Abstract
Introduction
Compared with European Americans, African Americans (AAs) are at higher risk for developing end-stage kidney disease (ESKD). Genome-wide association studies (GWAS) have identified >70 genetic variants associated with kidney function and chronic kidney disease (CKD) in patients with and without diabetes. However, these variants explain a small proportion of disease liability. This study examined the contribution of coding genetic variants for risk of type 2 diabetes (T2D)-attributed ESKD and advanced CKD in AAs.
Methods
Exome sequencing was performed in 456 AA T2D-ESKD cases, and 936 AA nondiabetic, non-nephropathy control individuals at the discovery stage. A mixed logistic regression model was used for association analysis. Nominal associations (P < 0.05) were replicated in an additional 2020 T2D-ESKD cases and 1121 nondiabetic, non-nephropathy control individuals. A meta-analysis combining 4533 discovery and replication samples was performed. Putative T2D-ESKD associations were tested in additional 1910 nondiabetic ESKD and 219 T2D-ESKD cases, as well as 912 AA nondiabetic non-nephropathy control individuals.
Results
A total of 11 suggestive T2D-ESKD associations (P < 1 x 10−4) from 8 loci (PLEKHN1, NADK, RAD51AP2, RREB1, PEX6, GRM8, PRX, APOL1) were apparent in the meta-analysis. Exclusion of APOL1 renal-risk genotype carriers identified 3 additional suggestive loci (OTUD7B, IFITM3, DLGAP5). Rs41302867 in RREB1 displayed consistent association with T2D-ESKD and nondiabetic ESKD (odds ratio: 0.47; P = 1.2 x 10−6 in 4605 all-cause ESKD and 2969 nondiabetic non-nephropathy control individuals).
Conclusion
Our findings suggest that coding genetic variants are implicated in predisposition to T2D-ESKD in AAs.
Keywords: African Americans, chronic kidney disease, end-stage kidney disease, exome sequencing, genetics, type 2 diabetes
African Americans (AAs) are disproportionately affected by end-stage kidney disease (ESKD); incidence rates of ESKD are 3.1-fold higher in AAs than European Americans.1 In 2014, 120,688 new cases of ESKD were diagnosed, of which 97.4% received dialysis and only 2.6% underwent kidney transplantation.1 The mortality rates for patients with ESKD on dialysis and after transplantation in 2014 were 166 and 30 per 1000 patient-years, respectively.1 Diabetes is one of the most common reported causes of ESKD, accounting for >44% of causes in the United States; of these, approximately 90% relate to type 2 diabetes (T2D).1 Even after adjustment for socioeconomic status and environmental factors, incidence rates and familial aggregation of diabetic kidney disease (DKD), including T2D-attributed ESKD (T2D-ESKD), remain significantly higher in AAs.2, 3 Several lines of evidence support genetic contributors to ESKD susceptibility.4, 5 Prior studies have shown that common genetic variants contribute to DKD susceptibility in multiple ethnic groups.6, 7, 8, 9, 10 Although the apolipoprotein L1 gene (APOL1) G1 and G2 alleles explain a substantial proportion (∼70%) of the disparity in nondiabetic ESKD in AAs versus European Americans, they fail to account for the excess risk of T2D-ESKD in AAs.11, 12, 13
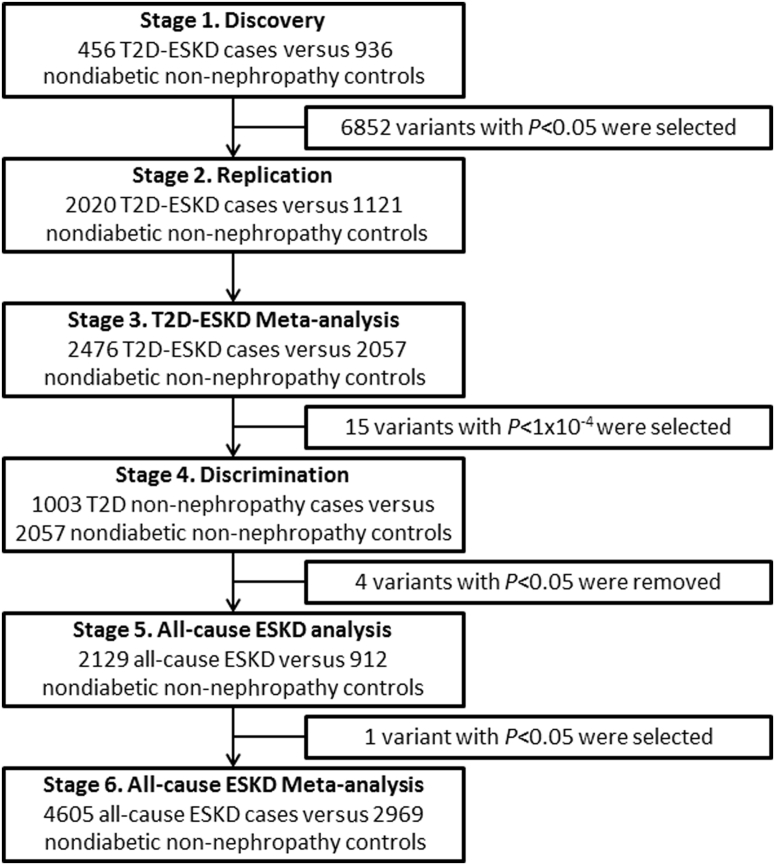
Although DKD-associated variants have been identified by genome-wide association studies (GWASs), their contributions to DKD risk are modest.6, 7, 8, 9, 14 GWASs are not efficient in testing low-frequency variants, and these may account for a portion of the missing heritability. In contrast, exome sequencing is a powerful tool to explore coding regions, particularly low-frequency variants, which are poorly imputed by GWASs. In our prior studies, several coding variants located in RREB1, NPHS1, CUBN, LRP2, COL4A3, and CLDN8 were shown to contribute to diabetic and/or nondiabetic ESKD in AAs.15, 16, 17, 18 Herein, we extend prior studies by performing a comprehensive association study of coding variants for T2D-ESKD risk in AAs. We examined 1392 AA individuals with or without T2D-ESKD by exome sequencing (stage 1), followed by replication (stage 2) in 3141 AA subjects, and a meta-analysis combining 4533 AAs from stage 1 and stage 2 (stage 3), and a discrimination analysis in 1003 subjects with T2D without nephropathy to exclude T2D-associated variants (stage 4). We also performed replication in an additional 3041 all-cause ESKD cases and controls (stage 5). Finally, all T2D-ESKD and nondiabetic ESKD cases as well as controls were meta-analyzed (n = 7574) to evaluate the impact of associated variants on risk of all-cause ESKD (stage 6).
Materials and Methods
Study Participants
Study participants were recruited by Wake Forest School of Medicine (stages 1–5) and Jackson Heart Study (stage 1). The studies were approved by the institutional review board of each participating center. All participants provided written informed consent. Description of a subset of study participants (1834 T2D-ESKD, 1295 nondiabetic non-nephropathy, 850 T2D-lacking nephropathy, 1705 nondiabetic ESKD) has previously been reported.15 Participants were considered to have T2D-ESKD when T2D was clinically diagnosed ≥5 years before the onset of ESKD (or with diabetic retinopathy to ensure long-duration T2D), and with at least 1 of the following: on renal replacement therapy, estimated glomerular filtration rate <30 ml/min per 1.73 m2 (chronic kidney disease [CKD4]), or urine albumin/creatinine ratio >300 mg/g (macroalbuminuria) if an estimated glomerular filtration rate was unavailable. Participants with CKD4 or macroalbuminuria were included as cases, given their high risk of developing ESKD (Supplementary Table S1). Nondiabetic ESKD cases lacked diabetes (or developed diabetes after initiating renal replacement therapy), and their kidney disease was attributed to chronic glomerular disease (e.g., focal segmental glomerulosclerosis), HIV-associated nephropathy, hypertension, or unknown cause. Those with ESKD attributed to surgical or urologic causes, polycystic kidney disease, autoimmune disease, hepatitis, IgA nephropathy, membranous glomerulonephritis, membranoproliferative glomerulonephritis, or monogenic kidney diseases were excluded. Nondiabetic non-nephropathy controls included those lacking diabetes and kidney disease (estimated glomerular filtration rate ≥ 60 ml/min per 1.73 m2 and urine albumin/creatinine ratio <30 mg/g [when urine albumin/creatinine ratio was available]). Individuals were considered to have T2D without nephropathy when their estimated glomerular filtration rate was ≥60 ml/min per 1.73 m2 and urine albumin/creatinine ratio was <30 mg/g in the presence of T2D.
Sequencing, Genotyping, and Quality Control
Discovery Cohort
A total of 456 AA T2D-ESKD cases (5 with CKD4), 936 AA nondiabetic non-nephropathy controls, and 338 AA T2D cases lacking nephropathy underwent exome sequencing as part of the T2D-GENES consortium project. Details of sequencing data generation have been described previously.19 Briefly, exome sequencing was performed using an Agilent V2 capture array platform (Agilent Technologies, Santa Clara, CA) at the Broad Institute (Cambridge, MA). Sequence data underwent multiple levels of quality control (QC) for both samples and sequence reads. A total of 357 samples with average coverage ≤20x, possible DNA contamination, showed non-AA ancestry in principal component analysis, or missing information was excluded. Aligned sequence reads were filtered based on multiple QC criteria, including number of mapped reads, fraction of properly paired reads, distribution of insertion sizes, distribution of average base quality, as well as guanine and cytosine bias. A total of 136,584 exome variants with minor allele frequency (MAF) ≥ 0.01 were included in this study.
Replication and Discrimination Cohort
The replication and discrimination cohorts included 2020 AA T2D-ESKD cases (143 had CKD4 or macroalbuminuria), 1121 AA nondiabetic non-nephropathy controls, and 665 AAs with T2D who lacked nephropathy. Samples were genotyped on a custom Affymetrix Axiom Biobank Genotyping Array (Affymetrix, Santa Clara, CA). Detailed variant information, custom content design, including fine mapping of candidate regions, genotyping methods, and QC have been reported.18 In brief, this array includes approximately 264K coding variants and insertions/deletions (indels), 70K loss-of-function variants, 2K pharmacogenomic variants, 23K expression quantitative trait loci markers, 246K multiethnic population-based genome-wide tag markers, and 115K custom content. A total of 724,530 variants were successfully called for downstream QC and analyses. No batch effect was observed. Variants with call rates <95%, departure from Hardy Weinberg Equilibrium (P < 0.0001), and monomorphic variants were removed. A total of 683,375 variants were kept for imputation. Sample QC was also performed to remove individuals with low call rate, gender discordance, DNA contamination, or non-AA ancestry. Duplicate samples were identified, and one of each duplicate pair removed. A total of 1039 samples were excluded from this study. Variants that passed QC were imputed to a combined haplotype reference panel including the 1000 Genomes phase 3 cosmopolitan reference panel (October 2014 version)20 and a version of the African Genome Variation Project reference panel including 640 African ancestry haplotypes kindly provided by the African Partnership for Chronic Disease Research and Wellcome Trust Sanger Institute.21 Prephasing was performed using SHAPEIT222 and imputation was performed using IMPUTE2.23 Variants with imputation info scores <0.4 were excluded from analysis.
All-Cause ESKD Replication Cohort
Stage 5 analyses included 1910 nondiabetic ESKD cases, 219 T2D-ESKD cases (7 with CKD4 or macroalbuminuria), and 912 nondiabetic non-nephropathy controls. These samples were genotyped on the Multi-Ethnic Genotyping Array (Illumina, San Diego, CA), designed to improve fine-mapping and functional variants discovery by increasing variant coverage across multiple ethnicities.24 In brief, the array includes variants from 2 major categories: (i) backbone content containing highly informative variants for GWAS and exome analyses in ancestrally diverse populations, and (ii) custom content used to replicate or generalize index GWAS associations, augment GWAS tagging variants in priority regions, enhance exome content in priority regions, fine-map GWAS loci, identify functional regulatory variants, explore medically important variants, and identify novel variant loci in candidate pathways. Genotyping was performed at Wake Forest School of Medicine. DNA from cases and controls was equally interleaved on 96-well plates to minimize artifactual errors during sample processing. A total of 48 samples sequenced as part of the 1000 Genomes Project20 at the Coriell Institute for Medical Research were included in genotyping and had a concordance rate of 98.6%. Genotype calling was performed using GenomeStudio (Illumina). A total of 1,705,970 variants were successfully called for downstream QC and analyses. No batch effect was observed. Variants with missing position or alleles, allele mismatch, call rates <95%, departure from Hardy Weinberg Equilibrium (P < 0.0001), frequency difference >0.2 compared with 1000 Genomes Project phase 3 reference panel, and monomorphic variants were removed. Multiple probe sets were compared and only one set with the highest call rate was kept. We kept 1,204,700 high-quality variants for subsequent imputation. Sample QC was performed to remove individuals with low call rates (<0.95), gender discordance, DNA contamination, or non-AA ancestry. Duplicate samples were compared, and one of each duplicate pair was removed. A total of 1519 samples were excluded from analysis. Variants and samples that passed QC were used to perform prephasing with SHAPEIT222 and imputation with IMPUTE223 using a combined haplotype reference panel from the 1000 Genomes Project phase 320 and the African Genome Variation Project,21 described previously. Variants with imputation info scores <0.4 were excluded from analysis.
Statistical Analysis
Discovery and Replication of Associations in T2D-ESKD (Baseline Model)
Single-variant association analyses in case-control samples (from all analysis stages) were performed using a logistic mixed model method implemented in the program GMMAT25 under an additive genetic model. This controlled for population structure and cryptic relatedness by incorporating a genetic relationship matrix estimated from a set of high-quality autosomal variants as a random effect. Principal component analysis was performed using EIGENSOFT (Broad Institute of Harvard and MIT, Cambridge, MA).26 The first eigenvector (PC1), representing African-European ancestry, along with age and sex, were used as covariates. Body mass index was not included because of changes after CKD onset.
In the stage 1 analysis, exome-wide analysis was performed in 456 T2D-ESKD cases and 936 nondiabetes non-nephropathy controls on 193,646 variants that passed QC and had MAF ≥ 0.01. Variants with nominal associations (P < 0.05) were further tested in a replication cohort with 2020 T2D-ESKD cases and 1121 nondiabetic non-nephropathy controls (stage 2 analysis). A P value of 0.05 was used as a cutoff because we had limited sample size and power in the discovery stage. Meta-analysis including the discovery and replication stages (2476 cases and 2057 controls) was performed using a fixed-effect inverse variance weighting method implemented in METAL27 in the stage 3 analysis. Variants with suggestive evidence of associations (P < 1 × 10–4) were selected for discrimination analysis.
Discrimination Analysis
To distinguish whether the meta-analysis association results were driven by T2D alone or T2D-ESKD, 1003 AAs with T2D-lacking nephropathy and 2057 nondiabetic non-nephropathy controls from analysis stages 1 and 2 were compared in a discrimination analysis (stage 4 analysis). Variants showing nominal association with T2D (P < 0.05) were excluded.
Replication of Associations in All-Cause ESKD
Variants that showed suggestive association (P < 1 × 10–4) in stage 3 T2D-ESKD meta-analysis and no evidence of association with T2D in the stage 4 discrimination analysis were tested for association with all-cause ESKD in 1910 nondiabetic ESKD cases, 219 additional T2D-ESKD cases, and 912 nondiabetic non-nephropathy controls (stage 5 analysis). Variants showing nominal association with all-cause ESKD (P < 0.05) were tested in a meta-analysis of all-cause ESKD that included all of the T2D-ESKD cases, nondiabetic ESKD cases, and controls from stages 1, 2, and 5 analyses (n = 7574). This meta-analysis evaluated whether T2D-ESKD associated gene variants contributed more broadly to other causes of ESKD. Subjects with stage 1 and 2 (n = 4533) were used to calculate the power to detect variants with a range of frequency (MAF = 0.01 to 0.4) and odds ratio (OR: 1.2 to 2) for a disease with 0.1% prevalence using CaTS (http://csg.sph.umich.edu/abecasis/cats/).
Association Analyses With Exclusion of APOL1 Risk Genotype Carriers (APOL1-Negative Model)
APOL1 G1 and G2 risk alleles explain a substantial proportion of genetic susceptibility to nondiabetic kidney disease in AAs.12 To minimize effects of misclassification, the same single-variant analyses were repeated (APOL1-negative model) by excluding APOL1 renal-risk-genotype carriers and those missing APOL1 genotype data from T2D-ESKD and nondiabetic ESKD samples. Individuals were considered APOL1 renal-risk-genotype carriers if they possessed 2 G1 alleles (rs60910145 G allele, rs73885319 G allele), 2 G2 alleles (6 base pair inframe deletion), or were compound heterozygotes (1 G1 and 1 G2 allele).12 This secondary analysis reduces heterogeneity in the T2D-ESKD case group, despite its smaller sample size. Specifically, 98 of 456 T2D-ESKD cases from stage 1 discovery and 386 of 2020 T2D-ESKD cases from stage 2 replication were removed. In addition, 936 of 2136 all-cause ESKD cases in the stage 5 analysis were removed, considering the impact of APOL1 in nondiabetic ESKD. This strategy may unmask effects of nondiabetic ESKD variants beyond APOL1.
Gene-based Analysis
Four gene-based tests implemented in RAREMETAL28 were applied: Sequence Kernel Association Test,29 Madsen-Browning test,30 Variable Threshold test,31 and Combined and Multivariate Collapsing test32 to test for the joint effect of rare variants (MAF < 0.05) within a gene on the stage 1 discovery analysis with exome sequencing data. Variants were categorized into 4 groups in all methods: (i) moderate to high-impact protein structure altering variants (transcript ablation, splice acceptor, splice donor, stop gained, frameshift, stop lost, start lost, transcript amplification, inframe insertion, inframe deletion, missense, and protein altering); (ii) a more restricted group with only high-impact protein structure–altering variants (transcript ablation, splice acceptor, splice donor, stop gained, frameshift, stop lost, start lost, transcript amplification); (iii) variants predicted to be deleterious by at least 1 of the 4 prediction methods, including SIFT (sorting intolerant from tolerant),33 LRT (likelihood ratio test),34 MutationTaster,35 and CADD (combined annotation–dependent depletion)36; and (iv) variants predicted to be deleterious by all 4 prediction methods. Age, gender, and PC1 were included as fixed-effect covariates. A P value <2.5 × 10−6 was considered exome-wide significant. Both baseline and APOL1-negative models were evaluated.
Functional Characterization
The publicly available expression quantitative trait loci database GTEx (http://www.gtexportal.org/home/) was used to determine potential influences of T2D-ESKD associated variants on nearby gene expression. We queried 14 top associations from either the baseline or APOL1-negative models in GTEx across multiple tissues. Additional functional annotation of genetic variants was performed with Variant Effect Predictor (VEP),37 SnpEff,38 and dbNSFP.39
Multiple Comparison Correction
We used Bonferroni correction to adjust for multiple comparisons problem for both single-variant analysis and gene-based analysis. Exome-wide significance was defined as P < 3.5 × 10−7 (0.05/136,584) and P < 2.5 × 10−6 (0.05/20,000) for single-variant analysis and gene-based analysis, respectively. We also calculated false discovery rate (Q-value) using the Benjamini-Hochberg algorithm for both discovery analysis and meta-analysis in single-variant analysis.
Results
Genetic variants located in coding regions were evaluated in 3 independent AA cohorts for association with T2D-ESKD and all-cause ESKD through a multistage study design (Figure 1). A total of 8577 individuals categorized as having T2D-ESKD, nondiabetic non-nephropathy, T2D-lacking nephropathy, and nondiabetic ESKD were included in a sequence of association analyses. Overall, 18 suggestive T2D-ESKD associations in 11 distinct regions were identified, including 2 missense variants, in either the baseline or the APOL1-negative models. Rs41302867 located in the RREB1 region revealed consistent association with T2D-ESKD and nondiabetic ESKD, which is consistent with our previous report.15 Additionally, gene-based associations identified 7 genes with suggestive evidence of association with T2D-ESKD.
Figure 1.
Analysis workflow of single-variant association analysis for T2D-ESKD exome sequencing study (baseline model). Stage 3 combines stage 1 and stage 2 samples; stage 6 combines stage 1, stage 2, and stage 5 samples. T2D, type 2 diabetes; ESKD, end-stage kidney disease.
Clinical Characteristics of Study Participants
Clinical characteristics of study participants from all analysis stages are presented in Table 1. Individuals with T2D-ESKD and T2D-lacking nephropathy were older than the nondiabetic non-nephropathy controls at recruitment. However, the average age of diagnosis of T2D in T2D-ESKD and of T2D-lacking nephropathy participants were similar or younger than healthy controls at recruitment. Overall, individuals with T2D-lacking nephropathy were more obese than individuals with T2D- or nondiabetic ESKD and healthy controls.
Table 1.
Clinical characteristics of study cohorts
| Characteristics | Discovery cohort |
Replication and discrimination cohort |
All-cause ESKD replication cohort |
||||||
|---|---|---|---|---|---|---|---|---|---|
| T2D-ESKD cases | Nondiabetic non-nephropathy controls | T2D-lacking nephropathy | T2D-ESKD cases | Nondiabetic non-nephropathy controls | T2D lacking nephropathy | Nondiabetic ESKD | T2D-ESKD | Nondiabetic non-nephropathy controls | |
| N | 456 | 936 | 338 | 2020 | 1121 | 665 | 1910 | 219 | 912 |
| Female (%) | 61.62 | 59.72 | 68.05 | 57.12 | 51.74 | 64.47 | 58.74a | 50.68 | 41.89a |
| Age (yr) | 64.61 ± 8.55a | 52.49 ± 11.25 | 57.42 ± 10.16a | 61.46 ± 10.88a | 46.50 ± 12.09 | 55.78 ± 11.61a | 55.35 ± 14.44a | 61.99 ± 10.99 | 44.82 ± 13.83a |
| Age at T2D onset (yr) | 46.2 ± 9.27 | — | — | 38.62 ± 12.76 | — | 46.20 ± 12.27 | — | 37.81 ± 9.60 | — |
| Duration of T2D for T2D-lacking nephropathy (yr) | — | — | — | — | — | 9.58 ± 9.12 | — | — | — |
| Duration of T2D before ESKD (yr) | 15.36 ± 8.80 | — | — | 19.66 ± 9.91 | — | — | — | 20.41 ± 9.55 | — |
| Duration of ESKD (yr) | 3.11 ± 3.73 | — | — | 3.61 ± 3.64 | — | — | 6.17 ± 5.80 | 4.04 ± 3.11 | — |
| Fasting serum glucose (mg/dl) | — | 84.75 ± 9.69 | — | — | 96.52 ± 20.93 | — | 87.00 ± 8.72a | — | 96.09 ± 10.46 |
| eGFR (ml/min per 1.73 m2) | — | 92.10 ± 17.33 | 94.44 ± 19.78 | — | 103.45 ± 19.08 | 95.71 ± 18.28a | — | — | 89.29 ± 16.72 |
| Body mass index (kg/m2) | 29.43 ± 6.52a | 30.86 ± 6.88 | 33.11 ± 6.22a | 30.74 ± 7.13a | 29.81 ± 7.36 | 33.10 ± 7.81a | 27.76 ± 7.21a | 29.73 ± 7.03 | 29.73 ± 6.60 |
eGFR, estimated glomerular filtration rate; ESKD, end-stage kidney disease; T2D, type 2 diabetes.
Categorical data expressed as percentage; continuous data as mean ± SD.
Significant difference (P < 0.05) compared with nondiabetic non-nephropathy controls.
Stage 1 Discovery Analysis in Exome Sequencing Cohort
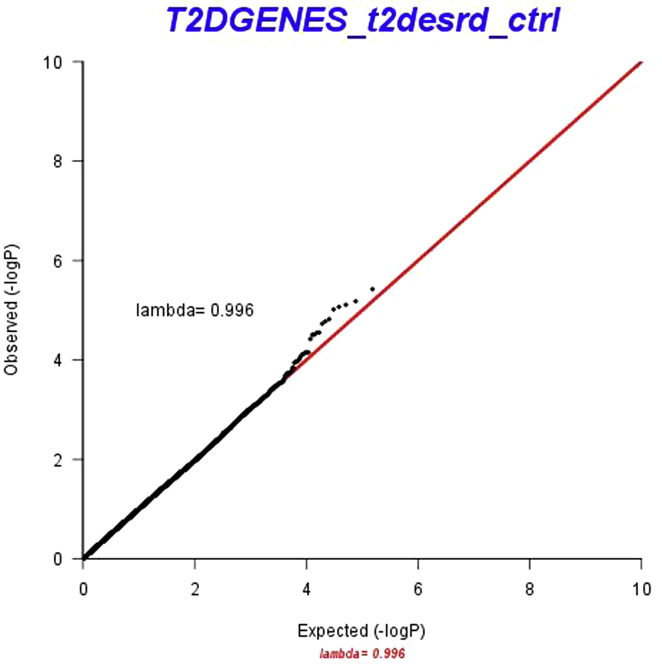
The stage 1 discovery analysis with exome sequencing data included 456 T2D-ESKD patients and 936 nondiabetic non-nephropathy controls (Figure 1). A total of 136,584 exome variants that passed QC and had MAF ≥ 0.01 were tested for association with T2D-ESKD using a logistic mixed model.25 Age, gender, and PC1 were included as fixed-effects in the model (baseline model). The association analysis yielded an inflation factor of 0.996 (Supplementary Figure S1), indicating that population structure and cryptic relatedness were sufficiently controlled. No exome-wide significant association (P < 3.5 × 10−7, 0.05/136,584) was observed. However, 20 variants showed suggestive significance at a P < 1 × 10−4. We selected 6852 variants with nominal significance (P < 0.05) for replication analysis, given the relatively small sample size and low power of the discovery cohort.
Stage 2 Replication Analysis
An independent cohort of 2020 AA T2D-ESKD cases and 1121 AA nondiabetic non-nephropathy controls with imputed dosage data was used to replicate the selected variants from the discovery stage (Figure 1). More than 93% of the variants were imputed with imputation quality info >0.4 for association analyses.
Stage 3 Meta-analysis
A total of 6852 variants with nominal association (P < 0.05) were selected for meta-analysis of the discovery and replication samples. Fifteen variants with consistent directions of association with T2D-ESKD at P < 1 × 10−4 were identified. However, no variant reached exome-wide significance (P < 3.5 × 10−7).
Stage 4 Discrimination Analysis
To differentiate T2D-ESKD associations identified in meta-analysis as putative T2D-ESKD or T2D-only (non-nephropathy) associated loci, we performed a T2D discrimination analysis comparing 1003 AA T2D cases lacking nephropathy with 2057 nondiabetic non-nephropathy controls from stages 1 and 2. Four of the 15 T2D-ESKD associated variants were nominally associated (P < 0.05) with T2D and were removed from subsequent analyses (Supplementary Table S2).
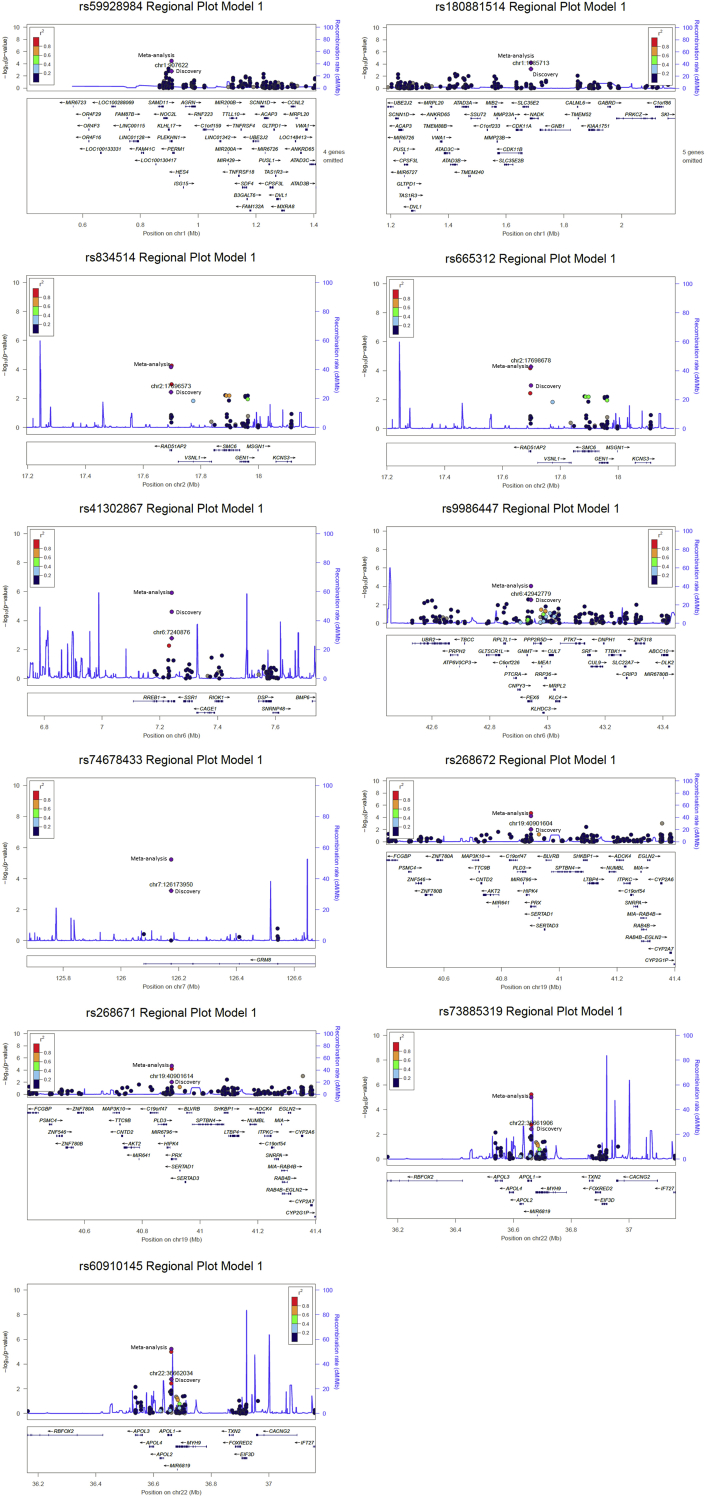
The remaining 11 T2D-ESKD associated variants were located in 8 loci, including PLEKHN1, NADK, RAD51AP2, RREB1, PEX6, GRM8, PRX, APOL1 (P < 1 × 10−4) (Table 2, Supplementary Figure S2). The appearance of 2 APOL1 G1 alleles, rs73885319 (OR: 1.32; 95% confidence interval [CI]: 1.17–1.50; P = 9.90 × 10−6) and rs60910145 (OR: 1.33; 95% CI: 1.18–1.51; P = 5.74 × 10−6), known to be associated with nondiabetic ESKD, may be due in part to misclassification of DKD cases.12(p1) T2D-ESKD associated variants located within RAD51AP2 and PRX are highly correlated (r2 > 0.9), and rs834514 at RAD51AP2 (OR: 0.76; 95% CI: 0.67–0.87; P = 6.56 × 10−5) and rs268671 at PRX (OR: 0.75; 95% CI: 0.65–0.86; P = 2.11 × 10−5) were missense variants. Moreover, variants from PLEKHN1, RAD51AP2, PEX6, and PRX were associated with the expression level of nearby genes across multiple tissues, suggesting potential regulatory roles (Supplementary Table S3).
Table 2.
T2D-ESKD associated variants in meta-analysis from discovery and replication cohorts (baseline model)
| Variant | Gene | Annotation | CHR | POS | EA | Discovery (456 T2D-ESKD cases vs. 936 nondiabetic non-nephropathy controls) |
Replication (2020 T2D-ESKD cases vs. 1121 nondiabetic non-nephropathy controls) |
Meta-analysis (2476 T2D-ESKD cases vs. 2057 nondiabetic non-nephropathy controls) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EAF | OR (95% CI) | P | EAF | OR (95% CI) | P | Info | EAF | OR (95% CI) | P | Q-value | ||||||
| rs59928984 | PLEKHN1 | Intron | 1 | 907622 | C | 0.22 | 1.46 (1.15–1.85) | 0.0017 | 0.21 | 1.27 (1.08–1.50) | 0.0041 | 0.91 | 0.21 | 1.33 (1.16–1.53) | 3.32E-05 | 0.023 |
| rs180881514 | NADK | Intron | 1 | 1685713 | T | 0.01 | 4.87 (1.96–12.06) | 0.00064 | 0.011 | 2.23 (1.20–4.13) | 0.011 | 0.91 | 0.011 | 2.85 (1.71–4.75) | 5.84E-05 | 0.031 |
| rs834514 | RAD51AP2 | Missense | 2 | 17696573 | T | 0.21 | 0.7 (0.55–0.89) | 0.0037 | 0.21 | 0.79 (0.68–0.93) | 0.0043 | 1.0 | 0.21 | 0.76 (0.67–0.87) | 6.56E-05 | 0.032 |
| rs665312 | RAD51AP2 | Synonymous | 2 | 17698678 | G | 0.24 | 0.69 (0.55–0.86) | 0.0011 | 0.24 | 0.81 (0.70–0.95) | 0.0077 | 1.0 | 0.24 | 0.77 (0.68–0.87) | 5.27E-05 | 0.031 |
| rs41302867 | RREB1 | Intron | 6 | 7240876 | A | 0.023 | 0.35 (0.18–0.67) | 0.0016 | 0.02 | 0.51 (0.33–0.80) | 0.003 | 1.0 | 0.021 | 0.45 (0.31–0.65) | 2.43E-05 | 0.019 |
| rs9986447 | PEX6 | Splice region | 6 | 42942779 | G | 0.22 | 0.71 (0.56–0.89) | 0.0028 | 0.21 | 0.8 (0.68–0.94) | 0.0074 | 0.98 | 0.21 | 0.77 (0.67–0.88) | 9.04E-05 | 0.039 |
| rs74678433 | GRM8 | Intron | 7 | 126173950 | G | 0.026 | 2.73 (1.54–4.84) | 0.0006 | 0.028 | 1.92 (1.27–2.91) | 0.0019 | 0.95 | 0.027 | 2.17 (1.55–3.03) | 5.96E-06 | 0.014 |
| rs268672 | PRX | Synonymous | 19 | 40901604 | A | 0.19 | 0.73 (0.58–0.93) | 0.0096 | 0.2 | 0.77 (0.66–0.91) | 0.002 | 1.0 | 0.19 | 0.76 (0.66–0.87) | 5.87E-05 | 0.031 |
| rs268671 | PRX | Missense | 19 | 40901614 | A | 0.19 | 0.73 (0.58–0.93) | 0.0096 | 0.2 | 0.76 (0.64–0.89) | 0.00072 | 1.0 | 0.19 | 0.75 (0.65–0.86) | 2.11E-05 | 0.018 |
| rs73885319 | APOL1 | Missense | 22 | 36661906 | G | 0.23 | 1.39 (1.11–1.73) | 0.0036 | 0.24 | 1.29 (1.11–1.50) | 0.00076 | 1.0 | 0.24 | 1.32 (1.17–1.50) | 9.90E-06 | 0.014 |
| rs60910145 | APOL1 | Missense | 22 | 36662034 | G | 0.23 | 1.42 (1.14–1.77) | 0.0017 | 0.24 | 1.29 (1.11–1.50) | 0.00081 | 1.0 | 0.24 | 1.33 (1.18–1.51) | 5.74E-06 | 0.014 |
CHR, chromosome; CI, confidence interval; EA, effect allele; EAF, effect allele frequency; ESKD, end-stage kidney disease; Info, information metric reported by IMPUTE2; OR, odds ratio; POS, position; T2D, type 2 diabetes.
Baseline model: adjusted for age, sex. Annotation, derived from Variant Effect Predictor (VEP). APOL1 renal-risk genotype carriers included.
Association Analysis Excluding APOL1 Renal-Risk Genotype Carriers
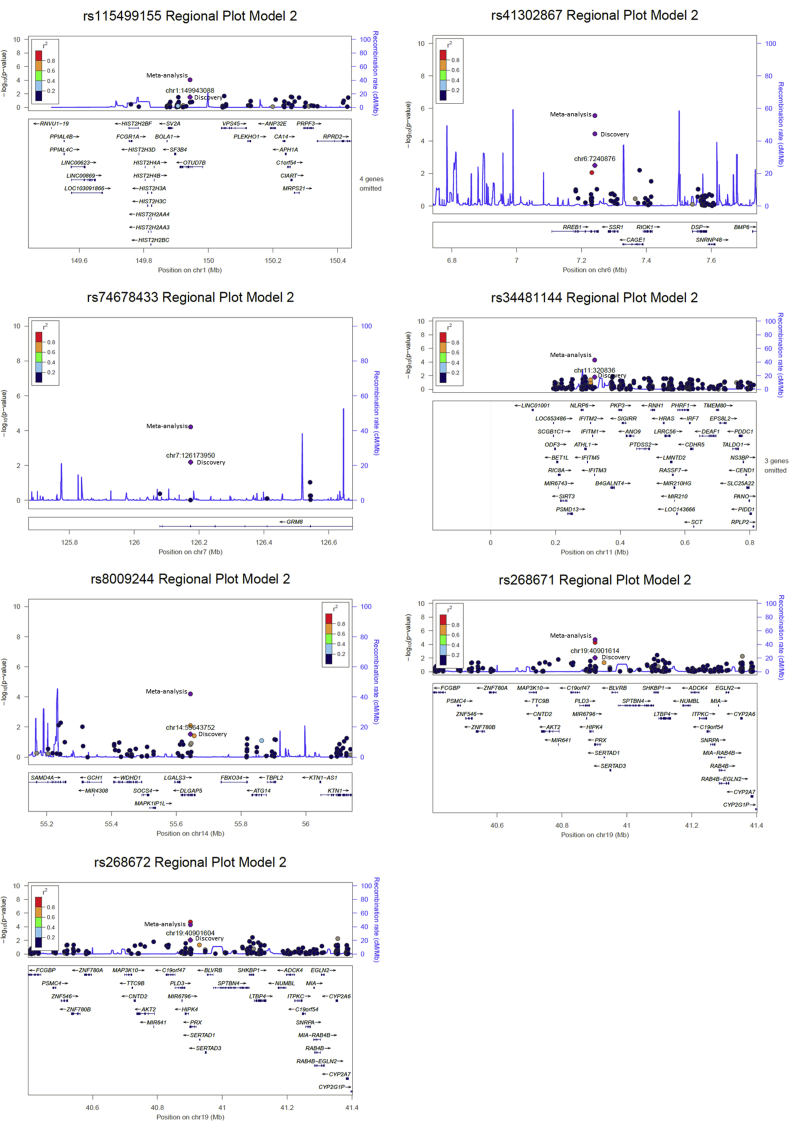
The primary analysis showed moderate association of APOL1 G1 alleles with T2D-ESKD, suggesting misclassification of some cases. A secondary analysis excluding APOL1 G1 and G2 risk genotype carriers (APOL1-negative model) from cases was performed. In the stage 1 discovery analysis, 358 T2D-ESKD cases were compared with 936 nondiabetic non-nephropathy controls. A total of 6896 variants showing nominal association (P < 0.05) were selected for replication in 1636 T2D-ESKD cases and 1121 nondiabetic non-nephropathy controls in a stage 2 analysis. Meta-analysis of samples from stages 1 and 2 revealed 10 suggestive signals with consistent association with T2D-ESKD at P < 1 × 10−4; no variant reached exome-wide significance (P = 3.5 × 10−7). Three variants were removed in a discrimination analysis due to their nominal association (P < 0.05) with T2D per se (Table 3, Supplementary Figure S3). Among the 7 top T2D-ESKD associated variants, 4 variants including rs41302867 at RREB1 (OR: 0.44; 95% CI: 0.30–0.65; P = 3.60 × 10−5), rs74678433 at GRM8 (OR: 2.11; 95% CI: 1.47–3.05; P = 6.24 × 10−5), and rs268672 and rs268671 at PRX (OR: 0.75; 95% CI: 0.65–0.86; P = 1.87 × 10−5; OR: 0.75; 95% CI: 0.65–0.86; P = 5.21 × 10−5) were identified in the baseline model. It is of note that the other 5 top associations (excluding 2 APOL1 G1 alleles) from the baseline model had moderate attenuation in significance, despite the similar effect size, due in part to the reduced sample size (Supplementary Table S4). In contrast, 3 additional associations were identified at OTUD7B (rs115499155; OR: 0.43; 95% CI: 0.28–0.65; P = 8.58 × 10–5), IFITM3 (rs34481144; OR: 0.67; 95% CI: 0.55–0.81; P = 5.25 × 10−5), and DLGAP5 (rs8009244; OR: 0.75; 95% CI: 0.65–0.86; P = 6.42 × 10−5). Variants in IFITM3 and DLGAP5 demonstrated association with transcript abundance of nearby genes based on GTEx (Supplementary Table S3). Excluding APOL1 risk genotype carriers from both cases and controls showed similar effect sizes despite slightly attenuated significance likely due to reduced sample size (Supplementary Table S5).
Table 3.
Top T2D-ESKD associations in meta-analysis after removing APOL1 renal-risk genotype carriers (APOL1-negative model)
| Variant | Gene | Annotation | CHR | POS | EA | Discovery (358 T2D-ESKD cases vs. 936 nondiabetic non-nephropathy controls) |
Replication (1636 T2D-ESKD cases vs. 1121 nondiabetic non-nephropathy controls) |
Meta-analysis (1994 T2D-ESKD cases vs. 2057 nondiabetic non-nephropathy controls) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EAF | OR (95% CI) | P | EAF | OR (95% CI) | P | Info | EAF | OR (95% CI) | P | Q-value | ||||||
| rs115499155 | OTUD7B | Synonymous | 1 | 149943088 | T | 0.02 | 0.44 (0.21–0.92) | 0.03 | 0.021 | 0.42 (0.25–0.71) | 0.00106 | 0.82 | 0.021 | 0.43 (0.28–0.65) | 8.58E-05 | 0.049 |
| rs41302867 | RREB1 | Intron | 6 | 7240876 | A | 0.025 | 0.36 (0.18–0.71) | 0.0032 | 0.021 | 0.49 (0.31–0.78) | 0.00282 | 1.0 | 0.022 | 0.44 (0.3–0.65) | 3.60E-05 | 0.040 |
| rs74678433 | GRM8 | Intron | 7 | 126173950 | G | 0.024 | 2.47 (1.29–4.73) | 0.0066 | 0.027 | 1.97 (1.26–3.07) | 0.00277 | 0.95 | 0.026 | 2.11 (1.47–3.05) | 6.24E-05 | 0.040 |
| rs34481144 | IFITM3 | 5’ UTR | 11 | 320836 | T | 0.1 | 0.66 (0.47–0.93) | 0.017 | 0.11 | 0.68 (0.54–0.86) | 0.00111 | 0.90 | 0.11 | 0.67 (0.55–0.81) | 5.25E-05 | 0.040 |
| rs8009244 | DLGAP5 | Intron | 14 | 55643752 | A | 0.23 | 0.75 (0.58–0.97) | 0.03 | 0.21 | 0.75 (0.63–0.89) | 0.0008 | 1.0 | 0.21 | 0.75 (0.65–0.86) | 6.42E-05 | 0.040 |
| rs268672 | PRX | Synonymous | 19 | 40901604 | A | 0.19 | 0.71 (0.55–0.92) | 0.0089 | 0.2 | 0.76 (0.64–0.9) | 0.00184 | 1.0 | 0.19 | 0.75 (0.65–0.86) | 5.21E-05 | 0.040 |
| rs268671 | PRX | Missense | 19 | 40901614 | A | 0.19 | 0.71 (0.55–0.92) | 0.0089 | 0.2 | 0.74 (0.63–0.88) | 0.00068 | 1.0 | 0.20 | 0.74 (0.64–0.85) | 1.87E-05 | 0.040 |
CHR, chromosome; CI, confidence interval; EA, effect allele; EAF, effect allele frequency; ESKD, end-stage kidney disease; Info, information metric reported by IMPUTE2; OR, odds ratio; POS, position; T2D, type 2 diabetes.
APOL1-negative model: adjusted for age, sex and PC1, APOL1 renal-risk genotype carriers removed. Annotation, derived from Variant Effect Predictor (VEP).
Replication Analysis in All-Cause ESKD
In the stage 5 analysis, an all-cause ESKD cohort consisting of 1910 nondiabetic ESKD cases, 219 T2D-ESKD cases, and 912 nondiabetic non-nephropathy controls was used to replicate the 14 T2D-ESKD associated signals identified from the baseline and the APOL1-negative models. This analysis evaluated the generalizability of T2D-ESKD loci in nondiabetic etiologies of ESKD. Initially, the 14 suggestive T2D-ESKD associations were tested in 2129 all-cause ESKD cases and 912 nondiabetic non-nephropathy controls. Only 1 variant, located in RREB1 (rs41302867; OR: 0.52; 95% CI: 0.31–0.88; P = 0.015), achieved P < 0.05. Similar results were obtained after excluding 219 T2D-ESKD cases from the association analysis; only rs41302867 reached P < 0.05. Consistent results were observed after excluding APOL1 renal-risk genotype carriers. Meta-analysis of rs41302867 in 4605 all-cause ESKD cases and 2969 nondiabetic non-nephropathy controls from stages 1, 2, and 5 analyses revealed strong evidence of association (OR: 0.47; 95% CI: 0.35–0.64; P = 1.2 × 10−6; Table 4), but did not reach exome-wide significance.
Table 4.
Meta-analysis combining T2D-ESKD and all-cause ESKD cohorts for rs41302867
| rs41302867 (A) RREB1 |
|||||
|---|---|---|---|---|---|
|
n Case/control |
EAF | OR (95% CI) | P | Info | |
| Discovery (T2D-ESKD cases versus nondiabetic non-nephropathy controls) | |||||
| 456/936 | 0.023 | 0.35 (0.18–0.67) | 0.0016 | — | |
| Replication (T2D-ESKD cases versus nondiabetic non-nephropathy controls) | |||||
| 2020/1121 | 0.020 | 0.51 (0.33–0.80) | 0.0030 | 1.0 | |
| All-cause ESKD cohort (nondiabetic ESKD and T2D-ESKD case versus nondiabetic non-nephropathy controls) | |||||
| 2129/912 | 0.020 | 0.52 (0.31–0.88) | 0.015 | 0.97 | |
| Meta-analysis of discovery and replication and all-cause ESKD cohort | |||||
| 4605/2969 | 0.021 | 0.47 (0.35–0.64) | 1.20E-06 | — | |
CI, confidence interval; EAF, effect allele frequency; ESKD, end-stage kidney disease; Info, information metric reported by IMPUTE2; OR, odds ratio; T2D, type 2 diabetes.
Gene-based Analysis
To increase power to detect association with low-frequency and rare variants, gene-based analyses were performed to aggregate effects of functional variants within each gene using only stage 1 exome sequencing data. Functional variants were categorized into 4 groups based on the level of deleterious impact. Both baseline and APOL1 risk genotype-negative models were examined, followed by a discrimination analysis comparing T2D-lacking nephropathy with nondiabetic non-nephropathy controls. Although no gene reached exome-wide significance (P < 2.5 × 10−6, adjusted for 20,000 genes), 8 revealed suggestive association with T2D-ESKD at P < 1 × 10−4 (Table 5) and no association was observed for T2D (P ≥ 0.05). TMEM5 showed the strongest association (P = 4.08 × 10−6 in the APOL1-negative model, P = 2.7 × 10−5 in the baseline model). Six additional genes, including SPATS2, ZIC4, HELZ3, ILDR2, LGALS3BP, and RSAD2, were nominally associated with T2D-ESKD (P < 1 × 10−4). We evaluated the T2D-ESKD associated loci from single-variant analyses in gene-based association results, but no strong gene-based associations were observed. The top overlapping association was from PEX6 (P = 6.3 × 10−3 in gene-based analysis) (Supplementary Table S6).
Table 5.
Top associations of gene-based analyses in baseline or APOL1-negative models
| Gene | Method | Model | Variant group | T2D-ESKD versus nondiabetic, non-nephropathy controls (n) |
T2D-lacking nephropathy versus controls (n) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
|
n case/control |
n variants | Average frequency | P |
n case/control |
n variants | P | ||||
| TMEM5 | SKAT | Baseline Model | Single predicted | 456/936 | 9 | 0.0056 | 2.70E-05 | 338/936 | 10 | 0.25 |
| TMEM5 | SKAT | APOL1-negative | Single predicted | 358/936 | 8 | 0.0062 | 4.08E-06 | 338/936 | 10 | 0.25 |
| SPATS2 | SKAT | APOL1-negative | Single predicted | 358/936 | 11 | 0.0019 | 9.22E-05 | 338/936 | 11 | 0.93 |
| ZIC4 | VT | APOL1-negative | Single predicted | 358/936 | 6 | 0.00039 | 4.68E-05 | 338/936 | 2 | 0.13 |
| HELZ2 | MB | APOL1-negative | H&M | 358/936 | 69 | 0.0035 | 8.12E-05 | 338/936 | 73 | 0.78 |
| ILDR2 | MB | APOL1-negative | H&M | 358/936 | 11 | 0.00070 | 9.31E-05 | 338/936 | 6 | 0.16 |
| LGALS3BP | MB | APOL1-negative | H&M | 358/936 | 19 | 0.0016 | 3.81E-05 | 338/936 | 3 | 0.93 |
| RSAD2 | MB | APOL1-negativel | H&M | 358/936 | 9 | 0.0074 | 8.31E-05 | 338/936 | 3 | 0.56 |
ESKD, end-stage kidney disease; MB, Madsen-Browning test; SKAT, Sequence Kernel Association Test; T2D, type 2 diabetes; VT, variable threshold test.
Single predicted variants predicted to be deleterious by at least 1 of the 4 prediction methods, including SIFT (sorting intolerant from tolerant), LRT (likelihood ratio test), MutationTaster, and CADD (combined annotation–dependent depletion); H&M, moderate to high-impact protein structure altering variants (transcript ablation, splice acceptor, splice donor, stop gained, frameshift, stop lost, start lost, transcript amplification, inframe insertion, inframe deletion, missense, and protein altering).
Discussion
This study presents results of an exome sequencing–based genetic association study evaluating the contribution of coding variants to risk of T2D-ESKD in AAs. We tested the top variants for replication in additional T2D-ESKD and control cohorts, and evaluated the generalizability of T2D-ESKD associations to common forms of nondiabetic ESKD. Evidence of nominal association with T2D-ESKD was observed for 11 loci, including PLEKHN1, NADK, RAD51AP2, RREB1, PEX6, APOL1, GRM8, PRX, OTUD7B, IFITM3, and DLGAP5 in single-variant analysis of either baseline or APOL1-negative models. Two associated variants from RAD51AP2 and PRX were missense mutations. Meta-analysis of all-cause ESKD revealed strong association at RREB1, previously identified as associated with ESKD.15 These results suggest it plays a role in AA diabetes- and nondiabetes-associated kidney disease. Gene-based analyses identified additional suggestive T2D-ESKD loci with multiple variants having cumulative effects: TMEM5, SPATS2, ZIC4, HELZ2, ILDR2, LGALS3BP, and RSAD2. However, there were minimal overlapping associations between single-variant and gene-based tests.
The only locus that revealed association with both T2D-ESKD and nondiabetic ESKD was an intronic variant at RREB1, a large complex gene encoding the ras responsive transcription factor. RREB1 has repeatedly been implicated in fasting glucose, T2D susceptibility, fat distribution, and adipocyte development in large-scale genetic studies.40, 41, 42, 43, 44 Relevant to kidney disease susceptibility, RREB1 variants had been reported to be associated with kidney function and interact with APOL1 renal-risk-alleles in nondiabetic nephropathy.45, 46 More recently, the same variant, rs41302867, was shown to be associated with all-cause ESKD in AAs and European Americans.15 Despite partially overlapping participants, the present results suggested a role for RREB1 in common forms of ESKD in AAs with a substantially expanded sample. In addition, it is noteworthy that the ESKD associated variant rs41302867 had no association with T2D alone in this study (P = 0.096); this suggests a pleiotropic effect of RREB1 in multiple traits including T2D and kidney disease.
Among the putative T2D-ESKD associated loci, an intronic variant at GRM8 (rs74678433) showed strong and consistent associations in baseline and APOL1-negative models (P < 5.96 × 10−6 and P < 6.24 × 10−5, respectively). Metabotropic glutamate receptor 8, the protein product encoded by GRM8, is associated with weight gain in mouse models.47, 48 Previous studies suggest that obesity is a major risk factor for kidney disease in patients with diabetes.49 This result may suggest potential genetic correlation between obesity and T2D-ESKD.
Two missense variants showing putative association with T2D-ESKD were identified, rs834514 and rs268671 located at RAD51AP2 and PRX, respectively. RAD51 Associated Protein 2, encoded by RAD51AP2, is a recombinase that plays a critical role in both DNA repair and meiotic recombination. Rs834514 may regulate the expression of RAD51AP2, GEN1, and VSNL1 in multiple tissues, including pancreas and adipose. The missense variant (rs268671) located in the periaxin gene (PRX), is a key myelination molecule forming tight junctions between myelin loops and axons. Mutations in PRX caused late-onset Charcot-Marie-Tooth neuropathy, a common inherited neurological disorder.50, 51 Charcot-Marie-Tooth neuropathy has been reported to be associated with kidney disease, mainly focal segmental glomerulosclerosis.52, 53, 54 The underlying mechanism linking these entities remains unknown.
A splice region variant (rs9986447) located in PEX6 was nominally associated with T2D-ESKD in the baseline model. Rs9986447 showed significant impact on the expression level of PEX6 in a number of tissues, including adipose and pancreas, indicating its regulatory role in this gene. This gene plays a direct role in the biosynthesis of peroxisome, a subcellular organelle involved in lipid metabolism. PEX6 may associate with DKD susceptibility, as dyslipidemia is thought to be a contributor.55
Analyses excluding APOL1 renal-risk genotype carriers provided an opportunity to uncover the genetic architecture in T2D-ESKD. In the APOL1-negative model, 4 additional variants revealing suggestive association with T2D-ESKD were identified in OTUD7B, IFITM3, and DLGAP5. OTU deubiquitinase 7B, encoded by OTUD7B, may be involved in the inflammatory response of DKD progression via regulating the nuclear factor kappa-B signaling pathway.56 However, IFITM3 (interferon induced transmembrane protein 3) and DLGAP5 (DLG associated protein 5) do not have literature to support their role in T2D-ESKD. Further studies are required to evaluate potential biological roles.
Gene-based methods provide an opportunity to evaluate the cumulative impact of low-frequency variants (MAF < 0.05) within a region on the disease of interest. Four types of gene-based analysis methods were used to test 4 categories of variants grouped by their functional impact. The top signal was found at a transmembrane protein gene TMEM5 showing consistent association in both the baseline and APOL1-negative models. For ILDR2 (Ig-like domain containing receptor 2), Ildr2 has been implicated in T2D susceptibility and hepatic lipid metabolism in mouse models.57, 58 Potential involvement of ILDR2 in T2D-ESKD disease remains to be assessed.
This study suggests that low-frequency and rare variants located at coding regions provide crucial information on understanding the genetic architecture of complex diseases such as DKD. We identified suggestive T2D-ESKD signals in 19 loci using multiple single-variant and gene-based analyses; however, additional replication is required to confirm findings given limited power in the current study. There was lack of enrichment between the single-variant analyses and gene-based tests. This may be partially due to the deleterious variants included in gene-based tests, which tend to have low frequencies and may be excluded (or underpowered) in single-variant analyses. On the other hand, gene-based associations were performed only in exome sequencing data; thus, with limited power.
The pathophysiology and pathology of DKD is heterogeneous, with effects of glycemia, blood pressure, albuminuria, diabetes duration, serum uric acid, dyslipidemia, obesity, and smoking.49, 59, 60 Thus, T2D-associated DKD may share a common genetic background with other phenotypes. It also has been proposed that genetic variation in Mendelian disease genes may in part account for common disease susceptibility.61, 62 Identification of GRM8, PEX6, ILDR2, RREB1, and PRX may indicate potential genetic correlations between T2D-ESKD with obesity, dyslipidemia, T2D, and Mendelian Charcot-Marie-Tooth neuropathy.
This study has similar limitations as in other reports. First, it is difficult to exclude all individuals misclassified as DKD due to the frequent lack of kidney biopsies. However, careful removal of patients with ESKD attributed to nondiabetic etiologies, and subsequently excluding APOL1 risk genotype carriers with high risk for nondiabetic kidney disease, should minimize misclassification. In addition, although the multistage study design brings together 8577 AA individuals, study power was moderate (Supplementary Tables S7 and S8). This is especially true in the discovery exome sequencing cohort, which may result in additional variants of modest effect and low frequency that were not captured. There are few other existing collections of appropriate AA samples; this limited possible replication studies. Finally, we used imputed data to replicate exome sequencing findings in stage 2 and to evaluate their association with nondiabetic ESKD in stage 5. Although we used info < 0.4 to filter out low-quality alleles, there might be some variants with low imputation quality included. However, the highlighted variants all showed high imputation quality (0.82 to 1).
In conclusion, an exome sequencing–based, multiphase study to identify T2D-ESKD susceptibility loci was performed in AAs, and 18 suggestive associations with T2D-ESKD were detected. RREB1 was consistently associated with diabetic and nondiabetic etiologies of ESKD in AAs. T2D-ESKD associated variants at GRM8, PEX6, ILDR2, RREB1, and PRX may support genetic correlation between T2D-ESKD and related phenotypes. Future efforts to confirm the newly identified associations and determine their potential impact on the biological processes related to DKD requires investment in additional sample recruitment and comprehensive functional evaluation.
Disclosure
All the authors declared no competing interests.
Acknowledgments
This work was supported by National Institutes of Health grants R01 DK53591 (DWB), R01 DK066358 (DWB), R01 DK087914 (MCYN), U01 DK105556 (MCYN), DK070941 (BIF), and DK084149 (BIF). The Jackson Heart Study is supported by contracts HHSN268201300046C, HHSN268201300047C, HHSN268201300048C, HHSN268201300049C, and HHSN268201300050C from the National Heart, Lung, and Blood Institute and the National Institute on Minority Health and Health Disparities. Dr. Wilson is supported by U54GM115428 from the National Institute of General Medical Sciences. We acknowledge the contributions of the study participants, coordinators, physicians, staff, and laboratory from the Wake Forest School of Medicine and the Jackson Heart Study.
Footnotes
Figure S1. QQ plot of T2D-ESKD versus controls in model 1 discovery stage (MAF ≥ 0.01).
Figure S2. Regional plot of top T2D-ESKD associations in baseline model.
Figure S3. Regional plot of top T2D-ESKD associations in APOL1-negative model.
Table S1. Breakdown of ESKD cases for each stage.
Table S2. Discrimination analysis for top associations of baseline model and APOL1-negative model.
Table S3. GTEx results of top associations (P < 1 × 10−4).
Table S4. Results of top associations from baseline model in APOL1-negative model.
Table S5. Top T2D-ESKD associations in meta-analysis after removing APOL1 renal-risk genotype carriers in both cases and controls.
Table S6. Gene-based analysis results for T2D-ESKD associated loci in single-variant association analysis.
Table S7. Power analysis of single-variant association analysis at significance level of α = 3.5 × 10−7.
Table S8. Power analysis of single-variant association analysis at significance level of α = 1.0 × 10−4.
Supplementary material is linked to the online version of the paper at http://www.kireports.org/.
Supplementary Material
Figure S1.
QQ plot of T2D-ESKD versus controls in model 1 discovery stage (MAF ≥ 0.01).
Figure S2.
Regional plot of top T2D-ESKD associations in baseline model.
Figure S3.
Regional plot of top T2D-ESKD associations in APOL1-negative model.
Breakdown of ESKD cases for each stage.
Discrimination analysis for top associations of baseline model and APOL1-negative model.
GTEx results of top associations (P < 1 × 10−4).
Results of top associations from baseline model in APOL1-negative model.
Top T2D-ESKD associations in meta-analysis after removing APOL1 renal-risk genotype carriers in both cases and controls.
Gene-based analysis results for T2D-ESKD associated loci in single-variant association analysis.
Power analysis of single-variant association analysis at significance level of α = 3.5 × 10−7.
Power analysis of single-variant association analysis at significance level of α = 1.0 × 10−4.
References
- 1.United States Renal Data System . National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda, MD: 2016. 2016 USRDS Annual Data Report: Epidemiology of Kidney Disease in the United States. [Google Scholar]
- 2.Spray B.J., Atassi N.G., Tuttle A.B., Freedman B.I. Familial risk, age at onset, and cause of end-stage renal disease in white Americans. J Am Soc Nephrol. 1995;5:1806–1810. doi: 10.1681/ASN.V5101806. [DOI] [PubMed] [Google Scholar]
- 3.Freedman B.I., Tuttle A.B., Spray B.J. Familial predisposition to nephropathy in African-Americans with non-insulin-dependent diabetes mellitus. Am J Kidney Dis. 1995;25:710–713. doi: 10.1016/0272-6386(95)90546-4. [DOI] [PubMed] [Google Scholar]
- 4.Köttgen A. Genome-wide association studies in nephrology research. Am J Kidney Dis. 2010;56:743–758. doi: 10.1053/j.ajkd.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 5.Friedman D.J., Pollak M.R. Genetics of kidney failure and the evolving story of APOL1. J Clin Invest. 2011;121:3367–3374. doi: 10.1172/JCI46263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeda S. Genome-wide search for susceptibility gene to diabetic nephropathy by gene-based SNP. Diabetes Res Clin Pract. 2004;66:S45–S47. doi: 10.1016/j.diabres.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 7.Pezzolesi M.G., Poznik G.D., Mychaleckyj J.C. Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes. 2009;58:1403–1410. doi: 10.2337/db08-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonough C.W., Palmer N.D., Hicks P.J. A genome wide association study for diabetic nephropathy genes in African Americans. Kidney Int. 2011;79:563–572. doi: 10.1038/ki.2010.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandholm N., Salem R.M., McKnight A.J. New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet. 2012;8:e1002921. doi: 10.1371/journal.pgen.1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandholm N., Zuydam N.V., Ahlqvist E. The genetic landscape of renal complications in type 1 diabetes. J Am Soc Nephrol. 2017;28:557–574. doi: 10.1681/ASN.2016020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tzur S., Rosset S., Shemer R. Missense mutations in the APOL1. Hum Genet. 2010;128:345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genovese G., Friedman D.J., Ross M.D. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopp J.B., Nelson G.W., Sampath K. APOL1 Genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22:2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyengar S.K., Sedor J.R., Freedman B.I. Genome-wide association and trans-ethnic meta-analysis for advanced diabetic kidney disease: Family Investigation of Nephropathy and Diabetes (FIND) PLoS Genet. 2015;11:e1005352. doi: 10.1371/journal.pgen.1005352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonomo J.A., Guan M., Ng M.C.Y. The ras responsive transcription factor RREB1 is a novel candidate gene for type 2 diabetes associated end-stage kidney disease. Hum Mol Genet. 2014;23:6441–6447. doi: 10.1093/hmg/ddu362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonomo J.A., Ng M.C.Y., Palmer N.D. Coding variants in nephrin (NPHS1) and susceptibility to nephropathy in African Americans. Clin J Am Soc Nephrol. 2014;9:1434–1440. doi: 10.2215/CJN.00290114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma J., Guan M., Bowden D.W. Association analysis of the cubilin (CUBN) and megalin (LRP2) genes with ESRD in African Americans. Clin J Am Soc Nephrol. 2016;11:1034–1043. doi: 10.2215/CJN.12971215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan M., Ma J., Keaton J.M. Association of kidney structure-related gene variants with type 2 diabetes-attributed end-stage kidney disease in African Americans. Hum Genet. 2016;135:1251–1262. doi: 10.1007/s00439-016-1714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchsberger C., Flannick J., Teslovich T.M. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurdasani D., Carstensen T., Tekola-Ayele F. The African Genome Variation Project shapes medical genetics in Africa. Nature. 2015;517:327–332. doi: 10.1038/nature13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delaneau O., Marchini J., Zagury J.-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 23.Marchini J., Howie B., Myers S. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 24.Bien S.A., Wojcik G.L., Zubair N. Strategies for enriching variant coverage in candidate disease loci on a multiethnic genotyping array. PLoS One. 2016;11:e0167758. doi: 10.1371/journal.pone.0167758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H., Wang C., Conomos M.P. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am J Hum Genet. 2016;98:653–666. doi: 10.1016/j.ajhg.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price A.L., Patterson N.J., Plenge R.M. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 27.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D.J., Peloso G.M., Zhan X. Meta-analysis of gene-level tests for rare variant association. Nat Genet. 2014;46:200–204. doi: 10.1038/ng.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu M.C., Lee S., Cai T. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madsen B.E., Browning S.R. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price A.L., Kryukov G.V., de Bakker P.I.W. Pooled association tests for rare variants in exon-resequencing studies. Am J Hum Genet. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B., Leal S.M. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chun S., Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 36.Kircher M., Witten D.M., Jain P. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLaren W., Pritchard B., Rios D. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cingolani P., Platts A., Wang L.L. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X., Jian X., Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Below J.E., Gamazon E.R., Morrison J.V. Genome-wide association and meta-analysis in populations from Starr County, Texas, and Mexico City identify type 2 diabetes susceptibility loci and enrichment for expression quantitative trait loci in top signals. Diabetologia. 2011;54:2047–2055. doi: 10.1007/s00125-011-2188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu C.-T., Monda K.L., Taylor K.C. Genome-wide association of body fat distribution in African ancestry populations suggests new loci. PLOS Genet. 2013;9:e1003681. doi: 10.1371/journal.pgen.1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahajan A., Sim X., Ng H.J. Identification and functional characterization of G6PC2 coding variants influencing glycemic traits define an effector transcript at the G6PC2-ABCB11 locus. PLoS Genet. 2015;11:e1004876. doi: 10.1371/journal.pgen.1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chu A.Y., Deng X., Fisher V.A. Multiethnic genome-wide meta-analysis of ectopic fat depots identifies loci associated with adipocyte development and differentiation. Nat Genet. 2017;49:125–130. doi: 10.1038/ng.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Q., Köttgen A., Dehghan A. Multiple genetic loci influence serum urate levels and their relationship with gout and cardiovascular disease risk factors. Circ Cardiovasc Genet. 2010;3:523–530. doi: 10.1161/CIRCGENETICS.109.934455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bostrom M.A., Kao W.H.L., Li M. Genetic association and gene-gene interaction analyses in African American dialysis patients with nondiabetic nephropathy. Am J Kidney Dis. 2012;59:210–221. doi: 10.1053/j.ajkd.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duvoisin R.M., Zhang C., Pfankuch T.F. Increased measures of anxiety and weight gain in mice lacking the group III metabotropic glutamate receptor mGluR8. Eur J Neurosci. 2005;22:425–436. doi: 10.1111/j.1460-9568.2005.04210.x. [DOI] [PubMed] [Google Scholar]
- 48.Davis M.J., Duvoisin R.M., Raber J. Related functions of mGlu4 and mGlu8. Pharmacol Biochem Behav. 2013;111:11–16. doi: 10.1016/j.pbb.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zoppini G., Targher G., Chonchol M. Predictors of estimated GFR decline in patients with type 2 diabetes and preserved kidney function. Clin J Am Soc Nephrol. 2012;7:401–408. doi: 10.2215/CJN.07650711. [DOI] [PubMed] [Google Scholar]
- 50.Tokunaga S., Hashiguchi A., Yoshimura A. Late-onset Charcot–Marie–Tooth disease 4F caused by periaxin gene mutation. Neurogenetics. 2012;13:359–365. doi: 10.1007/s10048-012-0338-5. [DOI] [PubMed] [Google Scholar]
- 51.Renouil M., Stojkovic T., Jacquemont M.L. [Charcot-Marie-Tooth disease associated with periaxin mutations (CMT4F): clinical, electrophysiological and genetic analysis of 24 patients] Rev Neurol (Paris) 2013;169:603–612. doi: 10.1016/j.neurol.2013.07.004. [in French] [DOI] [PubMed] [Google Scholar]
- 52.Nadal M.A., Lago N.R., Olivieri L.E. Fibrillary glomerulonephritis and Charcot-Marie-Tooth disease. Am J Kidney Dis. 1998;32:E3. doi: 10.1016/s0272-6386(98)70150-7. [DOI] [PubMed] [Google Scholar]
- 53.Boyer O., Nevo F., Plaisier E. INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. N Engl J Med. 2011;365:2377–2388. doi: 10.1056/NEJMoa1109122. [DOI] [PubMed] [Google Scholar]
- 54.De Rechter S., De Waele L., Levtchenko E., Mekahli D. Charcot-Marie-Tooth: Are you testing for proteinuria? Eur J Paediatr Neurol. 2015;19:1–5. doi: 10.1016/j.ejpn.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 55.Stadler K., Goldberg I.J., Susztak K. The evolving understanding of the contribution of lipid metabolism to diabetic kidney disease. Curr Diab Rep. 2015;15:40. doi: 10.1007/s11892-015-0611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao C., Huang W., Kanasaki K., Xu Y. The role of ubiquitination and sumoylation in diabetic nephropathy. BioMed Res Int. 2014;2014:e160692. doi: 10.1155/2014/160692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cooper J.N., Wei L., Fernandez S.A. Pre-operative prediction of surgical morbidity in children: comparison of five statistical models. Comput Biol Med. 2015;57:54–65. doi: 10.1016/j.compbiomed.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe K., Nakayama K., Ohta S. ZNF70, a novel ILDR2-interacting protein, contributes to the regulation of HES1 gene expression. Biochem Biophys Res Commun. 2016;477:712–716. doi: 10.1016/j.bbrc.2016.06.124. [DOI] [PubMed] [Google Scholar]
- 59.Macisaac R.J., Ekinci E.I., Jerums G. Markers of and risk factors for the development and progression of diabetic kidney disease. Am J Kidney Dis. 2014;63(2 Suppl 2):S39–S62. doi: 10.1053/j.ajkd.2013.10.048. [DOI] [PubMed] [Google Scholar]
- 60.Radcliffe N.J., Seah J.-M., Clarke M. Clinical predictive factors in diabetic kidney disease progression. J Diabetes Investig. 2017;8:6–18. doi: 10.1111/jdi.12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blair D.R., Lyttle C.S., Mortensen J.M. A nondegenerate code of deleterious variants in mendelian loci contributes to complex disease risk. Cell. 2013;155:70–80. doi: 10.1016/j.cell.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parsa A., Fuchsberger C., Köttgen A. Common variants in mendelian kidney disease genes and their association with renal function. J Am Soc Nephrol. 2013;24:2105–2117. doi: 10.1681/ASN.2012100983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Breakdown of ESKD cases for each stage.
Discrimination analysis for top associations of baseline model and APOL1-negative model.
GTEx results of top associations (P < 1 × 10−4).
Results of top associations from baseline model in APOL1-negative model.
Top T2D-ESKD associations in meta-analysis after removing APOL1 renal-risk genotype carriers in both cases and controls.
Gene-based analysis results for T2D-ESKD associated loci in single-variant association analysis.
Power analysis of single-variant association analysis at significance level of α = 3.5 × 10−7.
Power analysis of single-variant association analysis at significance level of α = 1.0 × 10−4.