Abstract
MYF5 is member of the Myc-like basic helix-loop-helix transcription factor family and, in cooperation with other myogenic regulatory factors MYOD and MYF5, is a key regulator of early stages of myogenesis. Here, we report three consanguineous families with biallelic homozygous loss-of-function mutations in MYF5 who define a clinical disorder characterized by congenital ophthalmoplegia with scoliosis and vertebral and rib anomalies. The clinical phenotype overlaps strikingly with that reported in several Myf5 knockout mouse models. Affected members of two families share a haploidentical region that contains a homozygous 10 bp frameshift mutation in exon 1 of MYF5 (c.23_32delAGTTCTCACC [p.Gln8Leufs∗86]) predicted to undergo nonsense-mediated decay. Affected members of the third family harbor a homozygous missense change in exon 1 of MYF5 (c.283C>T [p.Arg95Cys]). Using in vitro assays, we show that this missense mutation acts as a loss-of-function allele by impairing MYF5 DNA binding and nuclear localization. We performed whole-genome sequencing in one affected individual with the frameshift mutation and did not identify additional rare variants in the haploidentical region that might account for differences in severity among the families. These data support the direct role of MYF5 in rib, spine, and extraocular muscle formation in humans.
Keywords: MYF5, ophthalmaplegia, scoliosis, vertebral anomalies, rib anomalies, extraocular muscle, human genetics, exome sequencing, genome sequencing, myogenesis
Main Text
Myogenesis is determined by MYOD, MYF5, and MYF6 basic helix-loop-helix (bHLH) transcription factors (TFs).1, 2 Mouse knockout studies have revealed that any one of these factors can compensate for the other two during skeletal muscle development, and muscles of the body, neck, jaw, and face develop as long as any one of the three factors is expressed. Likely because of this redundancy, mutations in these genes have not been convincingly demonstrated to cause human disease. By contrast, extraocular muscles (EOMs) are composed of striated fibers distinct from skeletal muscle3 and lack a Myf5/Myf6-independent pathway to activate myogenesis through Myod. Thus, EOMs fail to form in mice lacking expression of both Myf5 and Myf6 and are hypoplastic with loss of either factor alone.4 In addition, expression of Myf5 and Myf6 in the hypaxial myotome promotes rib and vertebral formation in the adjacent sclerotome though a non-cell-autonomous effect mediated by PDGF and FGF signaling,5, 6 and mice lacking expression of both Myf5 and Myf6 can also develop rib and vertebral anomalies and die perinatally.2, 5, 6, 7, 8
Errors in development of EOMs or their innervating motor neurons can result in congenital ophthalmoplegia.9, 10, 11 In an effort to identify genetic causes of congenital ophthalmoplegia, we enrolled affected probands and their family members into an ongoing genetic study approved by the institutional review board of Boston Children’s Hospital. Written informed consent was obtained from participating family members or their parents, and investigations were conducted in accordance with the principles of the Declaration of Helsinki. Clinical data were ascertained through examinations and review of medical records, and DNA was extracted from blood or saliva from all participating family members for genetic analysis. Here, we report that members of three families affected with congenital ophthalmoplegia and scoliosis, with documented vertebral and rib anomalies in two, harbor homozygous recessive loss-of-function mutations in MYF5 (MIM: 159990) and define a previously undescribed human disorder we refer to as the “MYF5 syndrome” (Table 1).
Table 1.
Clinical Features of MYF5 Syndrome
|
Pedigree |
ALO |
ALO |
CHO |
BX |
B |
|---|---|---|---|---|---|
| Individual ID | 1 | 2 | 3 | 4 | 5 |
| Sex | F | M | M | F | F |
| Geographic location | Turkey (ALO:II-1) | Turkey (ALO:II-2) | Turkey (CHO:II-2) | Yemen (BX:IV-4) | Yemen (BX:IV-6) |
| Homozygous MYF5 variant | c.23_32delAGTTCTCACC | c.23_32delAGTTCTCACC | c.23_32delAGTTCTCACC | c.283C>T | c.283C>T |
| MYF5 protein | p.Gln8Leufs∗86 | p.Gln8Leufs∗86 | p.Gln8Leufs∗86 | p.Arg95Cys | p.Arg95Cys |
| Phenotype | |||||
| Strabismus | XT++, HT R+ | XT++ | HT++ | XT+++, HT L+ | XT + |
| External ophthalmoplegia | yes | yes | yes | yes | yes |
| Forced ductions | ND | ND | ND | ND | positive |
| Ptosis | ++R; +L | ++R; +L | +++ | no | +L |
| Torticollis | yes | yes | yes | ? | possibly |
| Scoliosis | yes | yes | yes | yes | ? |
| Motor development | normal | normal | delayed | normal | normal |
| Hypotonia/weakness | no | no | no | no | no |
| Cognition | normal | normal | normal | normal | normal |
| Imaging | |||||
| Orbital MRI | ND | ND | EOMs hypoplastic to absent | ND | ND |
| Ribs | dysmorphic, hypoplastic, fusion anomalies, fused sternum | dysmorphic, hypoplastic, shortened, fusion anomalies | dysmorphic, hypoplastic, shortened and missing ribs, fusion anomalies, pseudarthrosis | ND | ND |
| Spine | cervical scoliosis and fusions, clivus malformations, basilar invagination, narrow disc spaces | ND | cervical and thoracic scoliosis, cervical fusions, clivus malformations, basilar invagination, narrow disc spaces | ND | lumbar scoliosis |
Abbreviations: F, female; M, male; years, years; +, mild; ++, moderate; +++, severe, ND, no data; R, right; L, left; XT, exotropia; HT, hypotropia; ?, unknown.
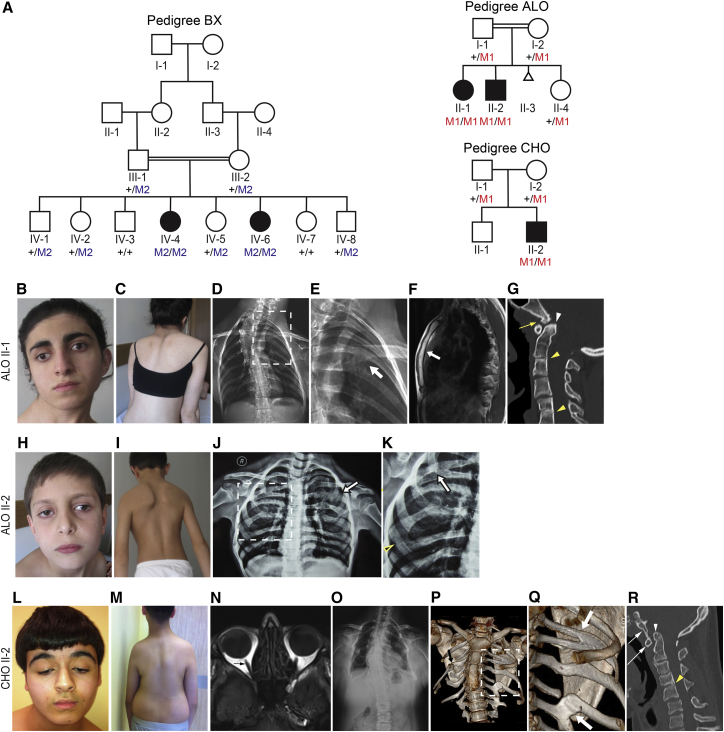
Pedigree BX is a previously published consanguineous Yemenite family12 with two affected (IV-4 and IV-6) and six unaffected siblings born to unaffected parents, all of whom participated in the research study (Figure 1A). The affected daughters were examined at 15 months (IV-6) and 4 years (IV-4) of age,12 and again as young adults. They had congenital bilateral nonprogressive external ophthalmoplegia with mild left-sided ptosis in IV-6 and absence of ptosis in IV-4. IV-4 developed scoliosis between age 4 and young adulthood. In photos, IV-6 was noted to have uneven shoulders suggestive of scoliosis and/or torticollis. Family history was noncontributory. On examination in primary position, IV-6 had a large bilateral exotropia and left hypotropia, while IV-4 had a small exotropia. Both had absence of elevation, marked limitation of adduction and depression, and moderate limitation of abduction. IV-6 underwent forced duction testing that revealed restriction to passive eye movement. Their cranial nerve exams were otherwise reported to be normal, as were their intellectual, social, and motor development. Further details of their general physical examinations were not available.
Figure 1.
MYF5-Affected Families and Phenotypes
(A) Pedigree schematics for pedigrees BX, ALO, and CHO as labeled. Double line denotes consanguinity. Filled symbol indicates affected. Pedigree position and mutation status is indicated under each enrolled participant. + denotes wild-type allele; M2 denotes p.Arg95Cys amino acid substitution; M1 denotes p.Gln8Leufs∗86 frameshift variant (refer to Figure 2).
(B–G) Phenotype of ALO II-1. Photographs show ptosis (R > L), exotropia, and torticollis (B) and scoliosis (C). Frontal radiograph (D) shows hypoplastic ribs, especially inferiorly, and left sided fusion anomalies highlighted by white arrow in magnification of boxed region (E). The rightward convex cervicothoracic spinal curvature is out of proportion to that expected exclusively due to the rib findings. Sagittal T1 weighted MR image (F) shows that the sternum is fused (white arrow), with absence of the normal cartilaginous articulations between manubrium, sternal body, and xiphoid process. Midline sagittal reformatted CT image (G) of the cervical spine demonstrates malformation of the clivus with a small bone projection that forms a pseudarthrosis with the anterior arch of C1 (yellow arrow), basilar invagination (white arrowhead), and fusion of C2-3 and C5-6 vertebral bodies (yellow arrowheads) and the C5-6 posterior elements. The posterior aspect of C3-4 disc space is narrowed with close approximation and possible fusion of C3-4 spinous processes.
(H–K) Phenotype of ALO II-2. Photographs show ptosis (R > L), exotropia, and torticollis (H) and scoliosis (I). Frontal radiograph (J) shows dysmorphic hypoplastic ribs with fusion anomalies (white arrow), several failing to extend anteriorly toward the sternum. Magnification (K) of boxed region in (J) highlighting fusion anomalies (white arrow) and hypoplasia of the rib with failure to extend anteriorly (yellow arrowhead).
(L–R) Phenotype of CHO II-2. (L) Photographs show bilateral ptosis, hypotropia, and torticollis (L) and (M) scoliosis. Axial T1 weighted MR image (N) shows absence of the extraocular muscles (EOMs), with only bilateral optic nerves seen (arrow). Frontal radiograph (O) shows that the ribs are few in number and hypoplastic with fusion anomalies on the left likely contributing to the rightward convex thoracic spinal curvature. CT 3D model (with soft tissues removed) (P) shows that the few ribs present are severely hypoplastic, with small posterior bones that do not extend anteriorly toward the sternum. Note the pseudarthrosis of the right 4th rib (black arrow). Magnification (Q) of boxed region in (P) highlights the left posterior rib fusion anomalies (white arrows). Midline sagittal reformatted CT image (R) of the cervical spine demonstrates fusion of the anterior arch of C1 (long white arrow) to the malformed clivus (short white arrow), basilar invagination (white arrowhead), and narrowing of the C2-3 and C3-4 disc spaces with a posterior fusion anomaly at C4-5 (yellow arrowhead). C2-3 also has early degenerative changes with anterior osteophyte formation.
Pedigree ALO is a consanguineous Turkish pedigree with two affected children (II-1 and II-2) and one unaffected healthy sibling (II-4) born to unaffected first-cousin parents, all of whom participated in the research study (Figure 1A). The parents also had one fetal loss of unknown gestational age and affection status (II-3). The affected children were born with nonprogressive ptosis and ophthalmoplegia and developed torticollis and scoliosis by 6 years of age. Both had normal intellectual, social, and motor development. Family history was noncontributory. On examination at ages 19 (II-1) and 9 (II-2) years, both had bilateral ptosis (R > L) and their primary eye positions were exotropic (Figures 1B and 1H). Residual eye movements were limited and no aberrant eye movements were noted. Cranial nerve testing was otherwise normal, and both had normal pupillary responses and full facial, jaw, neck, and skeletal muscle strength. Muscle tone, reflexes, coordination, sensation, and gait were also normal. No limb anomalies or dysmorphologies were noted.
Family CHO is a Turkish pedigree with one affected (II-2) and one unaffected (II-1) son born to unaffected parents who originated from the same village as each other and family ALO, but were not known to be related. Family history was noncontributory. The affected son was born after an uncomplicated pregnancy and delivery. He had nonprogressive bilateral ptosis and ophthalmoplegia from birth. He was noted to have torticollis in infancy and pectus carinatum as a child, and both progressed with age. He had normal intellectual and social development and mildly delayed motor milestones. When he came to medical attention at age 16, he had bilateral ptosis (R > L) with a chin-up and rotated head position, limited eye movements, down-slanting palpebral fissures, torticollis, narrow shoulders, thoracic scoliosis, and truncal obesity (Figures 1L and 1M). No limb anomalies or other dysmorphologies were noted, and his karyotype was normal.
CHO II-2 underwent magnetic resonance imaging (MRI) of the brain (which was normal) and of the orbits that revealed absence of the extraocular muscles (Figures 1N and S1E). Frontal radiographs and CT images of ALO II-1, ALO II-2, and CHO II-2 revealed a spectrum of dysmorphic ribs with hypoplasia, fusion anomalies, pseudarthrosis, missing ribs, and failure of some remaining ribs to extend anteriorly toward the sternum (Figures 1D, 1E, 1J, 1K, and 1O–1Q). The sternum of ALO II-1 was fused (Figure 1F). In addition, spine CT images of ALO II-1 and CHO II-2 revealed scoliosis, narrowed thoracic disc spaces, variable fusion anomalies of cervical and thoracic vertebral bodies and posterior elements, abnormally shaped and subluxed cervical facets, malformations of the clivus with basilar invagination and clefting of the anterior arch of C1, and abnormally shaped thoracic vertebrae. CHO II-2 also demonstrated atlantooccipital fusion (Figures 1G, 1R, and S1). BX IV-4 underwent MRI as a young adult; these images were not available for review, but were reported to reveal 25 degrees of dextroscoliosis centered at L1-L2 and mild nonspecific degenerative changes in the lower lumbar spine. No other imaging of the affected members of BX was performed.
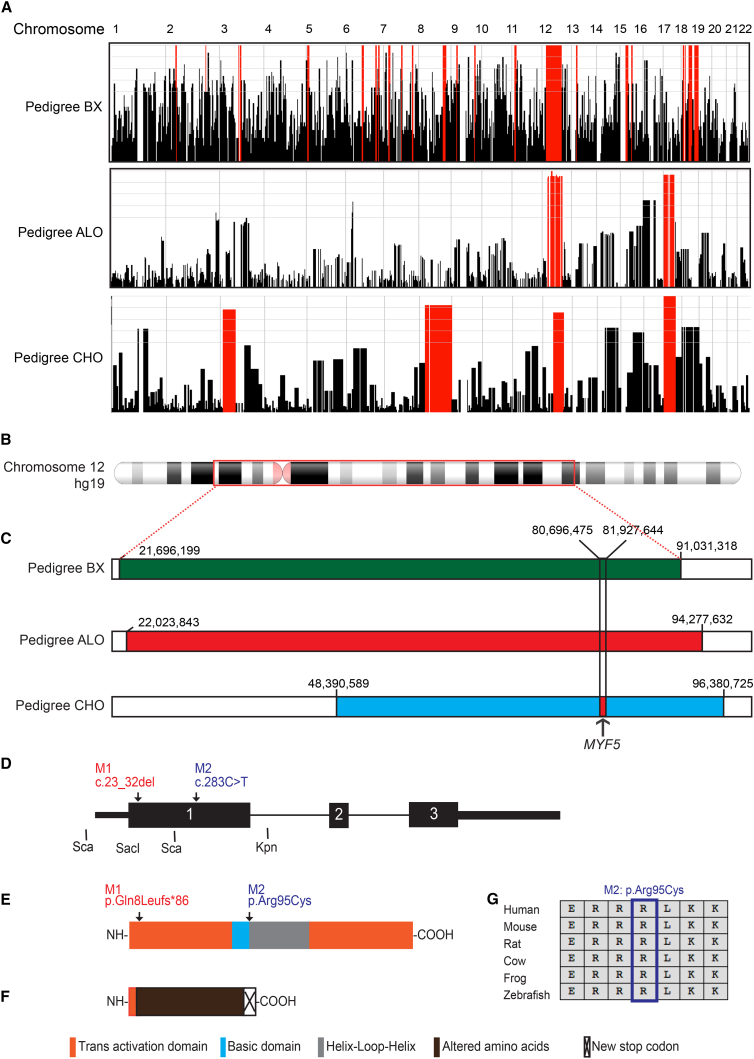
Genome-wide linkage analysis of pedigree BX, generated from Affymetrix Human Mapping 10K SNP array (ThermoFisher) data assuming recessive inheritance and a disease allele frequency of 0.001, yielded the maximum potential lod score of 2.1 at loci spanning 69 Mb and 1.4 Mb on chromosomes 12p12.1–12q21.31 and 18q12.3, respectively, and haplotype analysis revealed homozygosity across both regions (Figure 2A). Homozygosity mapping using Omni2.5-8 SNP array data from Illumina was performed for pedigrees ALO and CHO. Analysis of ALO revealed a 72 Mb region of homozygosity on chromosome 12p12.1–12q22 and a 50 Mb region on chromosome 17p13.2-17q22. Analysis of pedigree CHO revealed a 48 Mb region of homozygosity on chromosome 12q13.12–12q22, a 53 Mb region on 17p13.3–17q22 that overlapped with ALO but had an alternate haplotype, and 59 and 119 Mb regions on chromosome 3p25.3–3p14.1 and 8p21.1–8q24.3, respectively (Figure 2A). The regions of homozygosity on chromosome 12 overlapped in the three families for 42.6 Mb and, within this region, pedigrees ALO and CHO shared a 1.2 Mb region of haploidentity (chr12: 80,696,475–81,927,644) that encompasses PTPRQ (MIM: 613317), MYF5, MYF6 (MIM: 159991), LIN7A (MIM: 603380), ACSS3 (MIM: 614356), and portions of OTOGL (MIM: 614925) and PPFIA2 (MIM: 603143) (Figures 2B and 2C).
Figure 2.
MYF5 Genetics
(A) Homozygosity regions (red) in affected members of pedigrees BX (top), ALO (middle), and CHO (bottom) across chromosomes 1–22. Note overlapping regions of homozygosity on chromosome 12.
(B) Schematic of chromosome 12 with the region of linkage in pedigree BX boxed in red.
(C) Schematics of region of chromosome 12 linkage and homozygosity (green) in pedigree BX (top), and of homozygosity in pedigrees ALO (middle, red) and CHO (bottom, blue). hg19 chromosome 12 nucleotide positions are indicated over each homozygosity map. The region of haploidentity shared by ALO and CHO is denoted by vertical black lines and the red filled region in the blue CHO homozygosity map, and the location of MYF5 indicated beneath it.
(D) Human MYF5 schematic. Arrows denote nucleotide position of M1 and M2 mutations.
(E) Wild-type human MYF5 protein schematic. Arrows denote amino acid positions of M1 and M2, and key to colored domains is provided across the bottom of the figure.
(F) Predicted M1 mutant MYF5 protein schematic with 8 MYF5 amino acid residues followed by 86 altered residues prior to a new stop codon.
(G) Multispecies protein MacVector alignment of amino acids surrounding M2 substitution: all residues are conserved.
To identify causal variants in the three pedigrees, whole-exome sequencing (WES) was performed on DNA from BX III-1, III-2, IV-4, and IV-6, ALO I-2, II-1, and II-2, and CHO II-2. Libraries were prepared using Agilent SureSelect Human All Exon versions 4X, 5X, and 5+UTRs+MitoX for ALO, CHO, and BX, respectively, and sequenced on a HiSeq2000. Homozygous recessive variants of moderate to high impact and allele frequencies less than 0.01 were then identified using the seqr software platform (Table S1).
Evaluation of WES data from pedigree BX revealed nine rare coding variants that were homozygous in the affected daughters and heterozygous in the parents (Table S1). Of these, six were located within the pedigree’s 69 Mb region of homozygosity on chromosome 12, including two of particular interest: STAC3 (MIM: 615521) (hg19, GenBank: NM_145064.2; c.322C>G [p.Arg108Gly]) and MYF5 (hg19, GenBank: NM_005593.2; c.283C>T [p.Arg95Cys], M2), which are located outside of and within the 1.2 Mb region of shared haploidentity between ALO and CHO, respectively. A STAC3 biallelic missense (p.Trp284Ser) and truncating variants have been reported to cause a myopathic phenotype in four families distinct from BX: affected individuals had ptosis but not significant restriction of eye movements, and had facial weakness, generalized hypotonia and weakness, and susceptibility to malignant hyperthermia with or without cleft palate, scoliosis, and joint contractures.13, 14, 15 By contrast, mutations in MYF5 had not been reported, and the p.Arg95Cys variant had a CADD score of 34 and was predicted to be disease causing by MutationTaster, damaging by Fathmm, and probably damaging by PolyPhen.
Evaluation of WES data from pedigree ALO revealed ten rare coding variants that were homozygous in the two affected children and heterozygous in the mother (Table S1). Of these, nine were located within the homozygous region on chromosome 12, of which only one, a 10 base pair deletion in MYF5 (hg19, GenBank: NM_005593.2; c.23_32delAGTTCTCACC [p.Gln8Leufs∗86], M1) was in the haploidentical region shared with pedigree CHO. None was located in the homozygous region on chromosome 17.
Evaluation of WES data from the affected child in pedigree CHO revealed only six rare homozygous coding variants within the regions of homozygosity: one on chromosome 1, four on chromosome 3, and one on chromosome 12 (Table S1). The single homozygous variant on chromosome 12 fell within the 1.2 Mb region of shared haploidentity and was the identical MYF5 10 base pair deletion harbored by the affected members of ALO (Figures 2D–2F).
To exclude the presence of other homozygous structural variations (SVs) or non-coding variants not reported by exome sequencing within the haploidentical region of CHO and ALO, we performed PCR-free whole-genome sequencing in individual CHO-1 (30× average coverage, New York Genome Center, NY). After careful annotation of each non-coding variant and SV, we did not identify any novel homozygous rare events (cumulative MAF < 0.01) that might be linked to the observed phenotype (Table S2).
Both the MYF5 deletion and MYF5 missense variant were absent from the 1000 Genomes, GnomAD, and ExAC databases, and Sanger sequencing confirmed their appropriate segregation within each family (Figures 1A and S2; forward primer, 5′-TTACCGGAGCGACAGACTAG-3′; reverse primer, 5′-CCACACCAGGTTAAATGAGGT-3′). No other MYF5 variants or polymorphisms were present in any of the family members, and no other genes harbored rare coding variants in more than one family (Table S1). We also screened MYF5 sequence in probands from 28 recessive, 26 dominant, and 51 simplex unsolved pedigrees with congenital ophthalmoplegia and did not identify rare homozygous or compound heterozygous variants. While some probands had been diagnosed with “congenital fibrosis of the extraocular muscles” (CFEOM) similar to pedigree BX, none were known to have rib anomalies.
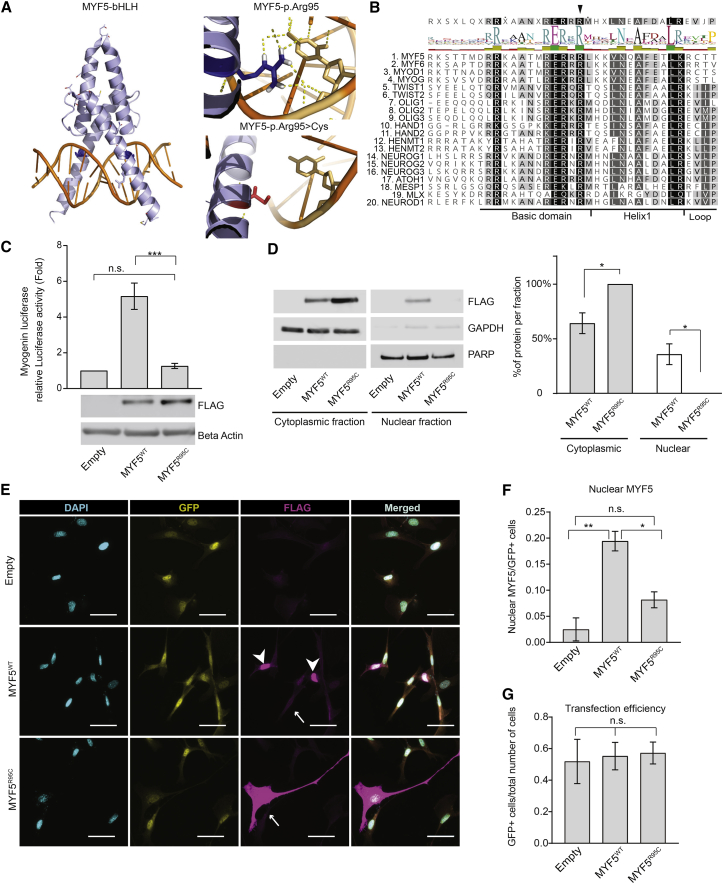
MYF5 contains a basic domain that interacts with DNA and a helix-loop-helix domain necessary for homo- and hetero-dimerization (Figure 3A).16 The frameshift deletion identified in families ALO and CHO is predicted to be the target of nonsense-mediated decay or, in case of escape, to generate a protein product containing only the first 7 MYF5 amino acids followed by 85 altered residues prior to a new stop codon (p.Gln8Leufs∗86), thus resulting in complete loss of MYF5 function (Figures 2F and S3). The missense mutation identified in family BX is predicted to alter an arginine at amino acid position 95, located at the end of the MYF5 basic domain. MYF5 Arg95 is highly conserved in other species (Figure 2G) as well as in 104 of the 113 human Myc-type bHLH TFs (Prosite: PS50888; Figure 3B), supporting its functional importance for TF binding activity. Moreover, missense mutations that alter this conserved arginine in the bHLH TF TWIST1 cause Saethre-Chotzen syndrome,17, 18, 19 and amino acid substitutions elsewhere in the basic domain of bHLH TF proteins are associated with additional human disorders.20, 21, 22, 23 Consistent with these data, in silico modeling of the Arg95 amino acid using PyMOL and the MYOD structure24 as a backbone highlights the direct interaction of this arginine residue with DNA, and loss of this DNA interaction when a cysteine is substituted for the arginine (Figure 3A).
Figure 3.
BX Family Mutation (p.Arg95Cys) Impairs MYF5 Nuclear Localization and Transcriptional Activity
(A) Left: 3D model of the bHLH domain (light blue) of MYF5 bound to DNA double helix (orange) generated using PyMol. MYF5 structural data were modeled over MYOD crystal structure.24 Residue Arg95 is indicated in dark blue. Right: Effect of p.Arg95Cys substitution on DNA binding. Arg95 (dark blue residue) forms strong hydrogen bonds (yellow dotted lines) with a cytosine (top). When mutated to Cys95 (red residue), these interactions are completely abolished (bottom).
(B) Top: Consensus sequence of 20 human bHLH proteins from Uniprot using ClustralW, with black arrowhead indicating the position of the conserved MYF5 Arg95 amino acid residue located at the end of the basic domain near its junction with Helix 1. Bottom: Alignment of 20 human bHLH proteins with degree of amino acid conservation denoted by shading from black (highly conserved) to white (not conserved). Above the alignment and under the consensus sequence is the amino acid logo and bar graph in which letter size, bar graph height, and color correlate with degree of conservation (larger/higher, more conserved; green, highly conserved; red, not conserved).
(C) Relative quantification of the activation (in fold increase on y axis) of myogenin-luciferase reporter following overexpression of equimolar quantities of WT and mutant (MYF5R95C) FLAG plasmids. Luciferase signals were normalized to relative Renilla control. The relative luciferase activity of the empty plasmid was set as 1. A representative western blot below the graph shows expression levels of the recombinant proteins detected with an anti-FLAG antibody with beta-actin antibody as a loading control. Statistical significance is calculated using unpaired Student’s t test. Data are presented as mean ± SEM of three independent triplicate experiments.
(D) Left: Subcellular fraction of total lysates of C3H10T1/2 cells overexpressed with empty, MYF5WT, MYF5R95C plasmid. Myf5 recombinant protein (FLAG) localizes to the cytoplasmic fraction in the mutant and WT lysates, but to the nuclear fraction only in the WT lysate. GAPDH (Santa Cruz, sc-25778, 1:50,000) and PARP (Cell Signaling, #9542, 1:1,000) signals are used as loading controls for the cytoplasmic and nuclear fractions, respectively. Right: quantification of the signal intensities of the western blot bands from three independent experiments. Signal was normalized to the cytoplasmic or nuclear loading control and calculated as percentage of the protein per fraction relative to the total signal (cytoplasmic + nuclear). Despite variability in the expression level between WT and mutant protein, MYF5 signal was never detected in the nuclear fraction of the mutant alleles at low exposure. At long exposure times, a faint band at the level of the mutant protein was visible (data not shown), suggesting that the nuclear transport is impaired but not completely abolished. Data are represented as mean ± SEM. Statistical significance is calculated using unpaired Student’s t test.
(E) Subcellular localization of MYF5 ectopic protein in C3H10T1/2 cells cotransfected with empty (top row), MYF5WT (wild-type, center row), or MYF5R95C (mutant, bottom row) FLAG and GFP plasmids. Both MYF5 wild-type and mutant proteins localize to the cytoplasm in transfected C3H10T1/2 cells (indicated by arrows), while by contrast, MYF5 protein localizes to the nucleus in ∼20% of wild-type and only ∼7% of mutant protein (not shown). Cyan, nuclear DAPI staining; yellow, GFP; magenta, FLAG. Merged images in right panels. Scale bar 50 μm.
(F) Percentage of cells with nuclear MYF5 expression relative to GFP-positive cells.
(G) Transfection efficiency calculated as total number of GFP-positive cells over total number of cells.
Quantitative data for (F) and (G) were obtained from double blinded counts from three independent experiments. Between 10 and 15 random fields per condition were counted for each experiment. Significance for (F) and (G) was calculated using one-way ANOVA with Turkey correction for multiple testing. Data are represented as mean ± SEM (∗p < 0.05; ∗∗p < 0.01; ns, p > 0.5).
To assess whether the MYF5 p.Arg95Cys amino acid substitution impairs MYF5 transcriptional function, we obtained a MYF5 expressing plasmid with a C-terminal Myc-FLAG tag (MYF5WT-FLAG, RC210156, OriGene) and mutagenized the plasmid to substitute Arg95 with a cysteine (MYF5R95C-FLAG). Mutagenesis was performed by GENEWIZ and confirmed by qualitative restriction digestion and sequencing. Both MYF5WT-FLAG and MYF5R95C-FLAG constructs were co-transfected with a previously generated myogenin-luciferase reporter25 in C3H10T1/2 cells (clone 8, ATCC CCL-226). Dual promoter luciferase assay (Promega) was then used to evaluate the relative activation of the luciferase compared to Renilla activation (Figure 3C). Cells transfected with MYF5WT-FLAG plasmid strongly activated the luciferase reporter while, by contrast, very minimal activation was observed in cells transfected with the same amount of MYF5R95C-FLAG plasmid, despite higher protein level of the latter as shown by western blot. These data establish that the mutant protein is deficient in promoting transcriptional activation in vitro.
To function as a TF, MYF5 must first translocate to the nucleus. MYF5 has a nuclear localization signal (NLS) in its basic domain26, 27 and, in zebrafish, a 23 amino acid sequence from the basic domain of Myf5 is sufficient and necessary to translocate a GFP protein to the nucleus and the nucleolus.27 Remarkably, Arg95 not only interacts with DNA but also is one of the residues within this 23 amino acid bipartite NLS ([KRKASTVDRRRAATMRERRRLKK] in which the two NLS are underlined and Arg96 is italicized).
To investigate whether p.Arg95Cys impairs nuclear localization as well as transcription, we first performed nuclear fractionation of transfected C3H10T1/2 cells and observed FLAG signal in the nuclear fraction of cells transfected with MYF5WT-FLAG but not MYF5R95C-FLAG (Figure 3D). Next, we examined and quantified the presence of the overexpressed proteins in the nucleus of fixed C3H10T1/2 cells (Figures 3E–3G). Both constructs showed cytoplasmic localization, but MYF5WT-FLAG localized to the nucleus more often (average ∼20% of nuclei) than MYF5R95C-FLAG (<10% of nuclei). While quantification of MYF5R95C-FLAG nuclear localization was not significantly different than that of the empty plasmid, we did observe MYF5R95C-FLAG in the nucleus of transfected cells (Figure 3F), suggesting that nuclear transport may have not been completely abolished by the mutation. Taken together, these data support the pathogenicity of p.Arg95Cys by demonstrating both impaired nuclear localization and transcriptional activity of the mutant MYF5 protein. Notably, Arg95 is located at the edge of the basic domain adjacent to helix 1 of the MYF5 protein (Figure 3B), and amino acid substitutions of resides in helix 1 of TWIST1 were reported to similarly both reduce nuclear localization and abolish DNA binding of the TWIST1 protein.21, 23
The pathogenicity of the human MYF5 variants harbored by these three families is supported by the remarkable phenotypic similarities of the human syndrome to that reported in a series of Myf5−/− mouse models generated using different constructs and cutting sites (Figure S4, Table S3). The reported phenotypes of Myf5−/− mice range from absent EOMs4, 28 and absent ribs5, 7, 8 with fusion of the sternum and the cervical and thoracic vertebrae resulting in perinatal death,2, 5, 7, 8, 29 to mildly hypoplastic EOMs4 and normal ribs30, 31 (Table S3). Over time, it has become evident that the more severe mouse phenotypes correlate with models in which large targeting vectors with ectopic promoters were inserted into Myf5 exon 1, with or without deletion of Myf5 exon 1 coding sequence, and it is now recognized that these constructs not only eliminate Myf5 expression but also reduce Myf6 expression, which is located ∼7 kb upstream of Myf5.2 Myf5 and Myf6 regulation is complex,32 and it has not yet been determined whether reduction of Myf6 expression in these models results from disruption of regulatory elements located within the Myf5 coding sequence, alteration of DNA secondary structure, or altered balance of Myf5 and Myf6 expression secondary to activity of the ectopic promoters included in the inserted cassettes. By contrast, milder phenotypes were reported in conditional Myf5 mouse models crossed to ubiquitous cre drivers. In the Myf5ΔloxP model, exon 1 was excised, leaving only a loxP sequence in its place;30 mutant mice survived and their ribs were normal in appearance, but EOMs were not examined.30, 31 In the Myf5loxP model, loxP was inserted into the coding sequence of exon 1;2 in these mutant mice, all EOM anlage were present although some appeared hypoplastic, but ribs were not examined.4 In both models, Myf5 functional activity was abolished and Myf6 expression was reported to be at normal or near normal levels.
Similar to the mouse models, phenotypic manifestations of the human MYF5 syndrome varies somewhat between pedigrees (Table 1). First, despite harboring the same disease allele predicted to result in complete loss of function of MYF5, CHO II-2 has more significant rib and spine anomalies than the affected members of ALO, supporting genetic and/or environmental modifiers. Second, affected members of pedigree BX who harbor the p.Arg95Cys substitution may have a milder phenotype, as they have mild or no ptosis, they have scoliosis that differs somewhat from that found in pedigrees ALO and CHO, and it is not known whether they have rib anomalies. Thus, the human MYF5 syndrome phenotype is, overall, less severe than mouse models with loss of both Myf5 and Myf6 expression. Indeed, the affected members of pedigree BX who harbor the missense variant most closely resemble the Myf5loxP model, suggesting that loss of MYF5 function alone is sufficient to cause the ocular phenotype. Due to the absence of tissue or cell lines from ALO and CHO, we could not evaluate the transcriptional level of MYF6 in these families to exclude the involvement of this gene in the rib phenotype. However, genome sequencing of CHO II-2 failed to identify any coding or non-coding changes or structural variants (such as a deletion/insertion into a regulatory element) within the haploidentical region that might affect MYF6 expression.
While the milder phenotype in pedigree BX could also arise from genetic or environmental modifiers similar to ALO, there are several alternative possibilities. First, given that in mouse models the degree of EOM and rib hypoplasia correlates with the degree to which expression of Myf6 is also reduced,2, 4 it is possible that the deletion, but not the missense variant, reduces MYF6 expression. Interestingly, based on ENCODE data, the 10 bp deletion falls in a region of DNase hypersensitivity. Although it is more likely that the open chromatin state is caused by the proximity to the Myf5 promoter, it is possible that the deletion affects transcriptional elements within coding sequence of MYF5 that regulate expression of nearby genes, such as MYF6. This phenomenon has been reported previously,33, 34 although it has not been studied at the MYF5-MYF6 locus. It is also possible that unlike the deletion, the missense mutation results in a MYF5 hypomorphic allele, leaving the mutant full-length protein with some residual activity. Indeed, we found that a minimal amount of mutant protein could translocate to the nucleus and might retain some transcriptional activity, although this observation did not reach statistical significance in vitro. Alternatively, the mutant protein, although incapable of binding to DNA, could retain its ability to complex with other molecules,35 thus permitting some specific cytoplasmic/complex function and partially rescuing the phenotype. Interestingly, a recent report has shown a role of MYF5 in binding the 3′ UTR to stabilize the mRNA of CCND1 (MIM: 168461), which encodes Cyclin D1, a key regulator of cell cycle.36 Future modeling of the specific human mutations in mice would permit these hypotheses to be tested.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the families for their participation; Eric Pierce and the HMS Ocular Genomics Institute and Daniel MacArthur and members of his laboratory for technical expertise and support; and Brenda Barry, Caroline Andrews, and Long Cheng for their assistance and enlightening discussions. Research was supported by Swiss National Science Foundation (P2LAP3_155081 and P300PA_164677) and Knights Templar Eye Foundation (S.A.D.G.); Boston Children’s Hospital IDDRC (U54 HD090255); and NEI (R01EY12498 and R01EY027421) (E.C.E.). E.C.E. is an investigator of the Howard Hughes Medical Institute.
Published: June 7, 2018
Footnotes
Supplemental Data include four figures and three tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.05.003.
Accession Numbers
The accession numbers for the sequence data reported in this paper are ClinVar: SCV000747041 and SCV000746968.
Web Resources
Combined Annotation Dependent Depletion (CADD), http://cadd.gs.washington.edu/
ENCODE, https://www.encodeproject.org/
Fathmm, http://fathmm.biocompute.org.uk/
gnomAD Browser, http://gnomad.broadinstitute.org/
MutationTaster, http://www.mutationtaster.org/
OMIM, http://www.omim.org/
PolyPhen, http://genetics.bwh.harvard.edu/pph2/
Prosite, http://prosite.expasy.org/
UCSC Genome Browser (February 2009), https://genome.ucsc.edu
UniProt, http://www.uniprot.org/
Supplemental Data
References
- 1.Rudnicki M.A., Schnegelsberg P.N., Stead R.H., Braun T., Arnold H.H., Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 2.Kassar-Duchossoy L., Gayraud-Morel B., Gomès D., Rocancourt D., Buckingham M., Shinin V., Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- 3.Porter J.D., Baker R.S. Muscles of a different ‘color’: the unusual properties of the extraocular muscles may predispose or protect them in neurogenic and myogenic disease. Neurology. 1996;46:30–37. doi: 10.1212/wnl.46.1.30. [DOI] [PubMed] [Google Scholar]
- 4.Sambasivan R., Gayraud-Morel B., Dumas G., Cimper C., Paisant S., Kelly R.G., Tajbakhsh S. Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev. Cell. 2009;16:810–821. doi: 10.1016/j.devcel.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Braun T., Rudnicki M.A., Arnold H.H., Jaenisch R. Targeted inactivation of the muscle regulatory gene Myf-5 results in abnormal rib development and perinatal death. Cell. 1992;71:369–382. doi: 10.1016/0092-8674(92)90507-9. [DOI] [PubMed] [Google Scholar]
- 6.Yoon J.K., Olson E.N., Arnold H.H., Wold B.J. Different MRF4 knockout alleles differentially disrupt Myf-5 expression: cis-regulatory interactions at the MRF4/Myf-5 locus. Dev. Biol. 1997;188:349–362. doi: 10.1006/dbio.1997.8670. [DOI] [PubMed] [Google Scholar]
- 7.Tajbakhsh S., Rocancourt D., Buckingham M. Muscle progenitor cells failing to respond to positional cues adopt non-myogenic fates in myf-5 null mice. Nature. 1996;384:266–270. doi: 10.1038/384266a0. [DOI] [PubMed] [Google Scholar]
- 8.Tallquist M.D., Weismann K.E., Hellström M., Soriano P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 2000;127:5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- 9.Engle E.C. Human genetic disorders of axon guidance. Cold Spring Harb. Perspect. Biol. 2010;2:a001784. doi: 10.1101/cshperspect.a001784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nugent A.A., Kolpak A.L., Engle E.C. Human disorders of axon guidance. Curr. Opin. Neurobiol. 2012;22:837–843. doi: 10.1016/j.conb.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones K.J., North K.N. External ophthalmoplegia in neuromuscular disorders: case report and review of the literature. Neuromuscul. Disord. 1997;7:143–151. doi: 10.1016/s0960-8966(97)00447-1. [DOI] [PubMed] [Google Scholar]
- 12.Traboulsi E.I., Lee B.A., Mousawi A., Khamis A.R., Engle E.C. Evidence of genetic heterogeneity in autosomal recessive congenital fibrosis of the extraocular muscles. Am. J. Ophthalmol. 2000;129:658–662. doi: 10.1016/s0002-9394(99)00467-5. [DOI] [PubMed] [Google Scholar]
- 13.Grzybowski M., Schänzer A., Pepler A., Heller C., Neubauer B.A., Hahn A. Novel STAC3 Mutations in the first non-Amerindian patient with Native American myopathy. Neuropediatrics. 2017;48:451–455. doi: 10.1055/s-0037-1601868. [DOI] [PubMed] [Google Scholar]
- 14.Horstick E.J., Linsley J.W., Dowling J.J., Hauser M.A., McDonald K.K., Ashley-Koch A., Saint-Amant L., Satish A., Cui W.W., Zhou W. Stac3 is a component of the excitation-contraction coupling machinery and mutated in Native American myopathy. Nat. Commun. 2013;4:1952. doi: 10.1038/ncomms2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Telegrafi A., Webb B.D., Robbins S.M., Speck-Martins C.E., FitzPatrick D., Fleming L., Redett R., Dufke A., Houge G., van Harssel J.J.T., Moebius Syndrome Research Consortium Identification of STAC3 variants in non-Native American families with overlapping features of Carey-Fineman-Ziter syndrome and Moebius syndrome. Am. J. Med. Genet. A. 2017;173:2763–2771. doi: 10.1002/ajmg.a.38375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun T., Winter B., Bober E., Arnold H.H. Transcriptional activation domain of the muscle-specific gene-regulatory protein myf5. Nature. 1990;346:663–665. doi: 10.1038/346663a0. [DOI] [PubMed] [Google Scholar]
- 17.Kress W., Schropp C., Lieb G., Petersen B., Büsse-Ratzka M., Kunz J., Reinhart E., Schäfer W.D., Sold J., Hoppe F. Saethre-Chotzen syndrome caused by TWIST 1 gene mutations: functional differentiation from Muenke coronal synostosis syndrome. Eur. J. Hum. Genet. 2006;14:39–48. doi: 10.1038/sj.ejhg.5201507. [DOI] [PubMed] [Google Scholar]
- 18.de Heer I.M., de Klein A., van den Ouweland A.M., Vermeij-Keers C., Wouters C.H., Vaandrager J.M., Hovius S.E., Hoogeboom J.M. Clinical and genetic analysis of patients with Saethre-Chotzen syndrome. Plast. Reconstr. Surg. 2005;115:1894–1902. doi: 10.1097/01.prs.0000165278.72168.51. discussion 1903–1895. [DOI] [PubMed] [Google Scholar]
- 19.Gripp K.W., Zackai E.H., Stolle C.A. Mutations in the human TWIST gene. Hum. Mutat. 2000;15:150–155. doi: 10.1002/(SICI)1098-1004(200002)15:2<150::AID-HUMU3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 20.Malecki M.T., Jhala U.S., Antonellis A., Fields L., Doria A., Orban T., Saad M., Warram J.H., Montminy M., Krolewski A.S. Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat. Genet. 1999;23:323–328. doi: 10.1038/15500. [DOI] [PubMed] [Google Scholar]
- 21.El Ghouzzi V., Legeai-Mallet L., Benoist-Lasselin C., Lajeunie E., Renier D., Munnich A., Bonaventure J. Mutations in the basic domain and the loop-helix II junction of TWIST abolish DNA binding in Saethre-Chotzen syndrome. FEBS Lett. 2001;492:112–118. doi: 10.1016/s0014-5793(01)02238-4. [DOI] [PubMed] [Google Scholar]
- 22.Marchegiani S., Davis T., Tessadori F., van Haaften G., Brancati F., Hoischen A., Huang H., Valkanas E., Pusey B., Schanze D. Recurrent mutations in the basic domain of TWIST2 cause ablepharon macrostomia and Barber-Say syndromes. Am. J. Hum. Genet. 2015;97:99–110. doi: 10.1016/j.ajhg.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Ghouzzi V., Legeai-Mallet L., Aresta S., Benoist C., Munnich A., de Gunzburg J., Bonaventure J. Saethre-Chotzen mutations cause TWIST protein degradation or impaired nuclear location. Hum. Mol. Genet. 2000;9:813–819. doi: 10.1093/hmg/9.5.813. [DOI] [PubMed] [Google Scholar]
- 24.Ma P.C., Rould M.A., Weintraub H., Pabo C.O. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 25.Parker M.H., Perry R.L.S., Fauteux M.C., Berkes C.A., Rudnicki M.A. MyoD synergizes with the E-protein HEB β to induce myogenic differentiation. Mol. Cell. Biol. 2006;26:5771–5783. doi: 10.1128/MCB.02404-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vandromme M., Cavadore J.C., Bonnieu A., Froeschlé A., Lamb N., Fernandez A. Two nuclear localization signals present in the basic-helix 1 domains of MyoD promote its active nuclear translocation and can function independently. Proc. Natl. Acad. Sci. USA. 1995;92:4646–4650. doi: 10.1073/pnas.92.10.4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y.H., Chen Y.H., Lu J.H., Tsai H.J. A 23-amino acid motif spanning the basic domain targets zebrafish myogenic regulatory factor myf5 into nucleolus. DNA Cell Biol. 2005;24:651–660. doi: 10.1089/dna.2005.24.651. [DOI] [PubMed] [Google Scholar]
- 28.Michalak S.M., Whitman M.C., Park J.G., Tischfield M.A., Nguyen E.H., Engle E.C. Ocular motor nerve development in the presence and absence of extraocular muscle. Invest. Ophthalmol. Vis. Sci. 2017;58:2388–2396. doi: 10.1167/iovs.16-21268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tajbakhsh S., Rocancourt D., Cossu G., Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–138. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- 30.Kaul A., Köster M., Neuhaus H., Braun T. Myf-5 revisited: loss of early myotome formation does not lead to a rib phenotype in homozygous Myf-5 mutant mice. Cell. 2000;102:17–19. doi: 10.1016/s0092-8674(00)00006-4. [DOI] [PubMed] [Google Scholar]
- 31.Vinagre T., Moncaut N., Carapuço M., Nóvoa A., Bom J., Mallo M. Evidence for a myotomal Hox/Myf cascade governing nonautonomous control of rib specification within global vertebral domains. Dev. Cell. 2010;18:655–661. doi: 10.1016/j.devcel.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 32.Chandra S., Terragni J., Zhang G., Pradhan S., Haushka S., Johnston D., Baribault C., Lacey M., Ehrlich M. Tissue-specific epigenetics in gene neighborhoods: myogenic transcription factor genes. Hum. Mol. Genet. 2015;24:4660–4673. doi: 10.1093/hmg/ddv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wenz P., Schwank S., Hoja U., Schüller H.J. A downstream regulatory element located within the coding sequence mediates autoregulated expression of the yeast fatty acid synthase gene FAS2 by the FAS1 gene product. Nucleic Acids Res. 2001;29:4625–4632. doi: 10.1093/nar/29.22.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lang G., Gombert W.M., Gould H.J. A transcriptional regulatory element in the coding sequence of the human Bcl-2 gene. Immunology. 2005;114:25–36. doi: 10.1111/j.1365-2567.2004.02073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamamori Y., Wu H.Y., Sartorelli V., Kedes L. The basic domain of myogenic basic helix-loop-helix (bHLH) proteins is the novel target for direct inhibition by another bHLH protein, Twist. Mol. Cell. Biol. 1997;17:6563–6573. doi: 10.1128/mcb.17.11.6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panda A.C., Abdelmohsen K., Martindale J.L., Di Germanio C., Yang X., Grammatikakis I., Noh J.H., Zhang Y., Lehrmann E., Dudekula D.B. Novel RNA-binding activity of MYF5 enhances Ccnd1/Cyclin D1 mRNA translation during myogenesis. Nucleic Acids Res. 2016;44:2393–2408. doi: 10.1093/nar/gkw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.