SUMMARY
Leukemia stem cells (LSCs) are thought to drive the genesis of acute myeloid leukemia (AML) as well as relapse following chemotherapy. Due to their unique biology, developing effective methods to eradicate LSCs has been a significant challenge. In the present study, we demonstrate that intrinsic over-expression of the mitochondrial dynamics regulator FIS1 mediates mitophagy activity that is essential for primitive AML cells. Depletion of FIS1 attenuates mitophagy and leads to inactivation of GSK3, myeloid differentiation, cell cycle arrest and a profound loss of LSC self-renewal potential. Further, we report the central metabolic stress regulator AMPK is also intrinsically activated in LSC populations and is upstream of FIS1. Inhibition of AMPK signaling recapitulates the biological effect of FIS1 loss. These data suggest a model in which LSCs co-opt AMPK/FIS1-mediated mitophagy as a means to maintain stem cell properties that may be otherwise compromised by the stresses induced by oncogenic transformation.
Keywords: Acute myeloid leukemia, Leukemia stem cells, Mitochondrial dynamics, Mitophagy, Differentiation, FIS1, GSK3, AMPK
eTOC
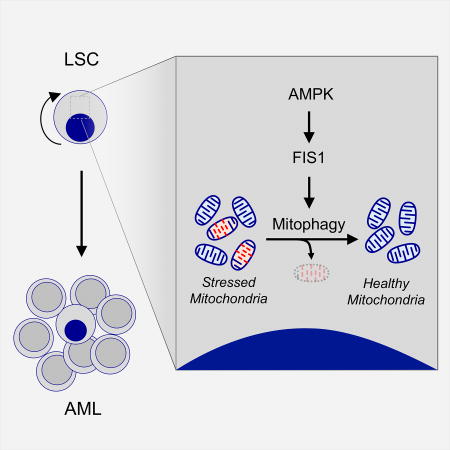
Human acute myeloid leukemia stem cells (LSCs) depend on FIS1-mediated mitophagy for self-renewal and survival. AMPK is constitutively active in human LSCs, is upstream of FIS1, and acts to stimulate mitophagy. Disruption of AMPK signaling or FIS1 activity results in eradication of LSCs.
INTRODUCTION
Despite decades of research, effective therapies for acute myeloid leukemia (AML) remain extremely limited. A majority of patients relapse following conventional chemotherapy, or in some cases never achieve a meaningful response (Dombret and Gardin, 2016). Analysis of AML biology has shown that disease pathogenesis is mediated by a subset of the tumor population termed leukemia stem cells (LSCs). Multiple studies have demonstrated that LSCs are biologically distinct from bulk tumor, have differential sensitivity to chemotherapy, and are likely a major cause of therapy resistance and relapse (Jordan et al., 2006; Mikkola et al., 2010). This concept is further supported by recent studies focusing on the clinical significance of LSCs. These studies demonstrate that the frequency and phenotypic diversity of LSCs increase substantially in relapse versus diagnosis AML (Ho et al., 2016). In addition, AML patients who have high expression of LSC gene expression signatures demonstrate significantly poorer treatment outcomes (Eppert et al., 2011; Gentles et al., 2010), suggesting that enrichment of LSC activities correlates with decreased efficacy of conventional therapy. Despite strong laboratory and clinical evidence emphasizing the need to improve anti-LSC regimens, the ability to effectively target such populations in patients remains a major challenge (Guzman and Allan, 2014; Pollyea and Jordan, 2017).
Although LSCs were first described as within the CD34+/CD38− subpopulation of human AML cells (Bonnet and Dick, 1997; Lapidot et al., 1994), later studies have shown significant inter- and intra-patient immunophenotypic heterogeneity in them, making CD34/CD38-based LSC isolation and characterization a significant challenge (Eppert et al., 2011; Ho et al., 2016; Ng et al., 2016; Sarry et al., 2011). As an alternative approach to characterization of cancer stem cell (CSC) populations, we and others have turned to the biochemical property of oxidative state as a means to identify and isolate primitive populations (Diehn et al., 2009; Lagadinou et al., 2013). As previously demonstrated for many types of stem cells, including normal hematopoietic stem cells (HSCs) (Jang and Sharkis, 2007), the level of reactive oxygen species (ROS) can be used to enrich for primitive populations. In all known instances, somatic stem cells reside in a relatively reduced condition, termed ROS-low, whereas more differentiated cells demonstrate a net increase in oxidative state termed ROS-high. The ROS-low physiological state is thought to be required by normal and malignant stem cell populations as a means to alleviate both intrinsic and extrinsic stresses, and to sustain their self-renewal and proliferation potential (Zhou et al., 2014). Thus, we have employed the ROS-low phenotype as an alternative means to enrich LSC populations for molecular, biochemical, and cellular analyses.
Using oxidative state to characterize primary human AML specimens, we previously compared the gene expression profile between ROS-low LSCs vs. ROS-high non-LSCs. These studies show that genes related to mitochondrial biology are up-regulated in the ROS-low LSC population, including FIS1 (Mitochondrial fission 1), a gene known to be critical for mitochondrial dynamics. The term mitochondrial dynamics describes the interconnected processes of mito-fusion, mito-fission and mitophagy (Chen and Chan, 2017). Briefly, the mito-fusion process functions to fuse individual mitochondrion together and is balanced by the opposite process of mito-fission to allow proper formation of mitochondrial network. When defective mitochondria accumulate upon stress, mito-fission and mitophagy can function together to isolate and degrade damaged mitochondria. These processes thus function to organize mitochondria into healthy networks and maintain a homeostatic balance required for proper cell function (Wai and Langer, 2016).
Despite a number of recent studies on the mechanisms that regulate mitochondrial dynamics, the functional role of processes such as mito-fusion, mito-fission and mitophagy in human cancers and/or cancer stem cells is largely unknown. In the present study, we dissect mitochondrial dynamics in human AML with a focus on LSC biology. We show that LSC-specific activation of the metabolic stress regulator AMPK induces up-regulation of FIS1, which in turn regulates mitophagy activity that functions to sustain AML LSCs. Inhibition of FIS1-mediated mitophagy induces myeloid differentiation, reduction in cell cycle activity, and loss of leukemic stem and progenitor cell potential.
RESULTS
LSCs have higher expression of FIS1 and distinct mitochondrial morphology
We previously utilized the ROS-low phenotype to enrich for LSCs, and then employed RNA-seq to identify molecular properties that are unique to the LSC population. Those studies show up-regulation of the mitochondrial dynamics regulator FIS1 (Lagadinou et al., 2013). Further analysis of those data identified a trend towards increased expression of many mitochondrial dynamics regulators in the ROS-low LSC population, including FIS1, TBC1D15, PINK1, MFF, MiD49 and MiD51 (Figures 1A, S1A and S1B). Among the genes known to regulate mitochondrial dynamics, FIS1 shows the clearest and most consistent differential expression in comparison to the ROS-high non-LSC population (Figure S1B). These data suggest FIS1 activity is elevated in primitive AML cells and may drive a distinct state of mitochondrial dynamics as a component of LSC growth and survival.
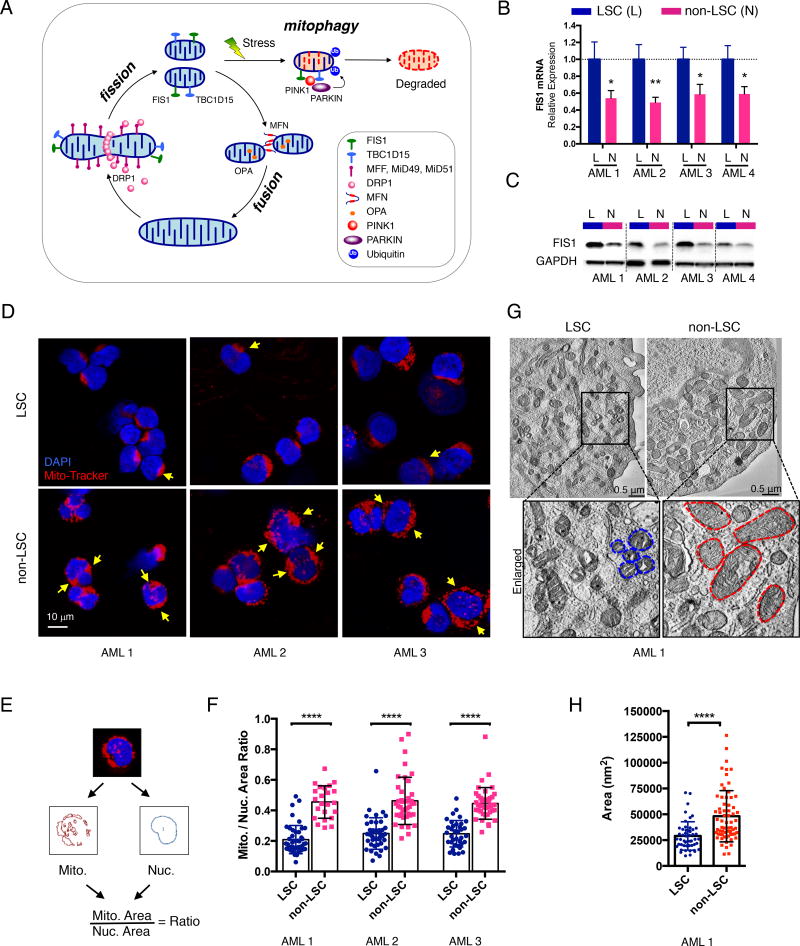
Figure 1. AML LSCs have higher expression of FIS1 and distinct mitochondrial morphology.
(A) A diagram showing regulation of healthy mitochondrial network through mito-fusion, mito-fission and mitophagy.
(B) qPCR results showing relative expression of FIS1 gene in sorted ROS-low LSCs (L) versus ROS-high non-LSCs (N). Mean ± SD (n=3). Type 2, two-tailed t-test.
(C) WB results showing expression of FIS1 protein in sorted ROS-low LSCs (L) versus ROS-high non-LSCs (N).
(D) Representative confocal images showing morphology of mitochondria in ROS-low LSCs versus ROS-high non-LSCs. Yellow arrows highlight distinct mitochondrial morphology.
(E) A diagram showing the method used to quantify the mitochondrial to nuclear area ratio.
(F) Mitochondrial to nuclear area ratio in LSCs vs. non-LSCs in 3 primary AML specimens. Each dot represents an individual cell. Mean ± SD. Type 3, two-tailed t-test.
(G) Representative TEM images showing morphology of mitochondria in ROS-low LSCs and ROS-high non-LSCs. Blue and red dotted lines outline mitochondrial shape.
(H) Quantification of mitochondrial cross-section area from the TEM images of AML 1. Each dot represents a single mitochondrion. Mean ± SD. Type 3, two-tailed t-test.
See also Figure S1.
To test this hypothesis, in the current study we isolated both ROS-low and ROS-high cells from an independent cohort of primary AML specimens by flow cytometric sorting (Figure S1C). We confirmed in multiple primary AML specimens that ROS-low AML cells are significantly enriched for both colony-forming and engraftment abilities in comparison to ROS-high AML cells (Figures S1D–S1G), demonstrating ROS-low as a reliable functional state to enrich for LSCs in AML, as we have previously reported (Lagadinou et al., 2013). Importantly, in this new cohort of primary AML specimens, both mRNA and protein expression of FIS1 are significantly higher in LSCs relative to non-LSCs (Figures 1B and 1C). Given the role of FIS1 in mitochondrial dynamics, we next examined mitochondrial morphology in LSCs versus non-LSCs using confocal and transmission electron microscopy (TEM). For confocal studies, we labeled mitochondria and nuclei of freshly sorted LSCs and non-LSCs with the mitochondrial dye Mito-Tracker and DNA dye DAPI. Compared to non-LSCs, the LSCs demonstrate a significantly lower mitochondrial to nuclear area ratio, suggesting they have less mitochondrial content (Figures 1D–1F and Figure S1H). In addition, the majority of mitochondria in LSCs are located in a single compact crescent-like shaped area while non-LSCs show a distinctly more diffuse labeling pattern (Figure 1D and Figure S1H). To further investigate mitochondrial morphology at individual mitochondrion resolution, TEM was performed on freshly isolated LSC and non-LSC populations. The TEM images revealed that the cross sections of mitochondria in LSCs consistently demonstrate a significantly smaller area compared to non-LSCs (Figures 1G and 1H), suggesting LSC mitochondria have smaller volume. Together, these data indicate that AML LSCs have unique mitochondrial morphology indicative of a distinct state of mitochondrial dynamics relative to non-LSCs.
Inhibition of FIS1 disrupts mitochondrial dynamics in AML
To test if FIS1 is responsible for the distinct mitochondrial morphology, we next performed shRNA-mediated knock-down studies in both the AML cell line MOLM-13 and primary AML cells to examine the impact of FIS1 loss on mitochondrial dynamics. We observed that the FIS1-depleted MOLM-13 cells have significantly increased mitochondrial content relative to the control cells (Figures 2A, 2B top row and 2C left). Notably, a recent study has shown FIS1 to be a regulator of mitophagy in mammalian cells (Shen et al., 2014). Thus, we hypothesized that increased mitochondrial content may be caused by a deficient state of mitophagy in FIS1-depleted AML cells. To test this hypothesis, we induced mitophagy using previously reported mitochondrial stress inducer valinomycin (Ashrafi and Schwarz, 2013). Valinomycin is a K+ inonphore that can permeabilize the mitochondrial membrane to K+, inducing mitochondrial membrane potential loss, thereby activating mitophagy (Rakovic et al., 2010; Seibler et al., 2011).
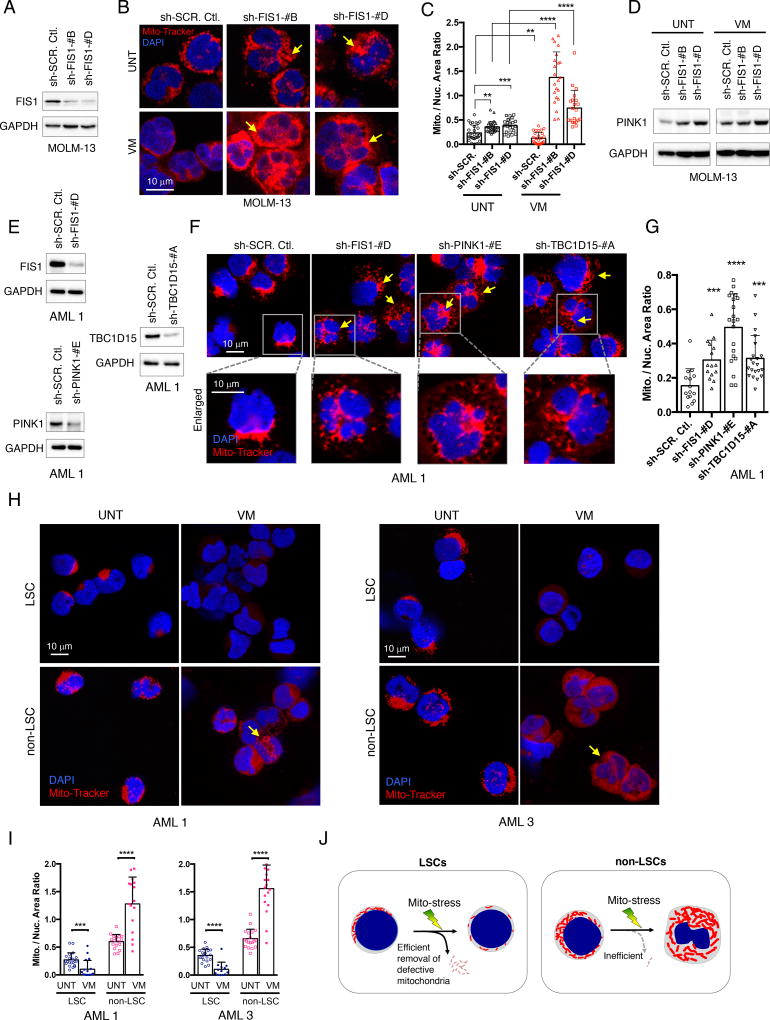
Figure 2. Loss of FIS1 induces mitophagy defects in AML.
(A) WB images showing knock-down efficiency of sh-FIS1 in MOLM-13 cells.
(B) Representative confocal images showing mitochondrial morphology of MOLM-13 cells at day 6 following knock-down with or without 5 uM valinomycin treatment for 3 hours. Arrows highlight distinct mitochondrial morphology.
(C) Mitochondrial to nuclear area ratio in MOLM-13 cells. Each dot represents an individual cell. Mean + SD. Type 3, two-tailed t-test.
(D) WB images showing expression of PINK1 in MOLM-13 cells at day 6 following shRNA-mediated knock-down of FIS1 with or without 5 uM valinomycin treatment for 3 hours.
(E) WB images showing knock-down efficiency of sh-FIS1, sh-PINK1 and sh-TBC1D15 in primary AML 1.
(F) Representative confocal images showing mitochondrial morphology of primary AML cells at day 6 following shRNA-mediated knock-down of FIS1, PINK1 and TBC1D15. Arrows highlight distinct mitochondrial morphology.
(G) Mitochondrial to nuclear area ratio in primary AML 1 with knock-down of FIS1, PINK1, or TBC1D15. Each dot represents an individual cell. Mean + SD. Type 3, two-tailed t-test.
(H) Representative confocal images showing mitochondrial morphology of ROS-low LSCs versus ROS-high non-LSCs, freshly sorted from primary AML specimens and then treated with or without 5uM valinomycin for 3 hours. Arrows highlight accumulated mitochondria induced by valinomycin treatment.
(I) Mitochondrial to nuclear area ratio in LSCs versus non-LSCs isolated from primary AML 1 and AML 3 at basal and valinomycin-treated conditions. Each dot represents an individual cell. Mean + SD. Type 3, two-tailed t-test.
(J) A diagram summarizing the mitochondrial morphology changes seen in LSCs versus non-LSCs with or without mitochondrial stress.
See also Figure S2.
We observed that while control cells responded to valinomycin treatment with loss of mitochondria, the shFIS1 cells further accumulated mitochondrial content, suggesting an inability to perform efficient mitophagy in response to mitochondrial stress (Figures 2B bottom row and 2C right). To further test this hypothesis, we probed for expression of PINK1, a kinase stabilized upon mitophagy activation and involved in the clearance of damaged mitochondria (Lazarou et al., 2015). We observed that while the control cells have increased PINK1 expression upon valinomycin treatment, the shFIS1 cells show no change in PINK1 level despite the same insult (Figure 2D). These data further suggest that the FIS1-depleted AML cells have inability to activate mitophagy upon mitochondrial stress increase.
To extend the above findings from MOLM-13 cells, shFIS1 knock-down studies were also performed in primary human AML cells (Figure 2E), where we observed that reduction of FIS1 also induced accumulation of mitochondria that led to a significant increase in the mitochondrial to nuclear area ratio in AML 1 (Figures 2F, 2G and S2A) and AML 7 (Figures S2B–S2E). In addition, depletion of known mitophagy regulators PINK1 and TBC1D15 (Nguyen et al., 2016; Yamano et al., 2014) reproduced the same mitochondrial accumulation phenotype in shFIS1 AML cells (Figures 2E–2G), further supporting the role of FIS1 in regulating mitophagy. Notably, we also observed that the mitophagy defect phenotype is distinctly different from a fission defect phenotype observed in shDRP1 MOLM-13 (Figures S2F–S2H) and primary AML cells (Figures S2I and S2J), where longer and more thread-like mitochondria are prevalent (Figure S2K). Lastly, the shFIS1 primary AML cells recapitulated the diffuse mitochondrial distribution phenotype seen in non-LSCs where FIS1 expression is low (Figure 2F, enlarged inserts).
To determine whether primary AML cells with higher expression of FIS1 have increased mitophagy activity, we performed valinomycin treatment experiments on freshly sorted LSCs and non-LSCs from human AML specimens. We found that, consistent with the knock-down experiments, LSCs also responded to valinomycin treatment with efficient clearance of mitochondria while non-LSCs showed further accumulation of mitochondria mass (Figures 2H and 2I).
Together, these data demonstrate FIS1 plays an important role in regulating the mitochondrial dynamics of leukemic cells largely through mitophagy and suggest the model illustrated in Figure 2J. LSCs or AML cells expressing higher levels of FIS1 possess fewer mitochondria and show localization to a defined region of the cell. In contrast, non-LSCs or AML cells engineered to reduce FIS1 expression have increased mitochondrial mass and more diffuse mitochondrial morphology. Elevated FIS1 expression in LSC populations mediates increased mitophagy activity, while non-LSCs with less FIS1 expression are inefficient in mitophagy, resulting in further accumulation of mitochondria under stress conditions (Figure 2J).
Loss of FIS1 impairs stem and progenitor potential of AML
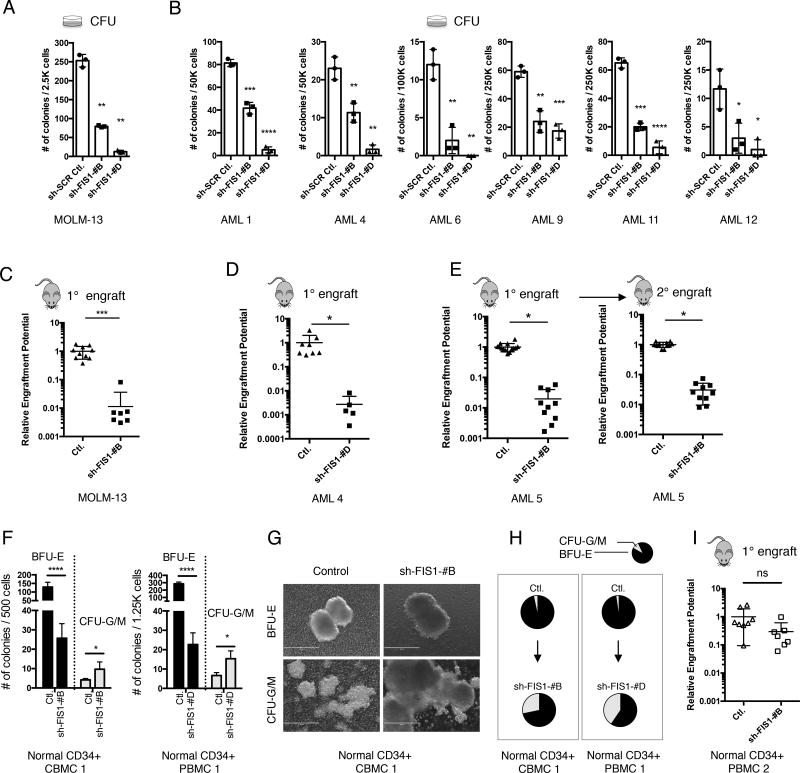
Given the profound impact of FIS1 loss on AML mitochondrial dynamics and the unique mitochondrial morphology present in AML LSCs, we next investigated the functional role of FIS1 and other mitophagy regulators in AML stem and progenitor cells. To this end, knock-down of FIS1, TBC1D15 and PINK1 were performed in both the MOLM-13 cell line and primary AML specimens (Figure S3A). Depletion of FIS1 using shRNA strongly reduced the colony-forming ability of MOLM-13 and primary AML cells (Figures 3A and 3B). In addition, shRNA-mediated knock-down of TBC1D15 and PINK1 also caused a significant loss of colony-forming ability in these specimens (Figure S3B), suggesting mitophagy in general might be required for stem and progenitor potential of AML. To directly assess the role of FIS1 on LSC growth and survival, we performed xenograft studies using shFIS1 transduced MOLM-13 and primary AML specimens. Importantly, given the known heterogeneity of LSCs, knock-down studies were performed using unfractionated primary AML specimens, so as to avoid any possible bias that may be introduced using a subpopulation of cells defined using a specific phenotype (e.g. CD34+, ROS-low, etc.). We observed that loss of FIS1 almost completely abolished the engraftment ability of MOLM-13 and AML specimens in immune-deficient mice (Figures 3C–3E and S3C). This depleting effect was evident in both primary and secondary transplantation assays (Figures 3E and S3C), indicating that FIS1 is required for LSC self-renewal potential. Finally, CRISPR/Cas9-mediated disruption of the FIS1 gene also significantly impaired both colony-forming and engraftment ability of MOLM-13 cells (Figures S3D–S3F), corroborating the results obtained with shRNA approaches. Together, these findings indicate a central role for FIS1 in the maintenance of LSC populations.
Fig. 3. Depletion of FIS1 impairs stem and progenitor potential of primary AML but relatively spares normal HSPCs.
(A) Colony-forming ability of MOLM-13 cells following shRNA-mediated knock-down of FIS1. Mean ± SD (n=3). Type 2, two-tailed t-test.
(B) Colony-forming ability of primary AML cells following shRNA-mediated knock-down of FIS1. Mean ± SD (n=3). Type 2, two-tailed t-test.
(C) Normalized relative engraftment potential of MOLM-13 cells with or without FIS1 knock-down.
(D) Normalized relative engraftment potential of primary AML.
(E) Normalized relative engraftment potential of primary AML 5 with or without FIS1 knock-down. 1° and 2° indicate primary and secondar y xenograft experiments, respectively.
(F) Number of BFU-E and CFU-G/M colonies produced by normal CD34+ CBMCs/PBMCs in methylcellulose. Mean ± SD, (n=3). Type 2, two-tailed t-test.
(G) Representative images showing morphology of BFU-E and CFU-G/M colonies.
(H) Pie charts showing changes in proportion of CFU-G/M and BFU-E colonies induced by FIS1 knock-down.
(I) Normalized relative engraftment potential of normal CD34+ PBMCs with or without FIS1 knock-down.
In C, D, E, and I, each dot represents an individual mouse and lines represent mean ± SD. Type 2, two-tailed t-test.
See also Figure S3.
FIS1 loss attenuates erythropoiesis without affecting the engraftment ability of normal hematopoietic stem and progenitor cells (HSPCs)
A recent study has shown that loss of MFN2 (mitofusin 2), another mitochondrial dynamics regulator, can impair the lymphoid potential of normal HSPCs (Luchsinger et al., 2016). To investigate the role of FIS1 in normal hematopoiesis, CD34+ normal cord/peripheral blood mononuclear cells (CD34+ CBMCs/PBMCs) were isolated from healthy donors and subjected to FIS1 knock-down using the same shRNA strategy employed for AML studies. We first investigated the impact of FIS1 loss on mitochondrial morphology of normal HSPCs. We found that the FIS1-depleted CD34+ cells had negligible change in their mitochondrial mass comparing to the control cells (Figures S3G and S3H), suggesting FIS1 is largely dispensable in the regulation of mitochondrial morphology in normal HSPCs. In addition, in contrast to the mitochondrial loss phenotype seen in valinomycin-treated AML, the same treatment induced mitochondrial accumulation in both control and shFIS1 CD34+ cells, suggesting a slower rate of mitophagy in normal HSPCs (Figure S3G and S3H). We next performed functional assays to evaluate the impact of FIS1 loss on stem/progenitor potential of normal HSPCs. Analysis of colony-forming ability showed two notable phenotypes. First, while the number of erythroid colonies (BFU-E) was markedly reduced (Figure 3F left), the size and morphology of BFU-Es were normal (Figure 3G top). In contrast, granulocyte/macrophage myeloid colonies (CFU-G/M) were more abundant and much larger upon FIS1 depletion (Figures 3F right and 3G bottom). As a result of this lineage skewing, FIS1 depletion substantially increased the proportion of CFU-G/M colonies compared to control (Figure 3H). Together, these data suggest that FIS1 loss predisposes normal hematopoietic progenitors to favor myelopoiesis over erythropoiesis. Consistent with this hypothesis, analysis of FIS1 expression during normal human hematopoiesis in the publically available data set GSE24759 (Bagger et al., 2016; Novershtern et al., 2011) shows that, among all stages of normal hematopoiesis, the expression of FIS1 is highest in the erythroid lineage (Figure S3I). This observation provides a possible explanation for the shFIS1-mediated selective impact on erythropoiesis.
To measure the impact of FIS1 loss in the HSC compartment, xenograft assays were performed to measure the engraftment ability of normal CD34+ PBMCs transduced with control vs. shFIS1 vectors. As shown in Figure S3J, we achieved a level of FIS1 knock-down comparable to AML experiments. Engraftment efficiency of total human cells in recipient mice was 14% ± 18% (n=15) at 12 weeks of post-transplant. As shown in Figure 3I and S3K, FIS1 knock-down caused a modest loss of engraftment potential (~3 fold) (Table S1). This is in sharp contrast to the ~50–350 fold drop in relative engraftment potential seen in primary AML cells (Figures 3D, 3E and S3C). Together, these data suggest that the functional role of FIS1 is critical to survival and self-renewal of AML LSCs, but that its role in normal mitochondrial morphology and self-renewal is much more subtle.
Loss of FIS1 induces GSK3 inhibition and myeloid differentiation in AML
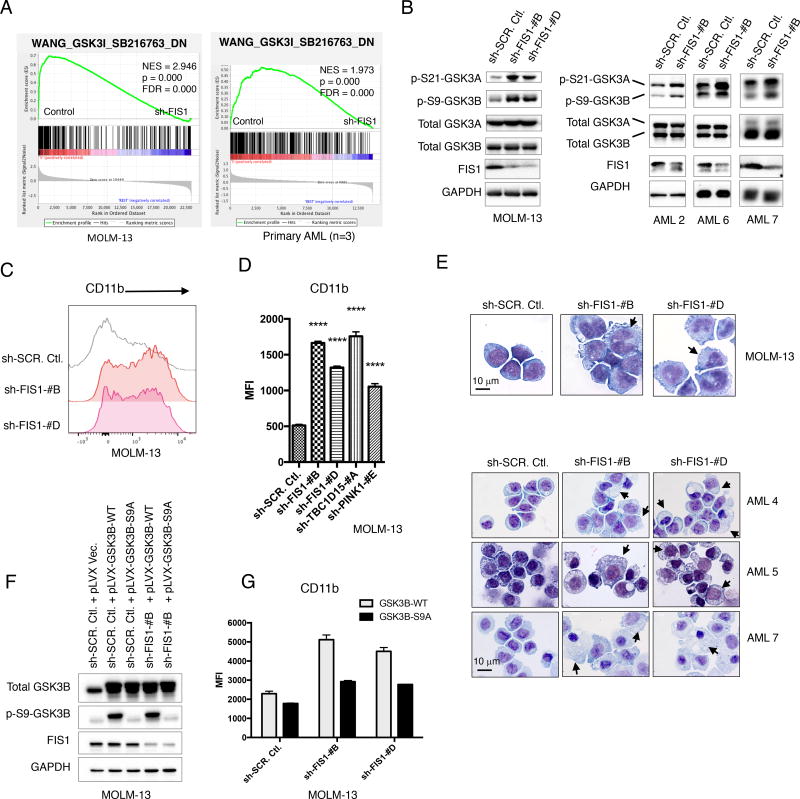
To investigate the mechanism underlying shFIS1-induced loss of stem and progenitor potential in AML, we performed two independent RNA-seq experiments to characterize global gene expression changes induced by shFIS1 in MOLM-13 and a cohort of 3 primary AML specimens (Figure S4A), respectively. Several previous studies have identified a link between GSK3B and DRP1 (Chou et al., 2012; Wu et al., 2013), suggesting the key stem/progenitor potential regulator GSK3 signaling and mitochondrial dynamics could be interconnected. Using gene set enrichment analysis (GSEA), we identified that FIS1 loss in both MOLM-13 and primary AML specimens strongly enriches a list of gene sets representing inactivation of GSK3 signaling in leukemic cells. These gene sets include the WANG_GSK3I_SB216763 gene set which is generated from treating human leukemia cell line RS4;11 cells with GSK3 inhibitor SB216763 (Wang et al., 2010) (Figure 4A). The list also includes the BANERJI_GSK3A_KD and BANERJI_GSK3B_KD gene sets which are produced by specifically knocking down GSK3A and GSK3B in leukemic cell line MOLM-14 cells (Banerji et al., 2012) (Figure S4B). GSK3A and GSK3B are inactivated by phosphorylation at S21 and S9, respectively (McCubrey et al., 2014). To confirm GSK3 inactivation, we probed phospho-S21-GSK3A and phospho-S9-GSK3B in AML cells following FIS1 depletion using both shRNA and CRISPR/Cas9 strategies. Both MOLM-13 and primary AML cells with depleted FIS1 expression showed elevated phospho-S21-GSK3A and phospho-S9-GSK3B (Figures 4B and S4E), indicating that GSK3 inactivation occurs as a result of losing FIS1.
Figure 4. Loss of FIS1 induces GSK3 inhibition and myeloid differentiation in primary AML.
(A) GSEA enrichment plots showing loss of FIS1 in MOLM-13 (3 technical replicates) and primary AML cells (3 biological replicates) results in down-regulation of the WANG_GSK3I_SB216732_DN gene set. “sh-FIS1” represents sh-FIS1-#B and sh-FIS1-#D together.
(B) WB images showing activity of GSK3 signaling in MOLM-13 and primary AML cells at day 6 following FIS1 knock down.
(C) Histogram showing flow cytometry analysis of CD11b expression in MOLM-13 cells at day 6 following FIS1 knock-down.
(D) Quantification of CD11b flow stain signal in MOLM-13 cells at day 6 following knockdown of each target gene. Mean ± SD, (n=3). Type 2, two-tailed t-test.
(E) Representative Giemsa staining images showing morphology of MOLM-13 and primary AML cells at day 6 following FIS1 knock-down. Arrows highlight distinct morphology indicative of differentiation.
(F) WB images showing expression of total GSK3B, p-S9-GSK3B and FIS1 in MOLM-13 cells engineered to express GSK3B-WT or GSK3B-S9A alleles with or without FIS1 knock-down.
(G) Quantification of CD11b flow stain signal in MOLM-13 cells at day 6 following simultaneous expression of GSK3B-WT or GSK3B-S9A alleles and knock-down of FIS1. Mean ± SD, (n=3).
In D and G, MFI stands for median fluorescent intensity.
See also Figure S4.
Inactivation of GSK3 signaling can lead to myeloid differentiation and impaired growth in various subtypes of primary AML (Banerji et al., 2012). Given that both enrichment of the GSK3 inhibition signature and inhibitory phosphorylation of GSK3 proteins are observed, we next investigated whether shFIS1-induced GSK3 inactivation would also result in a differentiation phenotype. First, we observed that shFIS1 enriches the KEGG_HEMATOPOIETIC_CELL_LINEAGE gene set and induces global upregulation of many hematopoietic lineage markers including CD33, CD22, CD14 and CD11b (Figures S4C and S4D). Next, flow cytometry analysis verified a strong increase of myeloid differentiation marker CD11b in FIS1-depleted MOLM-13 cells grown in culture (Figure 4C), methylcellulose (Figure S4F), and engrafted in immune-deficient mice (Figure S4G). Interestingly, knock-down of other mitophagy regulators such as TBC1D15 and PINK1 also strongly up-regulated CD11b expression (Figure 4D), suggesting inhibition of mitophagy activity in general can trigger myeloid differentiation in AML. Finally, using Giemsa staining, we observed that FIS1-depleted MOLM-13 and primary AML cells displayed crumpled cytoplasmic membranes, nuclear condensation and decreased nuclear-to-cytoplasmic area ratio, indicative of clear myeloid differentiation (Figures 4E and S4H). Lastly, despite a strong global upregulation of hematopoietic linage genes, we did not detect significant changes in histone H3K27ac marks (Figure S4I), an event often associated with activation of gene expression during stem cell differentiation (Atlasi and Stunnenberg, 2017).
To investigate whether GSK3 inhibition is required for the differentiation phenotype seen in the FIS1-depleted leukemic cells, we performed genetic rescue experiments. GSK3B-wt or GSK3B-S9A alleles were successfully expressed in combination with or without shFIS1 knock-down in MOLM13 cells (Figure 4F and Figure S4J). The GSK3B-S9A allele cannot be inhibited through phosphorylation of its Ser9 residue (Figure 4F), thus is commonly used as a constitutively active allele of GSK3B (Lang et al., 2013). As shown in Figure 4G, co-expression of the GSK3B-S9A but not the GSK3B-wt allele suppressed shFIS1-induced CD11b increase, suggesting that the shFIS1-induced differentiation phenotype is mediated at least in part by GSK3B inactivation. Expression of the GSK3B-S9A allele however did not rescue shFIS1-induced CFU loss (Figure S4K), suggesting multiple mechanisms exist downstream of FIS1 that control stem and progenitor potential of AML in addition to GSK3B. Lastly, expression of the constitutively active GSK3B-S9A allele failed to rescue shFIS1-induced mitochondrial accumulation phenotype (Figures S4L and S4M), suggesting GSK3B inactivation is likely downstream of the shFIS1-induced mitophagy defect event.
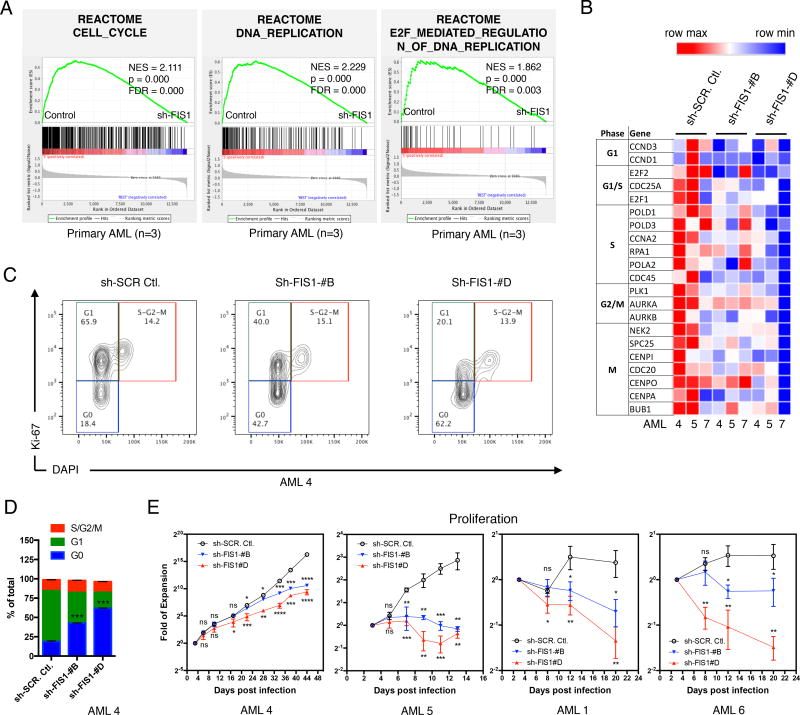
Depletion of FIS1 induces cell cycle arrest in AML
In addition to its role in maintaining leukemia transformation and suppression of differentiation (Banerji et al., 2012; Wang et al., 2010), active GSK3 signaling is also known to promote cell cycle activity in AML (Wang et al., 2008). Consistent with this observation, we also found that FIS1 loss in both primary AML specimens and MOLM-13 cells acted to strongly modulate cell cycle activity. As shown in Figures 5A and S5A, cell cycle inhibition is clearly evident from GSEA analysis of multiple cell cycle-related gene sets. In addition, comparative analysis of control vs. FIS1 knock-down showed a trend towards global inhibition of many representative cell cycle related genes at all major stages (G1, G1/S, S, G2/M and M) upon FIS1 loss (Figure 5B). The consequence was substantially reduced cell cycle activity shown by Ki-67/DAPI staining (Figures 5C and 5D) and significantly lower in vitro proliferation rate shown by cell count in primary AML cells (Figure 5E and Figure S5B). Together, the data in Figures 4 and 5 indicate that the strong impact of FIS1 loss on stem and progenitor potential of AML is related to its downstream GSK3 inactivation and cell cycle arrest effects.
Figure 5. Depletion of FIS1 induces cell cycle arrest in AML.
(A) GSEA enrichment plots showing loss of FIS1 in primary AML cells (3 biological replicates) results in down-regulation of 3 cell cycle-related gene sets from the GSEA Reactome collection. “sh-FIS1” represents sh-FIS1-#B and sh-FIS1-#D together.
(B) A heat map showing expression of representative cell cycle-related genes in primary AML cells with or without FIS1 knock down. Data are generated from RNA-seq analysis. Note that the genes included in this heat map do not necessarily meet the adjusted p-value of 0.05.
(C) Flow cytometry analysis of cell cycle status in primary AML cells at day 6 following FIS1 knock-down. Cell cycle profile is revealed by Ki-67 and DAPI staining.
(D) Quantification of the cell cycle profile shown in (C). Mean ± SD, (n=3). Type 2, two-tailed t-test was used to compare %G0 between control and knock-downs.
(E) Fold of expansion of primary AML cells cultured in complete serum-free media, plotted in log2 scale. Mean ± SD, (n=3). For each time point, knock-down clones are compared to the control using Type 2, two-tailed t-test.
See also Figure S5.
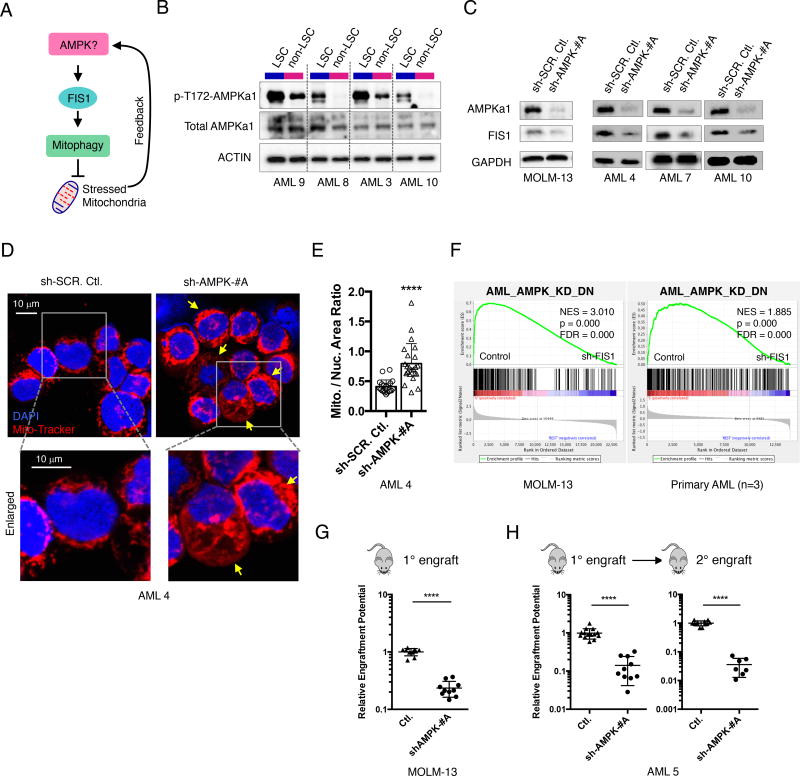
AMPK is upstream of FIS1
While the abovementioned studies demonstrated that FIS1-mediated mitophagy activity is important for the stem and progenitor potential of LSCs, the upstream signaling that maintains FIS1 activity in LSCs has not been characterized. A recent study in a mouse model reported that AMPK signaling is up-regulated in response to energetic stress in mouse LSCs (Saito et al., 2015). In addition, a separate study in osteosarcoma cells showed that AMPK is upstream of MFF (mitochondrial fission factor), another mitochondrial dynamics regulator (Toyama et al., 2016). Thus, we hypothesized that AMPK signaling might also regulate the activity of FIS1 in human LSCs as a means to manage mitochondrial stress (Figure 6A). To test this hypothesis, we first compared the activity of AMPK signaling in freshly sorted ROS-low LSCs versus ROS-high non-LSCs. Our results showed that AMPK is constitutively activated in LSC relative to non-LSC populations (Figure 6B), suggesting that AMPK may play an important role in human LSCs as well. To directly test if AMPK is upstream of FIS1, we knocked down AMPKa1(PRKAA1) subunit in both MOLM-13 and primary AML cells and measured its impact on FIS1 expression (Figures S6A–S6D). Our data clearly show that knock-down of AMPKa1 resulted in significant FIS1 mRNA and protein loss (Figures 6C and S6C), providing strong evidence that AMPK is upstream of FIS1. As a master regulator of energy metabolism, modulation of AMPK activity is expected to have a plethora of downstream effects. Thus, to measure how much the downstream effects of AMPK loss overlap with FIS1 loss, we examined if shAMPK also affects mitochondrial morphology in primary AML cells. Using confocal microscopy, we observed that the shAMPK primary AML cells also display a mitochondrial accumulation phenotype (Figures 6D and 6E) similar to that observed for FIS1 knock-down. These data further support AMPK as an upstream regulator of FIS1 in AML cells and suggest that inhibition of AMPK may largely recapitulate the biological effects of FIS1 loss in AML. To investigate this hypothesis, we first performed RNA-seq experiments to characterize global gene expression changes induced by AMPKa1 knock down in primary AML cells. Using a minimal p value of 0.05, we identified a list of significantly down-regulated genes induced by shAMPKa1. These genes were used to generate a gene set termed AML_AMPK_KD_DN (Table S2) to represent the biological effect of AMPK loss in AML. Interestingly, this gene set is significantly enriched by the shFIS1 gene expression profiles obtained from both primary AML cohort and MOLM-13 cells (Figure 6F), suggesting that AMPK loss indeed largely recapitulates the biological effects of FIS1 loss in AML.
Figure 6. Inhibition of AMPK signaling upstream of FIS1 depletes the LSC potential of primary AML.
(A) A diagram showing the rationale underlying AMPK hypothesis.
(B) WB images showing expression of p-T172-AMPKa1 and total AMPKa1 in ROS-low LSCs (L) versus ROS-high non-LSCs (N).
(C) WB images showing expression of FIS1 in MOLM-13 and primary AML cells at day 6 following AMPKa1(PRKAA1) knock-down.
(D) Representative confocal images showing mitochondrial morphology of primary AML cells at day 6 following shRNA-mediated knock down of AMPKa1. Arrows highlight mitochondrial accumulation.
(E) Mitochondrial to nuclear area ratio in control and shAMPK AML cells. Each dot represents a single cell. Mean + SD. Type 3, two-tailed t-test.
(F) GSEA enrichment plots showing loss of FIS1 in MOLM-13 (3 technical replicates) and primary AML cells (3 biological replicates) result in down-regulation of the AML_AMPK_KD_DN gene set. “sh-FIS1” represents the sh-FIS1-#B and sh-FIS1-#D together.
(G) Normalized relative engraftment potential of MOLM-13 cells with or without AMPK knock-down.
(H) Normalized relative engraftment potential of primary AML 5 with or without AMPK knock-down. 1° and 2° indicate primary and secondar y xenograft experiments, respectively.
In G and H, each dot represents an individual mouse and lines represent mean ± SD. Type 2, two-tailed t-test.
See also Figure S6.
Given that AMPK is upstream of FIS1, we next wanted to assess the impact of AMPK loss on AML LSC function. To this end, the engraftment ability of AML cells following shRNA-mediated depletion of AMPKa1 was measured in immune-deficient mice. Upon knock-down of AMPKa1, both MOLM-13 cells (Figure S6B) and primary AML specimens (Figures S6D) show greatly reduced engraftment potential (Figures 6G and 6H). Notably, the AMPK depleted primary AML cells show further loss of engraftment potential in secondary transplantation assays (Figure 6H, right), suggesting AMPK loss, like FIS1 depletion, can also impair long-term LSC potential. Together, these data demonstrate that AMPK regulates the activity of human AML LSCs, and indicates that the mechanism involves downstream modulation of FIS1 activity.
DISCUSSION
In AML, mitochondrial translation, mitochondrial DNA copy number, basal oxygen consumption rates and many other properties of mitochondria have been found to be differentially regulated in comparison to normal hematopoietic cells (Cole et al., 2015; Liyanage et al., 2017; Schimmer and Skrtic, 2012; Skrtic et al., 2011; Yeung et al., 2015), suggesting altered mitochondrial activity is important for leukemogenesis. Further, emerging studies have now described the highly dynamic nature of mitochondria, and their increasingly evident role in multiple cellular processes (Wai and Langer, 2016). Mito-fission, mito-fusion and mitophagy appear to co-exist in a homeostatic balance that is unique to varying cell types and physiological conditions. However, the biological significance of mitochondrial dynamics in cancer stem cells is as yet largely unknown (Chen and Chan, 2017). In the present study, we report that AML LSCs reside in a unique state of mitochondrial dynamics mediated by activation of AMPK and FIS1. This axis functions to inhibit differentiation via a GSK3-mediated downstream mechanism and supports self-renewal of primitive AML cells.
We show that the expression of FIS1 is significantly higher in AML LSCs versus non-LSCs. To date, only one previous report has linked FIS1 to AML biology. That study sought to identify poor prognosis markers of AML, and found that expression of FIS1 is higher in AML patients who are refractory to chemotherapy in comparison to drug responsive patients (Tian et al., 2014). Notably, several reports have indicated that LSC activity correlates with chemo-resistance in AML (Eppert et al., 2011; Gentles et al., 2010), suggesting a link between overall FIS1 expression, LSC biology and therapy resistance. Further, our studies have recently demonstrated that relapsed AML patients increase the frequency of LSCs by 9–90 fold (Ho et al., 2016). Thus, increased expression of FIS1 in refractory or relapsed AML may be a reflection of increased LSC burden.
The present study also observed a distinct mitochondrial morphology in AML LSCs. The compact crescent-like localization pattern of mitochondria in LSCs may suggest asymmetrical division, an activity that has been observed in HSCs (Ito et al., 2012). Upon depletion of FIS1 in AML cells, dramatic accumulation of mitochondria and more diffuse mitochondria localization were clearly evident, suggesting FIS1 is required to maintain the unique mitochondrial morphology seen in AML LSCs.
Importantly, our data clearly show that inhibition of FIS1 expression impairs the stem and progenitor potential of AML and this phenotype is largely recapitulated by inhibition of other mitophagy players PINK1 and TBC1D15, suggesting mitophagy activity is critical for sustaining AML LSCs. Our results also show that FIS1 activity is less important for normal HSPCs as evidenced by the limited impact of FIS1 knock-down on the mitochondrial morphology and the engraftment ability of HSPCs. These results indicate a possible therapeutic index between HSCs and LSCs when targeting the FIS1 axis. It is important to note however that mitophagy has been found to be required for normal HSC (Ito et al., 2016; Joshi and Kundu, 2013; Mortensen et al., 2011), suggesting the dependency of stem cell function on mitophagy is evolutionarily conserved. A more recent study shows that loss of autophagy in HSCs can cause mitochondrial accumulation and activate metabolic state, which impairs HSC self-renewal and regenerative potential (Ho et al., 2017). Thus, the possible therapeutic index may be specifically related to FIS1-mediated mitophagy. Given that multiple mitophagy pathways exist in addition to the FIS1/PINK1 axis described in the current study (Hamacher-Brady and Brady, 2016), we propose that the exact molecular mechanisms that maintain mitophagy activity might be different between HSCs and LSCs. In addition, our data suggest normal HSPCs may have a slower mitophagy rate compared to LSCs, suggesting they may better tolerate acute mitophagy inhibition. Lastly, our previous study showed that human AML LSCs preferentially rely on mitochondrial oxidative phosphorylation for energy production (Lagadinou et al., 2013). Given that mitophagy is a quality control mechanism for the health of mitochondria, and that oncogenic transformation increases cellular stress and potentially damages mitochondria, then it is possible that clearance of dysfunctional mitochondria through mitophagy would be very important for LSCs that require mitochondrial oxidative phosphorylation for survival. Thus, despite a role for mitophagy in normal HSCs, our data suggest that targeting specific mechanisms such as the AMPK/FIS1 axis can provide selective eradication of LSCs.
Notably, studies of CSCs in other cancer types including melanoma, glioblastoma and pancreatic cancer have also identified a dependence on oxidative phosphorylation (Roesch et al., 2013; Viale et al., 2014; Vlashi et al., 2011), suggesting targeting mitochondrial stress response mechanisms such as mitophagy could be applicable for selective eradication of CSCs in multiple types of cancer.
The GSK3B-SA9 rescue experiments support the concept that GSK3 inhibition is necessary for shFIS1-induced myeloid differentiation phenotype. The same allele however failed to rescue shFIS1-induced CFU loss and the mitochondrial accumulation phenotype. These data suggest: (1) multiple mechanisms exist downstream of FIS1 that control stem and progenitor potential of AML in addition to GSK3B. (2) GSK3B inactivation likely happens downstream of the shFIS1-induced mitophagy defect event. (3) GSK3A inactivation may also contribute to multiple phenotypes induced by FIS1 loss.
It is unclear how FIS1-mediated mitophagy impacts GSK3 phosphorylation. One possibility is through the PTEN/Akt/GSK3 axis. A recent study reported an N-terminally extended form of PTEN, known as PTENα, specifically localizes to mitochondria, where it forms a complex with canonical PTEN to increase PINK1 expression and promote energy production (Liang et al., 2014). This study raises the possibility that in the shFIS1 AML cells, the increased expression of PINK1 could be induced by formation of the PTENα/PTEN complex in the mitochondria. It would further suggest that shFIS1 can induce the translocation of canonical PTEN from cytosol to mitochondria, resulting in PTEN activity loss in the cytosol, subsequent Akt activation and GSK3 inhibition. This hypothesis is potentially interesting since it suggests that mitochondria have the ability to actively signal central metabolic pathways including the Akt pathway to “adapt” to differing physiological states.
In addition to GSK3 inhibition, we also observed strong cell cycle arrest in AML cells following FIS1 depletion. Cell cycle arrest often accompanies differentiation (Ruijtenberg and van den Heuvel, 2016), thus this observation may simply be a consequence of induced differentiation. Obviously, cell cycle arrest will block the growth of bulk leukemia, but it may also have implications in targeting LSCs. Several recent studies have suggested that LSCs may also display a more actively cycling phenotype. For example, in MLL-rearranged leukemia, CD93 marks a subset of actively cycling, non-quiescent AML cells enriched for LSC activity (Iwasaki et al., 2015). Similarly, human AML LSCs may not be enriched following chemotherapy treatment, suggesting some degree of cell cycle activity in them (Farge et al., 2017). Thus, we believe that the FIS1-induced cell cycle arrest may also contribute to the loss of LSC activity.
Lastly, although our data show AMPK can regulate FIS1 at both mRNA and protein levels, it remains a question whether the link between AMPK and FIS1 is direct or indirect. Importantly though, AMPK signaling is also preferentially active in ROS-low LSCs and inhibition of AMPK results in loss of LSC potential, recapitulating the anti-LSC effect of FIS1 loss in AML. These data suggest AMPK signaling could be a critical link between cellular/mitochondrial stress and activation of FIS1-mediated mitophagy (Figure 7). A recent study has convincingly demonstrated that AMPK activation is required to protect bone marrow LSCs from metabolic stress in a mouse model of MLL-AF9 induced leukemia (Saito et al., 2015). Others have shown that AMPK signaling can directly activate MFF, a critical regulator of mito-fission, in response to energy stress (Toyama et al., 2016). Our results suggest AMPK can also regulate mitophagy activity under mitochondrial stress. Given the fact that mito-fission and mitophagy are closely linked, it is reasonable to speculate that such dual abilities of AMPK in regulating both mito-fission and mitophagy make it an attractive target for therapies designed to selectively eradicate LSCs.
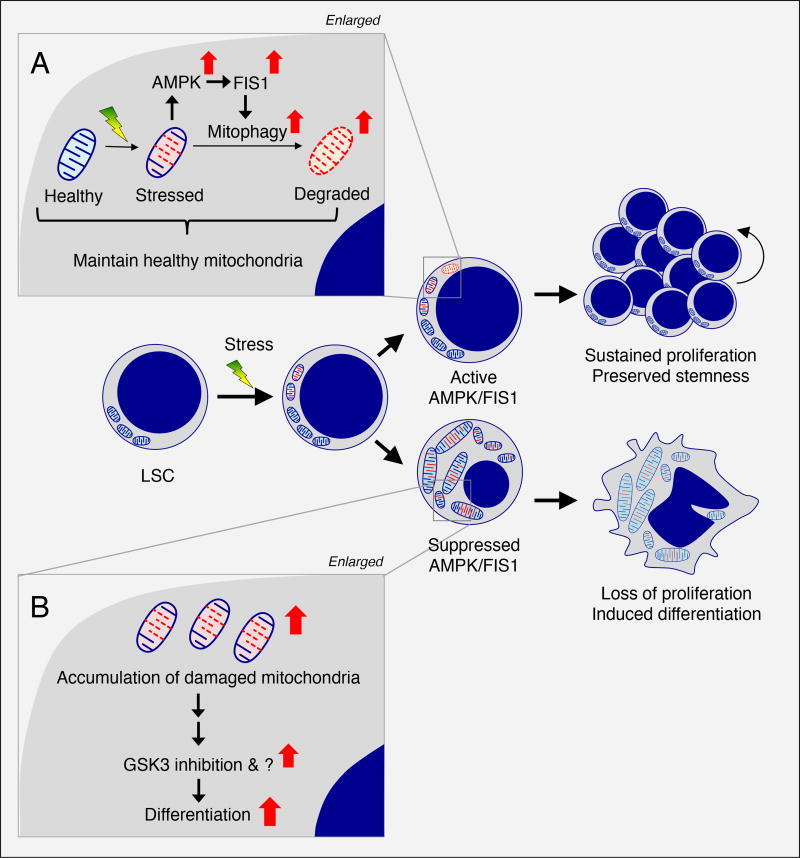
Figure 7. Proposed model for dependency of AML LSCs on AMPK/FIS1-mediated mitophagy.
(A) In LSCs, mitochondrial stress generated from oncogenic transformation may activate AMPK signaling which in turn drives FIS1-mediated mitophagy to degrade stressed mitochondria, maintaining a healthy mitochondrial network required for LSC potential.
(B) In LSCs with suppressed AMPK/FIS1 activity, accumulation of damaged mitochondria can lead to GSK3 inhibition and other unknown events, together inducing differentiation and impairing LSC function.
To date, the only other study that directly documents a role for mitochondrial dynamics in a cancer stem cell population was reported by Xie et al, who showed aberrant activation of DRP1 promotes increased mito-fission to support the survival of brain tumor-initiating cells (BTICs) in glioblastoma (Xie et al., 2015). Intriguingly, in the BTIC model, AMPK signaling is downstream of DRP1, and can rescue the loss of DRP1 signaling, suggesting that while mitochondrial dynamics may be broadly relevant to cancer initiating cells, the way in which mitochondria dynamics mediate “stemness” may be specific to different tumor types.
In conclusion, as outlined in Figure 7, human AML LSCs show intrinsic activation of AMPK, which we propose is a response to increased cellular and/or mitochondrial stress arising as a consequence of oncogenic transformation. AMPK activation in turn up-regulates FIS1-mediated mitophagy to facilitate the degradation of stressed mitochondria. This process likely functions as a quality control mechanism to maintain a healthy mitochondrial network, which is required to preserve stemness in LSCs (Figure 7A). Disruption of AMPK/FIS1-mediated mitophagy results in accumulation of defective mitochondria, inhibition of GSK3 and induction of differentiation (Figure 7B). This model suggests that targeting the AMPK/FIS1-mediated mitophagy axis may represent a promising strategy to eradicate AML LSCs.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Craig T. Jordan (craig.jordan@ucdenver.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human studies
Primary human AML specimens were obtained from apheresis product, peripheral blood, or bone marrow of AML patients who gave informed consent for sample procurement on the University of Colorado tissue procurement protocols (Colorado Multiple Institutional Review Board Protocol #12-0173 & #06-0720). Normal cord / peripheral blood specimens were obtained from volunteer donors who gave informed consent on a research subjects review board approved protocol at the University of Colorado (Colorado Multiple Institutional Review Board Protocol #12-0173). See Table S3 for age, sex, cytogenetics and mutation information on human specimens used in the current study.
In vivo animal studies
NSGS (NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ) mice (The Jackson Laboratory) were used for transplantation of MOLM-13, primary AML and normal CD34+ PBMC/CBMC cells in this study. Female mice ranging in age from 6 to 8 weeks were started on experiment with experiments lasting 6 to 12 weeks. Littermates of the same sex were randomly assigned to experimental groups. During all experiments, the weight of mice was approximately 20–30 grams with no animals losing greater than 10% body weight. The mice were kept in ventilated cages in the vivarium at University of Colorado. All animal work were performed in accordance with Institutional Animal Care and Use Committee protocol number 00308.
Cell lines and primary cultures
Primary human AML cells, normal cord/peripheral blood mononuclear cells (CBMCs/PBMCs) were resuspended at about 100–200 e6 cells per ml in freezing media composed of 50% FBS (GE Healthcare), 10% DMSO (Sigma) and 40% IMDM media (Gibco) and then cryo-preserved in liquid nitrogen. Cells were thawed in 37°C water bath, washed twice in thawing media composed of IMDM (Gibco), 2.5% FBS (GE Healthcare) and 10 ug/ml DNAse (Sigma). Normal CD34+ CBMCs/PBMCs were enriched from CBMCs/PBMCs using the CD34 MicroBead kit (Miltenyi Biotec). Cells were cultured in normal or complete serum-free media (SFM) in 37°C, 5% CO2 incubator. Normal SFM is composed of IMDM (Gibco), 20% BIT 9500 (STEMCELL Technologies), 10ug/ml LDL (Low Density Lipoprotein, Millipore), 55uM 2-Mercaptoethanol (Gibco) and 1% Pen/Strep (Gibco). Complete SFM were made by supplementing the normal SFM with FLT-3, IL-3 and SCF cytokines (PeproTech), each at 10 ng/ml. MOLM-13 cells were authenticated via STR profiling, cultured in MOLM-13 media composed of RPMI-1640 media (Gibco), 10% FBS (GE Healthcare) and 1% Pen/Strep (Gibco).
METHOD DETAILS
Sorting of LSCs and non-LSCs based on ROS profile
After thawing, about 100 e6 primary AML cells were stained with anti-CD45, anti-CD3 and anti-CD19 in FACS buffer (PBS + 0.5% FBS) at 4°C for 15 minutes. The cells were then washed one time in FACS buffer and stained with 5uM CellRox-DeepRed dye (Life Technologies) in ROS staining buffer (PBS + 2.5% FBS) at 37°C in CO2 incubator for 30 minutes. After CellRox staining, the cells were washed twice in ROS staining buffer and resuspended in FACS buffer containing 10uM DAPI to label late apoptotic cells. The BD FACSAria II Cell Sorter was used to analyze ROS profile and sort. The stained cells were gated through three serial gates including first DAPI-negative, then blast-positive (CD45 and SSC), and finally CD3 and CD19 double negative gates to enrich for viable lymphoid-excluded leukemic blasts. Within these cells, the ROS-low LSCs and ROS-high non-LSCs were identified and sorted at the bottom and top 10–15% of CellRox-DeepRed dye distribution, respectively (Figure S1C). A greater than 95% sorting purity was confirmed for all specimens using post-sort analysis (Figure S1C).
Transmission Electron Microscopy
Freshly sorted ROS-low LSCs and ROS-high LSCs were first embedded in 2% low-melting agarose in PBS, and then fixed in 4% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M cacodylate buffer, overnight at 4°C. This was followed by a secondary fixation step in 2% osmium tetraoxide for 1 hour and a tertiary fixation step in 2% uranyl acetate for 2 hours. The samples were then dehydrated in graded ethanol series: 15 minutes each in 25%, 50%, 75%, 90%, 95% and 3 times of 100% ethanol. After dehydration, samples were subjected to changes of propylene oxide for 3 times, 10 minutes each and followed by infiltration with resin : propylene oxide mix series: 2 hours in 1:2 resin : propylene oxide, 2 hours in 1:1 resin : propylene oxide and finally overnight in pure resin. Resin was made by mixing epon with araldite. After resin exchange, samples were put in embedding molds and cook overnight at 60°C. 85–200nm sections were cut, stained with uranyl acetate and lead citrate to enhance contrast, and then imaged on a FEI Tecnai F30, 300kV FEG- Transmission Electron Microscope equipped with a Gatan Ultrascan US-4000(4k-4k) CCD camera operated at 2k-2k to maximize efficiency and throughput. Quantification of mitochondrial volume was performed by first drawing contour of the cross-section of each mitochondrion and then calculating enclosed area in the ImageJ software version 1.50i.
Confocal Microscopy
MitoTracker-DeepRed dye (Thermo Scientific) was added into culture media at a final concentration of 100–500 nM to label the mitochondria of leukemic cells for 30 minutes in 37°C 5% CO2 incubator before harvesting cells for c onfocal studies. To prepare slides, labeled cells were washed 2 times with ice cold FACS buffer, 1 time with ice cold PBS and resuspended at a final concentration of ~500K cells /ml in ice cold PBS. 150 ul of cell suspension (~75K cells) was used for cytospin at 300rpm for 3 minutes at room temperature. The slides were air dried in dark box at room temperature for 5 minutes before fixed in −20°C 100% methanol for 10 minutes. The slides were then air dried in dark box at room temperature for 10 minutes and mounted in ProLong Gold Antifade Mountant solution (Thermo Scientific) before confocal imaging. The stained slides were imaged on the Zeiss LSM 780 confocal microscope using its Coherent Chameleon Ultra II laser to excite the DAPI dye and its 633nm laser to excite the MitoTracker-DeepRed dye.
Quantification of mitochondrial to nuclear area ratio
Quantification of mitochondrial to nuclear area ratio was performed in the ImageJ 1.51s software (Schneider et al., 2012). For each experimental condition, we analyzed 3 or more representative fields that pool together ~30–60 cells for the quantification. We then submit each cell to an automated macro function written in the ImageJ software, allowing segregation of DAPI and Mito-Tracker channels and subsequent quantification of the area of each channel (illustrated in Figure 1E). The macro function involves following major scripts in sequence: 1) For each cell: run("Split Channels"); 2) For each channel: setAutoThreshold("Default dark"); //run("Threshold…"); run("Convert to Mask"); run("Analyze Particles…", "size=8-Infinity show=Outlines clear include summarize in_situ"); 3) run(“Close”);. These scripts together allowed automated quantification of the area of each channel per cell. And finally, the ratio of mitochondrial to nuclear area was calculated in Excel and the results were plotted in Prism.
mRNA isolation and qPCR
Total mRNA was isolated using the RNeasy plus mini kit (Qiagen) according to manufacturer’s instructions. mRNA purity and quantity were determined with NanoDrop (Thermo Scientific) before qPCR and RNA-seq analysis. For qPCR, mRNA samples were reverse transcribed into cDNA using the iScript One-Step RT-PCR Kit (Bio-Rad). Quantitative Real-Time PCR was performed on LightCycler96 Real-Time PCR system using SYBR Green I Master Mix reagent (Roche). qPCR primers are listed in Table S4.
Protein lysates preparation and WB
Cells were harvested and washed in ice cold PBS before lysed in 1 × RIPA buffer (10mM Tris-Cl (pH 8.0), 1mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl and 1mM PMSF added fresh) or 1× Laemmli Sample Buffer (BIO-RAD) at 5 e6 / ml concentration. The RIPA-lysed samples were quantified using the DC Protein Assay Kit (BIO-RAD). For WB analysis, about 50–200K cells per lane was loaded and resolved on 6–12% SDS-PAGE gels, transferred to PVDF membranes, blocked in 5% milk in TBS with 0.1% Triton X-100 (Sigma). After incubation, the PVDF membranes were incubated with primary antibodies at 4°C overnight, washed, incubated with secondary antibodies at room temperature for 2 hours and subjected to imaging. The WB results were imaged on the ChemiDoc Imaging System and visualized in the Image Lab software (BIO-RAD). ACTIN and GAPDH expression were used as loading controls. The antibodies used are listed in the KEY RESOURCES TABLE.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CD45-Alexa700 | BD | Cat# 560566, RRID:AB_1645452 |
| Anti-CD3-PECY7 | BD | Cat# 557749, RRID:AB_396855 |
| Anti-CD19-PE | BD | Cat# 555413, RRID:AB_395813 |
| Anti-Ki67-Alexa647 | Biolegend | Cat# 350510, RRID:AB_10900821 |
| Anti-H3K27ac | Active Motif | Cat# 39133, RRID:AB_2561016 |
| In vivo anti-human CD3 | BioXCell | Cat# BE0001-2, RRID:AB_1107632 |
| Rabbit anti-FIS1 | Proteintech | Cat# 10956-1-AP, RRID:AB_2102532 |
| Rabbit anti-DRP1 | Cell Signaling | Cat# 8570, RRID:AB_10950498 |
| Rabbit anti-AMPKa1 total | Cell Signaling | Cat# 2793, RRID:AB_915794 |
| Rabbit anti-AMPKa1-p-T172 | Cell Signaling | Cat# 2535, RRID:AB_331250 |
| Rabbit anti-AMPKa1/2 | Cell Signaling | Cat# 2532, RRID:AB_330331 |
| Rabbit anti-GSK3-α/β total | Cell Signaling | Cat# 5676, RRID:AB_10547140 |
| Rabbit anti-GSK3-α/β-p-Ser21/9 | Cell Signaling | Cat# 8566S, RRID:AB_10860069 |
| Rabbit anti-GSK3-β total | Cell Signaling | Cat# 12456, RRID:AB_2636978 |
| Rabbit anti-GSK3-β-p-Ser9 | Cell Signaling | Cat# 5558, RRID:AB_10013750 |
| Rabbit anti-PINK1 | Cell Signaling | Cat# 6946S, RRID:AB_11179069 |
| Rabbit anti-TBC1D15 | Sigma | Cat# HPA013388, RRID:AB_2199233 |
| Mouse anti-GAPDH | Santa Cruz | Cat# sc-32233, RRID:AB_627679 |
| Goat anti-ACTIN | Santa Cruz | Cat# sc-1616, RRID:AB_630836 |
| Biological Samples | ||
| Primary AML | Craig Jordan Lab | n/a |
| Normal CBMC | Craig Jordan Lab | n/a |
| Normal PBMC | Craig Jordan Lab | n/a |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CellRox-DeepRed | Life Technologies | Cat# C10422 |
| MitoTracker-DeepRed FM | Life Technologies | Cat# M22426 |
| Polybrene (Hexadimethrine bromide) | Sigma | Cat# 107689-10G |
| Critical Commercial Assays | ||
| Mitochondria Isolation Kit for Cultured Cells | Thermo Scientific | Cat# 89874 |
| CD34 MicroBead Kit | Miltenyi Biotec | Cat# 130-046-702 |
| KWIK-DIFF Stain Kit | Thermo Scientific | Cat# 9990700 |
| ThruPLEX DNA-seq Kit | Rubicon Genomics | Cat# R400427 |
| Deposited Data | ||
| RNAseq of MOLM-13 cells with FIS1 knock-down | This paper | GSE114111 |
| RNAseq of primary AML with FIS1 knock-down | This paper | GSE114109 |
| RNAseq of primary AML with AMPK knock-down | This paper | GSE114105 |
| CHIPseq of MOLM-13 cells with FIS1 knock-down | This paper | GSE114106 |
| Experimental Models: Cell Lines | ||
| MOLM-13 | Tobias Neff Lab | n/a |
| 293T | Craig Jordan Lab | n/a |
| Experimental Models: Organisms/Strains | ||
| NSGS (NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3, CSF2,KITLG)1Eav/MloySzJ) | The Jackson Laboratory | Cat# JAX:013062, RRID:IMSR_JAX:013062 |
| Recombinant DNA | ||
| pLKO.1-GFP | Craig Jordan Lab | n/a |
| pLKO.1-Crimson | Craig Jordan Lab | n/a |
| pPax2 | Craig Jordan Lab | n/a |
| pMD2.G | Craig Jordan Lab | n/a |
| pLVX-EF1a-IRES-mCherry | Clontech | Cat# 631987 |
| LentiCRISPR-V2-GFP | Addgene | Cat# 82416 |
| HA GSK3 beta wt pcDNA3 | Addgene | Cat# 14753 |
| HA GSK3 beta S9A pcDNA3 | Addgene | Cat# 14754 |
| Software and Algorithms | ||
| bcl2fastq v1.8.4 | Illumina | n/a |
| Trimmomatic v0.32 | Usadel Lab | n/a |
| cufflinks v2.0.2 | Trapnell Lab | n/a |
| Bowtie2 v2.2.6 | Salzberg Lab | n/a |
| Flowjo v10.1 | Flowjo | n/a |
| Prism v6.0h | Prism | n/a |
| ImageJ v1.51s | Imagej.nih.gov | n/a |
Colony-forming unit assay
48 hours post lentiviral infection, freshly sorted DAPI− and GFP+ (healthy shRNA-expressing) MOLM-13, primary AML and CD34+ normal PBMC/CBMC cells were plated in triplicates in Human Methylcellulose Complete Media (R&D Systems) and grown in 37°C, 5% CO2 incubator for 2–4 weeks before imaging and counting. For MOLM-13, ~2–5K cells per ml were plated. For primary AML, ~50–250K cells per ml were plated. For CD34+ normal PBMC/CBMCs, ~1–5K cells per ml were plated.
Xenograft studies
NSGS mice were conditioned 24 hours prior to transplant with 25 mg/kg busulfan via intra-peritoneal (IP) injection. MOLM-13, primary AML and normal CD34+ PBMC cells were infected with shRNA lentivirus 48 hours prior to transplant. For MOLM-13, per mouse, 1.0 e6 cells in 0.1ml saline were tail vein injected. For primary AML specimens, per mouse, about 2.5 e6 cells in 0.1ml saline were tail vein injected. For normal CD34+ PBMCs experiments, CD34− cells were saved during CD34 enrichment, cultured in complete SFM side-by-side with infected CD34+ cells, and served as helper cells for transplant. The CD34+ and CD34− cells were mixed to achieve a final 5–10% CD34+ cells in the injected samples. The mixture was injected at about 0.5–1 e6 cells in 0.1ml saline per mouse via tail vein injection. For primary AML and normal CD34+ PBMC experiments, in vivo anti-human CD3 antibody (BioCell) was added at a final concentration of 1ug/e6 cells to prevent potential graft versus host disease. For all experiments, 10–12 mice were injected per experiment group. Mice engrafted with MOLM-13 cells were sacrificed after 2 weeks when tumor burden was at about 15–50% in the bone marrow. Mice engrafted with primary AML and normal CD34+ PBMC cells were sacrificed after 5–8 weeks when percentage of engraft was about 15–50%. Bone marrow was flushed from tibia and femur bones, subjected to red blood cell lysis, and stained with human CD45 antibody and DAPI to label live engrafted human cells.
Calculation of relative engraftment potential
Using flow cytometry, we first measured % of GFP+ cells within the DAPI− gate to determine the % of shRNA-expressing cells injected (%GFP-injected). After engraftment, we also measured % of GFP+ cells within the DAPI− and human CD45+ gate to determine the % of shRNA-expressing cells engrafted (%GFP-engrafted). For each experimental group (e.g. control as group x and shFIS1 as group y), we first calculated engraftment value x or y using the following formula: x or y = %GFP-engrafted ÷ %GFP-injected for each mouse. This calculation yielded an array of engraftment values for both control (x1, x2, x3, … x10) and shFIS1 (y1, y2, y3, … y10) groups. To calculate the final relative engraftment potential score, each engraftment value was normalized by the mean of (x1, x2, x3, …x10) from the control group. These scores were plotted to compare the relative engraftment potential between control and shFIS1.
Giemsa staining
Cells were cyto-spun to glass slides at about 0.1e6 per spot, air dried at room temperature for 1 hour and then stained using the KWIK DIFF Stain Kit (Thermo Scientific) according to manufacturer’s protocol. After staining, cells were dried at room temperature, mounted in Cytoseal 60 (Thermo Scientific) and submitted for imaging on Zeiss AX10 microscope (Zeiss).
Over-expression of GSK3B alleles
DNA sequences coding GSK3B-wt and GSK3B-S9A alleles were cut from the Addgene plasmids and inserted into the pLVX-EF1a-IRES-GFP vector (Clontech) to allow subsequent production of lentivirus from 293T cells. Lentiviral particles were harvested and used to infect MOLM-13 cells as described in the shRNA methods.
Cell cycle analysis
For each condition, 1–2 e6 cells were fixed in 1% Formaldehyde (Polysciences) on ice for 15 minutes, permeablized in 0.2% Triton X-100 (Sigma) on ice for another 15 minutes, washed in ice cold FACS buffer, and then stained with anti-Ki67 (BD) for 45 minutes on ice. After staining, cells were washed twice in FACS buffer and resuspended in FACS buffer containing 10uM DAPI. Cells were examined on the BD FACSCelesta Flow Cytometer (BD) and the results were analyzed in Flowjo version 9.0.
shRNA and lentiviral production
shRNA sequences were constructed into pLKO.1-GFP or pLKO.1-Crimson vectors, which contain a human U6 promoter to drive shRNA expression and a human PGK promoter to drive GFP or Crimson expression to label shRNA-expressing cells. The detailed methods for generating lentiviral particles, and infecting AML cells are described here (Ashton et al., 2012). Briefly, each pLKO.1-shRNA construct was co-transfected with pPax2 (provides packaging proteins) and pMD2.G (provides VSV-g envelope protein) plasmids into 293T cells (System Bioscience) to produce lentiviral particles. At about 48 hours post transfection, 293T culture supernatants containing lentiviral particles were harvested, filtered, aliquoted and stored in −80°C freezer for future use. At the day of infection, these lentivirus aliquots were thawed and used to infect cells. Primary AML, normal CD34+ CBMC/PBMCs were typically infected by mixing 1 e6 cells in 0.1ml complete SFM with 0.1ml of freshly thawed lentivirus and Polybrene at a final concentration of 10 ug/ml. MOLM-13 cells were typically infected by mixing 1 e6 cells in 0.5ml MOLM-13 media with 0.1ml of freshly thawed lentivirus and Polybrene at a final concentration of 10 ug/ml. These numbers were proportionally scaled when a different number of cells needed to be infected. At 3–6 days post infection, GFP-positive or Crimson-positive cells were sorted or analyzed. shRNA targeting sequences are listed in Table S5.
CRISPR/Cas9-mediated disruption of FIS1
The FIS1-targeting small guiding RNA (sg-FIS1) was designed according to the AVANA library (Doench et al., 2016) and inserted into the LentiCRISPR-V2-GFP vector (Sanjana et al., 2014). For control purpose, we also designed a sg-CTL sequence targeting a non-essential gene. Upon successful insertion of the sgRNA sequences, we generated the LentiCRISPR_V2_GFP_sg-CTL (sg-CTL) and the LentiCRISPR_V2 _GFP_sg-FIS1 (sg-FIS1) constructs that contain FIS1-targeting and control sgRNAs, a Cas9-coding gene, and a GFP marker allowing for sorting of successfully transduced cells. As shown in Figure S7A, we produced lentiviral particles using both constructs, infected MOLM-13 cells, and FACS sorted GFP-positive sg-CTL and sg-FIS1 cells. We then diluted the sorted GFP-positive cells to allow seeding these cells on 96-well plates at less than 1 cell per well. After about 3 weeks of culture, we obtained single cell-derived cell clones for both sg-CTL and sg-FIS1 genotypes. We sequenced and confirmed that we have successfully generated a sg-FIS1 clone that contains heterozygous mutations of the FIS1 gene in its genome (Figure S7B). The sequencing results showed that 5 out of 9 bacterial clones show a “T” insertion, while the other 4 show a “GC” deletion. In comparison, the genomic FIS1 locus is intact in the sg-CTL clone (Figure S7B). Importantly, both T-insertion and GC-deletion alleles create a same frame shift that essentially result in an early truncation of FIS1 protein after Lys58 with addition of a new peptide of 49 amino acids with unknown function (Figure S7C). In addition, it is known that the FIS1 protein contains an essential mitochondria-anchoring motif “SKSKS” at its extreme C-terminus (Stojanovski et al., 2004; Suzuki et al., 2003) (Figure S7C). Thus, an early truncation at the middle of the third alpha helix after Lys58 will result in a mutant FIS1 without the function of alpha helix 3–6 and lacking the ability to localize to mitochondrial membrane, essentially rendering it incompetent as a regulator of mitochondrial dynamics (Figure S7D). See also Figure S7. CRISPR/Cas9 targeting sequences are listed in Table S5.
RNA-seq
The total RNA concentration was determined with the NanopDrop 1000 spectrophotometer (NanoDrop) and RNA quality assessed with the Agilent Bioanalyzer (Agilent). The TruSeq Stranded mRNA Sample Preparation Kit (Illumina) was used for next generation sequencing library construction per manufacturer’s protocols. Briefly, mRNA was purified from 200ng total RNA with oligo-dT magnetic beads and fragmented. First-strand cDNA synthesis was performed with random hexamer priming followed by second-strand cDNA synthesis using dUTP incorporation for strand marking. End repair and 3’-adenylation was then performed on the double stranded cDNA. Illumina adaptors were ligated to both ends of the cDNA, purified by gel electrophoresis and amplified with PCR primers specific to the adaptor sequences to generate cDNA amplicons of approximately 200–500bp in size. The amplified libraries were hybridized to the Illumina single end flow cell and amplified using the cBot (Illumina). Single end reads of 100nt were generated for each sample using Illumina’s HiSeq2500v4. Raw reads generated from Illumina HiSeq2500 sequencing were de-multiplexed using bcl2fastq version 1.8.4. Quality filtering and adapter removal were performed using Trimmomatic version 0.32 (Bolger et al., 2014) with the following parameters: "SLIDINGWINDOW:4:20 TRAILING:13 LEADING:13 ILLUMINACLIP:adapters.fasta:2:30:10 MINLEN:25". Processed/cleaned reads were then mapped to the human reference sequence (hg19) using the SHRiMP version 2.2.3 alignment software given the following parameters: "--qv-offset 33 --all-contigs". Differential expression analyses and data normalization were performed using the CuffDiff tool from the cufflinks version 2.0.2 package given the following parameters: "--FDR 0.05 -u -b GENOME”.
GSEA analysis
GSEA analysis was performed using GSEA version 3.0 (Broad). FPKM values produced from CuffDiff analysis were formatted into GCT files containing expression values for genes in different biological states. CLS files were manually built to label biological states involved in each study. When performing GSEA analysis, several gene set databases were used including: c2.cp.kegg.v6.0.symbols.gmt, c2.cp.reactome.v6.0.symbols.gmt and an in-house database containing a collection of 860 gene sets of our interest. Following parameters were used: Number of permutations = 1000, permutation type = gene_set, chip platform = annotations_ALL.zip. Other parameters were left at default values.
ChIP-seq
The detailed methods for ChIP-seq are described here (Mack et al., 2014). Briefly, frozen cell pellets were resuspended and fixed in PBS with 1% PFA at room temperature for 10 min. Crosslinking was stopped by adding 125 mM glycine, followed by washing 2–3 times with ice cold PBS containing 1% BSA. Samples were then sonicated to ~200 bp fragments. Chromatin immunoprecipitation was then performed using 5 mg of H3K27ac antibody (Active Motif) overnight at 4°C. DNA was quantified using PicoGreen (Thermo Fisher) and libraries were prepared using ThruPLEX DNA-seq Kit (Rubicon Genomics). Samples were barcoded and pooled in equimolar amounts and sent out for nextgeneration sequencing on the Illumina HiSeq 2000 platform (illumina). ChIP-seq reads were mapped to hg19 genome using Bowtie2 v2.2.6. For data preprocessing, we collected 1k bp upstream and 2k bp downstream of the TSSs, which were calculated from merged gene lists obtained from GRCh37 GENCODE Gene annotation. Tables of bin counts at gene promoters were built for each experiment. Replicates of the same experiments were grouped together to derive technical variance. For testing, the edgeR version 3.18.1 package which uses the Negative Binomial Generalized Linear Model was utilized to test significance of fold change in R version 3.4.1. Using a relaxed p-value threshold of 0.05, a list of genes with significantly altered H3K27ac binding activity between scramble control and shFIS1-#B groups was created. This list was compared with the list of significantly altered genes from RNA-seq experiments (from CuffDiff analysis, see RNA-seq) using hypergeometric statistics. The comparison failed to reject null hypothesis of independence of two experiments with a p-value of 0.05, thus suggesting no significant link between H3K27ac events and gene expression changes seen in sh-FIS1 cells. For data visualization, ChIP-seq signals at the promoter regions were normalized before drawing heat maps using the built-in heatmap function in R version 3.4.1.
QUANTIFICATION AND STATISTICAL ANALYSIS
Unless otherwise specified, statistical analyses were performed using two-tailed (non-directional), type 2 (assume equal variance) or type 3 (assume unequal variance) student’s t-test. A p value of less than 0.05 indicates significance. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; ns, not significant. For RNA-seq experiments, the p values were adjusted by the Benjamini-Hochberg method. Detailed information of each test is in the figure legends.
DATA AND SOFTWARE AVAILABILITY
RNA-seq data of MOLM13 cells with FIS1 knock-down is available at GEO: GSE114111, RNA-seq data of primary AML cells with FIS1 knock-down is available at GEO: GSE114109, RNA-seq data of primary AML cells with AMPK (PRKAA1) knock-down is available at GEO: GSE114105, and H3K27Ac CHIP-seq data of MOLM-13 cells with FIS1 knock-down is available at GEO: GSE114106. Also see KEY RESOURCES TABLE.
Supplementary Material
Highlights.
Human AML LSCs display high FIS1 expression and unique mitochondrial morphology
FIS1 loss attenuates mitophagy and impairs AML LSC potential
FIS1 loss induces GSK3 inhibition, differentiation and cell cycle arrest in AML
AMPK is constitutively active in human AML LSCs and regulates FIS1 expression
Acknowledgments
We gratefully acknowledge our patients and their families for their participation in this study. Grant support: C.T.J. is generously supported by the Nancy Carroll Allen Chair in Hematology Research and NIH grants R01CA200707 and R01CA166265-05S1. B. A. was supported by F31CA196330-01. D.A.P. is supported by the University of Colorado Department of Medicine Outstanding Early Career Scholar Program. K.M.S. is supported by R01CA193994. A.C.T. is supported by University of Colorado Cancer Center Support Grant P30CA046934, Bioinformatics Core. A.I. was supported by NIH MSTP T32 GM008497. We are also grateful for funding from the University of Colorado Cancer Center Development Therapeutics Pilot Grant Award. We thank the Genomics Center at University of Rochester Medical Campus for their support in RNA-seq data analysis. We thank the Electron Microscopy Service core at University of Colorado-Boulder for their critical supports in TEM. We also thank the Advanced Light Microscopy Core at University of Colorado-Anschutz Medical Campus (supported in part by NIH/NCATS Colorado CTSI Grant Number UL1 TR001082) for their help with confocal studies. We greatly appreciate Drs. James DeGregori and Eric Pietras for their comments on our manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
S.P. and C.T.J. conceived the concept, designed the experiments, analyzed and interpreted the data and wrote the manuscript. M.M. designed and performed experiments related to AMPK studies. B.A. provided critical insights to manuscript writing. N.K. and B.A. assisted with animal studies. B.M.S. organized animal protocols, primary specimens and assisted with flow cytometry studies. J.R.M. and J.M.A. performed illumina sequencing and assisted with RNA-seq analysis. T.N. assisted with GSEA analysis of RNA-seq data. S.C.M., S.L. and J.N.R performed ChIP-seq experiment and initial data analysis. H.M.K. and A.T. performed complete analysis of the ChIP-seq data. A.I. and K.M.S. assisted with the CRISPR experiments. D.A.P. and C.A.S. obtained AML patient specimens and provided critical clinical insights to the current study.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton JM, Balys M, Neering SJ, Hassane DC, Cowley G, Root DE, Miller PG, Ebert BL, McMurray HR, Land H, et al. Gene sets identified with oncogene cooperativity analysis regulate in vivo growth and survival of leukemia stem cells. Cell stem cell. 2012;11:359–372. doi: 10.1016/j.stem.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlasi Y, Stunnenberg HG. The interplay of epigenetic marks during stem cell differentiation and development. Nat Rev Genet. 2017 doi: 10.1038/nrg.2017.57. [DOI] [PubMed] [Google Scholar]
- Bagger FO, Sasivarevic D, Sohi SH, Laursen LG, Pundhir S, Sonderby CK, Winther O, Rapin N, Porse BT. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 2016;44:D917–924. doi: 10.1093/nar/gkv1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji V, Frumm SM, Ross KN, Li LS, Schinzel AC, Hahn CK, Kakoza RM, Chow KT, Ross L, Alexe G, et al. The intersection of genetic and chemical genomic screens identifies GSK-3alpha as a target in human acute myeloid leukemia. The Journal of clinical investigation. 2012;122:935–947. doi: 10.1172/JCI46465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature medicine. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Chen H, Chan DC. Mitochondrial Dynamics in Regulating the Unique Phenotypes of Cancer and Stem Cells. Cell Metab. 2017;26:39–48. doi: 10.1016/j.cmet.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CH, Lin CC, Yang MC, Wei CC, Liao HD, Lin RC, Tu WY, Kao TC, Hsu CM, Cheng JT, et al. GSK3beta-mediated Drp1 phosphorylation induced elongated mitochondrial morphology against oxidative stress. PLoS One. 2012;7:e49112. doi: 10.1371/journal.pone.0049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole A, Wang Z, Coyaud E, Voisin V, Gronda M, Jitkova Y, Mattson R, Hurren R, Babovic S, Maclean N, et al. Inhibition of the Mitochondrial Protease ClpP as a Therapeutic Strategy for Human Acute Myeloid Leukemia. Cancer Cell. 2015;27:864–876. doi: 10.1016/j.ccell.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombret H, Gardin C. An update of current treatments for adult acute myeloid leukemia. Blood. 2016;127:53–61. doi: 10.1182/blood-2015-08-604520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, Metzeler KH, Poeppl A, Ling V, Beyene J, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nature medicine. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R, Bosc C, Sugita M, Stuani L, Fraisse M, et al. Chemotherapy-Resistant Human Acute Myeloid Leukemia Cells Are Not Enriched for Leukemic Stem Cells but Require Oxidative Metabolism. Cancer Discov. 2017;7:716–735. doi: 10.1158/2159-8290.CD-16-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles AJ, Plevritis SK, Majeti R, Alizadeh AA. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA. 2010;304:2706–2715. doi: 10.1001/jama.2010.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman ML, Allan JN. Concise review: Leukemia stem cells in personalized medicine. Stem Cells. 2014;32:844–851. doi: 10.1002/stem.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamacher-Brady A, Brady NR. Mitophagy programs: mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell Mol Life Sci. 2016;73:775–795. doi: 10.1007/s00018-015-2087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TC, LaMere M, Stevens BM, Ashton JM, Myers JR, O'Dwyer KM, Liesveld JL, Mendler JH, Guzman M, Morrissette JD, et al. Evolution of acute myelogenous leukemia stem cell properties after treatment and progression. Blood. 2016;128:1671–1678. doi: 10.1182/blood-2016-02-695312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TT, Warr MR, Adelman ER, Lansinger OM, Flach J, Verovskaya EV, Figueroa ME, Passegue E. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 2017;543:205–210. doi: 10.1038/nature21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, Schafer ZT, Evans RM, Suda T, Lee CH, et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nature medicine. 2012;18:1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Turcotte R, Cui J, Zimmerman SE, Pinho S, Mizoguchi T, Arai F, Runnels JM, Alt C, Teruya-Feldstein J, et al. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science. 2016;354:1156–1160. doi: 10.1126/science.aaf5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki M, Liedtke M, Gentles AJ, Cleary ML. CD93 Marks a Non-Quiescent Human Leukemia Stem Cell Population and Is Required for Development of MLL-Rearranged Acute Myeloid Leukemia. Cell stem cell. 2015;17:412–421. doi: 10.1016/j.stem.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang YY, Sharkis SJ. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood. 2007;110:3056–3063. doi: 10.1182/blood-2007-05-087759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan CT, Guzman ML, Noble M. Cancer stem cells. The New England journal of medicine. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- Joshi A, Kundu M. Mitophagy in hematopoietic stem cells: the case for exploration. Autophagy. 2013;9:1737–1749. doi: 10.4161/auto.26681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, Ashton JM, Pei S, Grose V, O'Dwyer KM, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell stem cell. 2013;12:329–341. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang UE, Kocabayoglu P, Cheng GZ, Ghiassi-Nejad Z, Munoz U, Vetter D, Eckstein DA, Hannivoort RA, Walsh MJ, Friedman SL. GSK3beta phosphorylation of the KLF6 tumor suppressor promotes its transactivation of p21. Oncogene. 2013;32:4557–4564. doi: 10.1038/onc.2012.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, He S, Yang J, Jia X, Wang P, Chen X, Zhang Z, Zou X, McNutt MA, Shen WH, et al. PTENalpha, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab. 2014;19:836–848. doi: 10.1016/j.cmet.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyanage SU, Hurren R, Voisin V, Bridon G, Wang X, Xu C, MacLean N, Siriwardena TP, Gronda M, Yehudai D, et al. Leveraging increased cytoplasmic nucleoside kinase activity to target mtDNA and oxidative phosphorylation in AML. Blood. 2017;129:2657–2666. doi: 10.1182/blood-2016-10-741207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchsinger LL, de Almeida MJ, Corrigan DJ, Mumau M, Snoeck HW. Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature. 2016;529:528–531. doi: 10.1038/nature16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn S, Stutz AM, Wang X, Gallo M, Garzia L, Zayne K, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506:445–450. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubrey JA, Steelman LS, Bertrand FE, Davis NM, Abrams SL, Montalto G, D'Assoro AB, Libra M, Nicoletti F, Maestro R, et al. Multifaceted roles of GSK-3 and Wnt/beta-catenin in hematopoiesis and leukemogenesis: opportunities for therapeutic intervention. Leukemia. 2014;28:15–33. doi: 10.1038/leu.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola HK, Radu CG, Witte ON. Targeting leukemia stem cells. Nat Biotechnol. 2010;28:237–238. doi: 10.1038/nbt0310-237. [DOI] [PubMed] [Google Scholar]
- Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, Stranks AJ, Glanville J, Knight S, Jacobsen SE, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, Arruda A, Popescu A, Gupta V, Schimmer AD, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540:433–437. doi: 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- Nguyen TN, Padman BS, Lazarou M. Deciphering the Molecular Signals of PINK1/Parkin Mitophagy. Trends in cell biology. 2016;26:733–744. doi: 10.1016/j.tcb.2016.05.008. [DOI] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollyea DA, Jordan CT. Therapeutic targeting of acute myeloid leukemia stem cells. Blood. 2017;129:1627–1635. doi: 10.1182/blood-2016-10-696039. [DOI] [PubMed] [Google Scholar]
- Rakovic A, Grunewald A, Seibler P, Ramirez A, Kock N, Orolicki S, Lohmann K, Klein C. Effect of endogenous mutant and wild-type PINK1 on Parkin in fibroblasts from Parkinson disease patients. Hum Mol Genet. 2010;19:3124–3137. doi: 10.1093/hmg/ddq215. [DOI] [PubMed] [Google Scholar]
- Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, Korbel C, Laschke MW, Gimotty PA, Philipp SE, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell. 2013;23:811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijtenberg S, van den Heuvel S. Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle. 2016;15:196–212. doi: 10.1080/15384101.2015.1120925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Chapple RH, Lin A, Kitano A, Nakada D. AMPK Protects Leukemia-Initiating Cells in Myeloid Leukemias from Metabolic Stress in the Bone Marrow. Cell stem cell. 2015;17:585–596. doi: 10.1016/j.stem.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarry JE, Murphy K, Perry R, Sanchez PV, Secreto A, Keefer C, Swider CR, Strzelecki AC, Cavelier C, Recher C, et al. Human acute myelogenous leukemia stem cells are rare and heterogeneous when assayed in NOD/SCID/IL2Rgammac-deficient mice. The Journal of clinical investigation. 2011;121:384–395. doi: 10.1172/JCI41495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmer AD, Skrtic M. Therapeutic potential of mitochondrial translation inhibition for treatment of acute myeloid leukemia. Expert Rev Hematol. 2012;5:117–119. doi: 10.1586/ehm.12.8. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibler P, Graziotto J, Jeong H, Simunovic F, Klein C, Krainc D. Mitochondrial Parkin recruitment is impaired in neurons derived from mutant PINK1 induced pluripotent stem cells. J Neurosci. 2011;31:5970–5976. doi: 10.1523/JNEUROSCI.4441-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Yamano K, Head BP, Kawajiri S, Cheung JT, Wang C, Cho JH, Hattori N, Youle RJ, van der Bliek AM. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol Biol Cell. 2014;25:145–159. doi: 10.1091/mbc.E13-09-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrtic M, Sriskanthadevan S, Jhas B, Gebbia M, Wang X, Wang Z, Hurren R, Jitkova Y, Gronda M, Maclean N, et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2011;20:674–688. doi: 10.1016/j.ccr.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D, Koutsopoulos OS, Okamoto K, Ryan MT. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J Cell Sci. 2004;117:1201–1210. doi: 10.1242/jcs.01058. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Jeong SY, Karbowski M, Youle RJ, Tjandra N. The solution structure of human mitochondria fission protein Fis1 reveals a novel TPR-like helix bundle. J Mol Biol. 2003;334:445–458. doi: 10.1016/j.jmb.2003.09.064. [DOI] [PubMed] [Google Scholar]
- Tian Y, Huang Z, Wang Z, Yin C, Zhou L, Zhang L, Huang K, Zhou H, Jiang X, Li J, et al. Identification of novel molecular markers for prognosis estimation of acute myeloid leukemia: over-expression of PDCD7, FIS1 and Ang2 may indicate poor prognosis in pretreatment patients with acute myeloid leukemia. PLoS One. 2014;9:e84150. doi: 10.1371/journal.pone.0084150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama EQ, Herzig S, Courchet J, Lewis TL, Jr, Loson OC, Hellberg K, Young NP, Chen H, Polleux F, Chan DC, et al. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science. 2016;351:275–281. doi: 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlashi E, Lagadec C, Vergnes L, Matsutani T, Masui K, Poulou M, Popescu R, Della Donna L, Evers P, Dekmezian C, et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16062–16067. doi: 10.1073/pnas.1106704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai T, Langer T. Mitochondrial Dynamics and Metabolic Regulation. Trends Endocrinol Metab. 2016;27:105–117. doi: 10.1016/j.tem.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Wang Z, Iwasaki M, Ficara F, Lin C, Matheny C, Wong SH, Smith KS, Cleary ML. GSK-3 promotes conditional association of CREB and its coactivators with MEIS1 to facilitate HOX-mediated transcription and oncogenesis. Cancer Cell. 2010;17:597–608. doi: 10.1016/j.ccr.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TC, Cleary ML. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455:1205–1209. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JH, Zhang SH, Gao FJ, Lei Y, Chen XY, Gao F, Zhang SJ, Sun XH. RNAi screening identifies GSK3beta as a regulator of DRP1 and the neuroprotection of lithium chloride against elevated pressure involved in downregulation of DRP1. Neurosci Lett. 2013;554:99–104. doi: 10.1016/j.neulet.2013.08.057. [DOI] [PubMed] [Google Scholar]
- Xie Q, Wu Q, Horbinski CM, Flavahan WA, Yang K, Zhou W, Dombrowski SM, Huang Z, Fang X, Shi Y, et al. Mitochondrial control by DRP1 in brain tumor initiating cells. Nat Neurosci. 2015;18:501–510. doi: 10.1038/nn.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Fogel AI, Wang C, van der Bliek AM, Youle RJ. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. Elife. 2014;3:e01612. doi: 10.7554/eLife.01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung M, Hurren R, Nemr C, Wang X, Hershenfeld S, Gronda M, Liyanage S, Wu Y, Augustine J, Lee EA, et al. Mitochondrial DNA damage by bleomycin induces AML cell death. Apoptosis. 2015;20:811–820. doi: 10.1007/s10495-015-1119-z. [DOI] [PubMed] [Google Scholar]
- Zhou D, Shao L, Spitz DR. Reactive oxygen species in normal and tumor stem cells. Adv Cancer Res. 2014;122:1–67. doi: 10.1016/B978-0-12-420117-0.00001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.