed] [Google Scholar]
- 15.Frangiskakis JM, Ewart AK, Morris CA, Mervis CB, Bertrand J, Robinson BF, Klein BP, Ensing GJ, Everett LA, Green ED, Proschel C, Gutowski NJ, Noble M, Atkinson DL, Odelberg SJ, Keating MT. LIM-kinase1 hemizygosity implicated in impaired visuospatial constructive cognition. Cell. 1996;86(1):59–69. doi: 10.1016/s0092-8674(00)80077-x. [DOI] [PubMed] [Google Scholar]
- 16.van Hagen JM, van der Geest JN, van der Giessen RS, Lagers-van Haselen GC, Eussen HJ, Gille JJ, Govaerts LC, Wouters CH, de Coo IF, Hoogenraad CC, Koekkoek SK, Frens MA, van Camp N, van der Linden A, Jansweijer MC, Thorgeirsson SS, De Zeeuw CI. Contribution of CYLN2 and GTF2IRD1 to neurological and cognitive symptoms in Williams Syndrome. Neurobiology of disease. 2007;26(1):112–24. doi: 10.1016/j.nbd.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 17.Wessel A, Gravenhorst V, Buchhorn R, Gosch A, Partsch CJ, Pankau R. Risk of sudden death in the Williams-Beuren syndrome. American journal of medical genetics Part A. 2004;127A(3):234–7. doi: 10.1002/ajmg.a.30012. [DOI] [PubMed] [Google Scholar]
- 18.Somerville MJ, Mervis CB, Young EJ, Seo EJ, del Campo M, Bamforth S, Peregrine E, Loo W, Lilley M, Perez-Jurado LA, Morris CA, Scherer SW, Osborne LR. Severe expressive-language delay related to duplication of the Williams-Beuren locus. The New England journal of medicine. 2005;353(16):1694–701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Depienne C, Heron D, Betancur C, Benyahia B, Trouillard O, Bouteiller D, Verloes A, LeGuern E, Leboyer M, Brice A. Autism, language delay and mental retardation in a patient with 7q11 duplication. Journal of medical genetics. 2007;44(7):452–8. doi: 10.1136/jmg.2006.047092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature genetics. 1999;23(2):185–8. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 21.O'Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annual review of neuroscience. 2002;25:315–38. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- 22.Phelan K, McDermid HE. The 22q13.3 Deletion Syndrome (Phelan-McDermid Syndrome) Molecular syndromology. 2012;2(3–5):186–201. doi: 10.1159/000334260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mari F, Azimonti S, Bertani I, Bolognese F, Colombo E, Caselli R, Scala E, Longo I, Grosso S, Pescucci C, Ariani F, Hayek G, Balestri P, Bergo A, Badaracco G, Zappella M, Broccoli V, Renieri A, Kilstrup-Nielsen C, Landsberger N. CDKL5 belongs to the same molecular pathway of MeCP2 and it is responsible for the early-onset seizure variant of Rett syndrome. Human molecular genetics. 2005;14(14):1935–46. doi: 10.1093/hmg/ddi198. [DOI] [PubMed] [Google Scholar]
- 24.Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, Napolitano C, Schwartz PJ, Joseph RM, Condouris K, Tager-Flusberg H, Priori SG, Sanguinetti MC, Keating MT. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119(1):19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Griesi-Oliveira K, Acab A, Gupta AR, Sunaga DY, Chailangkarn T, Nicol X, Nunez Y, Walker MF, Murdoch JD, Sanders SJ, Fernandez TV, Ji W, Lifton RP, Vadasz E, Dietrich A, Pradhan D, Song H, Ming GL, Gu X, Haddad G, Marchetto MC, Spitzer N, Passos-Bueno MR, State MW, Muotri AR. Modeling non-syndromic autism and the impact of TRPC6 disruption in human neurons. Molecular psychiatry. 2015;20(11):1350–65. doi: 10.1038/mp.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143(4):527–39. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Koscielska KA, Cao Z, Hulsizer S, Grace N, Mitchell G, Nacey C, Githinji J, McGee J, Garcia-Arocena D, Hagerman RJ, Nolta J, Pessah IN, Hagerman PJ. Signaling defects in iPSC-derived fragile X premutation neurons. Human molecular genetics. 2012;21(17):3795–805. doi: 10.1093/hmg/dds207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Pasca AM, Cord B, Palmer TD, Chikahisa S, Nishino S, Bernstein JA, Hallmayer J, Geschwind DH, Dolmetsch RE. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nature medicine. 2011;17(12):1657–62. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chailangkarn T, Trujillo CA, Freitas BC, Hrvoj-Mihic B, Herai RH, Yu DX, Brown TT, Marchetto MC, Bardy C, McHenry L, Stefanacci L, Jarvinen A, Searcy YM, DeWitt M, Wong W, Lai P, Ard MC, Hanson KL, Romero S, Jacobs B, Dale AM, Dai L, Korenberg JR, Gage FH, Bellugi U, Halgren E, Semendeferi K, Muotri AR. A human neurodevelopmental model for Williams syndrome. Nature. 2016;536(7616):338–43. doi: 10.1038/nature19067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy AL, Meisterernst M, Pognonec P, Roeder RG. Cooperative interaction of an initiator-binding transcription initiation factor and the helix-loop-helix activator USF. Nature. 1991;354(6350):245–8. doi: 10.1038/354245a0. [DOI] [PubMed] [Google Scholar]
- 31.Hirota H, Matsuoka R, Chen XN, Salandanan LS, Lincoln A, Rose FE, Sunahara M, Osawa M, Bellugi U, Korenberg JR. Williams syndrome deficits in visual spatial processing linked to GTF2IRD1 and GTF2I on chromosome 7q11.23. Genetics in medicine : official journal of the American College of Medical Genetics. 2003;5(4):311–21. doi: 10.1097/01.GIM.0000076975.10224.67. [DOI] [PubMed] [Google Scholar]
- 32.Morris CA, Mervis CB, Hobart HH, Gregg RG, Bertrand J, Ensing GJ, Sommer A, Moore CA, Hopkin RJ, Spallone PA, Keating MT, Osborne L, Kimberley KW, Stock AD. GTF2I hemizygosity implicated in mental retardation in Williams syndrome: genotype-phenotype analysis of five families with deletions in the Williams syndrome region. American journal of medical genetics Part A. 2003;123A(1):45–59. doi: 10.1002/ajmg.a.20496. [DOI] [PubMed] [Google Scholar]
- 33.Ferrero GB, Howald C, Micale L, Biamino E, Augello B, Fusco C, Turturo MG, Forzano S, Reymond A, Merla G. An atypical 7q11.23 deletion in a normal IQ Williams-Beuren syndrome patient. European journal of human genetics : EJHG. 2010;18(1):33–8. doi: 10.1038/ejhg.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai L, Bellugi U, Chen XN, Pulst-Korenberg AM, Jarvinen-Pasley A, Tirosh-Wagner T, Eis PS, Graham J, Mills D, Searcy Y, Korenberg JR. Is it Williams syndrome? GTF2IRD1 implicated in visual-spatial construction and GTF2I in sociability revealed by high resolution arrays. American journal of medical genetics Part A. 2009;149A(3):302–14. doi: 10.1002/ajmg.a.32652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antonell A, Del Campo M, Magano LF, Kaufmann L, de la Iglesia JM, Gallastegui F, Flores R, Schweigmann U, Fauth C, Kotzot D, Perez-Jurado LA. Partial 7q11.23 deletions further implicate GTF2I and GTF2IRD1 as the main genes responsible for the Williams-Beuren syndrome neurocognitive profile. Journal of medical genetics. 2010;47(5):312–20. doi: 10.1136/jmg.2009.071712. [DOI] [PubMed] [Google Scholar]
- 36.Edelmann L, Prosnitz A, Pardo S, Bhatt J, Cohen N, Lauriat T, Ouchanov L, Gonzalez PJ, Manghi ER, Bondy P, Esquivel M, Monge S, Delgado MF, Splendore A, Francke U, Burton BK, McInnes LA. An atypical deletion of the Williams-Beuren syndrome interval implicates genes associated with defective visuospatial processing and autism. Journal of medical genetics. 2007;44(2):136–43. doi: 10.1136/jmg.2006.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delgado LM, Gutierrez M, Augello B, Fusco C, Micale L, Merla G, Pastene EA. A 1.3-mb 7q11.23 atypical deletion identified in a cohort of patients with williams-beuren syndrome. Molecular syndromology. 2013;4(3):143–7. doi: 10.1159/000347167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jabbi M, Chen Q, Turner N, Kohn P, White M, Kippenhan JS, Dickinson D, Kolachana B, Mattay V, Weinberger DR, Berman KF. Variation in the Williams syndrome GTF2I gene and anxiety proneness interactively affect prefrontal cortical response to aversive stimuli. Translational psychiatry. 2015;5:e622. doi: 10.1038/tp.2015.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malenfant P, Liu X, Hudson ML, Qiao Y, Hrynchak M, Riendeau N, Hildebrand MJ, Cohen IL, Chudley AE, Forster-Gibson C, Mickelson EC, Rajcan-Separovic E, Lewis ME, Holden JJ. Association of GTF2i in the Williams-Beuren syndrome critical region with autism spectrum disorders. Journal of autism and developmental disorders. 2012;42(7):1459–69. doi: 10.1007/s10803-011-1389-4. [DOI] [PubMed] [Google Scholar]
- 40.Swartz JR, Waller R, Bogdan R, Knodt AR, Sabhlok A, Hyde LW, Hariri AR. A Common Polymorphism in a Williams Syndrome Gene Predicts Amygdala Reactivity and Extraversion in Healthy Adults. Biological psychiatry. 2017;81(3):203–210. doi: 10.1016/j.biopsych.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li HH, Roy M, Kuscuoglu U, Spencer CM, Halm B, Harrison KC, Bayle JH, Splendore A, Ding F, Meltzer LA, Wright E, Paylor R, Deisseroth K, Francke U. Induced chromosome deletions cause hypersociability and other features of Williams-Beuren syndrome in mice. EMBO molecular medicine. 2009;1(1):50–65. doi: 10.1002/emmm.200900003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakurai T, Dorr NP, Takahashi N, McInnes LA, Elder GA, Buxbaum JD. Haploinsufficiency of Gtf2i, a gene deleted in Williams Syndrome, leads to increases in social interactions. Autism research : official journal of the International Society for Autism Research. 2011;4(1):28–39. doi: 10.1002/aur.169. [DOI] [PubMed] [Google Scholar]
- 43.Lucena J, Pezzi S, Aso E, Valero MC, Carreiro C, Dubus P, Sampaio A, Segura M, Barthelemy I, Zindel MY, Sousa N, Barbero JL, Maldonado R, Perez-Jurado LA, Campuzano V. Essential role of the N-terminal region of TFII-I in viability and behavior. BMC medical genetics. 2010;11:61. doi: 10.1186/1471-2350-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mervis CB, Dida J, Lam E, Crawford-Zelli NA, Young EJ, Henderson DR, Onay T, Morris CA, Woodruff-Borden J, Yeomans J, Osborne LR. Duplication of GTF2I results in separation anxiety in mice and humans. American journal of human genetics. 2012;90(6):1064–70. doi: 10.1016/j.ajhg.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borralleras C, Sahun I, Perez-Jurado LA, Campuzano V. Intracisternal Gtf2i Gene Therapy Ameliorates Deficits in Cognition and Synaptic Plasticity of a Mouse Model of Williams-Beuren Syndrome. Molecular therapy : the journal of the American Society of Gene Therapy. 2015;23(11):1691–1699. doi: 10.1038/mt.2015.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheriyath V, Desgranges ZP, Roy AL. c-Src-dependent transcriptional activation of TFII-I. The Journal of biological chemistry. 2002;277(25):22798–805. doi: 10.1074/jbc.M202956200. [DOI] [PubMed] [Google Scholar]
- 47.Sinai L, Ivakine EA, Lam E, Deurloo M, Dida J, Zirngibl RA, Jung C, Aubin JE, Feng ZP, Yeomans J, McInnes RR, Osborne LR, Roder JC. Disruption of Src Is Associated with Phenotypes Related to Williams-Beuren Syndrome and Altered Cellular Localization of TFII-I(1,2) eNeuro. 2015;2(2) doi: 10.1523/ENEURO.0016-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tipney HJ, Hinsley TA, Brass A, Metcalfe K, Donnai D, Tassabehji M. Isolation and characterisation of GTF2IRD2, a novel fusion gene and member of the TFII-I family of transcription factors, deleted in Williams-Beuren syndrome. European journal of human genetics : EJHG. 2004;12(7):551–60. doi: 10.1038/sj.ejhg.5201174. [DOI] [PubMed] [Google Scholar]
- 49.Roy AL, Du H, Gregor PD, Novina CD, Martinez E, Roeder RG. Cloning of an inr- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. The EMBO journal. 1997;16(23):7091–104. doi: 10.1093/emboj/16.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheriyath V, Roy AL. Alternatively spliced isoforms of TFII-I. Complex formation, nuclear translocation, and differential gene regulation. The Journal of biological chemistry. 2000;275(34):26300–8. doi: 10.1074/jbc.M002980200. [DOI] [PubMed] [Google Scholar]
- 51.Perez Jurado LA, Wang YK, Peoples R, Coloma A, Cruces J, Francke U. A duplicated gene in the breakpoint regions of the 7q11.23 Williams-Beuren syndrome deletion encodes the initiator binding protein TFII-I and BAP-135, a phosphorylation target of BTK. Human molecular genetics. 1998;7(3):325–34. doi: 10.1093/hmg/7.3.325. [DOI] [PubMed] [Google Scholar]
- 52.Roy AL. Biochemistry and biology of the inducible multifunctional transcription factor TFII-I. Gene. 2001;274(1–2):1–13. doi: 10.1016/s0378-1119(01)00625-4. [DOI] [PubMed] [Google Scholar]
- 53.Roy AL. Biochemistry and biology of the inducible multifunctional transcription factor TFII-I: 10 years later. Gene. 2012;492(1):32–41. doi: 10.1016/j.gene.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grueneberg DA, Henry RW, Brauer A, Novina CD, Cheriyath V, Roy AL, Gilman M. A multifunctional DNA-binding protein that promotes the formation of serum response factor/homeodomain complexes: identity to TFII-I. Genes & development. 1997;11(19):2482–93. doi: 10.1101/gad.11.19.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hakre S, Tussie-Luna MI, Ashworth T, Novina CD, Settleman J, Sharp PA, Roy AL. Opposing functions of TFII-I spliced isoforms in growth factor-induced gene expression. Molecular cell. 2006;24(2):301–8. doi: 10.1016/j.molcel.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 56.Chimge NO, Makeyev AV, Ruddle FH, Bayarsaihan D. Identification of the TFII-I family target genes in the vertebrate genome. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(26):9006–10. doi: 10.1073/pnas.0803051105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Segura-Puimedon M, Borralleras C, Perez-Jurado LA, Campuzano V. TFII-I regulates target genes in the PI-3K and TGF-beta signaling pathways through a novel DNA binding motif. Gene. 2013;527(2):529–36. doi: 10.1016/j.gene.2013.06.050. [DOI] [PubMed] [Google Scholar]
- 58.Makeyev AV, Bayarsaihan D. Molecular basis of Williams-Beuren syndrome: TFII-I regulated targets involved in craniofacial development. The Cleft palate-craniofacial journal : official publication of the American Cleft Palate-Craniofacial Association. 2011;48(1):109–16. doi: 10.1597/09-093. [DOI] [PubMed] [Google Scholar]
- 59.Caraveo G, van Rossum DB, Patterson RL, Snyder SH, Desiderio S. Action of TFII-I outside the nucleus as an inhibitor of agonist-induced calcium entry. Science. 2006;314(5796):122–5. doi: 10.1126/science.1127815. [DOI] [PubMed] [Google Scholar]
- 60.Hartmann J, Dragicevic E, Adelsberger H, Henning HA, Sumser M, Abramowitz J, Blum R, Dietrich A, Freichel M, Flockerzi V, Birnbaumer L, Konnerth A. TRPC3 channels are required for synaptic transmission and motor coordination. Neuron. 2008;59(3):392–8. doi: 10.1016/j.neuron.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Letavernier E, Rodenas A, Guerrot D, Haymann JP. Williams-Beuren syndrome hypercalcemia: is TRPC3 a novel mediator in calcium homeostasis? Pediatrics. 2012;129(6):e1626–30. doi: 10.1542/peds.2011-2507. [DOI] [PubMed] [Google Scholar]
- 62.Adamo A, Atashpaz S, Germain PL, Zanella M, D'Agostino G, Albertin V, Chenoweth J, Micale L, Fusco C, Unger C, Augello B, Palumbo O, Hamilton B, Carella M, Donti E, Pruneri G, Selicorni A, Biamino E, Prontera P, McKay R, Merla G, Testa G. 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nature genetics. 2015;47(2):132–41. doi: 10.1038/ng.3169. [DOI] [PubMed] [Google Scholar]
- 63.Wang S, Wang B, Pan N, Fu L, Wang C, Song G, An J, Liu Z, Zhu W, Guan Y, Xu ZQ, Chan P, Chen Z, Zhang YA. Differentiation of human induced pluripotent stem cells to mature functional Purkinje neurons. Scientific reports. 2015;5:9232. doi: 10.1038/srep09232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao SY, Hu Y, Chen C, Yuan F, Xu M, Li Q, Fang KH, Chen Y, Liu Y. Enhanced derivation of human pluripotent stem cell-derived cortical glutamatergic neurons by a small molecule. Scientific reports. 2017;7(1):3282. doi: 10.1038/s41598-017-03519-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tcw J, Wang M, Pimenova AA, Bowles KR, Hartley BJ, Lacin E, Machlovi SI, Abdelaal R, Karch CM, Phatnani H, Slesinger PA, Zhang B, Goate AM, Brennand KJ. An Efficient Platform for Astrocyte Differentiation from Human Induced Pluripotent Stem Cells. Stem cell reports. 2017;9(2):600–614. doi: 10.1016/j.stemcr.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ehrlich M, Mozafari S, Glatza M, Starost L, Velychko S, Hallmann AL, Cui QL, Schambach A, Kim KP, Bachelin C, Marteyn A, Hargus G, Johnson RM, Antel J, Sterneckert J, Zaehres H, Scholer HR, Baron-Van Evercooren A, Kuhlmann T. Rapid and efficient generation of oligodendrocytes from human induced pluripotent stem cells using transcription factors. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(11):E2243–E2252. doi: 10.1073/pnas.1614412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimaraes KP, Benazzato C, Almeida N, Pignatari GC, Romero S, Polonio CM, Cunha I, Freitas CL, Brandao WN, Rossato C, Andrade DG, de Faria DP, Garcez AT, Buchpigel CA, Braconi CT, Mendes E, Sall AA, Zanotto PM, Peron JP, Muotri AR, Beltrao-Braga PC. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534(7606):267–71. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Yoon KJ, Jeang W, Lin L, Li Y, Thakor J, Berg DA, Zhang C, Kang E, Chickering M, Nauen D, Ho CY, Wen Z, Christian KM, Shi PY, Maher BJ, Wu H, Jin P, Tang H, Song H, Ming GL. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165(5):1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Danoff SK, Taylor HE, Blackshaw S, Desiderio S. TFII-I, a candidate gene for Williams syndrome cognitive profile: parallels between regional expression in mouse brain and human phenotype. Neuroscience. 2004;123(4):931–8. doi: 10.1016/j.neuroscience.2003.08.038. [DOI] [PubMed] [Google Scholar]
- 70.Huang WC, Young JS, Glitsch MD. Changes in TRPC channel expression during postnatal development of cerebellar neurons. Cell calcium. 2007;42(1):1–10. doi: 10.1016/j.ceca.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 71.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Novina CD, Kumar S, Bajpai U, Cheriyath V, Zhang K, Pillai S, Wortis HH, Roy AL. Regulation of nuclear localization and transcriptional activity of TFII-I by Bruton's tyrosine kinase. Molecular and cellular biology. 1999;19(7):5014–24. doi: 10.1128/mcb.19.7.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang W, Desiderio S. BAP-135, a target for Bruton's tyrosine kinase in response to B cell receptor engagement. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(2):604–9. doi: 10.1073/pnas.94.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sacristan C, Schattgen SA, Berg LJ, Bunnell SC, Roy AL, Rosenstein Y. Characterization of a novel interaction between transcription factor TFII-I and the inducible tyrosine kinase in T cells. European journal of immunology. 2009;39(9):2584–95. doi: 10.1002/eji.200839031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pena-Hernandez R, Marques M, Hilmi K, Zhao T, Saad A, Alaoui-Jamali MA, del Rincon SV, Ashworth T, Roy AL, Emerson BM, Witcher M. Genome-wide targeting of the epigenetic regulatory protein CTCF to gene promoters by the transcription factor TFII-I. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(7):E677–86. doi: 10.1073/pnas.1416674112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ku M, Sokol SY, Wu J, Tussie-Luna MI, Roy AL, Hata A. Positive and negative regulation of the transforming growth factor beta/activin target gene goosecoid by the TFII-I family of transcription factors. Molecular and cellular biology. 2005;25(16):7144–57. doi: 10.1128/MCB.25.16.7144-7157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim DW, Cochran BH. Extracellular signal-regulated kinase binds to TFII-I and regulates its activation of the c-fos promoter. Molecular and cellular biology. 2000;20(4):1140–8. doi: 10.1128/mcb.20.4.1140-1148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim DW, Cochran BH. JAK2 activates TFII-I and regulates its interaction with extracellular signal-regulated kinase. Molecular and cellular biology. 2001;21(10):3387–97. doi: 10.1128/MCB.21.10.3387-3397.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hong M, Lin MY, Huang JM, Baumeister P, Hakre S, Roy AL, Lee AS. Transcriptional regulation of the Grp78 promoter by endoplasmic reticulum stress: role of TFII-I and its tyrosine phosphorylation. The Journal of biological chemistry. 2005;280(17):16821–8. doi: 10.1074/jbc.M413753200. [DOI] [PubMed] [Google Scholar]
- 80.Parker R, Phan T, Baumeister P, Roy B, Cheriyath V, Roy AL, Lee AS. Identification of TFII-I as the endoplasmic reticulum stress response element binding factor ERSF: its autoregulation by stress and interaction with ATF6. Molecular and cellular biology. 2001;21(9):3220–33. doi: 10.1128/MCB.21.9.3220-3233.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Misra UK, Wang F, Pizzo SV. Transcription factor TFII-I causes transcriptional upregulation of GRP78 synthesis in prostate cancer cells. Journal of cellular biochemistry. 2009;106(3):381–9. doi: 10.1002/jcb.22016. [DOI] [PubMed] [Google Scholar]
- 82.Chen J, Malcolm T, Estable MC, Roeder RG, Sadowski I. TFII-I regulates induction of chromosomally integrated human immunodeficiency virus type 1 long terminal repeat in cooperation with USF. Journal of virology. 2005;79(7):4396–406. doi: 10.1128/JVI.79.7.4396-4406.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]