Abstract
Carbapenem resistant Enterobacteriaceae are a significant public health concern, and genes encoding the Klebsiella pneumoniae carbapenemase (KPC) have contributed to the global spread of carbapenem resistance. In the current study, we used whole-genome sequencing to investigate the diversity of blaKPC-containing plasmids and antimicrobial resistance mechanisms among 26 blaKPC-containing Escherichia coli, and 13 blaKPC-containing Enterobacter asburiae, Enterobacter hormaechei, K. pneumoniae, Klebsiella variicola, Klebsiella michiganensis, and Serratia marcescens strains, which were isolated from the same patients as the blaKPC-containing E. coli. A blaKPC-containing IncN and/or IncFIIK plasmid was identified in 77% (30/39) of the E. coli and other bacterial species analyzed. Complete genome sequencing and comparative analysis of a blaKPC-containing IncN plasmid from one of the E. coli strains demonstrated that this plasmid is present in the K. pneumoniae and S. marcescens strains from this patient, and is conserved among 13 of the E. coli and other bacterial species analyzed. Interestingly, while both IncFIIK and IncN plasmids were prevalent among the strains analyzed, the IncN plasmids were more often identified in multiple bacterial species from the same patients, demonstrating a contribution of this IncN plasmid to the inter-genera dissemination of the blaKPC genes between the E. coli and other bacterial species analyzed.
Introduction
Carbapenem-resistant Enterobacteriaceae (CRE) are a serious public health concern as they are often multidrug-resistant, thus making them exceedingly difficult to treat1–11. Also, carbapenem-resistant organisms have been associated with an increased likelihood of death compared with susceptible organisms and have been linked to deadly outbreaks1. The carbapenem antibiotics are a primary treatment approach for infections caused by extended-spectrum β-lactamases (ESBL)-producing bacteria, which are resistant to clinically important antibiotics including cephalosporins2,12. Thus, the global spread of ESBLs such as CTX-M, has made the increased prevalence of carbapenem resistance an even greater public health concern2,12,13.
Carbapenem resistance can result from multiple mechanisms including the activity of carbapenemases, contributions of mutations to β-lactamase function, and changes in outer membrane permeability14,15. A recent study using whole genome sequencing demonstrated there was considerable genomic diversity and numerous carbapenem resistance mechanisms among carbapenem-resistant organisms isolated from different healthcare facilities16.
Among the most prevalent mechanisms of carbapenem resistance is the Klebsiella pneumoniae carbapenemase (KPC), which is one of the class A β-lactamases that confer resistance to carbapenems, penicillins, and cephalosporins2,14. Following the initial description of KPC from a K. pneumoniae strain identified in 199617, KPCs have been identified in diverse bacteria including Enterobacter spp.18,19, Escherichia coli20–24, Salmonella spp.25, Acinetobacter spp.26 and Pseudomonas spp.27,28. The blaKPC gene is most often located on the Tn4401 transposon29–31; however, blaKPC genes have also been identified on other non-Tn4401 mobile elements including Tn172123,32,33. The transposons carrying the blaKPC gene have been identified on plasmids with a wide array of incompatibility (Inc) types including IncFIIK, IncFIA, IncN, IncP, ColE1, and IncA/C, and many of these plasmids have been detected in blaKPC-containing E. coli22–24,33–36.
In the current study, we used whole genome sequencing to characterize the diversity of all blaKPC-containing E. coli and any other blaKPC-containing bacterial species isolated from the same patients in a health system in Pennsylvania over time. This approach allowed us to investigate the diversity of mobile elements and antibiotic resistance genes carried by the blaKPC-containing E. coli among these patients, and also characterize the intergenera transmission of blaKPC genes among the E. coli and other co-occurring bacterial species. Additionally, long-read sequencing was used to generate a complete genome assembly of a diverse blaKPC-containing E. coli strain, and examine the distribution of the 70-kb blaKPC-3-containing IncN multidrug resistance plasmid and two additional antibiotic resistance plasmids from this E. coli strain.
Results
Characteristics of the blaKPC-harboring E. coli and other bacterial species
The 26 E. coli strains analyzed in this study were obtained from 26 different patients that received treatment at one of three hospitals in a large health system in Pennsylvania, United States (Table 1). The E. coli strains were isolated from at least seven different types of samples including Jackson-Pratt drainage (surgical site), urine, sputum, blood, abdominal drainage/abdominal fistula, tracheal aspirate, and bronchoalveolar lavage (BAL) (Table 1). These E. coli strains were selected for genome sequencing because they were initially identified as carbapenem-resistant in the clinical microbiology laboratories, and were later determined to be PCR-positive for a blaKPC gene21,22,37. The blaKPC-containing plasmid of E. coli strain YD626 was previously characterized by sequencing34; however, neither the whole genome of YD626 or any of the other E. coli strains included in this study have been previously analyzed using whole genome sequencing. Thus these results are meant to further examine the genomic and plasmid diversity of the blaKPC-containing E. coli. In addition to the E. coli strains, 10 of 26 patients had one or more non-E. coli cultures that were PCR-positive for a blaKPC gene (Table 1). The other bacterial species analyzed were identified as E. asburiae, E. hormaechei, K. pneumoniae, K. variicola, K. michiganensis, S. marcescens, or P. stuartii, and were cultured from the same or a subsequent sample from the patients that had the blaKPC-containing E. coli strains (Tables 1 and 2).
Table 1.
Features of patients and bacteria analyzed in this study.
| Patienta | Dateb | Samplec | blaKPC-containing E. coli strain | blaKPC gene | Other blaKPC-containing species from the patiente |
|---|---|---|---|---|---|
| 1 | 2008 | JP drainage | YDC107 | bla KPC-3 | K. pneumoniae YDC121 (blaKPC-3, 19 days after E. coli sample, abdominal abscess), S. marcescens YDC107-2 (blaKPC-3, 2009, sputum) |
| 2 | 2008 | urine | YDC134 | bla KPC-3 | |
| 3 | 2009 | urine | YDC304 | bla KPC-2 | |
| 4 | 2009 | abdominal drainage | YDC337 | bla KPC-3 | K. michiganensis YD358 (blaKPC-3, same day as E. coli sample, abdominal fistula), E. asburiae YDC337-2 (blaKPC-3, same day as E. coli sample, abdominal fistula) |
| 6 | 2009 | urine | YDC345 | bla KPC-2 | |
| 7 | 2009 | urine | YDC354 | bla KPC-2 | |
| 8 | 2009 | urine | YD439 | bla KPC-3 | |
| 9 | 2010 | BAL | YD509 | bla KPC-3 | S. marcescens YD509-2 (blaKPC-3, 2 days after E. coli sample, ascites) |
| 10 | 2010 | BAL | YDC419 | bla KPC-2 | |
| 11 | 2011 | blood | YDC462 | bla KPC-3 | |
| 12 | 2011 | JP drainage | YD648 | bla KPC-3 | K. pneumoniae YD648-2 (blaKPC-2, same day, blood) |
| 13 | 2011 | BAL | YD626 | bla KPC-2 | K. variicola YD626-2 (blaKPC-2, same sample) |
| 14 | 2011 | BAL | YD649 | bla KPC-3 | E. hormaechei YDC498 (KPC-3, 70 days after E. coli, BAL), E. hormaechei YDC518 (KPC-3, 107 days after E. coli, blood) |
| 15 | 2011 | urine | YD673 | bla KPC-2 | K. pneumoniae YDC465 (KPC-3, 153 days before E. coli, urine) |
| 16 | 2011 | urine | YD705 | bla KPC-2 | |
| 17 | 2012 | urine | YD736 | bla KPC-2 | |
| 18 | 2012 | urine | YD748 | bla KPC-2 | |
| 19 | 2012 | sputum | YD749 | bla KPC-3 | |
| 21 | 2012 | blood | YDC593 | bla KPC-2 | |
| 22 | 2013 | BAL | YDC595 | bla KPC-2 | |
| 23 | 2014 | blood | YD761 | bla KPC-3 | |
| 24 | 2014 | urine | YD762 | bla KPC-3 | K. pneumoniae YD762-3 (KPC-2, 78 days before E. coli, sputum), K. pneumoniae YD762-2 (KPC-2, 2 days after E. coli, tracheal aspirate) |
| 26 | 2015 | blood | YDC717 | bla KPC-2 | |
| 27 | 2015 | urine | YDC735 | bla KPC-2 | |
| 28 | 2015 | blood | YDC736-1 | bla KPC-2 | K. michiganensis YDC736-2 (KPC-2, same sample) |
| 29 | 2015 | BAL | YD789 | bla KPC-2 | Providencia stuartii YD789-2 (No KPC, 18 days before E. coli, BAL) |
aThe patient numbers 1-19 correspond with those in previous studies (Sidjabat et al.37, Kim et al.21, O’Hara et al.22).
bThe year the sample was collected.
cSample abbreviations: JP, Jackson-Pratt drain; BAL, bronchoalveolar lavage.
dThe blaKPC content, sample date, and sample type corresponding to each strain is indicated in parentheses. Providencia stuartii YD789-2 was PCR-positive for a blaKPC gene, but the blaKPC gene was not detected in the genome assembly.
Table 2.
Characteristics of the PacBio-sequenced genome assembly of E. coli strain YDC107.
| Contig | Description | Sequence length (bp) | GC% | Plasmid Inc-typesa | Resistance genesb | GenBank accession no. |
|---|---|---|---|---|---|---|
| YDC107 | chromosome | 5,198,311 | 50.63 | NA | none | CP025707 |
| pYDC107_184 | plasmid (complete) | 184,098 | 51.42 | IncFIA(AP001918), IncFIB(AP001918), IncFII(AY458016) | mrx, aadA5, tet(D), sul2, aac(3)-IIa, blaTEM-1, mphA, sul1, aph(6)-Id, tet(A), dfrA14, aph(3″)-Ib, ermB | CP025708 |
| pYDC107_85 | plasmid (incomplete) | 85,535 | 49.68 | IncI1 | bla CMY-44 | CP025709 |
| pYDC107_70 | plasmid (complete) | 70,372 | 53.34 | IncN | blaKPC-3, blaOXA-9, blaTEM-1, sul2, aadA, aac(6′)-Ib, aph(3″)-Ib, aph(6)-Id, dfrA14 | CP025710 |
| pYDC107_41 | plasmid (complete) | 41,544 | 45.34 | IncP-1-like trfA | none | CP025711 |
| YDC107_phage1 | phage | 46,701 | 49.89 | NA | none | CP025712 |
| YDC107_phage2 | phage | 18,617 | 47.27 | NA | none | CP025713 |
aThe incompatability type (Inc-type) of each plasmid. NA indicates the inc-typing was not applicable for the chromosome and phage contigs. pYDC107_41 was not predicted to have an inc-type using PlasmidFinder; however, the annotation contained an IncP-1-like trfA replication gene.
bThe antibiotic resistance genes identified on each plasmid. Efflux pumps or mutation-associated resistance genes were also identified on the chromosome that are not shown.
Antimicrobial susceptibility testing demonstrated that the blaKPC-containing E. coli and other bacterial species analyzed were non-susceptible (intermediate or resistant) to between four and 17 of the 21 antimicrobials examined (Supplementary Table S2). All of the blaKPC-containing E. coli and other bacterial species analyzed were resistant to aztreonam (ATM), and all but one of the strains exhibited resistance to ticarcillin-clavulanic acid (TIM) and cefotaxime (CTX) (Supplementary Table S2). All of the blaKPC-containing E. coli strains were susceptible to amikacin (AMK), tigecycline (TGC), colistin (CST), and polymyxin B (PMB) (Supplementary Table S2). In all but one example, the blaKPC-containing E. coli strains exhibited resistance to fewer antibiotics than the other bacterial species isolated from the same patient (Supplementary Table S2). On average, the blaKPC-containing E. coli exhibited resistance to nine antibiotics (range 4 to 14), while the other blaKPC-containing bacterial species isolated from the same patients had resistance to an average of 13 antibiotics (range 7 to 17) (Supplementary Table S2).
Genome characteristics of the E. coli and other bacterial species analyzed
Whole-genome sequencing was used to investigate the diversity of the mobile elements and antibiotic resistance genes carried by the 26 blaKPC-containing E. coli analyzed in this study. Among the blaKPC-containing E. coli in our study, 42% (11/26) had the blaKPC-3 gene, while 58% (15/26) had the blaKPC-2 gene (Supplementary Table S1, Supplementary Fig. S1). In silico multilocus sequence typing (MLST) demonstrated that the E. coli strains analyzed in this study had nine different STs (Supplementary Table S1). Of the 26 total E. coli strains analyzed, 65% (17/26) were identified as ST131, two were ST2521, and the remaining seven strains had different STs (Supplementary Table S1). In silico serotype prediction of each E. coli genome sequence demonstrated that the E. coli strains had 10 different serotypes (Supplementary Table S1). Phylogenomic analysis of the blaKPC-containing E. coli analyzed in this study together with all of the publicly-available blaKPC-containing E. coli in GenBank as of March 2017, highlighted the genomic diversity of blaKPC-containing E. coli analyzed in this study, which were isolated from at least seven different types of clinical samples (Supplementary Fig. S1, Supplementary Table S3). The blaKPC-containing E. coli analyzed in this study were identified in phylogroups B1, B2, D, and F (Supplementary Fig. S1). Although 50% (15/30) of the previously sequenced blaKPC-containing E. coli were in the ST648 MLST lineage, only one of the blaKPC-containing E. coli characterized in this study was identified in this lineage, suggesting that lineages of E. coli other than ST648 are involved in dissemination of the blaKPC genes among the patients analyzed in this study (Supplementary Fig. S1, Supplementary Table S3). More than half (65%, 17/26) of the blaKPC-containing E. coli characterized in this study were in the ST131 lineage (Supplementary Fig. S1, Supplementary Table S1). All of the previously described E. coli in the ST131 lineage possessed the blaKPC-3 gene, whereas 70% (12/17) of the E. coli ST131 genomes sequenced in our study contained the blaKPC-2 gene (Supplementary Fig. S1).
The 14 non-E. coli strains analyzed also had genomes sizes and a GC content consistent with other publicly-available genomes of the same species (Supplementary Table S1). In silico determination of the MLST STs of each of the K. pneumoniae genomes demonstrated that these strains belonged to the MLST lineages ST37, ST258, and ST454 (Supplementary Table S1). K. pneumoniae strains from these MLST lineages have previously been described carrying the blaKPC gene38,39. In particular, K. pneumoniae strains belonging to the ST258 lineage are among the most frequently identified KPC-producing strains, and have contributed to the global spread of blaKPC genes13,39,40.
Complete genome sequencing of blaKPC-3-containing E. coli strain YDC107
To investigate the plasmid diversity of a blaKPC-containing E. coli strain we generated a complete genome sequence for E. coli strain YDC107 (Table 2). E. coli strain YDC107 had a predicted serotype of O102:H6, belonged to the MLST lineage ST964, and was present in phylogroup D of the whole-genome phylogeny (Fig. 1). These characteristics make YDC107 a unique strain compared to the majority of the other blaKPC-containing E. coli strains, which primarily belong to the ST131 or ST648 lineages (Supplementary Fig. S1, Supplementary Table S3). The chromosome of E. coli strain YDC107 assembled into a single contig that was 5,198,311 bp in length and had a GC content of 50.63% (Table 2). Although the chromosome was a similar size to previously completed E. coli genomes it could not be circularized by the assembler, possibly due to the excision of phage regions (Table 2, phage 1 and phage 2) that could be placed both within, and independent from, the chromosome during the assembly process (Table 2). The four additional contigs of the YDC107 genome assembly contained predicted plasmid genes involved in replication, conjugative transfer, and stability (Supplementary Table S4). We have designated the plasmids as follows based on their sequence lengths: pYDC107_184 (184,098 bp), pYDC107_85 (85,535 bp), pYDC107_70 (70,372 bp), and pYDC107_41 (41,544 bp) (Table 2). Three of the plasmids (pYDC107_184, pYDC107_70, and pYDC107_41) circularized during assembly and thus represent complete plasmid sequences. The larger plasmids (pYDC107_184, pYDC107_85, and pYDC107_70) each contained one or more antibiotic resistance genes, whereas the smaller plasmid (pYDC107_41) did not have any predicted antibiotic resistance genes (Table 2, Supplementary Table S7).
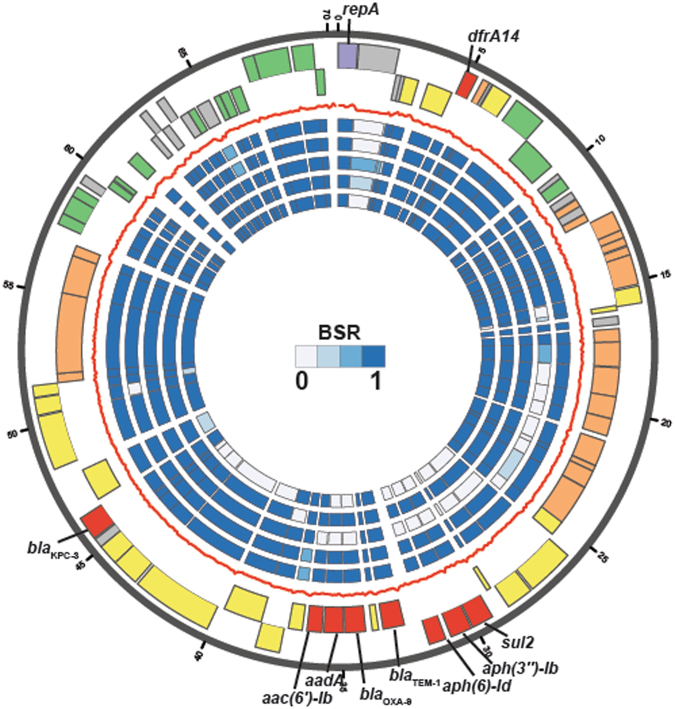
Figure 1.
Analysis of the blaKPC-3-containing plasmid pYDC107_70 from E. coli strain YDC107. The two outermost data tracks contain the predicted protein-coding genes on the forward (first track) or reverse (second track) strands of the plasmid. The gene colors indicate their predicted protein functions as follows: antibiotic resistance (red), plasmid replication (purple), plasmid stability (green), conjugative transfer (orange), transposition (yellow), and gray (unknown). The red line separating the gene tracks and the inner heat map tracks is the GC content of the plasmid, calculated on a 100 bp sliding window. The inner heat map tracks (numbered 1 to 5) contain BSR values from the in silico detection of each protein-coding gene in the genome sequences of K. pneumoniae strain YDC121 (track 1), S. marcescens strain YDC107-2 (track 2), both of which were isolated from the same patient as E. coli strain YDC107, as well as the previously sequenced blaKPC-containing IncN plasmids pYD626E (GenBank accession no. KJ933392.1) from E. coli strain YD62634 (track 3), pBK32602 (GenBank accession no. KU295134.1) from E. coli strain BK3260223 (track 4), and pECN580 (GenBank accession no. KF914891.1) from E. coli strain ECN58024 (track 5).
Mobile elements involved in dissemination of the blaKPC genes
Whole-genome sequencing also allowed us to investigate the diversity of mobile elements involved in dissemination of the blaKPC genes. All but one (P. stuartii YD789-2) of the genomes contained either the blaKPC-2 or blaKPC-3 gene, and all of the blaKPC genes were located on a Tn4401 transposon (Supplementary Table S1). Although the P. stuartii strain was initially PCR-positive for a blaKPC gene, the blaKPC gene was not detected in the genome assembly of this strain, suggesting the strain may have lost the plasmid or blaKPC gene prior to sequencing. All of the blaKPC-3 genes in the genomes sequenced in this study were located on the Tn4401b isoform (Supplementary Table S1). The blaKPC-2 genes were located on the Tn4401a isoform in 75% (15/20) of the blaKPC-2-containing strains analyzed (Supplementary Table S1). The blaKPC-2 genes in the E. coli and other bacterial species from patients 13, 22, and 28 instead carried blaKPC-2 located on Tn4401b (Supplementary Table S1). The genomes of the E. coli and non-E. coli strains from patients 1, 4, 9, 13, 14, and 28, contained the same blaKPC gene and Tn4401 isoform, while the E. coli and non-E. coli strains from patients 12, 15, and 24 had different blaKPC and Tn4401 isoform combinations (Supplementary Table S2).
By completing the genome of one of the blaKPC-containing E. coli strains (YDC107) we were able to investigate the similarity of one of the blaKPC-containing plasmids from an E. coli among the other blaKPC-containing E. coli and bacterial species analyzed in this study. The blaKPC-3 gene of E. coli strain YDC107 was identified on a 70-kb IncN plasmid pYDC107_70 along with additional resistance genes including the β-lactamase genes blaOXA-9 and blaTEM-1, which typically confer resistance to cephalosporins and aminopenicillins41,42 (Fig. 1, Table 2, Supplementary Table S6). The pYDC107_70 plasmid also contained dfrA14, sul2, aph(3″)-Ib, aph(6)-Id, aadA, and aac(6′)-Ib, which are known to confer resistance to trimethoprim, sulfonamides, and aminoglycosides, respectively (Fig. 1, Table 2, Supplementary Table S6). In addition to the resistance genes, pYDC107_70 carries genes for conjugative transfer and plasmid stability (Supplementary Table S6). Nearly all of the genes of pYDC107_70 were identified in K. pneumoniae strain YDC121 and S. marcescens strain YDC107-2 also from patient 1, suggesting that this IncN plasmid may have been transferred between these three species (heat map tracks 1 and 2 in Fig. 1). Comparison of the pYDC107_70 plasmid to three previously characterized blaKPC plasmids from E. coli demonstrated that two of the previously characterized blaKPC-containing plasmids, pYD626E (GenBank accession number KJ933392.1)34 and pECN580 (GenBank accession number KF914891.1)24, were missing the blaOXA-9 gene, and genes that confer resistance to aminoglycosides (aadA, aac(6′)-Ib, aph(6)-Id, and aph(3″)-Ib) or sulfonamides (sul2) (Fig. 1). In contrast, plasmid pBK32602 (GenBank accession number KU295134.1)23 contained nearly all of the protein-coding genes of pYDC107_70, including all of the antibiotic resistance genes (Fig. 1). The blaKPC gene of pYDC107 is located on a Tn4401b element that appears to be inserted within a Tn1331-like element, which is similar to the IncN plasmid pYD626E34; however, pYDC107_70 has the blaKPC-3 gene rather than the blaKPC-2 gene that was identified on pYD626E (Supplementary Table S6). Plasmids pYDC107_70 and pYD626E also differ among the other resistance genes they carry, with pYD626E carrying a blaLAP-1, qnrS1, and blaTEM-1 inserted adjacent to the blaKPC region, while pYDC107_70 has blaOXA-9, sul2, aadA, aac(6′)-Ib, aph(6)-Id, and aph(3″)-Ib in this same region with blaTEM-1 (Fig. 1).
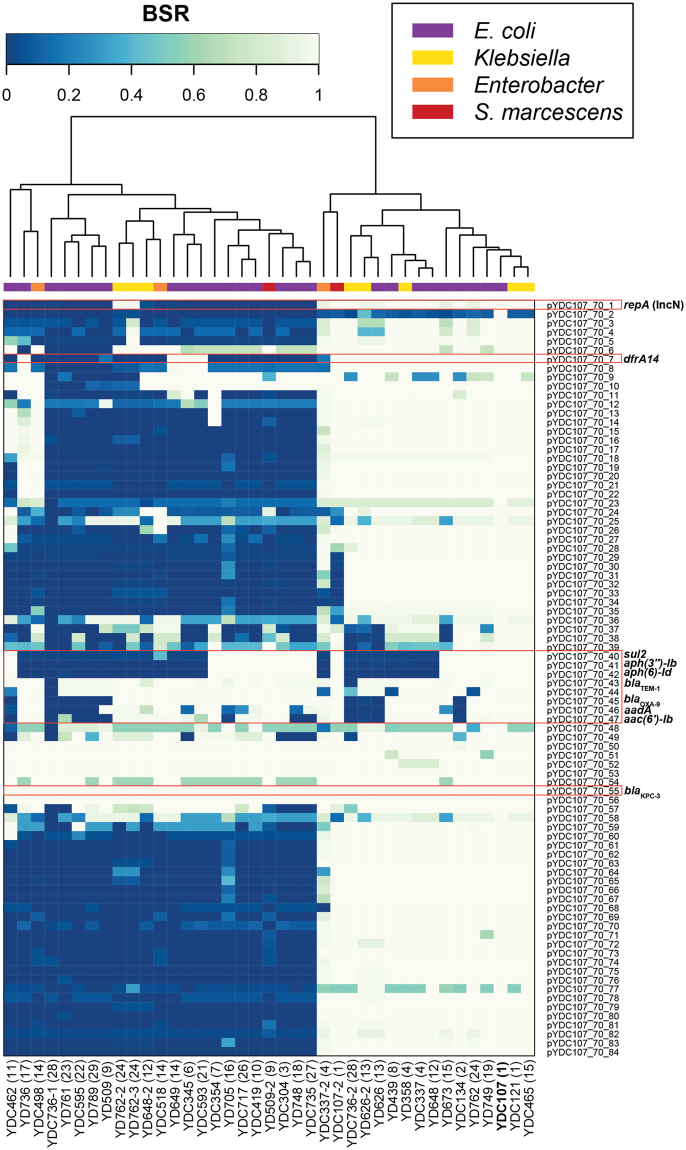
The blaKPC-containing contigs ranged in size from 6.5 to 78.8-kb, and 10 of the blaKPC-containing contigs were identified with markers of the IncFIIK plasmids (Supplementary Table S1). The blaKPC-containing contig of E. coli strain YD761 was most similar to an IncFII plasmid (GenBank accession no. CP000670), while the remaining blaKPC-containing contigs did not contain markers that could be used to identify them to a particular plasmid family (Supplementary Table S1). We further investigated the presence of genes associated with previously characterized blaKPC-containing plasmids in each of the E. coli and other bacterial species analyzed in this study, providing additional information regarding the diversity of potential blaKPC-containing plasmids in each genome. We used in silico analysis to identify genes of the blaKPC-3-containing IncN plasmid pYDC107_70 described in this study (Fig. 2), and also genes of the previously sequenced IncFIIK plasmid pKpQIL (GenBank accession no. GU595196.1) (Supplementary Fig. S4). In silico detection of the IncN plasmid pYDC107_70 demonstrated that this plasmid is present with significant similarity in 41% (16/39) of the blaKPC-containing E. coli and other bacterial species analyzed in this study, including nine of the E. coli and seven of the other bacterial species (Fig. 2). Detection of the IncFIIK plasmid pKpQIL43 demonstrated that 12 E. coli and one K. pneumoniae strain (YD648-2) had genes with significant similarity to this blaKPC-containing IncFIIK plasmid (Supplementary Fig. S4). The other four K. pneumoniae genomes analyzed (YDC121, YDC465, YD762-2, and YD762-3) exhibited similarity to genes from several of the regions of the plasmid, but were missing many of the genes of pKpQIL (Supplementary Fig. S4). Overall, 77% (30/39) of the blaKPC-containing E. coli and other bacterial species analyzed in this study have significant similarity to IncN and/or IncFIIK plasmids that carry the blaKPC gene (Supplementary Table S1, Figs 2 and S4).
Figure 2.
In silico detection of plasmid pYDC107_70 in the blaKPC-containing E. coli, and other bacterial species analyzed in this study. The heat map contains BSR values indicating the presence (very light green) or absence (dark blue) of each plasmid gene in each of the genomes. The heat map containing values clustered by column (genomes) was constructed with the heatmap.2 function of gplots using R v.3.4.1. Rows represent each of the protein-coding genes of pYDC107_70, while each column represents a different genome. The label of the E. coli YDC107 genome is indicated in bold. The species and/or genus of each genome is indicated by a square at the top of the heat map (see inset legend). The patient number (see Table S1) corresponding to each of the strains is indicated in parentheses next to the strain number.
Inter-genera dissemination of the blaKPC genes
Comparison of the blaKPC–containing plasmids among the different species isolated from the same or subsequent samples from the same patients demonstrated a contribution of IncN and IncFIIK plasmids to the dissemination of blaKPC among the species analyzed in this study. The IncN plasmid pYDC107_70 was present in all of the E. coli and other bacterial species isolated from patients 1, 4, 13, and 15 (Fig. 2). While the IncN plasmid was identified in both the E. coli and K. pneumoniae from patient 15, genes with similarity to those of the IncFIIK plasmid were also identified in these strains (Figs 2 and S4, Supplementary Table S1). Interestingly, the E. coli strains from patients 12 and 24 had genes with similarity to the IncN plasmid; however, the K. pneumoniae strains from each of these patients had genes with similarity to an IncFIIK plasmid (Figs 2 and S4, Supplementary Table S1). In contrast, an IncN plasmid was identified in K. michiganensis from patient 28, while the E. coli from this patient did not carry genes with similarity to the IncN plasmid or to the IncFIIK plasmid. The E. coli and other species characterized from patients 9 and 14 did not have genes with similarity to those of the IncN plasmid or to the IncFIIK plasmid (Supplementary Table S1), suggesting an uncharacterized plasmid may have been involved in the transfer of blaKPC among these strains. The blaKPC-3-containing contigs of E. coli strain YD509 and S. marcescens strain YD509-2 from patient 9 exhibited 100% nucleotide identity over 85 to 100% of the contig length compared to the 16.9 kb blaKPC-3-containing plasmid pBK28610 from E. coli strain BK28610, which was identified as a novel replicon (GenBank accession no. KU295136.1)23.
Additional antibiotic resistance genes and plasmids carried by the blaKPC-containing E. coli and other bacterial species
The E. coli and other bacterial species analyzed contained at least 37 different plasmid types, including members of the IncA/C, IncN, IncI1, IncFIIk, and IncFIB plasmid families (Supplementary S1), which have been previously described carrying antibiotic resistance genes44. Other types of resistance genes identified in the genomes included qnrS1, aadA, sul1, dfrA14, mphA, and cat, among many others, which confer resistance to fluoroquinolones, aminoglycosides, sulfonamides, trimethoprim, macrolides, and chloramphenicol, respectively (Supplementary Table S2). Comparison of the resistance gene content identified in strains from the same patient demonstrated that the strains contained many, but not all, of the same resistance genes (Supplementary Table S2). For example, among the three strains from Patient 14 (E. coli strain YD649, and E. hormaechei strains YDC498 and YDC518), all three of the genomes contained aadA, sul1, qnrA1, aac(6′)-Ib, ant(2″)-Ia (Supplementary Table S2). However, E. coli strain YD649 also contained a dihydrofolate reductase gene (dfrA14), which was absent from the E. hormaechei strains, while the E. hormaechei strains contained a chloramphenicol acetyltransferase gene (cat) that was absent from E. coli strain YD649 (Supplementary Table S2). In addition to having similar antibiotic resistance gene content, all three of the strains from Patient 14 also had an IncA/C2 plasmid (Supplementary Table S1, Supplementary Table S2), which have been previously characterized as large multidrug resistance plasmids44.
Completion genome sequencing of E. coli strain YDC107 allowed us to characterize and investigate the distribution of not only the blaKPC-containing plasmid from this E. coli strain, but also any additional plasmids harboring antibiotic resistance genes. In silico detection of the largest plasmid, pYDC107_184, among all of the blaKPC-containing sequenced in this study demonstrated that none of the other genomes analyzed had the entire plasmid; however, 21 of the other E. coli genomes had similarity to several regions of this plasmid (Supplementary Fig. S2, group II). The regions that were detected included genes from the antibiotic resistance region, conjugative transfer genes, and genes involved in plasmid stability including partitioning and toxin-antitoxin genes (Supplementary Fig. S2, Supplementary Table S4). The second largest plasmid of E. coli strain YDC107 is plasmid pYDC107_85, which is an IncI1 plasmid that has genes for conjugative transfer, but contains only a single antibiotic resistance gene (Supplementary Fig. S3, Supplementary Table S5). Detection of the pYDC107_85 genes in all of the E. coli and other bacterial species analyzed in this study demonstrated that many of the genes on this IncI1 plasmid are present in five of the other E. coli genomes (Supplementary Fig. S3). Several regions, including the region with the blaCMY-44 gene were absent from these five E. coli genomes; however, additional sequencing would be necessary to complete the IncI1 plasmids from these E. coli strains to determine whether the entire plasmid is present among these strains. Finally, detection of the smallest plasmid of E. coli strain YDC107, pYDC107_41, demonstrated that this plasmid is not present in any of the other blaKPC-containing E. coli or other bacterial species analyzed in this study.
Discussion
In the current study we used whole genome sequencing to gain insight into the mobile genetic elements and antibiotic resistance genes carried by blaKPC-containing E. coli and other bacterial species isolated from the same or subsequent patient samples. The blaKPC-containing E. coli strains analyzed in this study were isolated from at least seven different types of clinical samples and included not only members of the ST131 and ST648 lineages, which are well known to carry antibiotic resistance determinants13,16, but also included other genomically diverse E. coli strains. The majority (77%) of the blaKPC-containing genomes in the current study had genes with similarity to a blaKPC-containing IncN and/or IncFIIK plasmid. The IncFIIK plasmid family includes the pKpQIL-like plasmids, which have been implicated in the spread of blaKPC genes22,35,38,43,45,46. Also, a previous study demonstrated that IncN and IncFIIK plasmids were involved in the inter-genera transfer of blaKPC genes between K. pneumoniae and other bacterial species including E. coli, Enterobacter species, and Citrobacter species47. While the IncFIIK plasmids were prevalent among the E. coli analyzed in the current study22, the IncN plasmid was more frequently identified in all of the blaKPC-containing E. coli and other species from the same patient, suggesting this plasmid may have had a greater contribution to the inter-genera spread of blaKPC among the patients analyzed in this study. Interestingly, the E. coli from two patients carried an IncN plasmid while the other bacterial species from the same patients had an IncFIIK-like plasmid (patients 12 and 24), indicating multiple modes of acquisition of blaKPC among the bacterial species in these patients. Further functional studies are necessary to investigate whether the IncN plasmid may be more likely to be transferred between E. coli and other co-occurring bacterial species within a patient.
Complete genome sequencing of E. coli strain YDC107 allowed us to describe the blaKPC plasmid and other co-occurring plasmids in this strain. Interestingly, only the IncN blaKPC plasmid, pYDC107_70, from E. coli strain YDC107 appears to have been transferred and maintained among the two other blaKPC-containing bacterial species from this patient. Also, comparison of the blaKPC-3-containing IncN plasmid pYDC107_70 with the previously sequenced blaKPC-2-containing IncN plasmid from an E. coli that was isolated three years after the isolation of the E. coli strain YDC107 demonstrated that while these plasmids have a conserved backbone, they differ in their resistance gene content. This was similar to the comparison of pYDC107_70 among blaKPC-containing E. coli and other bacterial species analyzed in this study, which demonstrated that the plasmid was highly conserved, and most of the sequence variability was detected in the antibiotic resistance gene regions. Thus, the IncN plasmids involved in dissemination of blaKPC genes among patients in this health system have likely undergone changes in their resistance gene regions over time.
In summary, our findings demonstrate that blaKPC-containing IncN and IncFIIK plasmids are the most frequently identified plasmids among blaKPC-containing E. coli and other bacterial species from the same patients in a health system in Pennsylvania over 6 years. Whole-genome sequencing demonstrated the each of the blaKPC-containing E. coli and other species analyzed contained numerous antibiotic resistance genes that may be harbored on the same or a different plasmid than the blaKPC gene. Also, sequence analyses demonstrated that the blaKPC-containing IncN plasmid from patients in this health system has undergone modifications over time, which have occurred primarily in the resistance gene regions. Overall, our findings highlight the need for additional studies to investigate whether E. coli has an important role as a reservoir of blaKPC genes, which may be disseminated to co-occurring species that includes difficult-to-treat or outbreak-associated pathogens such as K. pneumoniae. Further studies are also necessary to better understand the dynamic nature of blaKPC-containing plasmids, and to determine how environmental and host factors can drive changes in resistance gene content and/or inter-genera plasmid dissemination, and whether these changes influence the clinical outcome of the patient.
Methods
Bacterial strains and antibiotic susceptibilities
Escherichia coli clinical strains that were reported as resistant to ertapenem were collected at the clinical microbiology laboratories at two teaching hospitals in Pittsburgh, PA between 2009 and 2015. Those that tested positive for blaKPC by conventional PCR were included in this study. When ertapenem-resistant strain(s) from other species were identified from the same patients from whom blaKPC-positive E. coli strains were identified and tested positive for blaKPC, these strains were also included. A limited number of the strains included in this study have been reported previously21,22,37. The blaKPC-containing strains analyzed in this study were tested for their susceptibility to 21 antibiotics by determining their minimum inhibitory concentrations (MIC) to each antibiotic using the Sensititre Gram-negative plates GNX2F (Thermo). The designations of susceptible, intermediate, or resistant were assigned based on the CLSI 2017 breakpoints for each species48.
Genome sequencing and assembly
Genomic DNA was extracted using the Sigma GenElute bacterial genomic DNA kit (Sigma-Aldrich; St. Louis, MO). All the genomes were sequenced using paired-end 500 bp insert libraries on the Illumina HiSeq. 4000 and the resulting 150 bp Illumina reads were assembled using SPAdes v.3.7.149. The final assemblies were filtered to contain only contigs that were ≥500 bp in length and had ≥5 × k-mer coverage. E. coli strain YDC107 was also sequenced using long-read sequencing to obtain a complete genome assembly, including any possible plasmids as previously described50.
In silico multilocus sequence typing, plasmid typing, serotyping, and antibiotic resistance gene detection
The MLST STs of each of the E. coli genome assemblies were determined based on the MLST scheme developed by Wirth et al.51. The sequences of the seven MLST loci (adk, gyrB, fumC, icd, mdh, purA, and recA) were located in each of the E. coli genomes using an in-house perl script. The sequences were queried against the BIGSdb database52 to obtain allele numbers, and the allelic profile of each strain was submitted to BIGSdb to obtain the ST for each of E. coli genomes analyzed (Supplementary Table S3). The STs of the K. pneumoniae genomes were determined by uploading each genome assembly to the BIGSdb whole-genome MLST prediction software on the Institut Pasteur website (http://bigsdb.pasteur.fr).
Plasmids were detected in each of the genome assemblies using PlasmidFinder v.1.353 using the default 95% nucleotide identity threshold. The molecular serotype of each E. coli genome was determined using SerotypeFinder v.1.1 (https://cge.cbs.dtu.dk/services/SerotypeFinder/) with the default settings of an 85% nucleotide identity threshold and 60% minimum alignment length54. Antibiotic resistance genes were detected in each of the genome assemblies using the resistance gene identifier (RGI) of the comprehensive antibiotic resistance database (CARD) v.1.1.8, with perfect or strict identification criteria55. The Tn4401 isoforms were determined by comparing the region surrounding each blaKPC gene to the sequences of the previously described Tn4401 isoforms and identifying the presence or absence of deletions that are characteristic of each isoform56.
Phylogenomic analysis
The 26 E. coli genomes sequenced in this study were compared with 30 publicly-available blaKPC-containing E. coli genomes as of March 2017, and a collection of 34 diverse E. coli and Shigella reference genomes using the single nucleotide polymorphism (SNP)-based In Silico Genotyper (ISG) as previously described57,58. The SNPs were predicted relative to the genome of E. coli strain IAI39 (GenBank accession number NC_011750.1) from phylogroup F. ISG identified 225,323 conserved SNP sites that were used to infer a maximum-likelihood phylogeny using RAxML v7.2.859, with the GTR model of nucleotide substitution, the GAMMA model of rate heterogeneity, and 100 bootstrap replicates.
In silico detection of plasmid genes
The predicted protein-coding genes on the E. coli strain YDC107 plasmids (pYDC107_184, pYDC107_85, pYDC107_70, and pYDC107_41) were identified in each of the genomes analyzed in this study using large-scale BLAST score ratio (LS-BSR) analysis as previously described60,61. The plasmid genes were compared to the E. coli genomes using BLASTN62.
Clustered heat maps were generated with the BSR values indicating the presence or absence of protein-coding genes of plasmids pYDC107_184, pYDC107_85, pYDC107_70, and the IncFIIK plasmid pKpQIL (Genome accession no. GU595196.1) in each of genomes analyzed. The heat maps were generated using the heatmap.2 function of gplots v. 3.0.1 in R v. 3.4.1, and the genomes in each heat map were clustered using the default complete linkage method with Euclidean distance estimation. The plasmid map of pYDC107_70 was generated using Circos 0.69-463. The heat map tracks of the circular plasmid plot contain the BLASTN BSR values of each of the protein-coding genes of pYDC107_70 compared with the plasmids and genomes described in the figure legend.
Data availability
The genome assemblies generated in this study are deposited in GenBank under the accession numbers listed in Supplementary Table S1.
Electronic supplementary material
Acknowledgements
The authors thank Jane Michalski for laboratory assistance. This project was funded in part by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under grant number U19 AI110820, R01AI104895, and R21AI123747.
Author Contributions
T.H.H., Y.D., and D.A.R. contributed to the study design and wrote the manuscript. T.H.H., R.M., C.L.M., S.L.B., and S.N. generated data. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-28085-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yohei Doi, Email: yod4@pitt.edu.
David A. Rasko, Email: drasko@som.umaryland.edu
References
- 1.Falagas ME, Tansarli GS, Karageorgopoulos DE, Vardakas KZ. Deaths attributable to carbapenem-resistant Enterobacteriaceae infections. Emerg Infect Dis. 2014;20:1170–1175. doi: 10.3201/eid2007.121004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9:228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 3.Woodford N, et al. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A beta-lactamase, KPC-3, in a New York Medical Center. Antimicrob Agents Chemother. 2004;48:4793–4799. doi: 10.1128/AAC.48.12.4793-4799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snitkin ES, et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agodi A, et al. Containment of an outbreak of KPC-3-producing Klebsiella pneumoniae in Italy. J Clin Microbiol. 2011;49:3986–3989. doi: 10.1128/JCM.01242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marsh JW, et al. Genomic Epidemiology of an Endoscope-Associated Outbreak of Klebsiella pneumoniae Carbapenemase (KPC)-Producing K. pneumoniae. PLoS One. 2015;10:e0144310. doi: 10.1371/journal.pone.0144310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang R, et al. Outbreak of Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae with high qnr prevalence in a Chinese hospital. J Med Microbiol. 2011;60:977–982. doi: 10.1099/jmm.0.015826-0. [DOI] [PubMed] [Google Scholar]
- 8.Cuzon G, Ouanich J, Gondret R, Naas T, Nordmann P. Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob Agents Chemother. 2011;55:2420–2423. doi: 10.1128/AAC.01452-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balkhy HH, et al. The epidemiology of the first described carbapenem-resistant Klebsiella pneumoniae outbreak in a tertiary care hospital in Saudi Arabia: how far do we go? Eur J Clin Microbiol Infect Dis. 2012;31:1901–1909. doi: 10.1007/s10096-011-1519-0. [DOI] [PubMed] [Google Scholar]
- 10.Mammina C, et al. Outbreak of infection with Klebsiella pneumoniae sequence type 258 producing Klebsiella pneumoniae Carbapenemase 3 in an intensive care unit in Italy. J Clin Microbiol. 2010;48:1506–1507. doi: 10.1128/JCM.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Logan LK, Weinstein RA. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J Infect Dis. 2017;215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 13.Woodford N, Turton JF, Livermore DM. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 14.Queenan, A. M. & Bush, K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev20, 440–458, table of contents, 10.1128/CMR.00001-07 (2007). [DOI] [PMC free article] [PubMed]
- 15.Davies TA, et al. Longitudinal survey of carbapenem resistance and resistance mechanisms in Enterobacteriaceae and non-fermenters from the USA in 2007-09. J Antimicrob Chemother. 2011;66:2298–2307. doi: 10.1093/jac/dkr290. [DOI] [PubMed] [Google Scholar]
- 16.Cerqueira GC, et al. Multi-institute analysis of carbapenem resistance reveals remarkable diversity, unexplained mechanisms, and limited clonal outbreaks. Proc Natl Acad Sci USA. 2017;114:1135–1140. doi: 10.1073/pnas.1616248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yigit H, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hargreaves ML, et al. Clonal Dissemination of Enterobacter cloacae Harboring blaKPC-3 in the Upper Midwestern United States. Antimicrob Agents Chemother. 2015;59:7723–7734. doi: 10.1128/AAC.01291-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hossain A, et al. Plasmid-mediated carbapenem-hydrolyzing enzyme KPC-2 in an Enterobacter sp. Antimicrob Agents Chemother. 2004;48:4438–4440. doi: 10.1128/AAC.48.11.4438-4440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris D, et al. Production of KPC-2 carbapenemase by an Escherichia coli clinical isolate belonging to the international ST131 clone. Antimicrob Agents Chemother. 2011;55:4935–4936. doi: 10.1128/AAC.05127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim YA, et al. Features of infections due to Klebsiella pneumoniae carbapenemase-producing Escherichia coli: emergence of sequence type 131. Clin Infect Dis. 2012;55:224–231. doi: 10.1093/cid/cis387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Hara JA, et al. Molecular epidemiology of KPC-producing Escherichia coli: occurrence of ST131-fimH30 subclone harboring pKpQIL-like IncFIIk plasmid. Antimicrob Agents Chemother. 2014;58:4234–4237. doi: 10.1128/AAC.02182-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chavda KD, Chen L, Jacobs MR, Bonomo RA, Kreiswirth BN. Molecular Diversity and Plasmid Analysis of KPC-Producing Escherichia coli. Antimicrob Agents Chemother. 2016;60:4073–4081. doi: 10.1128/AAC.00452-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, et al. Complete sequence of a KPC-producing IncN multidrug-resistant plasmid from an epidemic Escherichia coli sequence type 131 strain in China. Antimicrob Agents Chemother. 2014;58:2422–2425. doi: 10.1128/AAC.02587-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miriagou V, et al. Imipenem resistance in a Salmonella clinical strain due to plasmid-mediated class A carbapenemase KPC-2. Antimicrob Agents Chemother. 2003;47:1297–1300. doi: 10.1128/AAC.47.4.1297-1300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robledo IE, et al. Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob Agents Chemother. 2010;54:1354–1357. doi: 10.1128/AAC.00899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villegas MV, et al. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing beta-lactamase. Antimicrob Agents Chemother. 2007;51:1553–1555. doi: 10.1128/AAC.01405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirel L, Nordmann P, Lagrutta E, Cleary T, Munoz-Price LS. Emergence of KPC-producing Pseudomonas aeruginosa in the United States. Antimicrob Agents Chemother. 2010;54:3072. doi: 10.1128/AAC.00513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuzon G, Naas T, Nordmann P. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother. 2011;55:5370–5373. doi: 10.1128/AAC.05202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naas T, et al. Genetic structures at the origin of acquisition of the beta-lactamase blaKPC gene. Antimicrob Agents Chemother. 2008;52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathers, A. J. et al. Chromosomal Integration of the Klebsiella pneumoniae Carbapenemase Gene, blaKPC, In Klebsiella Species Is Elusive but Not Rare. Antimicrob Agents Chemother61, 10.1128/AAC.01823-16 (2017). [DOI] [PMC free article] [PubMed]
- 32.Shen P, et al. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob Agents Chemother. 2009;53:4333–4338. doi: 10.1128/AAC.00260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, et al. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li JJ, Lee CS, Sheng JF, Doi Y. Complete sequence of a conjugative Incn plasmid harboring blaKPC-2, blaSHV-12, and qnrS1 from an Escherichia coli sequence type 648 strain. Antimicrob Agents Chemother. 2014;58:6974–6977. doi: 10.1128/AAC.03632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, et al. Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York Hospitals. Antimicrob Agents Chemother. 2014;58:2871–2877. doi: 10.1128/AAC.00120-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoesser N, et al. Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli. Sci Rep. 2017;7:5917. doi: 10.1038/s41598-017-06256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sidjabat HE, et al. Interspecies spread of Klebsiella pneumoniae carbapenemase gene in a single patient. Clin Infect Dis. 2009;49:1736–1738. doi: 10.1086/648077. [DOI] [PubMed] [Google Scholar]
- 38.Ruiz-Garbajosa P, et al. Multiclonal dispersal of KPC genes following the emergence of non-ST258 KPC-producing Klebsiella pneumoniae clones in Madrid, Spain. J Antimicrob Chemother. 2013;68:2487–2492. doi: 10.1093/jac/dkt237. [DOI] [PubMed] [Google Scholar]
- 39.Cuzon G, et al. Worldwide diversity of Klebsiella pneumoniae that produce beta-lactamase blaKPC-2 gene. Emerg Infect Dis. 2010;16:1349–1356. doi: 10.3201/eid1609.091389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitchel B, et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother. 2009;53:3365–3370. doi: 10.1128/AAC.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob Agents Chemother. 2010;54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob Agents Chemother. 2010;54:4493–4496. doi: 10.1128/AAC.00175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leavitt A, Chmelnitsky I, Ofek I, Carmeli Y, Navon-Venezia S. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. J Antimicrob Chemother. 2010;65:243–248. doi: 10.1093/jac/dkp417. [DOI] [PubMed] [Google Scholar]
- 46.Doumith, M. et al. Major role of pKpQIL-like plasmids in the early dissemination of KPC-type carbapenemases in the UK. J Antimicrob Chemother, 10.1093/jac/dkx141 (2017). [DOI] [PubMed]
- 47.Adler A, Khabra E, Paikin S, Carmeli Y. Dissemination of the blaKPC gene by clonal spread and horizontal gene transfer: comparative study of incidence and molecular mechanisms. J Antimicrob Chemother. 2016;71:2143–2146. doi: 10.1093/jac/dkw106. [DOI] [PubMed] [Google Scholar]
- 48.Clinical and Laboratory Standards Institute In CLSI document M100-S21 (Clinical and Laboratory Standards Institute, Wayne, Pennsylvania, 2017).
- 49.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hazen, T. H., Michalski, J., Nagaraj, S., Okeke, I. N. & Rasko, D. A. Characterization of a Large Antibiotic Resistance Plasmid Found in Enteropathogenic Escherichia coli Strain B171 and Its Relatedness to Plasmids of Diverse E. coli and Shigella Strains. Antimicrob Agents Chemother61, 10.1128/AAC.00995-17 (2017). [DOI] [PMC free article] [PubMed]
- 51.Wirth T, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carattoli A, et al. In silico detection and typing of plasmids using Plasmid Finder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joensen KG, Tetzschner AM, Iguchi A, Aarestrup FM, Scheutz F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J Clin Microbiol. 2015;53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia B, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naas T, Cuzon G, Truong HV, Nordmann P. Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob Agents Chemother. 2012;56:4753–4759. doi: 10.1128/AAC.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hazen TH, et al. Refining the pathovar paradigm via phylogenomics of the attaching and effacing Escherichia coli. Proc Natl Acad Sci USA. 2013;110:12810–12815. doi: 10.1073/pnas.1306836110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sahl JW, et al. A comparative genomic analysis of diverse clonal types of enterotoxigenic Escherichia coli reveals pathovar-specific conservation. Infect Immun. 2011;79:950–960. doi: 10.1128/IAI.00932-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 60.Sahl JW, Caporaso JG, Rasko DA, Keim P. The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. PeerJ. 2014;2:e332. doi: 10.7717/peerj.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hazen TH, et al. Genomic diversity of EPEC associated with clinical presentations of differing severity. Nature Microbiology. 2016;1:15014. doi: 10.1038/nmicrobiol.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 63.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome assemblies generated in this study are deposited in GenBank under the accession numbers listed in Supplementary Table S1.