Abstract
Some patients with acute myeloid leukemia (AML) who are in complete remission after induction chemotherapy harbor persisting pre-leukemic clones, carrying a subset of leukemia-associated somatic mutations. There is conflicting evidence on the prognostic relevance of these clones for AML relapse. Here, we characterized paired pre-treatment and remission samples from 126 AML patients for mutations in 68 leukemia-associated genes. Fifty patients (40%) retained ≥1 mutation during remission at a VAF of ≥2%. Mutation persistence was most frequent in DNMT3A (65% of patients with mutations at diagnosis), SRSF2 (64%), TET2 (55%), and ASXL1 (46%), and significantly associated with older age (p < 0.0001) and, in multivariate analyses adjusting for age, genetic risk, and allogeneic transplantation, with inferior relapse-free survival (hazard ratio (HR), 2.34; p = 0.0039) and overall survival (HR, 2.14; p = 0.036). Patients with persisting mutations had a higher cumulative incidence of relapse before, but not after allogeneic stem cell transplantation. Our work underlines the relevance of mutation persistence during first remission as a novel risk factor in AML. Persistence of pre-leukemic clones may contribute to the inferior outcome of elderly AML patients. Allogeneic transplantation abrogated the increased relapse risk associated with persisting pre-leukemic clones, suggesting that mutation persistence may guide post-remission treatment.
Introduction
The development of acute myeloid leukemia (AML) from healthy hematopoietic stem or precursor cells (HSCs) is a multistep process [1]. Sequential acquisition of somatic mutations in AML driver genes can result in pre-leukemic HSCs capable of increased proliferation or self-renewal, but retaining normal multi-lineage differentiation potential. In some AML patients, pre-leukemic HSCs carrying early disease-associated “founder” mutations can be identified besides the fully transformed leukemia clone [1, 2]. Mutations in pre-leukemic HSCs frequently affect genes also implicated in age-associated clonal hematopoiesis, such as DNMT3A and TET2. Pre-leukemic clones may survive chemotherapy and persist in AML patients during complete remission (CR) [2–6]. Several groups have reported frequent persistence of DNMT3A mutations during CR, but found no association with AML relapse or survival [7–10]. In contrast, in another study analyzing a larger panel of genes in 50 AML patients, mutation persistence associated with shorter event-free and overall survival (OS) [11]. Of note, clonal cytogenetic alterations also remain detectable in some patients in morphological remission, and this has been linked to inferior outcomes [12, 13].
These conflicting results prompted us to genetically characterize paired samples, obtained at the time of diagnosis and during first remission, from a cohort of intensively treated AML patients [14]. We aimed to study associations between persistence of AML-associated somatic mutations during remission and patient outcomes in the context of other known prognostic factors, including the most recent European LeukemiaNet (ELN) classification of baseline genetic risk [15]. Since post-remission treatment is an important factor that may affect the ultimate outcome of patients harboring persisting pre-leukemic clones after the initial line of therapy, we also studied the impact of allogeneic transplantation on relapse risk in patients with and without mutation persistence.
Methods
Patients and study design
We performed a retrospective cohort study investigating the prevalence, spectrum, and prognostic relevance of persisting pre-leukemic mutations in 129 adult AML patients in first remission (median age, 54 years [y]; range, 20–80 y). Patients were enrolled in the German AML Cooperative Group AMLCG-2008 phase III trial (NTC01382147; n = 99) [16] or a patient registry (n = 27; Supplementary Table 1). Treatment regimens are described in the Supplementary Information. All patients in this analysis achieved CR or CR with incomplete blood count recovery (CRi), defined according to standard criteria [17], after first-line induction chemotherapy. Patient selection for this analysis was based solely on the availability of paired specimens for genetic analysis, obtained before treatment and during first remission. Genetic risk groups were defined according to the 2010 Medical Research Council (MRC) cytogenetic classification and the 2017 ELN risk stratification system [15, 18]. All study protocols were in accordance with the Declaration of Helsinki and approved by the institutional review boards of participating centers. Patients provided written informed consent for inclusion into the clinical trial or registry and genetic analyses.
Genetic analyses
We studied bone marrow or peripheral blood samples, obtained before treatment and during first remission, from 129 patients. For 36/54 patients with documented recurrence, a relapse specimen was also available. Mutations in 68 recurrently mutated genes were analyzed by targeted sequencing (Haloplex, Agilent, Boeblingen, Germany/Miseq, Illumina, San Diego, CA, USA) using genomic DNA from Ficoll-enriched mononuclear cells, as reported previously [14]. We sequenced the entire coding sequence of 37 genes, and recurrently mutated regions in the remaining 31 (Supplementary Table 2). The mean sequencing depth was 570×, and >99% of the target region was covered ≥30-fold.
We identified sequence alterations with a variant allele frequency (VAF) of ≥2%. This threshold was based on the specificity of our assay (Supplementary Results) and is comparable to previous analyses [11] and in agreement with a working definition of clonal hematopoiesis [19]. Variants were classified as known/putative driver mutations, variants of unknown significance, or germline polymorphisms based on published data [20–22], as reported in detail previously [14]. Additionally, NPM1 and CEBPA mutations and FLT3 internal tandem duplications (FLT3-ITDs) were tested by standard methods [23–27]. Genotyping of T lymphocytes was performed for selected patients as described in the Supplementary Information.
Minimal residual disease assessment
Minimal residual disease (MRD) levels were measured in the remission samples by quantitative real-time PCR (qPCR) for NPM1 mutations (types A/B/D) or by flow cytometry in 46 and 81 patients, respectively, using published methods [23, 28]. Eleven additional patients had other types of NPM1 mutations, and no qPCR assay was available for these. Flow cytometric MRD assessment, based on detection of leukemia-associated immunophenotypes, was performed using FACSCalibur (BD Biosciences, San Jose, CA, USA; 60 patients) or NAVIOS (Beckman Coulter, Brea, CA, USA; 21 patients) instruments.
Statistical analyses
Associations between mutation persistence and clinical parameters were studied using the Wilcoxon rank-sum test for continuous and Fisher’s exact test for categorical variables. For analyses of treatment outcomes, endpoints (relapse-free survival (RFS) and OS) were defined according to standard criteria [17]. The Kaplan–Meier method and log-rank test were used for unadjusted analyses of time-to-event endpoints. To evaluate the risk of disease recurrence, we studied cumulative incidence of relapse (CIR) while treating allogeneic stem cell transplantation (alloSCT) and death in remission (whichever occurred first) as competing events; patients with and without mutation persistence were compared using Gray’s test [29]. Multivariate Cox proportional hazards models included alloSCT as a time-dependent covariate [30] and were stratified with separate strata for each of the AMLCG-2008 trial arms and the patient registry. HRs, 95% confidence intervals (CIs), and Wald-test p values are reported. Two-sided p values ≤0.05 were interpreted as significant and are considered hypothesis generating. Statistical analyses were performed using R, version 3.2.3 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Patients
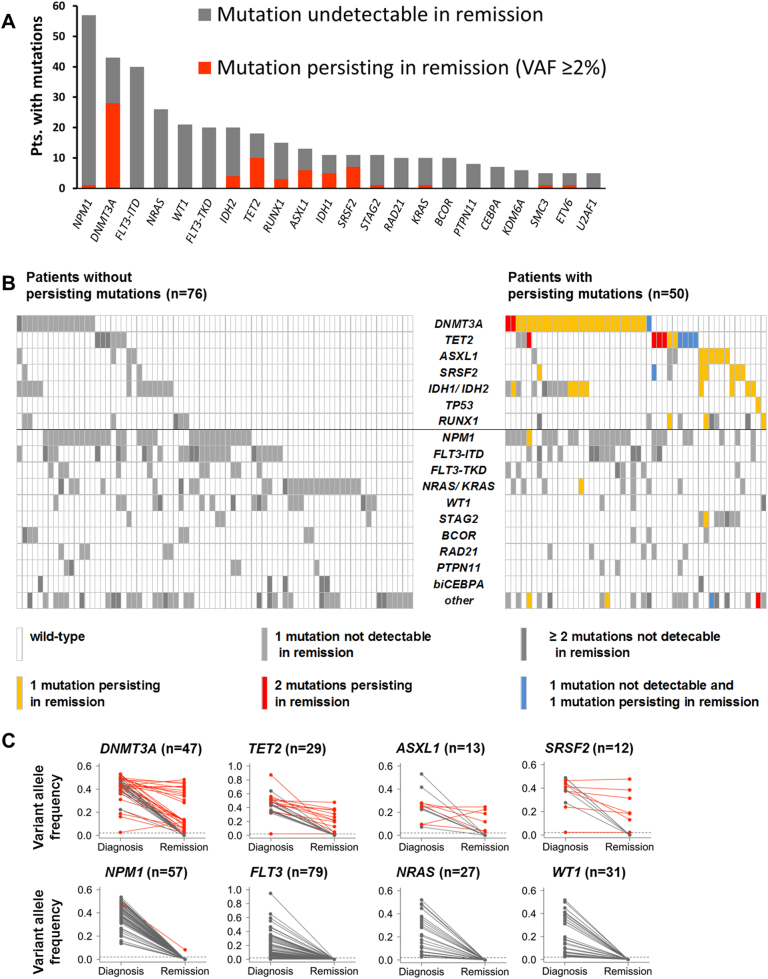
Targeted sequencing of pre-treatment specimens from 129 AML patients identified a total of 481 variants affecting 41 genes that were classified as putative driver mutations (median, 4 mutations/patient; range, 0–10; Supplementary Table 3). In 126/129 (98%) patients, we detected ≥1 driver mutation at diagnosis. Three patients harbored balanced chromosomal translocations (KMT2A-MLLT1, KMT2A-MLLT3, and CBFB-MYH11) but no detectable gene mutations, and were excluded from further analyses. Detailed characteristics of the remaining 126 patients are listed in Table 1 and Supplementary Table 1. The most commonly mutated genes were NPM1 (57 mutations/57 patients), DNMT3A (47 mutations/43 patients), and FLT3 (54 internal tandem duplications [ITDs]/40 patients, 21 tyrosine kinase domain mutations/20 patients) (Fig. 1a).
Table 1.
Pre-treatment patient characteristics and baseline parameters
| Variable | Total cohort | Patients with ≥1 persisting mutation in remission | Patients without persisting mutation in remission | p |
|---|---|---|---|---|
| Patient number | n = 126 | n = 50 | n = 76 | |
| Age [years], median (range) | 54 (20–80) | 63.5 (21–78) | 48 (20–80) | <0.0001 |
| Pts. ≤60 years/>60 years | 84/42 | 21/29 | 63/13 | |
| Male gender | 62 (49%) | 21 (42%) | 41 (54%) | 0.21 |
| Disease type | ||||
| De novo AML | 108 (86%) | 40 (80%) | 68 (89%) | 0.079 |
| Secondary AML | 9 (7%) | 3 (6%) | 6 (8%) | |
| Therapy-related AML | 6 (5%) | 5 (10%) | 1 (1%) | |
| ECOG performance status | 0.44 | |||
| 0 | 45 (36%) | 15 (30%) | 30 (39%) | |
| 1 | 50 (40%) | 20 (40%) | 30 (39%) | |
| 2 | 7 (6%) | 4 (8%) | 3 (4%) | |
| ≥3 | 6 (5%) | 2 (4%) | 4 (5%) | |
| WBC [ × 109/l], median (range) | 20.3 (0.5–406) | 26.8 (0.5–406) | 15.2 (0.9–364) | 0.41 |
| Blast count at diagnosis | ||||
| % Bone marrow blasts, median (range) | 72 (17–97) | 73.5 (17–97) | 71 (17–97) | 0.68 |
| % Peripheral blood blasts, median (range) | 37 (0–96) | 42 (0–96) | 31 (0–96) | 0.73 |
| MRC cytogenetic risk category | 0.029 | |||
| Favorable | 8 (6%) | 0 (0%) | 8 (11%) | |
| Intermediate | 102 (81%) | 42 (84%) | 60 (79%) | |
| Adverse | 11 (9%) | 6 (12%) | 5 (7%) | |
| Missing | 5 (4%) | 2 (4%) | 3 (4%) | |
| ELN 2017 classification | 0.92 | |||
| Favorable | 49 (39%) | 19 (38%) | 30 (39%) | |
| Intermediate | 37 (29%) | 14 (28%) | 23 (39%) | |
| Adverse | 40 (32%) | 17 (34%) | 23 (30%) | |
| Patients with gene mutations detected in pretreatment sample | ||||
| NPM1 | 57 (45%) | 26 (52%) | 31 (41%) | 0.20 |
| DNMT3A | 43 (34%) | 28 (56%) | 15 (20%) | <0.0001 |
| FLT3-ITD | 40 (32%) | 15 (30%) | 25 (33%) | 0.85 |
| TET2 | 18 (14%) | 12 (24%) | 6 (8%) | 0.018 |
| RUNX1 | 15 (12%) | 9 (18%) | 6 (8%) | 0.10 |
| ASXL1 | 13 (10%) | 9 (18%) | 4 (5%) | 0.034 |
| SRSF2 | 11 (9%) | 8 (16%) | 3 (4%) | 0.025 |
| CEBPA | ||||
| -Mono-allelic | 3 (2%) | 3 (6%) | 0 (0%) | 0.060 |
| -Bi-allelic | 6 (5%) | 1 (2%) | 5 (7%) | 0.40 |
| Remission status post induction | 0.12 | |||
| CR | 98 (78%) | 35 (70%) | 63 (83%) | |
| CRi | 28 (22%) | 15 (30%) | 13 (17%) | |
| Allogeneic transplantation | 80 (64%) | 30 (60%) | 50 (66%) | 0.57 |
| In first CR/CRi | 53 | 19 | 34 | |
| After relapse | 27 | 11 | 16 | |
| Material studied (diagnosis/remission/relapse) | – | |||
| Bone marrow | 115/117/35 | 70/72/17 | 45/45/18 | |
| Peripheral blood | 11/9/1 | 6/4/0 | 5/5/1 | |
ECOG Eastern Cooperative Oncology Group, MRC British Medical Research Council, ELN European LeukemiaNet, CR complete remission, CRi complete remission with incomplete blood count recovery, ITD internal tandem duplication.
Fig. 1.
Overview of mutations at diagnosis and in remission. a Frequency of mutations identified at diagnosis in ≥4 patients. b Heatmap depicting gene mutations occurring in ≥4 patients. Each column represents one patient. The left panel represents patients without, and the right panel shows patients with persisting mutations in remission. c Allele frequencies of mutations in commonly mutated genes in paired diagnosis and remission samples from individual patients. Persisting mutations are shown in red, and mutations undetectable in remission in gray
All 126 patients achieved CR (98/126, 78%) or CRi (28/126, 22%) after induction chemotherapy. The median time from diagnosis until documented first CR/CRi was 64 days (range: 26–314 days). Most remission samples (103/126, 87%) were collected within 60 days of the documented CR/CRi date, while 23 samples were collected >60 days after remission was first achieved (Supplementary Table 4). Twelve patients died while in first remission, and relapse was observed in 55 patients, 34 of whom subsequently died. The median follow-up, calculated according to the reverse Kaplan–Meier method [31], was 51.9 months (m). The median RFS for the entire cohort was 24.8 m (95% CI; 15.9 m—not reached), and the median OS was not reached. Since by definition, mutation persistence can only be studied in patients who achieved CR/CRi, outcomes of our cohort compare favorably to unselected populations of newly diagnosed AML patients.
Persistence of mutations in remission
In the samples obtained during first remission, we re-identified 78 mutations in 16 different genes that had already been present in the matched diagnostic specimens (persisting mutations). Persistence of ≥1 mutation was found in 50/126 patients (40%; median, 1 mutation/patient; range, 1–5). In the remaining 76 patients, all driver mutations were undetectable in the remission specimen at the level of sensitivity of our assay (VAF, ≥2%; Fig. 1b). The frequency of mutation persistence was similar in samples obtained within 60 days of the documented CR/CRi date (43/102) and in samples obtained later during first remission (7/24) (p = 0.35). Mutation persistence was most frequently observed in DNMT3A (28/43 patients with mutations at diagnosis; 65%), TET2 (10/18; 56%), SRSF2 (7/11; 64%), and ASXL1 (6/13; 46%). In contrast, persistence of driver mutations was very rare in other AML-associated genes such as NPM1 (persisting in 1/57 patients, 2%), FLT3 (0/79), NRAS (0/27), and WT1 (0/31) (Fig. 1a–c).
Most persisting mutations were recurrent somatic alterations found in age-associated clonal hematopoiesis and myeloid neoplasms (Supplementary Table 3). The allele frequencies of these variants at diagnosis and/or during remission were mostly incompatible with heterozygous or homozygous germline polymorphisms (expected VAFs, 50% or 100%, respectively), supporting their somatic origin (Fig. 1c, Supplementary Fig. 1). Additionally, we genotyped T cells from 5 of the 18 patients who had mutations persisting at VAFs >25%, to prove their somatic origin (Supplementary Information).
Allele frequencies of variants detected in remission were ≥10% in most patients (35/50; 70%; Supplementary Table 3). In the remaining patients, VAFs of persisting mutations were between 10% and our limit of detection of 2%. Assuming heterozygosity, this indicates that in most patients persisting mutations were carried by ≥20% of cells in the remission specimen, while the morphological bone marrow blast count was <5%. Mutations that persisted in remission had a higher median VAF at diagnosis (43%, range: 2–94%), and thus were present in a larger fraction of cells, than mutations that were subsequently cleared in remission (31%, range: 2–95%, p < 0.0001; Supplementary Fig.1). Furthermore, 47/50 patients with a persisting mutation had ≥1 additional mutation at diagnosis that was cleared in remission (Fig. 1b). Specimens obtained at AML relapse were available for 36 patients (Supplementary Results). In 35 patients, the initial and relapse clones were genetically related, as indicated by common driver mutations. Eighteen patients had persisting mutations during remission that were also detected at the time of relapse. In 14 of them, additional mutations present at diagnosis became undetectable during remission and re-emerged at relapse. The other four patients relapsed with a clone carrying the persisting mutations, but none of the other mutations present in the initial specimen. In these patients, relapse may have originated from the pre-leukemic clone.
Mutation persistence, patient characteristics, and treatment outcomes
Patients with mutation persistence during remission were significantly older than patients who cleared all mutations (median, 63.5 y vs. 48 y; p < 0.0001; Table 1). The median age of patients with persisting DNMT3A mutations was 58 y, compared to 67 y for patients with persistent mutations in other genes only (p = 0.0004; Fig. 2). Other pre-treatment clinical characteristics were not associated with mutation persistence (Table 1). Persisting mutations were observed in 42/102 patients with intermediate-risk cytogenetics (41%) and in 6/11 patients (55%) with adverse risk cytogenetics, but in 0/8 patients with a favorable karyotype (p = 0.029). The frequency of persisting mutations was similar in patients with CR or CRi (p = 0.12).
Fig. 2.
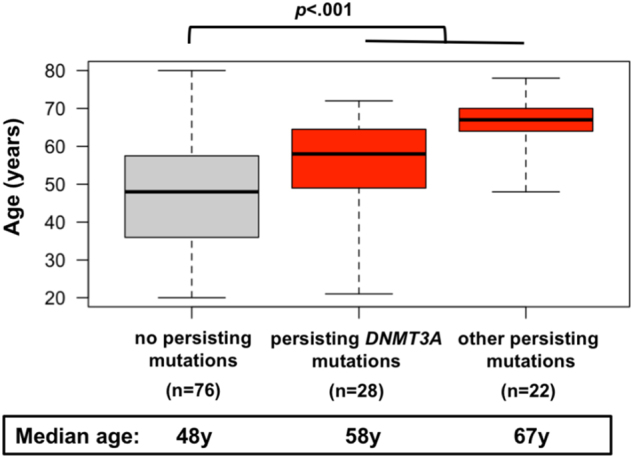
Age distribution of patients without persisting mutations. Age distribution of patients without persisting mutations in remission, patients with persisting mutation in DNMT3A, and patients with persisting mutations only in genes other than DNMT3A. Boxplots depict median, 25th and 75th percentile, and minimum/maximum age in each cohort
Persistence of ≥1 mutation in remission, in contrast to complete mutation clearance, significantly associated with shorter RFS (median, 55.7 m for patients without vs. 11.7 m for patients with persisting mutations; p = 0.0005; Fig. 3a). Likewise, patients with persisting mutations had significantly shorter OS than those without (median, 31.3 m vs. not reached; p = 0.0019; Fig. 3b). When this analysis was restricted to clinically defined de novo AML, persistence of mutations during remission still associated with shorter RFS and OS (Supplementary Fig. 2).
Fig. 3.
Outcomes of patients with and without persisting mutations in remission. a RFS and b OS for patients with and without persisting mutations in the remission sample. c CIR, with alloSCT or death in first CR/CRi treated as competing events, for patients with and without persisting mutations. d CIR after alloSCT in first CR/CRi
Since alloSCT performed in first remission may eliminate persisting pre-leukemic clones, we analyzed CIR while considering transplantation and death in first remission as competing events. Patients with persisting mutations had a higher incidence of relapse before allogeneic transplantation (3 y CIR, 53.0%; 95% CI, 38.9–67.2%) than those with complete mutation clearance (3 y CIR; 25.8%; 95% CI, 15.8–35.8%; p = 0.028; Fig. 3c). In the 53 patients (42%) who underwent alloSCT in first remission, CIR after transplantation was low, and no significant difference was observed between patients who did or did not have mutation persistence before alloSCT (5/34 patients without and 2/19 patients with persisting mutations relapsed after alloSCT; p = 0.67; Fig. 3d).
In more than half of the patients with mutation persistence (28/50), DNMT3A mutations at codon 882 (16/28) or other DNMT3A mutations (12/28) were detectable in the remission sample, whereas 22 patients had persisting mutations only in genes other than DNMT3A. We observed no survival differences between these groups (Supplementary Fig. 3a,b). Outcomes were also similar for patients with small or large persisting clones (i.e., remission VAF, <10% or ≥10%; RFS, p = 0.75; OS, p = 0.79; Supplementary Fig. 3c,d), and for patients with one or >1 persisting mutation (RFS, p = 0.72; OS, p = 0.47; Supplementary Fig. 3e,f), however, patient numbers in these subgroup analyses were small.
Supplementary Table 5 shows the association of clinical and genetic factors with treatment outcomes in our cohort. We used multivariate analyses to study the impact of persisting mutations while adjusting for other risk factors (Table 2). In a model for RFS adjusting for age, ELN 2017 genetic risk groups, leukocyte count, remission status (CRi vs. CR), and alloSCT in first remission, patients with ≥1 persisting mutation had an over twofold increased risk of relapse or death (HR, 2.34; 95% CI, 1.31–4.17; p = 0.0039). Adverse baseline genetic risk, higher leukocyte counts, and alloSCT, treated as a time-dependent covariate, were the only other factors significantly associated with RFS. There was no significant interaction between mutation persistence and age, the ELN risk groups, or allogeneic transplantation. Similarly, in a multivariate model for OS, persistence of ≥1 mutation associated with increased risk of death compared to patients who cleared all mutations (HR, 2.14; 95% CI, 1.05–4.35; p = 0.036). In this model, patients with adverse genetics also had inferior survival. Multivariate models using the MRC cytogenetic risk classification yielded similar results (Supplementary Table 6).
Table 2.
Multivariate analyses
| RFS | OS | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | p | HR (95% CI) | p |
| ≥1 persisting mutation | 2.34 (1.31–4.17) | 0.0039 | 2.14 (1.05–4.35) | 0.036 |
| Age (continuous, hazard ratio per 10 y increase) | 0.98 (0.80–1.20) | 0.62 | 1.03 (0.80–1.32) | 0.85 |
| Leukocyte count (continuous, hazard ratio per 10 y increase) | 1.04 (1.01–1.08) | 0.0042 | 1.00 (1.00–1.00) | 0.44 |
| ELN 2017 genetic risk group | ||||
| Favorable | 1.07 (0.51–2.24) | 0.85 | 1.42 (0.55–3.68) | 0.47 |
| Intermediate | 1 (reference) | – | 1 (reference) | |
| Adverse | 3.23 (1.58–6.61) | 0.0013 | 3.80 (1.61–8.94) | 0.0023 |
| CRi vs. CR | 1.01 (0.53–1.93) | 0.97 | 0.97 (0.46–2.03) | 0.94 |
| AlloSCT in first CR/CRia | 0.28 (0.14–0.57) | 0.0004 | 0.69 (0.33–1.45) | 0.32 |
The Akaike Information Criterion (AIC) was 412.00 for the RFS model shown here, compared to 418.47 for the same model but without persisting mutations, with a lower value indicating a better fit of the model. The AIC was 308.54 for the OS model, compared to 311.01 for the same model but without persisting mutations. Both multivariate regression models were stratified according to trial arm.
aAllogeneic stem cell transplantation was included as a time-dependent covariate. In either model, there was no statistically significant interaction between alloSCT and mutation persistence.
RFS relapse-free survival, OS overall survival, HR hazard ratio, CI confidence interval, ELN European LeukemiaNet, CR complete remission, CRi complete remission with incomplete blood count recovery, alloSCT allogeneic stem cell transplantation.
Mutation persistence and MRD
We questioned whether detection of persisting mutations in remission specimens was indicative of higher levels of MRD as determined by established molecular and flow cytometry-based methods (Supplementary Table 7). Among 46 patients with NPM1 mutation, there was a nonsignificant trend towards higher molecular MRD levels in patients with (n = 22) compared to those without persisting pre-leukemic mutations (n = 24; p = 0.11; Fig. 4a). Flow cytometric MRD assessment in the remission sample was available for 81/126 patients. There was no significant difference in flow MRD levels between the groups with (n = 30) and without persisting mutations (n = 51; p = 0.38; Fig. 4b).
Fig. 4.
Persisting pre-leukemic mutations and minimal residual disease. MRD levels were assessed a by qPCR in 46 NPM1-mutated patients and b by flow cytometry in 82 patients. Boxplots show the median, 25th/75th percentile, and minimum/maximum level of a NPM1mut/ABL1 ratio and b LAIP-positive cells, for patients with and without persisting mutations in the remission sample
Discussion
In our cohort of intensively treated AML patients in first CR or CRi, persistence of at least one leukemia-associated mutation was observed in 40%. In addition to persisting DNMT3A, TET2, IDH1/IDH2, ASXL1, and RUNX1 mutations, which have been previously reported [2, 11], we also identified frequent persistence of SRSF2 mutations. Persistence of leukemia driver mutations associated with an over twofold increased risk of relapse or death in multivariate analyses adjusting for other known prognostic factors, including the most recent ELN classification of pre-treatment genetic risk. By its design, our study only included patients who had achieved a remission after first-line induction therapy. Therefore, patients with adverse genetic characteristics were underrepresented due to their lower response rates, and outcomes in general were relatively favorable compared to cohorts of untreated AML patients [14]. However, RFS and OS of our cohort were similar to the outcomes of all patients treated on the AMLCG-2008 trial who reached CR/CRi, suggesting that our results can be generalized to patients in CR/CRi after intensive induction chemotherapy.
Several groups reported frequent persistence of DNMT3A codon R882 mutations in AML patients in CR, using assays with sensitivities comparable to our study [6–10, 32]. In these analyses, no prognostic significance of DNMT3A mutation persistence was found. These discrepant results might be explained by the relatively small cohort sizes in some studies, and the focus on a single hotspot codon reflecting only a subset of mutations in one gene, as opposed to a broader panel of driver genes in our study. On the other hand, our results are in agreement with a seminal report by Klco et al., who identified persisting mutations in samples obtained ~30 days after the start of induction therapy in 24/50 AML patients [11]. In this cohort, which was partially pre-selected based on patient outcomes, mutation persistence associated with shorter EFS, but not OS, in multivariate analyses. Our work and the study by Klco and colleagues used similar VAF thresholds for the detection of persisting mutations (2% and 2.5%, respectively). Variants exceeding these allele frequencies are present in a substantial fraction of cells, thereby supporting clonal hematopoiesis with a persisting pre-leukemic clone rather than detection of a small number of residual leukemia cells (i.e., MRD). In our analysis, persisting mutations associated with inferior outcomes irrespective of their allele frequency in remission. It remains unclear whether very small preleukemic clones below our limit of detection carry a similar prognostic weight. In another recently published study, an NGS assay with a sensitivity of 0.2% was used to study persistence of mutations in 122 genes during CR in 59 patients. Persistence of ≥2 lesions, but not persistence of a single lesion, associated with significantly reduced leukemia-free survival and OS [5]. In the same study, patients with ≥1 mutation persisting at a VAF of ≥3.33% had shorter leukemia-free survival when censoring for alloSCT. These results are consistent with our findings and suggest that persistence of a single pre-leukemic variant at a low allele frequency may not carry the same adverse prognostic implications as persistence of larger clones or multiple mutations.
Almost half of the patients in our analysis underwent alloSCT in first remission, while the others received consolidation chemotherapy. To shed some light on the impact of post-remission therapy in patients with and without mutation persistence, we analyzed CIR before and after transplantation. In an analysis that treated alloSCT as a competing risk, mutation persistence associated with a significantly increased risk of relapse among patients who had not, or not yet, undergone transplantation. This difference was no longer observed after alloSCT in first remission, although the low number of recurrences after transplantation limited the power of this analysis. Since most allografted patients reach full hematopoietic chimerism, alloSCT may reducing the risk of relapse by not only eradicating residual leukemic cells, but also pre-leukemic clones. We could not directly verify this hypothesis by testing for persisting mutations in post-transplantation samples, due to lack of consent for genetic testing from SCT donors.
Mutation persistence was observed across the entire age range of our cohort. It affected ~25% of patients aged ≤60 y, by far exceeding the reported incidence of clonal hematopoiesis in healthy persons in this age group. Among patients >60 y, over two thirds had persisting mutations in remission. Older age is one of the strongest predictors of inferior treatment outcomes in AML, yet the reasons for this association still are incompletely understood. Pre-treatment genetics and patient-related factors such as performance status and comorbidities only partially explain the worse prognosis of elderly patients. In our cohort, older age by itself showed a significant association with shorter RFS and OS. However, mutation persistence was more closely associated with both endpoints than age in bivariate models (Supplementary Table 8). These data suggest that a higher frequency of pre-leukemic clones surviving chemotherapy is an additional factor contributing to the poor prognosis of older AML patients. Our analysis of relapse specimens shows that pre-leukemic clones can act as a source of AML relapse harboring the same pre-leukemic variants, but a different set of additional mutations, compared to the initial leukemia clone. In other cases, the genotype at relapse indicated a close relationship to the original clone, suggesting that persistence of a pre-leukemic clone may also be a surrogate for a more difficult-to-eradicate phenotype of the leukemia clone itself.
Our study adds to a growing body of data showing that monitoring of leukemic and pre-leukemic clones after therapy (through MRD assays and analyses of persisting mutations, respectively) may complement or even supersede pre-treatment genetic factors for prognostic stratification [33]. MRD measurements by flow cytometry or qPCR rely on informative immunophenotypes or molecular markers. Consequently, MRD levels by either method were only available in a subset of our patients, precluding assessment of their prognostic value alone or relative to persistence of pre-leukemic clones. However, mutation persistence and MRD levels were not closely correlated, suggesting that both variables might ultimately provide complementary prognostic information.
In summary, our study provides further evidence that persistence of pre-leukemic clones during first remission after induction chemotherapy is an adverse risk factor in AML. Mutation persistence is common in elderly patients and may contribute to the inferior outcomes in this age group. While the prognostic value of mutation persistence was independent of other known risk factors, including the current ELN genetic risk classification of AML, patients with persisting mutations who did not undergo alloSCT had a particularly high relapse risk. Future studies should evaluate the predictive value of mutation persistence for post-remission treatment decisions.
Electronic supplementary material
Acknowledgements
This study was supported by an European Hematology Association (EHA) Clinical Research Fellowship, and by grant support from Deutsche Forschungsgemeinschaft (DFG SFB 1243) and the German Cancer Consortium (DKTK, Heidelberg, Germany). The AMLCG-2008 clinical trial was supported by Deutsche Krebshilfe. SKB gratefully acknowledges support from Leukemia & Blood Cancer New Zealand and the family of Marijanna Kumerich.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41375-018-0034-z) contains supplementary material, which is available to authorized users.
References
- 1.Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 2012;4:149ra118. doi: 10.1126/scitranslmed.3004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci USA. 2014;111:2548–53. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–33. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirsch P, Zhang Y, Tang R, Joulin V, Boutroux H, Pronier E, et al. Genetic hierarchy and temporal variegation in the clonal history of acute myeloid leukaemia. Nat Commun. 2016;7:12475. doi: 10.1038/ncomms12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirsch P, Tang R, Abermil N, Flandrin P, Moatti H, Favale F, et al. Precision and prognostic value of clone-specific minimal residual disease in acute myeloid leukemia. Haematologica. 2017;102:1227–37. doi: 10.3324/haematol.2016.159681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thol F, Klesse S, Köhler L, Gabdoulline R, Kloos A, Liebich A, et al. Acute myeloid leukemia derived from lympho-myeloid clonal hematopoiesis. Leukemia. 2017;31:1286–95. doi: 10.1038/leu.2016.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pløen GG, Nederby L, Guldberg P, Hansen M, Ebbesen LH, Jensen UB, et al. Persistence of DNMT3A mutations at long-term remission in adult patients with AML. Br J Haematol. 2014;167:478–86. doi: 10.1111/bjh.13062. [DOI] [PubMed] [Google Scholar]
- 8.Debarri H, Lebon D, Roumier C, Cheok M, Marceau-Renaut A, Nibourel O, et al. IDH1/2 but not DNMT3A mutations are suitable targets for minimal residual disease monitoring in acute myeloid leukemia patients: a study by the Acute Leukemia French Association. Oncotarget. 2015;6:42345–53. doi: 10.18632/oncotarget.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatnagar B, Eisfeld AK, Nicolet D, Mrózek K, Blachly JS, Orwick S, et al. Persistence of DNMT3AR882 mutations during remission does not adversely affect outcomes of patients with acute myeloid leukaemia. Br J Haematol. 2016;175:226–36. doi: 10.1111/bjh.14254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaidzik VI, Weber D, Paschka P, Kaumanns A, Krieger S, Corbacioglu A, et al. DNMT3A mutant transcript levels persist in remission and do not predict outcome in patients with acute myeloid leukemia. Leukemia. 2017;32:30–37. doi: 10.1038/leu.2017.200. [DOI] [PubMed] [Google Scholar]
- 11.Klco JM, Miller CA, Griffith M, Petti A, Spencer DH, Ketkar-Kulkarni S, et al. Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA. 2015;314:811–22. doi: 10.1001/jama.2015.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcucci G, Mrózek K, Ruppert AS, Archer KJ, Pettenati MJ, Heerema NA, et al. Abnormal cytogenetics at date of morphologic complete remission predicts short overall and disease-free survival, and higher relapse rate in adult acute myeloid leukemia: results from cancer and leukemia group B study 8461. J Clin Oncol. 2004;22:2410–8. doi: 10.1200/JCO.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Cortes J, Estrov Z, Faderl S, Qiao W, Abruzzo L, et al. Persistence of cytogenetic abnormalities at complete remission after induction in patients with acute myeloid leukemia: prognostic significance and the potential role of allogeneic stem-cell transplantation. J Clin Oncol. 2011;29:2507–13. doi: 10.1200/JCO.2010.34.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metzeler K, Herold T, Rothenberg-Thurley M, Amler S, Sauerland MC, Görlich D, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128:686–98. doi: 10.1182/blood-2016-01-693879. [DOI] [PubMed] [Google Scholar]
- 15.Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–47. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braess J, Kreuzer KA, Spiekermann K, Lindemann HW, Lengfelder E, Graeven U, et al. High efficacy and significantly shortened neutropenia of dose-dense S-HAM as compared to standard double induction: first results of a prospective randomized trial (AML-CG 2008) Blood. 2013;122:619. doi: 10.1182/blood-2013-06-508481. [DOI] [PubMed] [Google Scholar]
- 17.Cheson BD, Bennett JM, Kopecky KJ, Büchner T, Willman CL, Estey EH, et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol. 2003;21:4642–9. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 18.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–65. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 19.Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015;43:D6–17. doi: 10.1093/nar/gku1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hubmann M, Köhnke T, Hoster E, Schneider S, Dufour A, Zellmeier E, et al. Molecular response assessment by quantitative real-time polymerase chain reaction after induction therapy in NPM1-mutated patients identifies those at high risk of relapse. Haematologica. 2014;99:1317–25. doi: 10.3324/haematol.2014.104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benthaus T, Schneider F, Mellert G, Zellmeier E, Schneider S, Kakadia PM, et al. Rapid and sensitive screening for CEBPA mutations in acute myeloid leukaemia. Br J Haematol. 2008;143:230–9. doi: 10.1111/j.1365-2141.2008.07328.x. [DOI] [PubMed] [Google Scholar]
- 25.Kiyoi H, Naoe T, Nakano Y, Yokota S, Minami S, Miyawaki S, et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood. 1999;93:3074–80. [PubMed] [Google Scholar]
- 26.Thiede C, Steudel C, Mohr B, Schaich M, Schäkel U, Platzbecker U, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–35. doi: 10.1182/blood.V99.12.4326. [DOI] [PubMed] [Google Scholar]
- 27.Papadaki C, Dufour A, Seibl M, Schneider S, Bohlander SK, Zellmeier E, et al. Monitoring minimal residual disease in acute myeloid leukaemia with NPM1 mutations by quantitative PCR: clonal evolution is a limiting factor. Br J Haematol. 2009;144:517–23. doi: 10.1111/j.1365-2141.2008.07488.x. [DOI] [PubMed] [Google Scholar]
- 28.Köhnke T, Sauter D, Ringel K, Hoster E, Laubender RP, Hubmann M, et al. Early assessment of minimal residual disease in AML by flow cytometry during aplasia identifies patients at increased risk of relapse. Leukemia. 2015;29:377–86. doi: 10.1038/leu.2014.186. [DOI] [PubMed] [Google Scholar]
- 29.Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988;16:1141–54. doi: 10.1214/aos/1176350951. [DOI] [Google Scholar]
- 30.Altman DG, De Stavola BL. Practical problems in fitting a proportional hazards model to data with udated measurements of the covariates. Stat Med. 1994;13:301–41. doi: 10.1002/sim.4780130402. [DOI] [PubMed] [Google Scholar]
- 31.Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Control Clin Trials. 1996;17:343–6. doi: 10.1016/0197-2456(96)00075-X. [DOI] [PubMed] [Google Scholar]
- 32.Jezísková I, Musilova M, Culen M, Foltankova V, Dvoráková D, Mayer J, et al. Distribution of mutations in DNMT3A gene and the suitability of mutations in R882 codon for MRD monitoring in patients with AML. Int J Hematol. 2015;102:553–7. doi: 10.1007/s12185-015-1856-3. [DOI] [PubMed] [Google Scholar]
- 33.Ivey A, Hills RK, Simpson MA, Jovanovic JV, Gilkes A, Grech A, et al. Assessment of minimal residual disease in standard-risk AML. N Engl J Med. 2016;374:422–33. doi: 10.1056/NEJMoa1507471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.