Abstract
Precise control over microbial cell growth conditions could enable detection of minute phenotypic changes, which would improve our understanding of how genotypes are shaped by adaptive selection. Although automated cell-culture systems such as bioreactors offer strict control over liquid culture conditions, they often do not scale to high-throughput or require cumbersome redesign to alter growth conditions. We report the design and validation of eVOLVER, a scalable DIY framework that can be configured to carry out high-throughput growth experiments in molecular evolution, systems biology, and microbiology. We perform high-throughput evolution of yeast across systematically varied population density niches to show how eVOLVER can precisely characterize adaptive niches. We describe growth selection using time-varying temperature programs on a genome-wide yeast knockout library to identify strains with altered sensitivity to changes in temperature magnitude or frequency. Inspired by large-scale integration of electronics and microfluidics, we also demonstrate millifluidic multiplexing modules that enable multiplexed media routing, cleaning, vial-to-vial transfers and automated yeast mating.
INTRODUCTION
Living organisms exist in complex environments that select for evolved phenotypes1–3 (Fig. 1a). In the laboratory, experiments using growth selection in liquid culture have been integral to studying the relationship between genotype and phenotype. These include functional genomic library screening and selection4,5, characterization of natural and synthetic cellular systems6–8, and directed evolution to either study evolutionary processes9–11 or evolve new functions12–15. Development of technologies for precise control of growth conditions has not kept pace with improvements in genotype characterization using sequencing technologies. For example, bioreactors are highly controllable but are low-throughput whereas batch cultures are difficult to control but can be high-throughput. This has hampered understanding how genotypes evolve under selective pressure16 and constrained experimental designs.
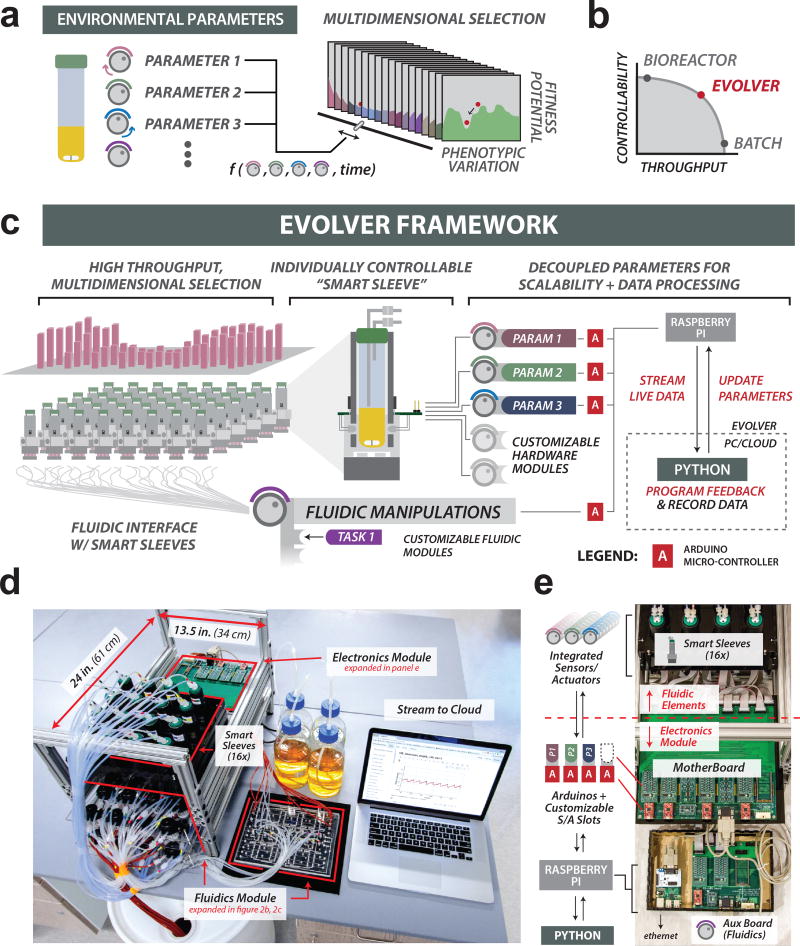
Figure 1. eVOLVER: an integrated framework for high-throughput, automated cell culture.
(a) Understanding how cellular phenotypes arise from multidimensional selection gradients requires multi-parameter control of culture conditions. (b) Growth fitness experiments face a tradeoff between precision control of culture conditions and throughput. eVOLVER enables reliable scaling along both axes. (c) eVOLVER hardware, fluidic, and software modules. System design is modular and synergistic. Left: eVOLVER is designed to scale to high-throughput. Center Top: Smart Sleeve unit. Smart Sleeves integrate sensors and actuators needed to measure and control parameters of individual cultures. Center Bottom: eVOLVER fluidic manipulation system (peristaltic pumps or millifluidic devices) controls movement of media and culture within the system. Right: A modular, scalable hardware architecture interfaces with Smart Sleeve and fluidic modules to achieve individually addressable, real-time culture control. The hardware functions as a bidirectional relay, streaming live data (via Raspberry Pi) collected from each Smart Sleeve to the external computing infrastructure running control software (written in Python). This software records and processes data and returns commands to the hardware in order to update culture parameters. System customization can be achieved by swapping fluidic handling devices, adding new parameter control modules, or programming new feedback control routines between culture and software. (d) 16-culture eVOLVER base unit. Fluidics (media input, waste output) are physically separated from the electronics. The base unit can be cloned and parallelized to increase experimental throughput. (e) eVOLVER hardware architecture. Smart Sleeves communicate with electronics module via a motherboard. Control modules, which control single parameters across for all Smart Sleeves within a 16-culture unit, are composed of Arduino-connected control boards occupying motherboard S/A slots. Arduinos are programmed to interpret and respond to serial commands from the Raspberry Pi, which communicates with software run on a user’s computer or server.
The main challenge to designing growth selection experiments is balancing the tradeoff between control and throughput (Fig. 1b). Batch cultures maintained by serial passage permit parallel testing of many strains or conditions, but are inherently discontinuous and offer limited temporal control17. Although automated cell growth systems can maintain constant growth rates under precisely defined conditions, they are difficult to parallelize due to cost, space and design complexity16–18. The convergence of several open-source technologies—inexpensive additive manufacturing, do-it-yourself (DIY) software/hardware interfaces and cloud computing—has enabled facile fabrication of custom laboratory platforms19–22. Various examples of automated cell culture include devices for automated dilution routines to investigate acquisition of antibiotic resistance23, real-time monitoring of bulk fluorescence7, light-based feedback control of synthetic gene circuits24, and chemostat parallelization25. Unfortunately, the ad hoc design of these systems limits their scalability and restricts their reconfiguration for other experimental purposes.
We present eVOLVER, a DIY framework that offers users the freedom to define the parameters of automated culture growth experiments e.g. temperature, culture density, media composition, and scale them to any size. eVOLVER is constructed using highly modular, open-source wetware, hardware, electronics and web-based software that can be rapidly reconfigured for virtually any type of automated growth experiment. eVOLVER can continuously control and monitor hundreds of individual cultures, collecting, measuring and recording experimental data in real-time, for any timescale. Facile programming of algorithmic culture ‘routines’ is possible, such that feedback between the growing culture and the system couple the status of a culture e.g. high optical density (OD) to its automated manipulation e.g. dilution with fresh media. By combining this programmability with arbitrary throughput scaling, eVOLVER can be used for fine resolution of fitness landscapes, or determination of phenotypes that arise during selection.
We apply eVOLVER to carry out diverse growth and selection experiments. First, we evolve yeast populations in multiple selection conditions at high throughput and measure evolved fitness in several conditions to assess adaptive outcomes. Next, by performing growth selection for a yeast knockout (YKO) library under temporally variable temperature stress conditions, we show that eVOLVER can be used to explore the relationship between environmental fluctuations and adaptive phenotypes. Finally, by integrating millifluidic multiplexing modules, we show that eVOLVER can carry out complex fluidic manipulations, thereby extending the scope and range of possible growth and selection experiments.
RESULTS
Design of an automated cell culture framework
Most automated culture systems are designed to address specialized needs and control just one or more pre-determined culture parameters. We designed eVOLVER so that it could be configured to measure and control an arbitrary set of user-defined parameters. In order to accomplish this, the hardware design enables rapid, cost-effective scaling and customization, including with future technology (Fig. 1c, Supplementary Figs. 1, 2 and 3). eVOLVER hardware comprises three modules (Fig. 1d). First, a customizable Smart Sleeve houses and interfaces with individual culture vessels. Second, a fluidic module controls movement of liquid in and out of each culture vessel, and third, the modular hardware infrastructure simplifies high-volume bidirectional data flow by decoupling each parameter into individual microcontrollers (Fig. 1e). A detailed description of eVOLVER design and construction can be found in Supplementary Notes 1–11 (available online at fynchbio.com).
The Smart Sleeve mediates monitoring and control of growing cultures (Fig. 2a). Each sleeve is composed of a machined aluminum tube (for temperature control), printed circuit board (PCB) mounted sensors, actuators, and other electronic components, all attached to a custom 3D printed mount. The Smart Sleeve can be readily mass-produced for high-throughput experiments, or reconfigured to meet custom experimental needs, such as larger culture volumes, pH or oxygen sensors for bioprocess applications, or light emitting diodes (LEDs) for optogenetic studies. In this article we use a Smart Sleeve configuration that accommodates 40 mL glass vials, and is configured to control three experimental parameters: stirring, temperature, and culture density (Fig. 2a, Supplementary Figs. 4, 5, 6, 7 and 8).
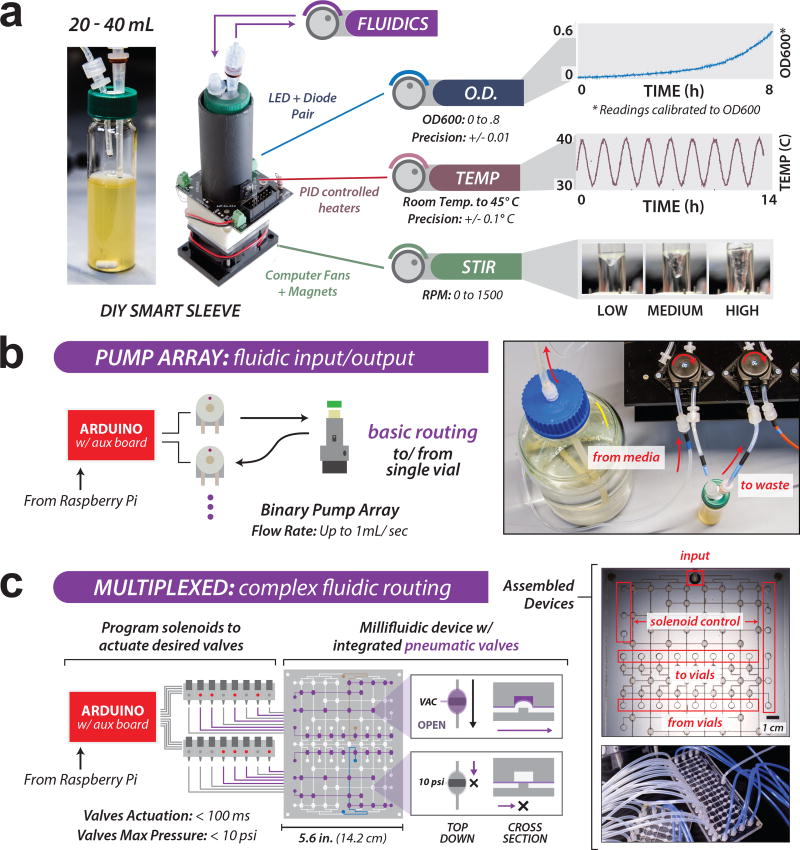
Figure 2. Design and performance of eVOLVER modules.
(a) Generalizable configuration of Smart Sleeves for continuous culture; control of fluidic input/output, optical density, temperature, and stir rate. Left: Smart Sleeves are designed to accommodate 40 mL autoclavable borosilicate glass vials. Efflux straw length determines culture volume. Center: Smart Sleeve integrated electronic components. LED/photodiode sensor pairs perform OD900 readings. Thermistors and heaters attached to a machined aluminum tube maintain PID temperature control. Magnet-attached computer fans rotate stir bars inside the vials. Components are wired to a PCB and mounted on an inexpensive 3D printed chassis. Individual sleeves cost ~$25 and can be assembled in ~10 minutes. Right: Specifications of Smart Sleeve parameters: optical density, temperature, and stirring. Device measurement precision varies with experimental conditions (e.g. cell type, room temperature) but can be adjusted to achieve necessary precision and range (e.g. tuning temperature PID constants, or filtering OD measurements) (see Supplementary Note 4). Reported values are typical for experiments described in Figs. 3–4. Calibration may be performed as often as desired, though settings are largely invariant over thousands of hours of use. (b) "Basic" fluidic handling in eVOLVER utilizes pumps with fixed flow rates of ~1 mL/sec and can be actuated with a precision of ~100 ms. (c) Millifluidic multiplexing devices enable novel, customized liquid routing. Devices are fabricated by bonding a silicone membrane between two plastic layers with laser-etched flow channels. Integrated pneumatic valves actuate on the membrane to direct fluidic routing from media input to output ports (to or from vials).
eVOLVER’s fluidic module controls movement of media, culture, and liquid reagents in the system (Figs. 2b, 2c and Supplementary Fig. 9). It can be configured to two fluidic handling modes: either a basic mode, which uses peristaltic pumping to control media influx and efflux for each culture26 (Fig. 2b), or a complex mode in which multiplexed routing enables sophisticated fluidic manipulations (Fig. 2c). User-actuated peristaltic pumps are robust, simple to use, and can be scaled to carry out media dilution routines for multiple parallel continuous cultures25,26 (Supplementary Fig. 10). However, they scale poorly to more sophisticated routing where the number of required connections and control elements expands non-linearly with the number of cultures. Our complex fluidic mode applies principles of large-scale integration (LSI) to overcome this limitation. Originally developed for electronic devices, and then adopted in microfluidics27,28, LSI uses combinatorial multiplexing to expand the number of input-output paths per control channel29,30. Inspired by LSI we created physically-compact millifluidic multiplexing devices by adhering a silicone rubber membrane between two clear sheets of laser-etched plastic, each patterned with desired channel geometries and aligned to form an intact device (Fig. 2c, Supplementary Figs. 11). Devices can be designed as needed to carry out custom fluidic protocols, including complex media dispensing routines, transfer of liquid between cultures, or periodic cleaning protocols to maintain sterility. Multiplex device design and fabrication protocols are presented in Supplementary Note 6.
We developed a hardware and software infrastructure for eVOLVER that complements the Smart Sleeve and fluidic modules (Fig. 1c). Our design uses a central organizing PCB (motherboard) that controls core functionalities e.g. serial communication, signal routing, and contains slots for plug-in boards that manage specialized features e.g. graphics cards (Fig. 1e, Supplementary Figs. 1, 2 and 3). The control modules—custom PCBs wired to Arduino microcontrollers—read and power Smart Sleeve-mounted components. During an experiment, individual control modules manage each culture parameter e.g. temperature, stirring. A motherboard distributes control of up to 4 independent parameters across a set of sixteen Smart Sleeves, comprising a single eVOLVER base unit (Fig. 1d). Microcontrollers associated with each base unit are coordinated by a single Raspberry Pi, which is a small, low-cost, single-board computer that serves as a bidirectional relay to a user’s computer or a cloud server (Figs. 1c, 1e). The modularity of this design facilitates repurposing and scaling; users can easily modify or augment eVOLVER’s experimental capability by connecting new control modules, while additional base-units can easily be cloned to achieve higher throughput (Figs. 1d and Supplementary Fig. 12).
An important feature of eVOLVER’s design is the use of network connectivity to coordinate and run experiments over the internet. eVOLVER’s distributed hardware architecture enables efficient transmission of large packets of high-dimensional, real-time data (Figs. 1c, 1e) such that a single computer, located anywhere with an internet connection, can monitor hundreds of cultures in real time (Supplementary Fig. 3). Python scripts manage the acquired data and execute the control algorithms that define a selection scheme.
For example, during a typical data acquisition/control protocol, a scripted routine can query the Raspberry Pi every 30 seconds for Smart Sleeve-acquired culture status data e.g. temperature or optical density (Figs. 1c, 1e). Recorded data is converted into higher-order data e.g. growth rate by an algorithm and used to update individual Smart Sleeves or fluidic channels with new settings e.g. adjusted temperature or media dilution. By modifying scripts, a user can change selection criteria (Supplementary Fig. 13) or specify a selection pressure design by subtle iterations of the same algorithm across multiple Smart Sleeves.
The eVOLVER system presented in this article can run long-term (250+ h) experiments without electronic or software failure (Fig. 3 and Supplementary Fig. 14). Most components have a lifetime of >4000 h, so hardware replacement is unlikely during most experiments. Hardware calibration can be done as much as needed but we found that settings were unaltered during dozens of experiments that comprised more than 1700 h of operation (Supplementary Figs. 4, 6, 8, and 10). eVOLVER is designed to withstand liquid spills, with Smart Sleeves and fluidic components physically separated from control hardware (Fig. 1e). Additionally, the system can be set up to avoid microbial contamination. In a control experiment, we found that the system remained sterile for a ~10 day experiment while incubating uninoculated, antibiotic-free media alongside passaged Escherichia coli cultures (Supplementary Fig. 14).
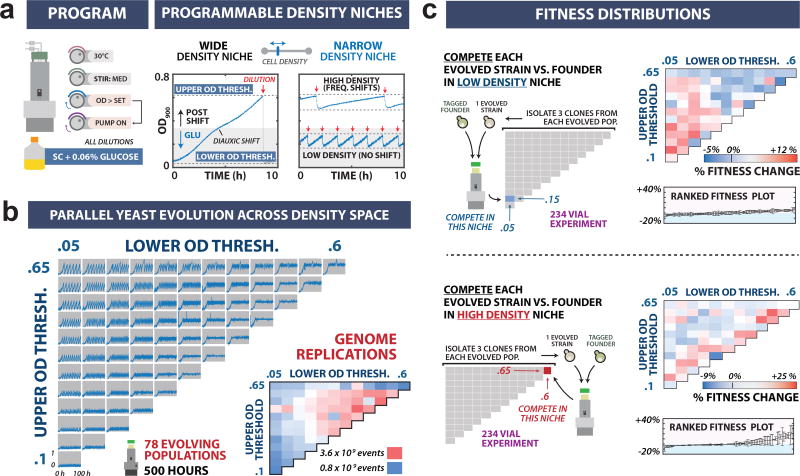
Figure 3. High-throughput experimental evolution across a multidimensional selection space.
(a) Programming eVOLVER to maintain culture density selection routines during yeast evolution. Left: eVOLVER was configured to maintain cultures within defined density niches using a feedback between OD measurements and dilution events (turbidostat function). Right: Representative growth traces for yeast (Saccharomyces cerevisiae FL100) cultures growing under wide and narrow density niches. For each culture, the programmed OD window determines population size, and the consequent dilution rate and diauxic shift frequency. (b) Parallel evolution of 78 yeast populations in distinct density niches. Culture OD traces are shown for populations evolved for 500 h in density windows with varied lower (0.05–0.6) and upper (0.1–0.65) OD thresholds. Lower right: Heat map of the estimated genome replication events for the 78 populations. Values were calculated by multiplying average number of cells by the number of doublings, both estimated through segmentation of the OD trace. (c) Fitness distributions of evolved strains. Three clones from each evolved population were competed against the ancestral strain under low-density (OD 0.05–0.15, top) and high-density (OD 0.60–0.65, bottom) growth regimes. Right: Heat maps for mean fitness change relative to the ancestor (top) and ranked fitness with standard error bars representing competitive fitness for each clone (bottom).
Experimental yeast evolution
We first configured eVOLVER to function as a turbidostat in order to study the relationship between culture density and fitness in yeast populations. Culture density was maintained in a constant, defined window bounded by lower and upper OD thresholds (ODlower threshold – ODupper threshold). Using continuously recorded OD data (Supplementary Figs. 7 and 8), an algorithm activates dilution when the upper threshold is exceeded. Dilution continues until the lower threshold is reached (Fig. 3a). Population growth rates are calculated in real-time by segmenting and fitting the OD trace between dilution events. We varied the upper and lower OD thresholds across 78 yeast populations grown in parallel, thereby defining a two-dimensional selection space based on minimum and maximum culture density (Fig. 3b) (Online Methods, Supplementary Note 12). Cultures were grown in glucose-limited media for 500 h (40–280 generations depending on the culture), resulting in diauxic shifts with OD windows that exceed ~0.35 (Supplementary Fig. 15). The frequency with which the culture experiences the shift is a function of density-dependent selection prescribed to individual cultures (Fig. 3b). OD measurements allowed us to generate maps of population and evolution parameters across the selection space, including population growth rates (Supplementary Fig. 16) and average genome replication events as a function of OD and growth rate (Fig. 3b and Supplementary Fig. 17).
We used eVOLVER to evaluate fitness parameters of isolates from each of the evolved populations by competing isolated strains against a fluorescently-labelled ancestor strain at low- or high-density continuous growth conditions (468 total cultures), assaying for population ratio over time using flow cytometry31 (Online Methods). The fitness distributions in low- and high-density growth conditions were distinct (Fig 3c). K-means analysis of the fitness phenotypes yielded three distinct groups: low-density specialists, high-density specialists, and the remainder which have low fitness in both measured niches (Supplementary Fig. 18). Low-density specialists evolved from most of the cultures maintained at lower OD thresholds (Fig. 3c top, Supplementary Fig. 18). In contrast, high-density specialists were isolated only from cultures maintained within narrow, high-OD windows (Fig. 3c bottom, Supplementary Fig. 18). These results demonstrate that by systematically varying of the features of a selective environment, eVOLVER can be used to precisely resolve adaptive niches.
Selection under temporally varying pressure regimes
There is growing interest in understanding how temporal changes in parameters of an environment shape adaptation32. Fluctuating conditions may yield adaptations distinct from those selected under monotonic pressure33,34. Prior studies often either use highly controlled microfluidic systems35 to study a few strains at a time, or tedious bulk techniques36 for limited perturbation on a pooled population of thousands of strains. eVOLVER can provide both control and throughput, enabling temporal changes of one selective pressure on a large population (>107 cells) while holding other culture conditions constant. To demonstrate this, we carried out growth selection experiments on a pooled YKO library4,5,37 (5,149 unique members) under conditions in which a single environmental variable—temperature—was temporally varied. A two-dimensional selection space was programmed by varying the magnitude and period of square wave temperature oscillations (Fig. 4a, 4b). Cultures were maintained for 6 days in turbidostat mode, and samples collected every 48 h (Online Methods and Supplementary Note 13). At the conclusion of the experiment, next-generation sequencing was performed on each selected population to determine library member frequency38,39, which was used to calculate the fitness of each member31 (Online Methods, Supplementary Figs. 19–21, and Supplementary Note 13).
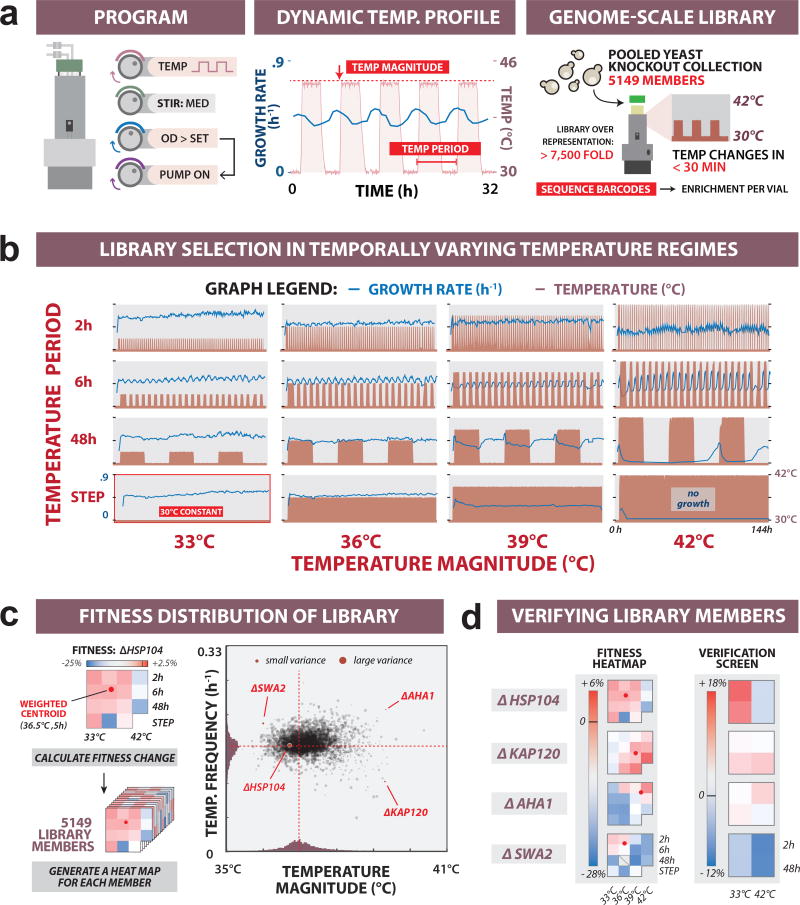
Figure 4. Genome scale library fitness under temporally varying selection pressure.
(a) Programming temporally varying temperature regimes. Left: eVOLVER configuration for conducting turbidostat experiments (OD window: 0.15–0.2) under fluctuating temperature stress. Middle: Snapshot of temperature waveform (red) alternating between 30°C and 39°C on a 6 h period, and corresponding culture growth rate (blue). Right: Parallel cultures of pooled YKO collection were grown. Selection-based enrichment of library members was quantified at various timepoints using next-generation sequencing. (b) Full set of dynamic temperature regimes. Temperature magnitudes (33°C, 36°C, 39°C or 42°C) were varied against periods (2h, 6h, or 48h, or a constant step), and run against a 30°C control culture. Recorded temperature (red) is plotted with culture growth rates calculated between dilutions (blue). (c) Mapping fitness of library members to dynamic selection space. Left: For each library member, fitness heat maps were generated in each selection regime, and used to calculate weighted fitness centroids within temperature magnitude/frequency coordinate space. Right: Scatter plot of fitness centroids for the full library. (d) Validation of library selection. Four strains with distinct profiles were chosen for verification and competed against a neutral control strain (ΔHO) under four different temporal selection regimes. Population ratios were measured using quantitative PCR.
We computed a 2D weighted centroid for each library member by transforming its measured fitness into coordinates of temperature magnitude and frequency, which allowed us to compare the library phenotypes across the 2D temporal selection space (Fig. 4c, Supplementary Fig. 22 and Supplementary Note 13). To justify the centroid as a representative phenotypic metric, we picked four library members from different regions of the distribution (ΔHSP104, ΔKAP120, ΔAHA1, and ΔSWA2) for experimental validation in four temperature selection conditions. We also chose a control strain (ΔHO) whose centroid is near the population mean, suggesting the knockout had no adaptive value. Fitness measurements for these strains in pairwise competition against ΔHO were consistent with their centroid positions from the pooled library experiment (Fig. 4d), supporting our use of the fitness centroid as an appropriate phenotypic metric. To compare with prior literature, fitness centroids of previously annotated gene sets40 were then statistically compared to the fitness centroid distribution of all KO members. We expected and observed gene sets associated with relevant annotations, like heat sensitivity, to be significantly skewed from the population mean (Supplementary Fig. 23). Similar analysis on individual KO strains identified several chaperone and chaperone cofactor genes, which are known to have a role in response to thermal stress41 (Supplementary Fig. 24).
To characterize similarities between environments, we used cross-correlation and principle component analysis on the library composition over time to quantify the degree to which different conditions bring about the same fitness changes in the KO library. We observed three distinct groupings: First, a high-temperature, high-frequency group was composed of 42°C fluctuations with a period of ≤ 6 h. Second, a high-temperature low-frequency group was composed of 42°C fluctuation with 24 h period and constant 39°C conditions. Finally, a mild temperature group was composed of all remaining conditions, with temperatures ≤ 39°C (Supplementary Figs. 25 and 26). We identified gene ontology (GO) term annotations that are linked to fitness defects in one or more of these groups (Supplementary Fig. 26). We observed that genes known to affect growth rate e.g. mitochondrial function, ribosome biogenesis, significantly altered fitness in the mild temperature group. Interestingly, ribosome components and processing factors also showed high-frequency sensitivity at 42°C, supporting a potential role for ribosome biogenesis in transitions in and out of stress42. We further analyzed potential sources of frequency-dependence. We found that the high- and low-frequency groups were characterized by annotations associated with cell cycle checkpoints e.g. DNA damage response, organelle fission which regulate cellular processes over time and therefore might be expected to affect cellular response to fluctuating stresses at different frequencies.
Automated Culturing with Fluidic Multiplexing Devices
In order to show the potential of the fluidic multiplexing framework to automate movement of reagents and cells in eVOLVER, we designed devices for three experiments (Supplementary Note 14). These included dynamic media mixing during continuous culture to track yeast response to ratios of sugars, preventing bacterial biofilm formation by automated passaging, and programming sexual reproduction between adapting yeast populations.
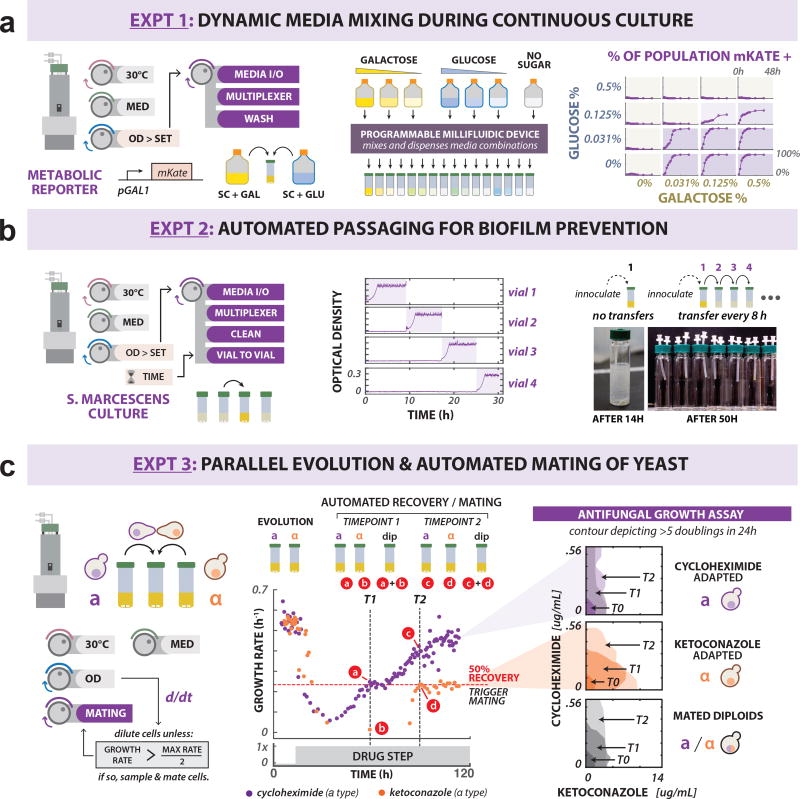
We show that fluidic multiplexing can be used to manage media composition for multiple cultures maintained by eVOLVER by constructing an 8-channel media selector device that draws media from multiple input sources and dispenses a defined mixture to a culture of choice (Fig. 2c, Supplementary Figs. 27 and 28). Galactose utilization in yeast is regulated by the ratiometric sensing of available galactose and glucose43. We maintained and tracked a yeast strain harboring a galactose-inducible fluorescent reporter (pGAL1-mKate2) in continuous cultures featuring 16 different sugar compositions (Supplementary Figs. 29, 30, and Supplementary Note 15). Dyed media, tracking relative sugar levels in each culture, confirmed that the device can dynamically mix and dispense in correct ratios for extended periods of time (Supplementary Fig. 29). We used flow cytometry measurements to track galactose-induced population fractions, confirming maintenance of sugar-dependent gene induction (Fig. 5a).
Figure 5. Integrated millifluidic devices enable scaling of complex fluidic manipulation.
(a) Demonstrating dynamic media mixing in continuous culture. Left: eVOLVER program for maintaining cells in turbidostat mode using millifluidic device to mix and dispense appropriate dilution volumes. A yeast galactose-inducible reporter (pGAL1-mKate2) was used to validate the device by maintaining cultures in turbidostat mode at different ratios of glucose and galactose. Center: Any combination of seven media inputs can be mixed and dispensed into any of the 16 culture vessels. Right: Reporter induction (by population percentage) for 16 cultures containing different glucose: galactose ratios, as measured by flow cytometry. (b) Preventing biofilm formation with automated vial-to-vial transfers. Left: A millifluidic device can enable inter-culture transfers between any of the 16 cultures. Center: Serratia marcescens cultures were maintained in turbidostat mode, with culture transfer events triggered every 8 h. Right: A culture maintained in a single vessel forms a thick biofilm after 14 h, while automated transfer prevents visible biofilm formation. (c) Using millifluidic devices to automate yeast mating. Left: Haploid strains containing opposite mating types are maintained as turbidostat cultures under antifungal selection. Vial-to-vial transfers are triggered by growth rate feedback control, used to sample haploids and form diploids within the device using an automated mating protocol. Center: Growth rate of haploid cells evolved under cyclohexamide (CHX, 0.2 ug/mL, purple) or ketoconazole (KETO, 6 ug/mL, orange) selection was monitored continuously following drug exposure. Once growth rates of either drug-evolved culture equal 50% of the wild-type growth rate under no selection, automated mating and transfer is carried out. This was performed at two timepoints: t1=68.7 h and t2 = 98.1 h. Right: Antifungal resistance was assayed for recovered haploid and diploid populations. Contours correspond to an antifungal concentration range in which at least five generations of growth were observed in 24 h (on average).
The utility of the millifluidic system for mediating liquid transfer between cultures was demonstrated using a device that overcomes biofilm formation during long-term continuous growth experiments (Supplementary Figs. 28, 31, and Supplementary Note 16). Serratia marcescens44, a bacterial species that readily forms biofilm on glass surfaces, was grown in turbidostat mode. Using a vial-to-vial transfer device, inoculation of a fresh vial was performed at 8 h intervals, followed by automated sterilization of culture-exposed fluidic paths (Fig. 5b). While thick biofilm deposition was observed after only 14 h in a culture grown continuously in a single vial (no transfers), no deposition was observed in vials maintained under the transfer routine (Fig. 5b). With daily replacement of used vials, four Smart Sleeves were used to passage a culture, biofilm-free, for an indefinite period of time (Supplementary Fig. 32).
Finally, we demonstrate that multiple functionalities can be combined to automate complex fluidic handling routines. We designed a fluidic routine for automated sexual reproduction of adapting yeast populations by integrating multiplexed media selection, vial-to-vial transfer, and post-transfer device cleaning (Supplementary Figs. 27, 28, 31 and 33). This routine was used to conduct mating between separate, opposite mating-type haploid yeast evolving under different antifungal selections: cyclohexamide (CHX) and ketoconazole (KETO) (Fig. 5c and Supplementary Note 17). Rather than manually sampling at arbitrary timepoints, once haploid cultures reached user-defined growth rate milestones, automatic sampling and mating were carried out on the device (Supplementary Figs. 33 and 34). A minimum inhibitory concentration (MIC) assay performed on both diploid and parental populations indicated that the haploid-evolved CHX resistance was transferred to diploids in a dominant manner (Fig. 5c, Supplementary Figs. 35 and 36). Conversely, the general antifungal resistance that emerged under KETO selection appeared to be recessive. Further sequencing of KETO-evolved haploid strains revealed nonsense mutations in ERG3 (Supplementary Fig. 37), which has been shown to confer recessive resistance to azole antifungals45.
DISCUSSION
We report the development of eVOLVER, which is a DIY framework for automated cell growth experiments. Our design is customizable and provides researchers with the ability to design, build and share both experimental configurations and data (Figs. 1–2). With straightforward modifications, eVOLVER can be reconfigured to conduct any of the recently reported continuous growth studies (Supplementary Table 1), and can replace batch culture techniques used in several recent experimental evolution studies9,11,32,46. It is straightforward to add hardware components to the platform as they become available. For example, integration with open-source pipetting robots would automate culture sampling, thereby enabling fluorescence-activated cell sorting (FACS) for assaying gene expression, or droplet microfluidics for single-cell studies. The configuration we report in this article is designed for well-mixed liquid cultures, but eVOLVER can be adapted for the coordination of multiple arrayed sensors to capture spatial distributions in static liquid cultures or phototrophic cultures. We have reported use of eVOLVER for growing lab-adapted suspension cultures of bacteria and yeast. We note that whilst it is feasible to use our system for growing mammalian cell lines, additional attention to sterility and removal of residual cleaning agents would be needed, and bead/matrix systems might be required for adherent cells.
The multiple possible configurations enable precise specification of individual culture environments. By systematically co-varying parameters, eVOLVER can be used to investigate cellular fitness along multidimensional environmental gradients, potentially allowing for experimental decoupling of overlapping selection pressures. The ability to arbitrarily program feedback control between culture conditions and fluidic functions allows the user to algorithmically define highly specialized environmental niches.
We showcased the versatility of our system using an experimental evolution study (Fig. 3) of yeast in 78 different culture density windows. We then generated fitness distributions by testing fitness of evolved clones in low- and high-density niches, identifying low- and high-density specialists. Interestingly, high-density specialists were most often derived from evolution in narrow OD windows. Since the prescribed culture density windows are related to the frequency of diauxic shift at limiting glucose concentrations47, it is interesting to speculate that these strains selected for metabolic programs that facilitate rapid metabolic shifts. Further work is needed to confirm that the differences observed in the fitness distributions are relevant. For example, a baseline fitness distribution generated from a large number of replicate evolutions could be used to rule out the possibility that stochastic events dominate the observed fitness differences. Additionally, comparing the fitness of whole evolved populations in each condition could help isolate true adaptation from variation observed due to clonal differences. Finally, assaying fitness in additional niches is required to determine how well fitness distributions correlate with the assayed niche, as well as to ascertain the existence of generalists.
In a second experiment, we assessed growth fitness of a yeast genome-wide KO library under systematically varied temperature fluctuations, demonstrating that eVOLVER could be used to extract unique fitness information across a temporally varied selection surface (Fig. 4). In a similar approach—using sub-lethal stress administered under temporally diverse selection conditions—selection experiments could be conducted for other types of libraries at a resolution beyond what is available for standard growth selection schemes4,38,39. Using this approach, algorithms with additional parameters could be used to perform fitness and selection experiments investigating biological phenomenon associated with temporal adaptation, such as bet-hedging, noise and transcriptional feedback.
Accurate fluidic manipulation is a requirement of continuous culture automation, but past approaches to fluidic routing have been tedious to customize and difficult to control, imposing limitations on experimental design. We presented a method to build and control millifluidic devices that plugs into the eVOLVER framework for programmatic routing of fluids during continuous culture. To highlight the utility and robustness of the devices, we performed three experimental demonstrations: sophisticated fluidic mixing and dispensation, vial-to-vial transfers, and integration of multiple devices for more complex culture routines (Fig. 5). These experiments illustrate the potential of custom millifluidics with this platform.
In the future we hope that our framework could be applied in studying contributions of individual species to community fitness in microbial consortia, designing synthetic circuits that minimize fitness costs to the host cell3, identifying circuit designs for producer strains to maximize stability over time in industrial bioreactors48, or in optimization of synthetic microbial genomes49,50. We hope that eVOLVER will serve as a democratic platform for research by a broad community of users to build, execute and share experiments.
ONLINE METHODS
General methods and analysis used throughout the study are described here. A detailed description of eVOLVER device design can be found in Supplementary Notes 1–11. For a more complete description of experiments depicted in Figs. 3–5, we refer the reader to Supplementary Notes 12–17, which includes detailed methods, results, and data analysis.
Strain Construction
Genotypes for yeast and bacterial strains used in this study are listed in Supplementary Table 2. Plasmids were constructed using standard molecular biology techniques. Strains were generated using standard lithium acetate transformation. When appropriate, non-isogenic pooled population samples were given unique designations for clarity (Supplementary Table 2).
Routine Cell Culture Techniques
Culture conditions varied according to the needs of particular experiments (see methods in Supplementary Notes 10 – 17). Cultures used to seed eVOLVER experiments were prepared as follows: Saccharomyces cerevisiae (obtained from frozen stock or single colonies) was grown in 2 mL of YPD media (2% glucose) at 30°C in a shaking incubator (300 rpm) for at least 36 h. For routine overnight culture of Escherichia coli or Serratia marcescens, cells obtained from frozen stock were used to inoculate 2 mL of LB Miller broth grown at 37°C for 12 h in a shaking incubator (300 rpm).
Flow Cytometry
Flow cytometry was used to measure single-cell fluorescence throughout the study. Prior to measurement, 200 uL of yeast culture (see methods for experiment-specific growth conditions) was diluted with 100 uL of filter-sterilized PBS supplemented with cyclohexamide to a final concentration of 20 ug/mL, then incubated at 4°C in the dark for no less than 3 h to allow for fluorophore maturation. An Attune NxT Flow Cytometer (Invitrogen) equipped with an autosampler was used to acquire data. For a typical experiment, at least 10,000 events were acquired. Cells were analyzed using FlowJo (Treestar Software). Intact cells were gated using forward and side scatter, followed by gating on fluorescence channels (green and/or red, as appropriate) to determine the fractional distribution of each population.
Fitness Calculations
Competitive fitness, in which a strain of interest is co-cultured in competition with a reference strain, was assayed in the same fashion throughout the study. The ratio of the two strains was determined at multiple timepoints – generally at the beginning and end of an experiment – either by flow cytometry or qPCR. Fitness values, F, were calculated using the following equation31:
where t is number of generations, and n and nr are cell counts for the strains of interest and reference strain, respectively.
Setup Procedure for eVOLVER Experiments
Prior to each eVOLVER experiment, 40 mL borosilicate glass vials outfitted with a stir bar and capped with an influx port and an efflux port were sterilized by autoclave. Media and waste lines were sterilized by pumping 10% bleach (20 mL), followed by 70% ethanol (20 mL). Lines were cleared by pumping air for 20 s, followed by media (20 mL). Media and waste lines were then attached to the influx and efflux ports of each vial, and each vial filled by pumping 25 mL of the appropriate media through the influx port. At this point, Python control code was initiated, triggering a blank media measurement, and activating Smart sleeve heating elements. Prior to seeding with cells, fresh media was incubated in the device for a length of time sufficient for the first 15 optical density recordings to be taken (~2.5 min) and for media to reach the programmed temperature.
Extracting Yeast Genomic DNA
To extract genomic DNA, ~2×106 yeast cells (roughly 30 µL of overnight culture) were pelleted by centrifugation (5 min, 1000 rcf). Supernatant was removed, and pellets were resuspended in 30 µL 0.2% sodium dodecyl sulfate (SDS), followed by vortexing for 15 s. Suspensions were transferred to PCR tubes and heated in a thermal cycler (37°C) for 5 min, followed by 98°C for 5 min before cooling to 4°C. Extracts were diluted with H2O to a final volume of 75 µL prior to being used as PCR template. Primers used in this study are listed in Supplementary Table 3.
Library Preparation and Barcode Sequencing
Libraries for next-generation sequencing were prepared by PCR-amplification of genomic DNA (LightCycler 480 Instrument II, Roche), purification (Zymo Research). Primers used for indexing (culture, timepoint) and sequencing adapters were added via PCR. NextSeq sequencing (Harvard Biopolymers Facility) was used to sequence the culture index, the timepoint index, and a 55 bp single end read of the barcode construct. PhiX was added at 50% to increase sequence diversity. Alignment was performed using custom code harnessing MATLAB's Bioinformatics Toolbox and Boston University's parallel computing cluster. Alignment scores were calculated using the Smith-Waterman algorithm (swalign function) and assigned based on best score above a minimum threshold. In total, we assigned more than 244 million reads to 5149 unique library members to track frequency across the 4 time points for each of the 16 conditions. A more detailed description of the library preparation and sequencing can be found in Supplementary Note 13.
DATA AVALIBILITY
Data supporting these studies are available upon reasonable request.
Supplementary Material
Acknowledgments
We are grateful to B. Stafford for his assistance in design architecture of the system. We thank H. Khalil, A. Soltanianzadeh, A. Sun, S. Pipe, and A. Cavale for help on construction of the system. We are indebted to the Electronics Design Facility (EDF) and Engineering Product Innovation Center (EPIC) at Boston University for their services. We also thank D. Segrè, J. Ngo, J. Tytell, W. Wong, and members of the Khalil Lab for insightful comments on the manuscript. This work was supported by a NSF CAREER Award (MCB-1350949 to A.S.K.) and a DARPA grant (HR0011-15-C-0091 to A.S.K.). A.S.K. also acknowledges funding from the NIH Director’s New Innovator Award (1DP2AI131083-01), DARPA Young Faculty Award (D16AP00142), and NSF Expeditions in Computing (CCF-1522074).
Footnotes
AUTHOR CONTRIBUTIONS
B.G.W, C.J.B, and A.S.K. conceived the study. B.G.W. developed the system with guidance and input from all authors. B.G.W and C.P.M. performed and analyzed experiments. S.K. generated reagents. C.J.B. and A.S.K. oversaw the study. All authors wrote the paper.
COMPETING FINANCIAL INTERESTS STATEMENT
Authors have declared that no competing interests exist.
References
- 1.Elena SF, Lenski RE. Microbial genetics: Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 2.Nichols RJ, et al. Phenotypic Landscape of a Bacterial Cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nevozhay D, Adams RM, Van Itallie E, Bennett MR, Balázsi G. Mapping the Environmental Fitness Landscape of a Synthetic Gene Circuit. PLoS Comput. Biol. 2012;8:e1002480. doi: 10.1371/journal.pcbi.1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 5.Li Z, et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat. Biotechnol. 2011;29:361–7. doi: 10.1038/nbt.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuleta IA, Aranda-Díaz A, Li H, El-Samad H. Dynamic characterization of growth and gene expression using high-throughput automated flow cytometry. Nat. Methods. 2014;11:443–8. doi: 10.1038/nmeth.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi CN, Miller AW, Ekness F, Dunham MJ, Klavins E. A low cost, customizable turbidostat for use in synthetic circuit characterization. ACS Synth. Biol. 2015;4:32–38. doi: 10.1021/sb500165g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feist AM, Herrgård MJ, Thiele I, Reed JL, Palsson BØ. Reconstruction of biochemical networks in microorganisms. Nat. Rev. Microbiol. 2008;7:129–143. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lang GI, et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 2013;500:571–574. doi: 10.1038/nature12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maddamsetti R, et al. Adaptation, Clonal Interference, and Frequency-Dependent Interactions in a Long-Term Evolution Experiment with Escherichia coli. Genetics. 2015;200:619–31. doi: 10.1534/genetics.115.176677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yona AH, et al. Chromosomal duplication is a transient evolutionary solution to stress. Proc. Natl. Acad. Sci. U.S.A. 2012;109:21010–5. doi: 10.1073/pnas.1211150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499–503. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravikumar A, Arrieta A, Liu CC. An orthogonal DNA replication system in yeast. Nat. Chem. Biol. 2014;10:175–177. doi: 10.1038/nchembio.1439. [DOI] [PubMed] [Google Scholar]
- 15.Crook N, et al. In vivo continuous evolution of genes and pathways in yeast. Nat. Commun. 2016;7:13051. doi: 10.1038/ncomms13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bull AT. The renaissance of continuous culture in the post-genomics age. J. Ind. Microbiol. Biotechnol. 2010;37:993–1021. doi: 10.1007/s10295-010-0816-4. [DOI] [PubMed] [Google Scholar]
- 17.Gresham D, Dunham MJ. The enduring utility of continuous culturing in experimental evolution. Genomics. 2014;104:399–405. doi: 10.1016/j.ygeno.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piper MDW, et al. Reproducibility of oligonucleotide microarray transcriptome analyses. An interlaboratory comparison using chemostat cultures of Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:37001–37008. doi: 10.1074/jbc.M204490200. [DOI] [PubMed] [Google Scholar]
- 19.Cressey D. The DIY electronics transforming research. Nature. 2017;544:125–126. doi: 10.1038/544125a. [DOI] [PubMed] [Google Scholar]
- 20.Kong DS, et al. Open-source, community-driven microfluidics with Metafluidics. Nat. Biotechnol. 2017;35:523–529. doi: 10.1038/nbt.3873. [DOI] [PubMed] [Google Scholar]
- 21.Adamo A, et al. On-demand continuous-flow production of pharmaceuticals in a compact, reconfigurable system. Science. 2016;352:61–7. doi: 10.1126/science.aaf1337. [DOI] [PubMed] [Google Scholar]
- 22.Perkel JM. The Internet of Things comes to the lab. Nature. 2017;542:125–126. doi: 10.1038/542125a. [DOI] [PubMed] [Google Scholar]
- 23.Toprak E, et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet. 2011;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milias-Argeitis A, Rullan M, Aoki SK, Buchmann P, Khammash M. Automated optogenetic feedback control for precise and robust regulation of gene expression and cell growth. Nat. Commun. 2016;7:12546. doi: 10.1038/ncomms12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hope EA, et al. Experimental Evolution Reveals Favored Adaptive Routes to Cell Aggregation in Yeast. Genetics. 2017;206:1153–1167. doi: 10.1534/genetics.116.198895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toprak E, et al. Building a morbidostat: an automated continuous-culture device for studying bacterial drug resistance under dynamically sustained drug inhibition. Nat. Protoc. 2013;8:555–567. doi: 10.1038/nprot.nprot.2013.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002;298:580–4. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 28.Melin J, Quake SR. Microfluidic Large-Scale Integration: The Evolution of Design Rules for Biological Automation. Annu. Rev. Biophys. Biomol. Struct. 2007;36:213–231. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- 29.Unger MA. Monolithic Microfabricated Valves and Pumps by Multilayer Soft Lithography. Science (80-.) 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 30.Grover WH, Skelley AM, Liu CN, Lagally ET, Mathies RA. Monolithic membrane valves and diaphragm pumps for practical large-scale integration into glass microfluidic devices. Sensors Actuators, B Chem. 2003;89:315–323. [Google Scholar]
- 31.Kryazhimskiy S, Rice DP, Jerison ER, Desai MM. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science (80-.) 2014;344:1519–1522. doi: 10.1126/science.1250939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell A, et al. Adaptive prediction of environmental changes by microorganisms. Nature. 2009;460:220–224. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- 33.Ketola T, et al. Fluctuating Temperature Leads To Evolution Of Thermal Generalism And Preadaptation To Novel Environments. Evolution (N. Y) 2013;67:2936–2944. doi: 10.1111/evo.12148. [DOI] [PubMed] [Google Scholar]
- 34.Sæther B-E, Engen S. The concept of fitness in fluctuating environments. Trends Ecol. Evol. 2015;30:273–281. doi: 10.1016/j.tree.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Bennett MR, et al. Metabolic gene regulation in a dynamically changing environment. Nature. 2008;454:1119–1122. doi: 10.1038/nature07211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salignon J, Richard M, Fulcrand E, Yvert G. Genomics of cellular proliferation under periodic stress. 2017:1–17. doi: 10.15252/msb.20177823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giaever G, Nislow C. The yeast deletion collection: A decade of functional genomics. Genetics. 2014;197:451–465. doi: 10.1534/genetics.114.161620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith AM, et al. Quantitative phenotyping via deep barcode sequencing. Genome Res. 2009;19:1836–1842. doi: 10.1101/gr.093955.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibney PA, Lu C, Caudy AA, Hess DC, Botstein D. Yeast metabolic and signaling genes are required for heat-shock survival and have little overlap with the heat-induced genes. Proc. Natl. Acad. Sci. 2013;110:E4393–E4402. doi: 10.1073/pnas.1318100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cherry JM, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morano KA, et al. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics. 2012;190:1157–95. doi: 10.1534/genetics.111.128033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loar JW, et al. Genetic and biochemical interactions among Yar1, Ltv1 and RpS3 define novel links between environmental stress and ribosome biogenesis in Saccharomyces cerevisiae. Genetics. 2004;168:1877–1889. doi: 10.1534/genetics.104.032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Escalante-Chong R, et al. Galactose metabolic genes in yeast respond to a ratio of galactose and glucose. Proc. Natl. Acad. Sci. U.S.A. 2015;112:1636–41. doi: 10.1073/pnas.1418058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rice S, et al. Biofilm Formation and Sloughing in Serratia marcescens Are Controlled by Quorum Sensing and Nutrient Cues. J. Bacteriol. 2005;187:3477–3485. doi: 10.1128/JB.187.10.3477-3485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson JB, Sirjusingh C, Ricker N. Haploidy, diploidy and evolution of antifungal drug resistance in Saccharomyces cerevisiae. Genetics. 2004;168:1915–23. doi: 10.1534/genetics.104.033266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.González C, et al. Stress-response balance drives the evolution of a network module and its host genome. Mol. Syst. Biol. 2015;11:827. doi: 10.15252/msb.20156185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brauer MJ, Saldanha AJ, Dolinski K, Botstein D. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol. Biol. Cell. 2005;16:2503–17. doi: 10.1091/mbc.E04-11-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12678–83. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hutchison CA, et al. Design and synthesis of a minimal bacterial genome. Science (80-.) 2016;351:aad6253–aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 50.Richardson SM, et al. Design of a synthetic yeast genome. Science (80-.) 2017;355:1040–1044. doi: 10.1126/science.aaf4557. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.