Abstract
Background & Aims
Barrett’s esophagus (BE) is the greatest risk factor for esophageal adenocarcinoma, but only a small proportion of patients with BE develop cancer. Biomarkers might be able to identify patients at highest risk of progression. We investigated genomic differences in surveillance biopsies collected from patients whose BE subsequently progressed compared to patients whose disease did not progress.
Methods
We performed a retrospective case–control study of 24 patients with BE that progressed to high-grade dysplasia (HGD, n=14) or esophageal adenocarcinoma (EAC, n=10). The control group (n=73, called non-progressors) comprised patients with BE and least 5 years of total endoscopic biopsy surveillance without progression to HGD or EAC. From each patient, we selected a single tissue sample obtained more than 1 year before progression (cases) or more than 2 years before the end of follow up (controls). Pathogenic mutations, gene copy numbers, and ploidy were compared between samples from progressors and non-progressors.
Results
TP53 mutations were detected in 46% of samples from progressors and 5% of non-progressors. In this case–control sample set, TP53 mutations in BE tissues increased the adjusted risk of progression 13.8-fold (95% CI, 3.2–61.0) (P<.001). We did not observe significant differences in ploidy or copy number profile between groups. We identified 147 pathogenic mutations in 57 distinct genes—the average number of pathogenic mutations was higher in samples from progressors (2.5) than non-progressors (1.2) (P<.001). TP53 and other somatic mutations were recurrently detected in samples with limited copy number changes (aneuploidy).
Conclusions
In genomic analyses of BE tissues from patients with or without later progression to HGD or EAC, we found significantly higher numbers of TP53 mutations in BE from patients with subsequent progression. These mutations were frequently detected before the onset of dysplasia or substantial changes in copy number.
Keywords: esophageal cancer, preneoplastic, esophagus
INTRODUCTION
Esophageal adenocarcinoma (EAC) is a devastating disease with an average five-year survival of only 15%.1 Moreover, rates of EAC in the U.S. population have increased >450% in recent decades.2 EAC is believed to arise from Barrett’s esophagus (BE), intestinal metaplasia of the lower esophagus that forms in response to reflux injury. The prevalence of BE at the gastroesophageal junction has been estimated to be around 1.6 to 11% of adults, putting millions at heightened risk of EAC.3,4 However, the annual incidence of progression among patients with BE remains low, estimated at ≤0.33% in a recent meta-analysis.5 This disparity between the large number of patients with BE and the modest risk of progression to advanced disease poses challenges to screening and surveillance paradigms. Guidelines recommend those with non-dysplastic BE (NDBE) undergo surveillance endoscopy and biopsies every three to five years.6 Standard screening is reliant upon identification of histologic dysplasia, leading to selection of patients for more intense screening, ablation or even surgical esophagectomy. One limitation of this approach is substantial inter-observer variability between pathologists in histological grading of BE, especially to diagnose low-grade dysplasia (LGD).7,8 Current strategies also require a large population to undergo frequent, invasive and costly endoscopic procedures. Given all of these challenges, there is a clear need for improved understanding of the biology of disease progression and development of predictive markers that can identify which among the large population of patients with BE are at elevated risk of progression.
Previous studies have detected chromosomal alterations and aneuploidy9,10 or methylation of key tumor suppressor genes11 in BE samples obtained before the development of high-grade dysplasia (HGD) or EAC, suggesting that acquisition of aneuploidy or methylation to be a potential biomarkers for risk stratification. In addition, there have been previous comparisons of the genomic features of NDBE to dysplasia and cancer.12–15 Many of these studies, however, did not examine the progression history of their BE patients or even excluded NDBE patients who later progressed12,13,16,17, making it impossible to distinguish early high-risk genomic features that may be key drivers and early biomarkers of progression.
The tumor suppressor p53 has been identified as a risk factor for progression in BE, most commonly when aberrant p53 expression is detected via immunohistochemistry (IHC).18–21 p53 testing has often been evaluated as a marker to assist with the pathologic diagnosis of HGD. While several of these studies have suggested promising results, p53 IHC is not recommended for routine use in the evaluation of BE biopsies owing in part to limited prospective data, the lack of clear guidelines for the interpretation of p53 staining, and lack of guidelines for integrating the results with dysplasia grading.6,22 There have also been a multitude of studies looking at TP53 mutations in BE and EAC.10,17,23–26 However, most of these studies either looked at BE adjacent to an already formed EAC or BE biopsies without clinical follow-up. Although results have not always been consistent, most studies have suggested TP53 mutations are absent or rare in NDBE.16,17,23,24 For example, Weaver et al performed targeted next-generation sequencing on a collection of NDBE samples from patients with stable, non-progressive disease and found TP53 mutations in only 2.5%.16 No studies have looked at potential differences in specific mutations of TP53 found in BE patients or the broader genomic context (other genomic alterations) in which the TP53 mutations arose. Compared to many previously used techniques to study genomics of BE, modern next-generation sequencing technologies now have the ability to detect somatic mutations at lower allelic fraction than previously feasible and to jointly assess mutations and somatic copy-number aberrations. These technologies allow a more thorough and accurate analysis of BE tissues, especially when using small clinical FFPE biopsies that may have a limited amount of BE epithelium present.
Here we report a case–control study evaluating genomic alterations that are associated with subsequent progression to HGD or EAC. We utilize routine clinical samples obtained at least one year prior to progression in cases and controls that are matched on clinical risk factors (BE segment length and pre-progression dysplasia history). We identified several genomic alterations in BE samples that are enriched in patients who later progress to HGD or EAC. In addition, by performing paired analysis of mutations and copy-number alterations in these preneoplastic samples, we demonstrated that these mutations commonly occur in the absence of substantial aneuploidy. Together, these lead to revisions of the current models of progression of BE to EAC and underscore the ultimate potential for genomic testing to facilitate risk stratification of patients with non-dysplastic BE.
METHODS
Patient and Sample Selection
After IRB approval, patients for our study set were identified from a retrospective cohort with endoscopic biopsies for surveillance of BE performed at one of 4 endoscopy centers within the University of Pittsburgh Medical Center (UPMC) system. All patients had histologically confirmed intestinal metaplasia. For detailed patient and sample selection process and comparison to reference populations, please see the Supplementary Methods, Supplementary Table 1 and 2.
A formalin-fixed, paraffin-embedded (FFPE) block was chosen that represented the highest grade of neoplasia (NDBE, IFD, or LGD) originally diagnosed up to that point in surveillance for 83/97 patients. In the remaining 14 patients, a secondary block had to be chosen due to lack of availability of the initially selected block (sample selection is detailed in Supplementary Table 2 and Supplementary Figure 1 and 2).
For our validation cohort who were subjected to p53 IHC validation, a collection of NDBE biopsies from 16 patients (29 samples) with no history of dysplasia but subsequently progressed to HGD or EAC and 28 consecutive NDBE patients (44 samples) with no known subsequent dysplasia were collected from the archives of Miraca Life Sciences.
Histopathologic Review
In our study set, ten to 15 serial 5 micron sections were cut from each paraffin block. One was stained with hematoxylin and eosin and jointly reviewed by three gastrointestinal pathologists (RO, JH, and AA) with expertise in BE using a multi-headed microscope to provide a histologic diagnosis for each sample. In the 3 cases where there was disagreement between the three pathologists, the majority (two out of three) diagnosis was taken (Supplementary Table 2). Pathologists were blinded to original diagnosis, progression status, and sequencing results.
Clinical Data
Patient’s age, sex, BE segment length, surveillance biopsy and resection diagnoses, history of ablative and surgical treatment and clinical follow-up after the diagnosis of EAC was recorded. After selection, individual patients were given a unique study number and patient demographics were blinded to all investigators with JD holding the key. For this study, the follow up interval was calculated from the date of the endoscopic procedure on which the sample was originally obtained to the date of first diagnosis of HGD or EAC (progressors) or date of last surveillance biopsy or first ablation procedure (non-progressors).
P53 Immunohistochemistry
On samples with available tissue sections, one was prepared for p53 immunohistochemistry. A single pathologist (JD (study set) and MR (validation set)) reviewed all p53 immunostains in conjunction with a hematoxylin and eosin stained slide blinded to mutation and outcome data. Aberrant expression was defined as strong nuclear overexpression or complete lack of expression in at least 1 glandular complex (pit or crypt and associated glands) with or without surface expression (Supplementary Methods).
Statistical Analysis
Differences in categorical variables were analyzed with Fisher’s exact test (two-tailed P value) while differences in continuous variables were evaluated with the Mann-Whitney U Test. P<0.05 was considered significant. Statistical analysis was performed with IBM SPSS (version 22).
Sequencing
For full sequencing and analysis details, see supplemental methods. After DNA isolation and fragmentation, libraries were constructed and underwent hybrid capture for the target genes (Supplementary Table 3) and sequencing. Mutations, copy number variants, ploidy, and genomic doubling was determined in each sample. To identify any possible TP53 mutations missed by MuTect, sequence reads (blinded to outcome status) were manually reviewed around the region of TP53. To remove known germline polymorphisms and likely passenger events, all called variants were filtered to retain only likely pathogenic events (see Supplementary Methods). A similar approach has been successfully used in a clinical setting when testing tumor samples for somatic alterations.27,28
Data Availability
All sequencing results analyzed during this study are included in this published article. As all samples were only sequenced on a small targeted platform without normal control, mutation calls from MuTect and GATK are listed in Supplementary Table 4.
RESULTS
Patient selection
For our study set, we collected archival samples from 24 patients who were under routine BE surveillance and later progressed to HGD (n=14) or EAC (n=10) more than 1 year after their index BE diagnosis. The included cases were representative of the population of incident progressors as demonstrated by their similarity to progressors not used in the study with respect to age, sex, BE segment length and history of IFD and LGD (Supplementary Table 1).
We collected samples from a 73-patient control group with > 2 years follow-up after the tested sample, and at least 5 years of total endoscopic biopsy surveillance without progression to HGD or EAC. Because IFD and LGD is prevalent among progressors, we selected controls matched with cases on history dysplasia based upon the original clinical diagnosis. Consequently, selected controls were more likely to have a diagnosis of IFD or LGD, more likely to have long segment BE and more often male than a typical unselected reference population of BE patients with at least 5 years of total endoscopic surveillance and no evidence of progression (Supplementary Table 1).
Clinical and histological characteristics
Selected cases and controls were similar with respect to clinical diagnosis of IFD or LGD during surveillance, BE segment length and sex, but cases were almost 6 years older (67.5 vs. 61.6 years) on average than controls (Table 1). As expected, in the non-progressor control group there was a significantly longer mean duration of follow-up (6.7 vs. 3.3 years, P<0.001) and greater mean number of follow-up endoscopies (5 vs 3, P<0.001).
Table 1.
Patient/Sample characteristics
| Non-progressor N=73 | Progressor N=24 | p-value6 | |
|---|---|---|---|
| Patient age, mean (range) | 61.6 (41.1–81.3) | 67.5 (52.1–85.3) | 0.002 |
| Sex, n (%) | 0.172 | ||
| Female | 21 (28.8) | 3 (12.5) | |
| Male | 52 (71.2) | 21 (87.5) | |
| Barrett’s segment length, n (%) | 0.811 | ||
| Irregular z-line | 4 (5.5) | 0 (0) | |
| Short segment (<3 cm) | 35 (47.9) | 11 (45.8) | |
| Long segment (≥3 cm) | 29 (39.7) | 11 (45.8) | |
| Unknown | 5 (6.8) | 2 (8.3) | |
| Highest originally diagnosed dysplasia grade1 | 0.293 | ||
| Negative | 21 (28.8) | 10 (41.7) | |
| Indefinite | 31 (42.5) | 6 (25) | |
| Low grade | 21 (28.8) | 8 (33.3) | |
| Consensus sample diagnosis, n (%)2 | 1.0 | ||
| Negative | 66 (90.4) | 21 (87.5) | |
| Indefinite | 2 (2.7) | 1 (4.2) | |
| Low grade | 5 (6.8) | 2 (8.3) | |
| Pre-progression endoscopic ablation treatment3 | N/A | ||
| No | 65 (89.0) | 24 (100) | |
| Yes | 8 (11.0) | 0 (0) | |
| Years of follow-up, mean (range)4 | 6.7 (2.4–13.8) | 3.3 (1.4–9.0) | <0.001 |
| No. of follow-up surveillance exams, mean (range)5 | 5 (1–18) | 3 (1–7) | <0.001 |
Highest grade of dysplasia originally diagnosed in the UPMC system during surveillance prior to progression up to and including the sample submitted for sequencing.
Consensus diagnosis of 3 gastrointestinal pathologists on review of the analyzed biopsy sample (blinded to outcomes and NGS analysis)
Ablation treatment prior to a diagnosis of HGD or EAC (end of follow up was time of ablation)
Time from sample biopsy date to end of follow up
Number of endoscopic exams with biopsy after the sample biopsy date to end of follow-up
Mann-Whitney U Test for continuous variables; Fisher’s exact test for categorical variables.
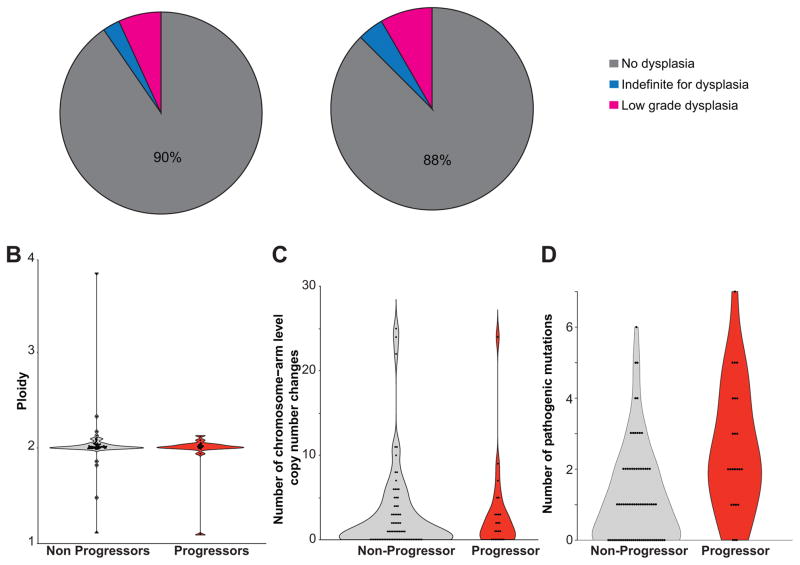
We selected a single archival paraffin block for each patient, representing the highest grade of dysplasia (NDBE, IFD, or LGD) originally diagnosed at that pre-progression time point (see Methods, Supplementary Table 2 and Supplementary Figure 1). We subjected all samples to blinded expert review by three gastrointestinal pathologists to obtain a consensus or (2/3) majority diagnosis for each tissue sample. Consistent with other studies where cases of IFD/LGD are subjected to pathologic review, our review panel downgraded the histopathologic diagnosis of the majority of samples with initial diagnosis of IFD/LGD, both among the progressing and non-progressing patients.8,29 Of the 97 samples, all 42 originally diagnosed as NDBE were confirmed on review. Of the 55 samples originally diagnosed as IFD or LGD, 45 were downgraded to NDBE and 10 were confirmed as IFD/LGD (Supplementary Table 2 and Supplementary Figure 2). In sum, 87.5% of progressor samples and 90.4% of control samples were assessed by consensus review to be NDBE and there was no difference in the proportion of samples with consensus IFD or LGD (P=1.000; Fig. 1a and Table 1). The estimated mean fraction of BE cells in the tissue samples was identical between progressors (39.8%) and non-progressors (39.9%).
Figure 1.
Comparison of Barrett’s esophagus samples with and without subsequent progression. (a) Pie charts displaying the histologic diagnosis of each sample after blinded, consensus histologic review by three gastrointestinal pathologists. (b) Violin plot showing the inferred ploidy of each sample. (c) Violin plot showing number of chromosomal or chromosomal arm level copy number changes. (d) Violin plot showing the number of pathogenic mutations.
Sequencing
We performed NGS of 243 genes commonly altered in EAC (Supplementary Table 3, Supplementary Figure 3) and additional loci throughout the genome to facilitate copy-number analysis. Average mean targeted coverage for this cohort was ~160x. All samples used for analysis contained at least 30x coverage over at least 80% of the targeted genome. The number of identified mutations did not correlate with the percentage of BE cells in the non-progressing samples (Spearman R=0.20). Additionally, pathogenic mutations were identified in samples with the lowest percentage of BE cells, as gauged by pathology review and computational analysis, confirming the ability to perform genomic analysis in clinical BE biopsies.
Ploidy and copy number alterations
As others have reported that major changes in ploidy and chromosomal copy-number can be identified in Barrett’s tissue within 24–48 months before the development of EAC 10,14, we first evaluated somatic copy number alterations using the ABSOLUTE computational algorithm.30 We found no significant difference in the average ploidy of progressors vs. non-progressors (Fig. 1b, Supplementary Table 5). Whole chromosome or arm level copy number changes were found in 16 (67%) progressors and 42 (58%) of non-progressors (P=0.48) (Fig. 1c, Supplementary Figure 4, Supplementary Table 5, 6). We identified four samples with evidence of genome doubling (two non-progressors and two progressors), Supplementary Table 5. Excepting these samples, the number of arm or chromosomal copy number events (mean 2.2 for progressors and 2.4 for non-progressors, P=0.81) and total copy number events (mean 5.3 for progressors and 5.7 for non-progressors, P=0.77) in each sample was low. Focal homozygous deletions in tumor suppressor genes were limited to the CDKN2A/CDKN2B locus, present in 14 (19%) non-progressors and 2 (8%) progressors. We identified no evidence of focal amplifications in EAC oncogenes. Overall, the ploidy and copy number profile of our samples showed no discernable differences between progressors and non-progressors, suggesting these mainly NDBE samples from progressors were taken before the development of large-scale copy number alterations shown to occur shortly before progression to EAC.14
Pathogenic mutation burden
We next asked if there was a difference in the relative burden of pathogenic mutations in histologically identical samples from progressors and non-progressors. As matched germline samples were not available, we filtered the data to enrich for likely pathogenic alterations by removing common germline variants and mutations not identified in other cancer sequencing studies (see supplementary methods). Following filtering, we identified 147 pathogenic mutations across 57 distinct genes with the average burden of pathogenic mutations higher in progressors than in non-progressors (2.5 vs. 1.2, P<0.001; Fig. 1d, Supplementary Table 7).
Differentially altered genes
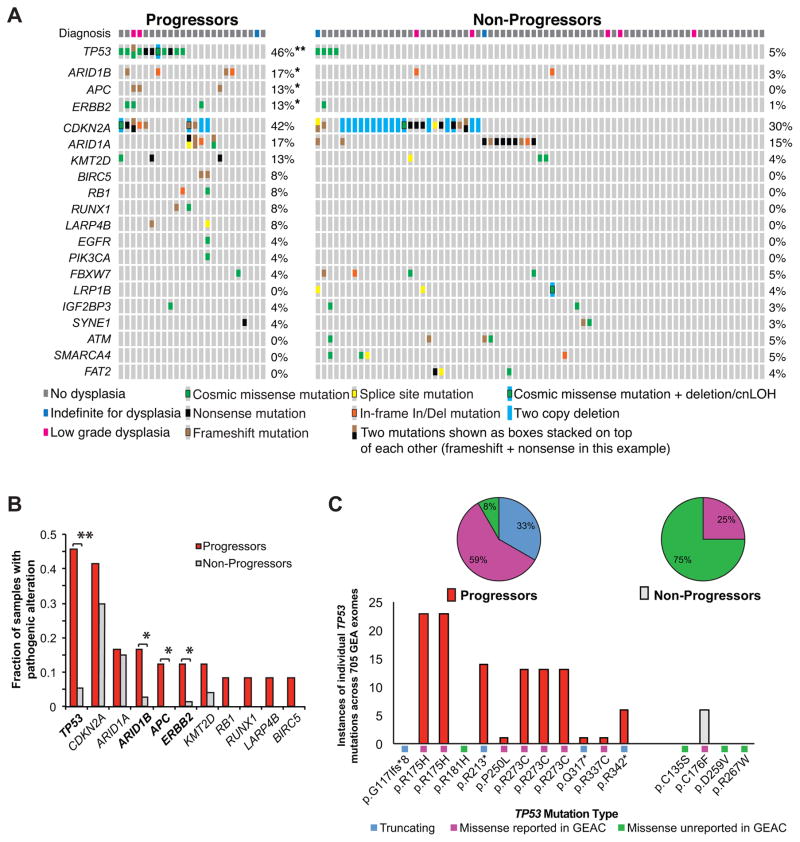
We next asked if individual genes were differentially altered in the progressors and non-progressors (Fig. 2a, b, Supplementary Table 7). Both populations harbored high rates of CDKN2A alterations and ARID1A mutations. Three of four progressors with ARID1A mutations had two mutations, suggesting biallelic inactivation, while none of the controls with ARID1A mutations had more than one alteration identified. We also identified several cancer-associated genes with higher somatic mutation rates in progressing patients. Most strikingly, we identified TP53 mutations in 11 of 24 progressors (46%) but only 4 (5%) in the non-progressors (P<0.0001), adjusted OR 13.8 (95% CI 3.2–60.5). Of the 15 samples with TP53 mutations, the expert histologic diagnosis was NDBE in 12 (80%), IFD in 1 (7%), and LGD in 2 (13%). There was no significant association between dysplasia grade (NDBE, IFD, LGD) and TP53 mutation status (Supplementary Table 8).
Figure 2.
Recurrent alterations in progressors and non-progressors. (a) Diagram displaying the identified pathogenic alterations in each sample along with the histologic diagnosis (top). Each patient is represented by a column and genes with 3 or more alterations or genes with enriched alterations within progressors are shown. (b) Fraction of samples within the progressor and non-progressor cohorts that contained a pathogenic alteration in the given gene. **:P<0.0001 *:P<0.05. (c) Breakdown of identified TP53 mutations. Bar graph representing the specific number of times the identified TP53 mutation was previously reported in 705 gastroesophageal adenocarcinomas (GEA) of the TCGA (CBioPortal) database (bottom). The percentage of TP53 mutations that were truncating, missense mutations previously reported in GEA sequencing studies, and missense mutations not previously reported in GEA (top).
As histologic state was downgraded for many samples, we also evaluated TP53 mutation status using the original diagnosis (Supplementary Table 2). In the 13 progressors whose original diagnosis was NDBE, 6 (46%) contained a TP53 mutation compared to 1 of 29 (3.4%) of the non-progressors originally diagnosed as NDBE (Fisher exact, 2 tail P=0.0019). There was also a significant difference in the samples originally diagnosed as LGD, 67% vs. 7% (P=0.014) and a trend for IFD samples (20% vs. 7%) for progressors and non-progressors, respectively. Additionally, in a few cases, the highest-grade block (LGD or IFD) was unavailable (see methods). We therefore evaluated the rates of TP53 mutation based upon the patient’s highest-grade diagnosis up to the time the tested sample was taken, regardless of the sample analyzed (progressors by definition had a HGD or EAC diagnosis at a later time point). Among patients diagnosed exclusively with NDBE during clinical pathology evaluation, 5 of 10 (50%) progressors but only 1 of 21 (4.8%) non-progressors possessed TP53 mutations, P= 0.008. Among those patients with a diagnosis of IFD, TP53 mutations were found in 2 of 6 (33.3%) of progressors and 2 of 31 (6.5%) of non-progressors; P=0.115. Among patients with a diagnosis of LGD, TP53 mutations were found in 4 of 8 (50%) progressors and 1 of 21 (4.8%) non-progressors; p =0.0123.
Further inspection indicated that TP53 amino acid changes induced by mutations in progressors may be more pathogenic than those in the non-progressors. Four of 11 progressors possessed truncating TP53 mutations and 2 of 11 progressors either had a second TP53 mutation or loss of heterozygosity (LOH), while none of the controls had TP53 LOH or truncating events. In addition, we compared the TP53 mutations found in our cohort to those detected across exome sequencing of 705 gastroesophageal adenocarcinomas (GEA). Only one of the four mutations in the non-progressors was previously reported in gastroesophageal adenocarcinoma. In contrast 10 of 11 progressors with a TP53 mutation had mutations previously reported. Among the progressing patients we identified two R175H and three R273C mutations, both highly recurrent across human cancer (Fig. 2c).
Beyond TP53, several known GEA tumor suppressors and oncogenes (ARID1B, APC, ERBB2, RB1, RUNX1, LARP4B, and BIRC5) had more frequent mutations in progressors (Fig 2b, Supplementary Table 7). Among these, ARID1B, APC, and ERBB2 were significantly enriched in the progressors (P<0.05). In addition, when we compared the pre-progression samples (cases) from those who progressed to a HGD endpoint and those who progressed to a cancer endpoint, there was no difference in ploidy, number of chromosomal changes, percent of samples with TP53 mutations, or number of pathogenic mutations (Supplementary Table 9).
Analysis of subsequent progression samples
In 6 patients who progressed to EAC or HDG, the subsequent progression lesion was also available for sequencing. In 5 of these patients, the pre-progression sample contained pathogenic mutations. Overall, 11/20 mutations found in the pre-progression sample were also identified in the progression lesion (Supplementary Table 10). Two of the 5 patients had a single TP53 mutation in the pre-progression BE sample, with both mutations also found at progression. One pre-progression BE sample, patient #20 with LGD, had two TP53 mutations, p.G117Ifs*8 and R175H. The progression sample had the p.G117Ifs*8 mutation but not the R175H, suggesting that sampled area of LGD had additional genomic evolution following the branching off of the region of BE that led directly to subsequent progression. More broadly, our finding that the same TP53 mutations in pre-progression samples were also found at progression indicates that these mutations were not merely a sign of a field of tissue at higher risk of somatic mutations. Instead, these mutations directly mark the precursor lesions to more advanced disease.
p53 immunohistochemistry
We also evaluated p53 IHC on 57 sequenced samples from our study set with sufficient tissue for evaluation. Five of 7 (71.4%) TP53 mutant samples compared to 3 of 50 (6%) samples without a detectable TP53 mutation had abnormal p53 overexpression by IHC (Supplementary Fig. 5), indicating a strong but imperfect correlation between the two assays (P<0.001). Similar to TP53 sequencing, abnormal p53 IHC was enriched in the progressor group, detected in 4 of 9 (44.4%) evaluable progressors compared to 4 of 47 (8.5%) evaluable non-progressors. In patients with results from both assays, 5 of 9 (56%) evaluable progressors had either TP53 mutation or abnormal p53 expression (n=3, mutation+ IHC+; n=1, mutation+ IHC−; n=1, mutation− IHC+), compared to 5 of 47 (10.6%) evaluable non-progressors (n=2, mutation+ IHC+; n=1, mutation+ IHC−; n=2, mutation− IHC+).
p53 immunohistochemistry of an independent cohort
To evaluate our findings in an independent validation cohort, we collected another set of NDBE biopsies in patients with no history of dysplasia but subsequent progression to cancer or high-grade dysplasia (29 blocks from 16 patients) and 28 consecutive NDBE patients (44 total blocks) with no subsequent dysplasia as controls. As many of these samples’ tissue blocks were older and had limited amount of tissue sections, we could not obtain sufficient quality DNA for genomic sequencing and instead focused on evaluating p53 status via IHC. In the progressors, 11/16 (69%) patients had an abnormal p53 IHC pattern. When we evaluated all NDBE biopsies from these 16 patients, 14/29 (48%) had an abnormal p53 IHC result. In the controls samples, by contrast, only 2/28 (7.1%) patients and 3/44(6.8%) biopsies had a mutant IHC pattern (Supplementary Table 11). This discrepancy (P<0.0001 (patients) and P=0.0001 (total biopsies)) in p53 aberrations is consistent with our genomic results, validating that TP53 abnormalities in NDBE can mark patients who subsequently progress.
DISCUSSION
Our finding of highly recurrent, pathogenic TP53 mutations in predominantly non-dysplastic surveillance biopsies taken up to 9 years prior to a diagnosis of HGD or EAC challenges the prevailing models of BE progression in which these mutations are thought to occur contemporaneously with high-grade dysplasia or cancer. These results likely differ from those in the past as studies that failed to identify recurrent TP53 mutations in NDBE either did not selectively evaluate patients with subsequent progression or used older, less sensitive genomic assays.16
There are however, previous reports that support our findings of TP53 mutations in BE from progressors yet being rare in similar tissue from non-progressors. Several studies that performed multi-region sequencing noted TP53 mutations in NDBE adjacent to invasive EAC in surgical resection specimens26,31,32 and in biopsies adjacent to HGD or EAC.33,34 Del Portillo et al, utilized a targeted sequencing panel without paired normal, similar to our technique, to identify mutations in NDBE from biopsies or endoscopic mucosal resections in eight patients with concurrent HGD/EAC or in 13 patients with no history of progression. They found TP53 mutations in 5 of 8 (63%) NDBE samples with concurrent HGD or EAC but none in the non-progressors. Similarly, we previous reported TP53 mutations in 5 of 7 (71%) NDBE samples adjacent to EAC.31,34 Additional p53 IHC and FISH studies are also supportive of our data. Timmer et al showed that 13/22 patients that progressed had a p53 abnormality by IHC or FISH, though they also found 97/406 non-progressing patients to have an abnormality by these technologies.35 Importantly, this study specifically excluded LGD samples, thus also showing TP53 alterations can occur in non-dysplastic samples. For non-progressors, Weaver et al sequenced 40 NDBE samples and only identified one TP53 mutation, which supports our finding of a very low rate of TP53 mutations in NDBE from non-progressors.16 Additionally, rereview of data from Dr. Reid’s group show that 15/27 (56%) of progressors and 18/216 (8%) of non-progressing BE patients had a detectable TP53 mutation. A limitation of these latter data is a lack of refined histological breakdown of the BE samples. Finally, a recently published meta-analysis of biomarkers predicting risk of malignant progression in Barrett’s esophagus calculated TP53 mutation had a sensitivity of 60% and specificity of 82%, diagnostic odds ratio of 10.23.36 Together, even though many of these prior studies used less sensitive assays, the results are consistent with our data.
Among the most notable of our findings are the clear presence of TP53 mutations in NDBE prior to progression. The fact that a number of the cases had their histologic state downgraded during pathologic review can complicate the relationship between grade and mutation status. In routine clinical practice, diagnosis of IFD and LGD is commonly downgraded after re-review, as suggested by numerous studies.37–39 However, our rate of downgrading samples was higher than what has been seen in some studies. Here, 30/35 (86%) of IFD samples and 16/20 (80%) LGD samples were downgraded. While most studies only looked at LGD, Sonwalker et al, found that over 80% of IFD diagnoses were downgraded.37 For low grade dysplasia, Curvers et al found 85% of LGD samples were downgraded (a rate actually higher than ours) and Duits et al found that 73% of LGD was downgraded by a panel of expert GI pathologists.8,38 However, when we looked exclusively at the consistently called NDBE (concordant NDBE in UPMC system and expert review diagnosis) samples from 31 patients, TP53 mutation remained highly significantly associated with progression. These results indicate that our conclusions are not due to excessive downgrading dysplastic samples on expert pathologic review. Additionally, there were 2 cases and 7 controls for which we tested a biopsy with a diagnosis of IFD or NDBE when there was an antecedent biopsy with an original diagnosis of LGD that was unable to be used. None of these samples had a TP53 mutation, indicating that these cases did not sway our results. Our findings indicate that TP53 mutations can be detected in NDBE and, when detected, identify patients at increased risk for progression. However, given that BE surveillance relies on endoscopy with random biopsies, we cannot exclude the possibility that regions not sampled by the endoscopist had dysplasia.
Our results are also notable for the presence of TP53 mutations despite the lack of marked aneuploidy. Previous studies have shown increased copy number changes as patients approach progression,10,14 especially within the 24 months preceding the development of EAC.14 As with any genomic study, technical considerations must be considered for the differences in these data. However, while the methods to query copy number in these prior studies may be more sensitive than our methods, these prior studies evaluated copy-number across 1 Mb segments of the genome that we are readily able to detect with our assays. Alternatively, we posit a biological explanation for the quieter copy-number profiles in our NDBE samples. Specifically, the majority of our samples of NDBE were obtained >24 months before progression, possibly before the development of significant aneuploidy. Indeed, as our TP53 mutant NDBE did not have a different ploidy and had a similar mean number of chromosomal or total copy number changes as patients with TP53 wild type NDBE, these data suggest TP53 mutations precede aneuploidy, a logical sequence as TP53 loss may allow cells to tolerate the structural genome alterations that may more directly promote dysplasia and cancer.9,10,14,40 Given the known association of TP53 mutations, genome doubling, and aneuploidy, this would support a model of progression where some patients with BE develop an early TP53 mutation, which allows the development and tolerance of genome doubling and increased copy number changes.31,41,42 Recent pan-cancer studies from both the TCGA and ICGC are consistent with this supposition regarding TP53 as an early event as they have shown that TP53 mutations often occur as a clonal driver in cancer.30,43,44 Our data are in contrast to prior models suggesting that BE progression is initiated by early loss of CDKN2A.45 While we found frequent alterations in CDKN2A, rates were comparable in both progressors and non-progressors, suggesting that these alterations do not play a major role in driving progression but may be important in Barrett’s epithelial growth or expansion.
The study was designed to compare progressors with clinically well-characterized, non-progressive controls possessing similar risk factors (history of dysplasia and BE segment length). In this context, while limited due to the relatively low number of samples, we detected a nearly 13.8-fold (95% CI, 3.2–60.5) increase in the odds of progression with TP53 mutation (Supplementary Table 7) after adjusting for differences in age and sex. For progression to HGD or EAC in this patient population, TP53 mutation had a sensitivity of 45.8% (95% CI: 26.1–66.7%) and a specificity of 94.5% (95% CI: 85.8–98.2%), positive predictive value of 73.3% (95% CI: 44.8–91.1%) and negative predictive value of 84.1% (95% CI: 74.0–91.0%). This held when we only included patients with an original NDBE diagnosis (samples not downgraded by expert review), Supplementary Table 12. Furthermore, our finding that TP53 mutations from BE were also identified in the subsequent progression samples adds support to our belief that these mutations were pathogenic. However, our findings of TP53 mutations in 5% of our control population, similar to the 2.5% rate in non-progressive NDBE using similar sequencing methods16, demonstrates how even TP53 mutation does not guarantee progression. Moreover, additional studies have found TP53 mutations in histologically normal squamous tissue, including the esophagus, findings not associated with clear cancer risk, again reinforcing that TP53 mutations are not always associated with future cancer.46,47 As somatic genomic analytics are increasingly introduced into the assessment of pre-neoplastic conditions, it will be critical to comprehensively assess the degree that different mutations impact risk of neoplastic progression across distinct lineages.
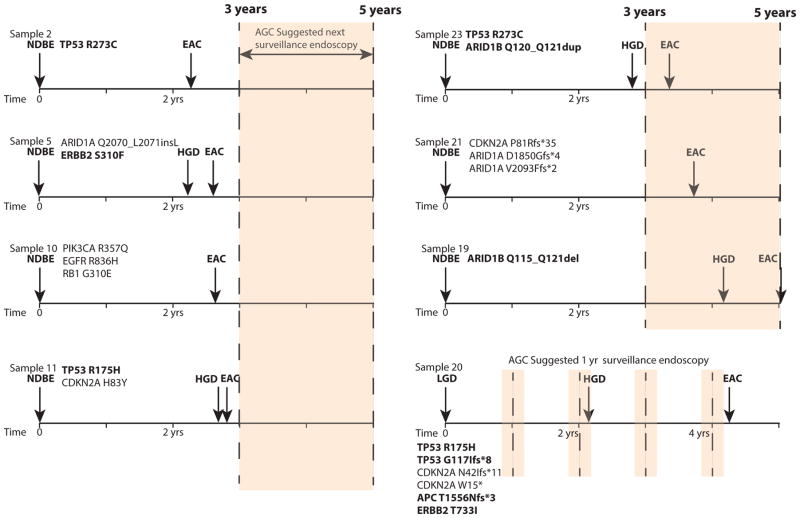
Additionally, although our results suggest that TP53 mutation is the most substantial genomic marker of progression risk, it is also notable that mutations in other pathogenic cancer-associated genes were also enriched in our progressor population. These data indicate the potential to construct panels with a combination of biomarkers for risk stratification. Biomarker panels may include a variety of genomic/structural alterations and even epigenetic biomarkers, as methylation has been shown to predict progression risk.11,48 Furthermore, as biomarkers are refined, it will be important to recognize that the nature of the mutations within individual genes may impact progression risk, as suggested by the distinct class of TP53 mutations found in the progressors. Future studies will need to evaluate the best combination of genomic aberrations that may predict a patient’s risk of progression in standard clinical tissue samples. The ability to detect copy number alterations, ploidy, and mutations from clinical FFPE biopsies using a single targeted assay as was performed in this study suggests such testing is feasible. As an example, in our cohort of progressors in which 20 of 24 samples were obtained 2–7 years before documented progression, we identified four patients with NDBE who subsequently progressed to invasive EAC within three years (the time of the next standard surveillance endoscopy) and another three with progression to cancer within five years (Fig. 3). We observed pathogenic mutations in each NDBE sample. These data suggest that integrating risk stratification into BE screening may be able to identify patients with greatest need for close surveillance or, perhaps, therapeutic ablation. Notably, these somatic alterations included genes other than TP53, again highlighting the potential for multi-plexed genomic assay to assess BE progression risk even when dysplasia is not detected.
Figure 3.
Genomic features of patients with subsequent progression to EAC within 5 years. Timeline showing when the tested sample was taken in relation to the patient’s diagnosis of progression in patients who developed EAC within ~5 years of the sample biopsy. Potential pathogenic mutations identified in each sample are listed. Mutations in genes that were more commonly mutated in progressors are bolded. The next suggested surveillance endoscopy interval from the tested sample is shaded.
Limitations of this study include the analysis of only a single BE sample from each patient and the relatively low number of progression patients. Future studies will need to determine the degree of spatial and temporal heterogeneity of distinct genomic alterations across the field of BE in patients with or without subsequent progression. Such studies will refine our understanding of how long before a cancer diagnosis specific risk-predicting features can be found. Larger studies in these populations will be needed to refine development of a panel of genomic markers, especially those other than TP53 mutation, that mark high-risk patients. Furthermore, future studies should evaluate the potential to integrate key coding mutations with the specific candidate structural alterations suggested by others to be highly enriched in progressors.10,35 Additionally, while multiple lines of evidence (allele fraction, likely pathogenic nature) suggest the identified mutations are likely somatic, without a paired germline control we cannot definitively exclude the potential for any of the mutations we identified to be germline variants.
In sum, these data show that TP53 mutations are often present in NDBE before the onset of progression and argue for the potential to shift the screening paradigm in BE towards the use of molecular and genomic markers to assess risk in this premalignant tissue. In addition, this and other studies have shown that such testing can be accomplished using a clinically relevant, targeted sequencing panel with standard, formalin-fixed biopsies.34 If coupled with more effective screening procedures, biomarker-guided approaches may help prevent EAC or detect it at early, curable stages and may ultimately limit the need for frequent surveillance of the large population of patients who never ultimately progress to invasive cancer thus decreasing the under and over diagnosis common in this disease.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases grant K08 DK109209 and the Prevent Cancer Foundation Grant (M.D.S.), the National Cancer Institute P01 CA098101, pilot project grant, and grant from the Broad Next10 Initiative (A.J.B.).
We thank the Dana-Farber Center for Cancer Genome Discovery for their assistance. This work was supported by the National Institute Health K08 DK109209 and the Prevent Cancer Foundation Grant (M.D.S.), P01CA098101 (A.J.B.) and the Broad Institute Next10 Program.
Abbreviations
- BE
Barrett’s esophagus
- EAC
esophageal adenocarcinoma
- FFPE
formalin fixed paraffin embedded
- IFD
indefinite for dysplasia
- IHC
immunohistochemistry
- LGD
low grade dysplasia
- NDBE
non-dysplastic Barrett’s esophagus
Footnotes
Author Contributions: Data analysis: MDS, AN, ART, JMD, AJB; Computational analysis: NDC, MD, SLC; Patient selection and clinical data collection: CD, JMD, MR; Slide cutting and DNA isolation: CD, AK; Histologic review: ATA, RDO, JLH, AN, MR, RHL; Conceived of study: MDS, JMD, AJB; Manuscript preparation: MDS, NDC, JMD, AJB; Manuscript review: MDS, ATA, RDO, JLH, AN, ART, SLC, JMD, AJB
Disclosures: The authors declare no competing financial, personal, other interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Author names in bold designate shared co-first authorship
- 1.Holmes RS, Vaughan TL. Epidemiology and pathogenesis of esophageal cancer. Semin Radiat Oncol. 2007;17:2–9. doi: 10.1016/j.semradonc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Brown LM, Devesa SS, Chow W-H. Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. J Natl Cancer Inst. 2008;100:1184–7. doi: 10.1093/jnci/djn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ormsby AH, Kilgore SP, Goldblum JR, et al. The Location and Frequency of Intestinal Metaplasia at the Esophagogastric Junction in 223 Consecutive Autopsies: Implications for Patient Treatment and Preventive Strategies in Barrett ’ s Esophagus. Mod Pathol. 2000;6:614–620. doi: 10.1038/modpathol.3880106. [DOI] [PubMed] [Google Scholar]
- 4.Ronkainen J, Aro P, Storskrubb T, et al. Prevalence of Barrett’s esophagus in the general population: an endoscopic study. Gastroenterology. 2005;129:1825–31. doi: 10.1053/j.gastro.2005.08.053. [DOI] [PubMed] [Google Scholar]
- 5.Desai TK, Krishnan K, Samala N, et al. The incidence of oesophageal adenocarcinoma in non-dysplastic Barrett’s oesophagus: a meta-analysis. Gut. 2012;61:970–6. doi: 10.1136/gutjnl-2011-300730. [DOI] [PubMed] [Google Scholar]
- 6.Shaheen NJ, Falk GW, Iyer PG, et al. ACG Clinical Guideline: Diagnosis and Management of Barrett’s Esophagus. Am J Gastroenterol. 2016;111:30–50. doi: 10.1038/ajg.2015.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reid BJ, Haggitt RC, Rubin CE, et al. Observer variation in the diagnosis of dysplasia in Barrett’s esophagus. Hum Pathol. 1988;19:166–78. doi: 10.1016/s0046-8177(88)80344-7. [DOI] [PubMed] [Google Scholar]
- 8.Curvers WL, ten Kate FJ, Krishnadath KK, et al. Low-grade dysplasia in Barrett’s esophagus: overdiagnosed and underestimated. Am J Gastroenterol. 2010;105:1523–30. doi: 10.1038/ajg.2010.171. [DOI] [PubMed] [Google Scholar]
- 9.Eluri S, Brugge WR, Daglilar ES, et al. The Presence of Genetic Mutations at Key Loci Predicts Progression to Esophageal Adenocarcinoma in Barrett’s Esophagus. Am J Gastroenterol. 2015;110:828–34. doi: 10.1038/ajg.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Paulson TG, Galipeau PC, et al. Assessment of Esophageal Adenocarcinoma Risk Using Somatic Chromosome Alterations in Longitudinal Samples in Barrett’s Esophagus. Cancer Prev Res. 2015;8:845–56. doi: 10.1158/1940-6207.CAPR-15-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Z, Cheng Y, Gu W, et al. A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett’s esophagus. Cancer Res. 2009;69:4112–4115. doi: 10.1158/0008-5472.CAN-09-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paulson TG, Galipeau PC, Xu L, et al. p16 mutation spectrum in the premalignant condition Barrett’s esophagus. PLoS One. 2008;3:e3809. doi: 10.1371/journal.pone.0003809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong DJ, Paulson TG, Prevo LJ, et al. p16 INK4a Lesions Are Common , Early Abnormalities that Undergo Clonal Expansion in Barrett ’ s Metaplastic Epithelium p16 INK4a Lesions Are Common , Early Abnormalities that Undergo Clonal Expansion. Cancer Res. 2001;61:8284–8289. [PubMed] [Google Scholar]
- 14.Li X, Galipeau PC, Paulson TG, et al. Temporal and Spatial Evolution of Somatic Chromosomal Alterations: A Case-Cohort Study of Barrett’s Esophagus. Cancer Prev Res. 2014;7:114–127. doi: 10.1158/1940-6207.CAPR-13-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu J, Ajani Ja, Hawk ET, et al. Genome-wide catalogue of chromosomal aberrations in barrett’s esophagus and esophageal adenocarcinoma: a high-density single nucleotide polymorphism array analysis. Cancer Prev Res. 2010;3:1176–86. doi: 10.1158/1940-6207.CAPR-09-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weaver JMJ, Ross-Innes CS, Shannon N, Lynch AG, et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat Genet. 2014;46:837–843. doi: 10.1038/ng.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novotna K, Trkova M, Pazdro A, et al. TP53 gene mutations are rare in nondysplastic Barrett’s esophagus. Dig Dis Sci. 2006;51:110–113. doi: 10.1007/s10620-006-3093-3. [DOI] [PubMed] [Google Scholar]
- 18.Younes M, Brown K, Lauwers GY, et al. p53 protein accumulation predicts malignant progression in Barrett’s metaplasia: a prospective study of 275 patients. Histopathology. 2017;38:42–49. doi: 10.1111/his.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaye PV, Ilyas M, Soomro I, et al. Dysplasia in Barrett’s oesophagus: p53 immunostaining is more reproducible than haematoxylin and eosin diagnosis and improves overall reliability, while grading is poorly reproducible. Histopathology. 2016;69:431–440. doi: 10.1111/his.12956. [DOI] [PubMed] [Google Scholar]
- 20.Davelaar AL, Calpe S, Lau L, et al. Aberrant TP53 detected by combining immunohistochemistry and DNA-FISH improves Barrett’s esophagus progression prediction: A prospective follow-up study. Genes Chromosomes Cancer. 2015;54:82–90. doi: 10.1002/gcc.22220. [DOI] [PubMed] [Google Scholar]
- 21.Kastelein F, Biermann K, Steyerberg EW, et al. Aberrant p53 protein expression is associated with an increased risk of neoplastic progression in patients with Barrett’s oesophagus. Gut. 2013;62:1676–83. doi: 10.1136/gutjnl-2012-303594. [DOI] [PubMed] [Google Scholar]
- 22.Srivastava A, Appelman H, Goldsmith JD, et al. The Use of Ancillary Stains in the Diagnosis of Barrett Esophagus and Barrett Esophagus-associated Dysplasia: Recommendations From the Rodger C. Haggitt Gastrointestinal Pathology Society Am J Surg Pathol. 2017;41:e8–e21. doi: 10.1097/PAS.0000000000000819. [DOI] [PubMed] [Google Scholar]
- 23.Djalilvand A, Pal R, Goldman H, et al. Evaluation of p53 mutations in premalignant esophageal lesions and esophageal adenocarcinoma using laser capture microdissection. Mod Pathol. 2004;17:1323–7. doi: 10.1038/modpathol.3800231. [DOI] [PubMed] [Google Scholar]
- 24.Dolan K, Walker S, Gosney J, et al. TP53 mutations in malignant and premalignant Barrett’s esophagus. Dis Esophagus. 2003;16:83–9. doi: 10.1046/j.1442-2050.2003.00302.x. [DOI] [PubMed] [Google Scholar]
- 25.Jenkins GJS, Doak SH, Griffiths aP, et al. Early p53 mutations in nondysplastic Barrett’s tissue detected by the restriction site mutation (RSM) methodology. Br J Cancer. 2003;88:1271–6. doi: 10.1038/sj.bjc.6600891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider PM, Casson AG, Levin B, et al. Mutations of p53 in Barrett’s esophagus and Barrett’s cancer: a prospective study of ninety-eight cases. J Thorac Cardiovasc Surg. 1996;111:323–31. 3. doi: 10.1016/s0022-5223(96)70441-5. [DOI] [PubMed] [Google Scholar]
- 27.Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI insight. 2016;1:e87062. doi: 10.1172/jci.insight.87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia EP, Minkovsky A, Jia Y, et al. Validation of OncoPanel: A Targeted Next-Generation Sequencing Assay for the Detection of Somatic Variants in Cancer. Arch Pathol Lab Med. 2017;141:751–758. doi: 10.5858/arpa.2016-0527-OA. [DOI] [PubMed] [Google Scholar]
- 29.Lash RH, Deas TM, Wians FH. Healthcare Cost of Over-Diagnosis of Low-Grade Dysplasia in Barrett’s Esophagus. Adv Ther. 2016;33:684–697. doi: 10.1007/s12325-016-0308-7. [DOI] [PubMed] [Google Scholar]
- 30.Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stachler MD, Taylor-Weiner A, Peng S, et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat Genet. 2015;47:1047–55. doi: 10.1038/ng.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavery DL, Martinez P, Gay LJ, et al. Evolution of oesophageal adenocarcinoma from metaplastic columnar epithelium without goblet cells in Barrett’s oesophagus. Gut. 2016;65:907–913. doi: 10.1136/gutjnl-2015-310748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross-Innes CS, Becq J, Warren A, et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett’s esophagus and esophageal adenocarcinoma. Nat Genet. 2015;47:1038–1046. doi: 10.1038/ng.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Del Portillo A, Lagana SM, Yao Y, et al. Evaluation of Mutational Testing of Preneoplastic Barrett’s Mucosa by Next-Generation Sequencing of Formalin-Fixed, Paraffin-Embedded Endoscopic Samples for Detection of Concurrent Dysplasia and Adenocarcinoma in Barrett’s Esophagus. J Mol Diagn. 2015;17:412–9. doi: 10.1016/j.jmoldx.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Timmer MR, Martinez P, Lau CT, et al. Derivation of genetic biomarkers for cancer risk stratification in Barrett’s oesophagus: a prospective cohort study. Gut. 2016;65:1602–10. doi: 10.1136/gutjnl-2015-309642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altaf K, Xiong J-J, De la Iglesia D, et al. Meta-analysis of biomarkers predicting risk of malignant progression in Barrett’s oesophagus. Br J Surg. 2017;104:493–502. doi: 10.1002/bjs.10484. [DOI] [PubMed] [Google Scholar]
- 37.Sonwalkar SA, Rotimi O, Scott N, et al. A study of indefinite for dysplasia in Barrett’s oesophagus: reproducibility of diagnosis, clinical outcomes and predicting progression with AMACR (alpha-methylacyl-CoA-racemase) Histopathology. 2010;56:900–7. doi: 10.1111/j.1365-2559.2010.03571.x. [DOI] [PubMed] [Google Scholar]
- 38.Duits LC, Phoa KN, Curvers WL, et al. Barrett’s oesophagus patients with low-grade dysplasia can be accurately risk-stratified after histological review by an expert pathology panel. Gut. 2015;64:700–6. doi: 10.1136/gutjnl-2014-307278. [DOI] [PubMed] [Google Scholar]
- 39.Duits LC, van der Wel MJ, Cotton CC, et al. Patients With Barrett’s Esophagus and Confirmed Persistent Low-Grade Dysplasia Are at Increased Risk for Progression to Neoplasia. Gastroenterology. 2017;152:993–1001.e1. doi: 10.1053/j.gastro.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Maley CC, Galipeau PC, Li X, et al. The Combination of Genetic Instability and Clonal Expansion Predicts Progression to Esophageal Adenocarcinoma. Cancer Res. 2004;64:7629–7633. doi: 10.1158/0008-5472.CAN-04-1738. [DOI] [PubMed] [Google Scholar]
- 41.Fujiwara T, Bandi M, Nitta M, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–7. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 42.Dewhurst SM, McGranahan N, Burrell Ra, et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov. 2014;4:175–85. doi: 10.1158/2159-8290.CD-13-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerstung M, Jolly C, Leshchiner I, Dentro S, et al. The evolutionary history of 2 ,658 cancers. BioRxiv. 2017 [Google Scholar]
- 44.Campbell PJ, Getz G, Stuart JM, Korbel JO, et al. Pan-cancer analysis of whole genomes. bioRxiv. 2017;3:162784. [Google Scholar]
- 45.Contino G, Vaughan TL, Whiteman D, et al. The Evolving Genomic Landscape of Barrett’s Esophagus and Esophageal Adenocarcinoma. Gastroenterology. 2017;153:657–673e1. doi: 10.1053/j.gastro.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martincorena I, Roshan A, Gerstung M, et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science (80- ) 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jonason AS, Kunala S, Price GJ, et al. Frequent clones of p53-mutated keratinocytes in normal human skin. Proc Natl Acad Sci U S A. 1996;93:14025–9. doi: 10.1073/pnas.93.24.14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alvi MA, Liu X, O’Donovan M, et al. DNA methylation as an adjunct to histopathology to detect prevalent, inconspicuous dysplasia and early-stage neoplasia in Barrett’s esophagus. Clin Cancer Res. 2013;19:878–888. doi: 10.1158/1078-0432.CCR-12-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing results analyzed during this study are included in this published article. As all samples were only sequenced on a small targeted platform without normal control, mutation calls from MuTect and GATK are listed in Supplementary Table 4.