Abstract
Multiple protozoans produce homologs of the cytokine MIF which play a role in immune evasion, invasion and pathogenesis. However, how parasite-encoded MIF activity is controlled remains poorly understood. Cytokine activity can be inhibited by intracellular binding partners that are released in the extracellular space during cell death. We investigated the presence of an endogenous parasite protein that was capable of interacting and interfering with MIF activity. A screen for protein-protein interaction was performed using immunoaffinity purification of amebic cell lysate with specific anti-Entamoeba histolytica MIF (EhMIF) antibody followed by mass spectrometry analysis, which revealed an E. histolytica-produced JAB1 protein (EhJAB1) as a potential binding partner. JAB1 was found to be highly conserved in protozoans. Direct interaction between the EhMIF and EhJAB1 was confirmed by several independent approaches with GST pull-down, co-immunoprecipitation, and Biolayer interferometry (BLI) assays. Furthermore, the C-terminal region outside the functional JAMM deneddylase motif was required for EhMIF binding, which was consistent with the top in silico predictions. In addition, EhJAB1 binding blocked EhMIF-induced IL-8 production by human epithelial cells. We report the initial characterization of a parasite-encoded JAB1 and uncover a new binding partner for a protozoan-produced MIF protein, acting as a possible negative regulator of EhMIF.
Introduction
Protozoan parasites represent a major threat to health and contribute significantly to morbidity and mortality worldwide. For example, Entamoeba histolytica is a protozoan parasite that causes colitis1. Severe forms of amebic colitis are associated with high case fatality rates ranging from 40% to 89%2. There is neither an effective vaccine nor have there been advancements in therapies for amebic colitis for over fifty years3. Therefore, there remains an ongoing need to find new drug and vaccine targets through a better understanding of parasite biology.
Macrophage migration inhibitory factor (MIF), one of the first cytokines to be discovered, is a pleiotropic inflammatory cytokine and a critical upstream mediator of innate immunity. Many of the inflammatory effects of MIF are mediated through direct binding to the CD74 cell surface receptor, causing the secretion of proinflammatory cytokines such as IL-84–6. An increase in MIF expression contributes to excessive inflammation and immunopathology. Hence, MIF has been reported to have a role in the pathogenesis of several inflammatory diseases such as inflammatory bowel disease and rheumatoid arthritis7,8. MIF proinflammatory properties also make it a crucial mediator in the immune response against a wide range of pathogens9–11.
Counterintuitively, MIF homologs have been characterized in several pathogenic protozoans including Entamoeba, Plasmodium, Toxoplasma, and Leishmania. These protozoan MIF homologs have demonstrated similar proinflammatory activities to that of human MIF, and play a role in immune evasion, invasion and pathogenesis12–19. Despite the growing literature on protozoan-encoded MIF proinflammatory activity, very little is known about how it is regulated. Here, we uncover a parasite-encoded JAB1 (c-Jun activation domain binding protein 1), which is highly conserved throughout protozoan parasites, as a novel binding partner and potential regulator of the MIF homolog of Entamoeba histolytica.
Results
Identification and characterization of a parasite-produced JAB1
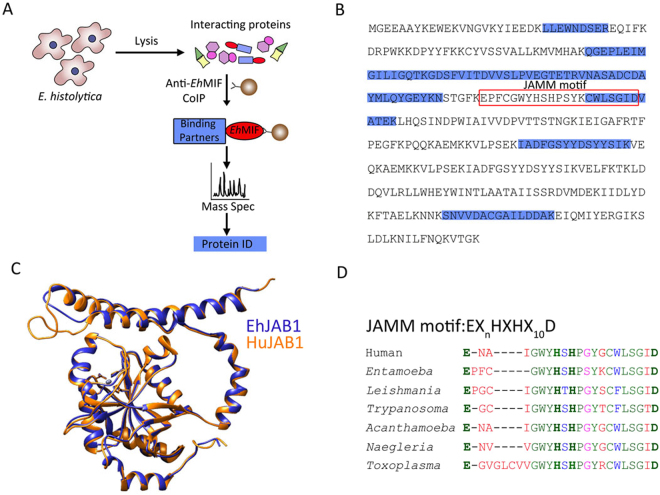
Co-immunoprecipitation (co-IP) of proteins followed by mass spectrometric identification is a standard approach for identifying novel protein-protein interactions20,21. Parasite cell lysates were incubated with a specific antibody against Entamoeba histolytica MIF (EhMIF) or IgG control antibody. The immunoprecipitates were subjected to mass spectrometry analysis. An Entamoeba histolytica-encoded JAB1 (EhJAB1, EHI_050500) was identified as a potential EhMIF binding partner (Fig. 1A,B). While a number of putative interacting proteins were co-precipitated with anti-EhMIF antibody, EhJAB1 was selected for further analysis given: (i) EhJAB1 was detected in the anti-EhMIF co-IP sample and not detected in the control IP; (ii) EhJAB1 was among the highest percent coverage (Supp. Fig. S1); and (iii) previous report of the intracellular interaction between human MIF and human JAB1 (HuJAB1)22–24. EhJAB1 gene consists of 957 base pairs with no intron and a GC content of 30%. The gene encodes for a protein of a predicted molecular weight of 36.5 kDa.EhJAB1 has 39% identity and 61% similarity with HuJAB1 and appears structurally conserved (Figs 1C and S2). JAB1 is conserved in multiple parasites that cause disease in humans, all containing the MPN domain with the JAMM (JAB1/MPN/Mov34 metalloenzyme) motif (Fig. 1D). JAMM motif is a metalloprotease motif, consisting of the amino acid sequence EXnHXHX10D, present in the deubiquitinating enzymes RPN11 and JAB125. RPN11 was also found is these parasites (Supp. Fig. S3).
Figure 1.
Characterization of EhJAB1. (A) Schematic of procedure for identifying novel protein-protein interaction with co-immunoprecipitation followed by mass spectrometric analysis. (B) E. histolytica JAB1 (EhJAB1) amino acid sequence. Peptides unique to the EhJAB1 protein identified by mass spectrometry are highlighted (blue). JAMM motif (EXnHXHX10D) shown in red box. (C) Structural homology between Human JAB1 (HuJAB1) and EhJAB1. HuJAB1 (orange) was superimposed with the predicted structure of EhJAB1 (blue). (D) Multiple sequence alignment of the conserved JAMM motif of JAB1 from pathogenic parasites. Identical (green), conserved (blue), semi-conserved (pink), and non-conserved residues (red).
EhMIF directly interacts with EhJAB1
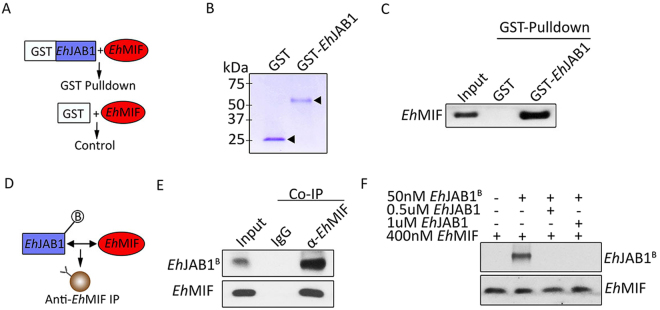
We investigated whether the interaction between EhMIF and EhJAB1 was direct or indirect. First, we performed GST-pulldown assays (Fig. 2A). EhMIF and EhJAB1 fused to GST were expressed and purified (Fig. 2B). GST-EhJAB1 immobilized on magnetic beads was incubated with EhMIF. GST alone immobilized on magnetic beads and incubated with the same concentration of EhMIF was used as control. As shown in Fig. 2C, the GST-EhJAB1 fusion protein readily pulled down EhMIF relative to the GST control, demonstrating direct interaction. In an independent approach, we used co-immunoprecipitation experiments to further verify direct interaction. EhJAB1 immunoprecipitated with anti-EhMIF antibody, but not with the IgG antibody control (Fig. 2D–F). Together, these findings suggest a direct binding between EhMIF and EhJAB1 proteins.
Figure 2.
Direct interaction between EhMIF and EhJAB1. (A) Schematic of GST pull-down assay. (B) Purified recombinant GST and GST-EhJAB1 proteins (arrowhead) used in the GST pull-down assay were separated by SDS/PAGE and stained with Coomassie Blue. (C) Interaction between EhMIF and EhJAB1 by GST pull-down assay. GST-EhJAB1 and GST control were mixed with EhMIF. Input (10%) and pull-down material were separated by SDS/PAGE, and EhMIF was detected by immunoblot analysis using anti-EhMIF antibody. (D) Schematic diagram of co-immunoprecipitation assay. (E,F) Co-immunoprecipitation of EhMIF and biotinylated EhJAB1 (EhJAB1B) mixture using specific anti-EhMIF antibody bound beads. EhJAB1B was detected by immunoblot analysis using goat anti-biotin HRP conjugated antibody.
High-affinity binding between EhMIF and EhJAB1 proteins
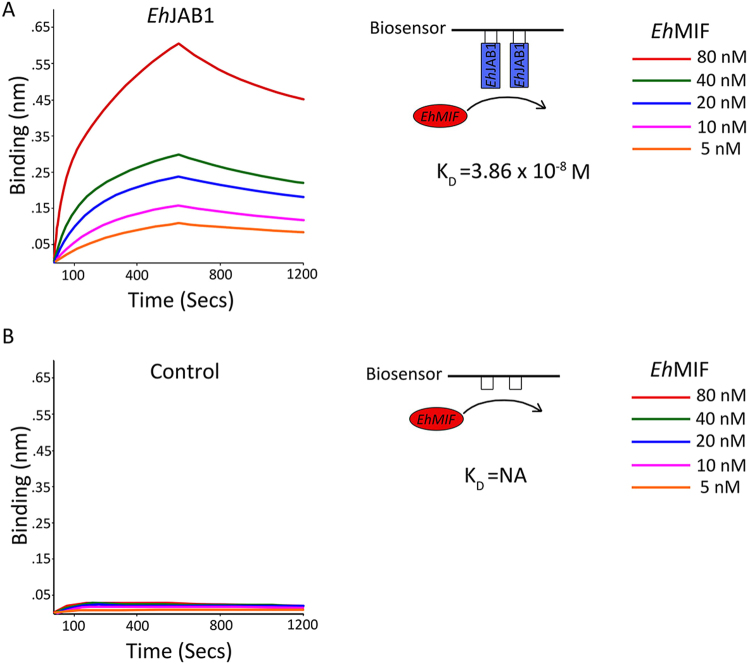
Biolayer interferometry (BLI) is a useful technique for measuring interactions between proteins in real time26. We determined the equilibrium dissociation constant for EhMIF binding to EhJAB1 using BLI. GST-tagged EhJAB1 was coupled to the surface of anti-GST antibody coated BLI sensors, followed by binding measurements in different concentrations of EhMIF. Analysis revealed a dissociation constant of KD of 3.86 × 10−8 M (Fig. 3A). GST alone coupled to anti-GST antibody coated BLI sensors was used as control. BLI measurements demonstrate that EhMIF did not bind to the GST control, KD not applicable (Fig. 3B). These findings suggest EhMIF binds to EhJAB1 within the range considered biologically relevant27,28.
Figure 3.
Characterization of EhMIF binding to EhJAB1 using the Biolayer interferometry. (A) Representative binding and dissociation curves for EhMIF binding to EhJAB1. Anti-GST antibody-coated biosensors were loaded with GST-EhJAB1. Sensors were placed into solutions with EhMIF, concentrations range from 5 to 80 nM (association analysis: 0 to 600 secs). Subsequently, the sensors were transferred to buffer without EhMIF for dissociation analysis (from 600 to 1200 secs). Analysis revealed a dissociation constant of KD of 3.86 × 10−8 M. (B) Biosensors loaded with GST only were used as controls.
EhJAB1 interacts with EhMIF via its C-terminal domain
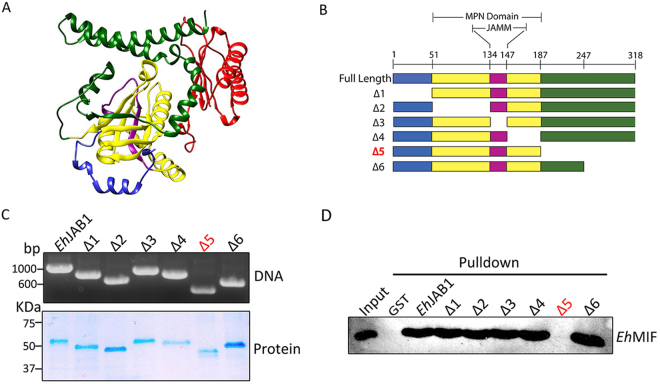
We investigated the binding domain of EhJAB1 responsible for interacting with EhMIF. First, an in silico approach using pyDockWEB was applied to assess the binding of EhJAB1 to EhMIF. pyDockWEB allows the best rigid-body docking orientations generated by pyDock scoring function, which consists of electrostatics, desolvation energy and limited van der Waals contribution29. The C-terminal domain of EhJAB1 was revealed to be the EhMIF binding region in all top ten predictions (Figs 4A and S6). Next, we further examine the region of EhJAB1 that is responsible for the formation of the complex with EhMIF using deletion mutant analysis. GST-tagged full-length EhJAB1 and various GST-tagged deletion mutants of EhJAB1, with either N-terminal, MPN, JAMM or C-terminal domain deletion (Fig. 4B,C), were immobilized on magnetic beads and incubated with EhMIF protein. EhMIF specifically interacted with GST–EhJAB1, except for the mutant lacking the C-terminal domain. Interaction was regained when the 187–246 amino acid sequence of the C-terminal was restored (Fig. 4D). These results indicate binding of EhJAB1 to EhMIF is mediated through the C-terminal region outside of the MPN domain.
Figure 4.
Mapping the EhMIF-interacting domain of EhJAB1. (A) Predicted interaction between EhMIF and EhJAB1. EhMIF (red) and EhJAB1 N-terminal (blue), MPN domain (yellow), JAMM motif (purple) and C-terminal (green). (B) Schematic representations of full-length and deletion mutants of EhJAB1. Δ 1 (deletion of 3–50 aa), Δ 2 (deletion of 51–133 aa), Δ 3 (deletion of 134–147 aa), Δ 4 (deletion of 148–187 aa), Δ 5 (deletion of 187–318 aa), Δ 6 (deletion of 247–318 aa). (C) PCR amplicons and protein expression of GST-tagged full-length EhJAB1 and GST-tagged deletion mutants of EhJAB1. (D) Interactions of EhMIF with deletion mutants were analyzed by immunoblotting.
EhJAB1 inhibits EhMIF interaction with the CD74 receptor and EhMIF-induced cytokine production
CD74 is a cell surface receptor for human MIF, which mediates many of its inflammatory effects30. Several parasite MIF homologs, including EhMIF, were previously shown to interact with the CD74 receptor13,17,18,31. Here, we were able to reproduce this finding and found that EhMIF-CD74 interaction was blocked by preincubating EhMIF with EhJAB1 (Fig. 5A). The epithelial surfaces of the skin, nasal, intestinal, respiratory, and genitourinary tracts are the first points of contact for many protozoans. Epithelial cells express CD74 and are a rich source of IL-832. Stimulation of IL-8 production has been the most reproducible activity of protozoan MIF homologs. EhMIF was recently shown to stimulate IL-8 secretion from human intestinal epithelial cells19. IL-8 is a potent neutrophil chemoattractant that contributes to inflammation in various infectious and inflammatory diseases. We proceeded to examine if EhJAB1 binding to EhMIF attenuates its proinflammatory function using two different colonic epithelial cell lines. EhJAB1 blocked EhMIF-induced IL-8 secretion by HCT116 and Caco2 colonic cells (Fig. 5B,C). These data indicate that when bound to EhJAB1, EhMIF is no longer capable of carrying out its inflammatory functions.
Figure 5.
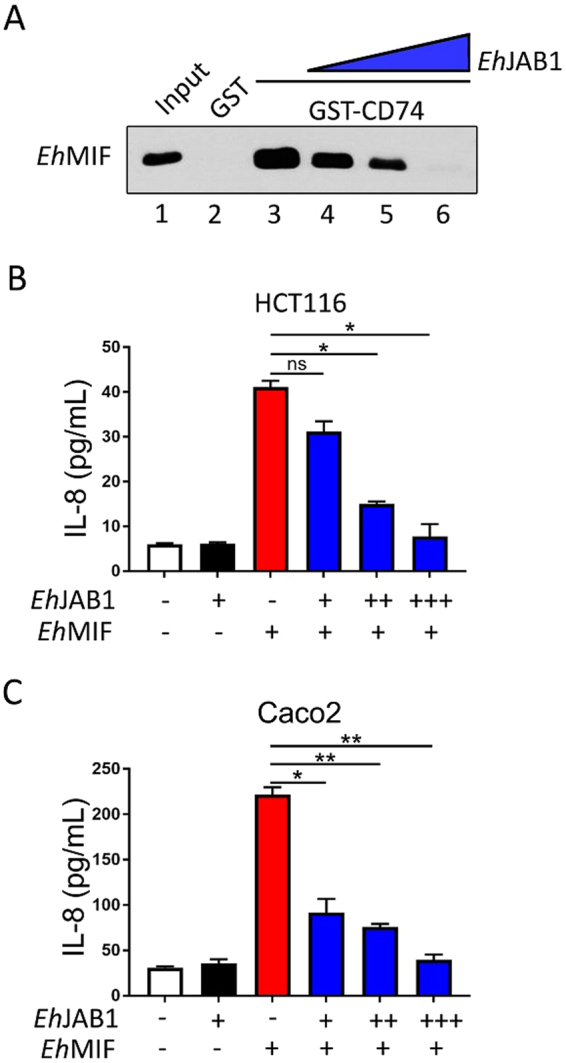
Effect of EhJAB1 binding on EhMIF activity. (A) EhJAB1 inhibits EhMIF interaction with the human MIF receptor CD74 by GST pull-down competition assay. Input (lane 1) and GST control (lane 2). EhMIF was preincubated with or without EhJAB1 at increasing doses before mixing with GST-CD74 (lanes 3–6). (B,C) EhJAB1 inhibits EhMIF-induced IL-8 production in a dose-dependent manner. Human colonic epithelial cells HCT116 and Caco2 were incubated with EhMIF with or without EhJAB1 at increasing doses. Culture supernatants were collected after 8 h and IL-8 was quantified by ELISA. Data represent mean and SD of triplicates from 1 experiment and are representative of 3 independent experiments. *P < 0.01; **P < 0.001. ns, not significant.
Discussion
Parasites can exploit the host inflammatory response to promote tissue invasion1. EhMIF-stimulated human intestinal epithelial cells secrete IL-8, a potent promoter of inflammation19. One of the downstream effects of E. histolytica MIF-induced inflammation is an increase in matrix metalloproteinases (MMPs) production, which was shown recently to promote E. histolytica tissue invasion in human colon19,33. That said, parasites are likely to have developed mechanisms to regulate their MIF actions especially in situations where they fail to evade the host inflammatory response induced by producing such a molecule.
Identifying a binding partner that inhibits MIF function provides insight into how MIF’s actions are regulated27. In this study, a proteomics approach was used to identify parasite-encoded protein interaction partners of a homolog of MIF from Entameoba histolytica. We found that EhJAB1 protein interacts with EhMIF. This interaction was validated by multiple independent approaches with GST-pulldowns, co-immunoprecipitation, in silico experiments and Biolayer interferometry.
JAB1, also known as COP9 signalosome subunit 5 (CSN5), is well characterized in non-parasitic eukaryotes34. It constitutes the catalytic center of the large multi-protein COP9 signalosome complex, since it harbors the JAMM/MPN+ metalloprotease motif35–37. It carries out a Zn dependent reaction called deneddylation, equivalent to de-ubiquitylation, where it removes a ubiquitin (Ub) like protein Nedd8 from the Cullin-RING E3 Ub-ligases (CRL)35,38,39. Regulation of CRLs, such as the Skp/Cullin/F-box (SCF) complex, by CSN mediated deneddylation is critical for proper cell division, cell cycle control and DNA damage response40,41. Here, we report the initial characterization of a parasite-encoded JAB1. Given parasites including E. histolytica express cullins and Nedd8 proteins42–45, it is plausible that JAB1 in parasites regulate cullins by deneddylation and modulate ubiquitin proteasomal system (UPS) activity, however, further studies to confirm this are needed. Surprisingly, we did not find a JAB1/CSN5 homolog in Plasmodium genome. However, the incomplete genome assembly and annotation in Plasmodium spp. could limit our in-silico analysis and explain why JAB1 was not identified in Plasmodium spp.
JAB1 function is not limited to CSN dependent deneddylation. JAB1 is also stable and functional in a free, monomeric, CSN independent form46–48. JAB1 monomers are catalytically inactive, but bind to certain proteins and alter their activity34,48. We showed that the JAMM catalytic motif of EhJAB1 was not required for its interaction with EhMIF, which is consistent with the previous report on mammalian proteins23. The dissociation constant for EhJAB1-EhMIF complex was well within the range considered physiologically relevant, suggesting a biologically significant interaction27,28.
Intracellular molecules released from damaged cells can have profound effects on the immune response. There is growing evidence that not all of these immunomodulatory molecules are pro-inflammatory, and interestingly, some have immunosuppressive or regulatory effects. These anti-inflammatory actions occur either directly or indirectly. For example, in the case of human MIF, an endogenous binding partner that is released in the extracellular space during cell death interacts and negatively interferes with MIF activity27,49–51. JAB1 is an intracellular protein22, which is consistent with our mass spectrometric analysis of the parasite cytosolic fraction; while MIF homologs, such as EhMIF are secreted proteins18,19. E. histolytica parasite has developed a number of mechanisms to evade the host immune response1,52. However, amebic parasites could become damaged if they fail to evade the inflammatory response triggered by EhMIF. It would be reasonable to speculate that the free monomeric EhJAB1 released from injured cells into the extracellular environment would then form complexes with EhMIF, preventing interaction with the host receptor CD74 and reducing EhMIF-induced inflammation, as a negative feedback mechanism. In addition, given the structural and functional similarity between human and E. histolytica MIF and JAB1 proteins, we do postulate that EhJAB1 could also interact with human MIF. It would be interesting to determine whether an EhJAB1/human MIF interaction functions to evade the host immune response in future studies.
In conclusion, our data suggest that E. histolytica homolog of JAB1 interacts with the cytokine EhMIF and negatively regulates its pro-inflammatory function. Our study also generates the hypothesis that targeting EhMIF and its interaction with EhJAB1 may disrupt the parasite’s ability to exploit the host immune response and ultimately serve as a therapeutic target against this devastating parasitic disease.
Methods
Plasmids, cloning and PCR
The EhJAB1 gene, codon optimized for expression in E.coli BL21(DE3) cells, was cloned within pDEST15 vector. Deletion constructs were prepared by inverse PCR on the pDEST15-full length JAB1 clone using the primers listed in the Supp. Fig. S4. A schematic of the mutation strategy is provided in the Supp. Fig. S5. Clones were screened by PCR across the gene boundaries within the vector followed by confirmation with sequencing. The EhJAB1 gene was amplified from the pDEST15 vector with primers carrying 5′BamHI and 3′XhoI sites and sub-cloned within BamHI and XhoI sites of pGEX-4T1 vector to utilize the thrombin site for cleaving off the GST tag from the GST-EhJAB1 protein. Also, the previously described codon optimized EhMIF gene cloned within pJexpress414 vector (DNA2.0)17 was used in this study.
Protein expression and purification
Protein expression of the recombinant EhMIF and EhJAB1was done following the previously described protocol17 except that the induction with isopropyl β- D –thiogalactoside (IPTG) was done for 18 hours at 15 °C. Cells were pelleted and lysed in CelLyticTM B Cell Lysis Reagent (Sigma) at room temperature for 15 minutes and lysate was collected following 30 min spin at maximum speed at 4 °C. The purification of GST-fusion protein and His-tagged fusion protein was done as previously described17 using glutathione-sepharose (GE-Healthcare) and Ni-NTA agarose beads (Qiagen) respectively. Polymyxin B (Sigma) was used in the purification procedures for the removal of endotoxin. On-column cleavage of the GST-EhJAB1 protein was done with the Thrombin Cleavage Capture Kit (Millipore) that utilizes biotinylated thrombin for its streptavidin agarose based removal.
GST-Pulldown Assay
GST pull down assays were done using the MagneGSTTM Protein Purification System kit (Promega). Bacterial lysate expressing GST fused- full length or deletion mutant EhJAB1 protein, diluted in MagneGST binding/wash buffer to a concentration 500 ng/ml in 500 µl, was mixed with 25 µl magnetic beads and was incubated overnight at 4 °C with rotation. Beads were washed 3 times with 500 µl binding/wash buffer followed by incubation with 1 µg purified EhMIF at 4 °C for overnight. The GST-CD74 pulldown assays were performed as previously described17,31. Briefly, EhMIF protein was pre-incubated with or without 1X, 2X or 5X concentration of EhJAB1 protein for 30 minutes in 500 µl binding/wash buffer prior to incubation with the GST-CD74 bound magnetic beads at 4 °C for overnight. The bound complexes were eluted in 40 µl elution buffer after washing the beads 5 times with 500 µl binding/wash buffer. In a control experiment equivalent amount of GST protein was used in place of GST-EhJAB1 or GST-CD74 proteins. Twenty five percent of the eluted samples were assayed by immunoblots.
Immunoprecipitation
Approximately 1.25 × 107 trophozoites of E. histolytica, grown at 37 °C in TYI-S-33 medium, were harvested in 3 ml non-denaturing lysis buffer (20 mM Tris HCl pH 8, 137 mM NaCl, 1% TritonX-100, 2 mM EDTA, 1x Protease inhibitor freshly added). Lysis was done with sonication followed by centrifugation at high speed for 10 minutes at 4 °C. Immunoprecipitations were performed with anti-EhMIF rabbit serum17 and rabbit IgG (Santa Cruz Biotechnology). For each, 100 µl DynabeadsTM Protein A (Invitrogen) was chemically conjugated with 10 µg antibodies using 5 mM Bis[Sulfosuccinimidyl] suberate or BS3 (Thermo Scientific) following the manufacturer’s manual. Antibody conjugated beads were added to 500 µl amebic lysate and incubated overnight at 4 °C with rotation. Following binding, beads were washed 5 times with 300 µl wash buffer (10 mM Tris HCl pH 7.4, 150 mM NaCl, 1% TritonX-100, 1 mM EDTA, 1x Protease inhibitor freshly added). The bound complexes were eluted in 40 µl 1x Laemmli sample buffer (Bio-Rad) by boiling. Co-precipitated endogenous proteins were analyzed by Mass-Spectrometry. For direct binding experiment, tag free EhJAB1protein was biotinylated using the EZ-LinkR Sulfo-NHS-LC-Biotinylation kit (Thermo Scientific). Immunoprecipitation was performed with 50 µl dynabeads bound to 10 µg anti-EhMIF rabbit serum or rabbit IgG. For each set, 1.5 µg of EhMIF mixed with 500 ng biotinylated EhJAB1protein was incubated with the antibody coated beads in 300 µl binding buffer (as described above) for overnight at 4 °C with rotation. Wash and elution of the bound complexes were done the same way as above. The immunoprecipitation was repeated in the presence of 5 µg and 10 µg non-biotinylated tag free EhJAB1 protein. Twenty-five percent of the eluted samples were assayed by immunoblots.
Mass Spectrometry
The entire eluted fraction from anti-EhMIF and rabbit IgG immunoprecipitations were subjected to electrophoresis on SDS polyacrylamide gel. Proteins were separated via SDS-PAGE for a length of 1 cm53. About 1 cm × 1 cm section of the gel, spanning all of the resolved proteins in each well, were excised. The gel samples were submitted to the W. M. Keck Biomedical Mass Spectrometry Laboratory for mass spectrometry analysis.
Immunoblotting
Protein samples obtained from GST-pulldown and immunoprecipitation experiments were resolved by SDS-PAGE followed by transfer onto polyvinylidene difluoride membranes (Millipore). The membranes were incubated overnight with primary antibodies at 4 °C. For EhMIF protein detection, rabbit anti-EhMIF antibody was used followed by anti-rabbit IgG HRP conjugate (Sigma) secondary antibody. For biotinylated EhJAB1, goat anti-biotin HRP conjugated antibody (Cell Signaling Technology) was used. Enhanced chemiluminescence (Thermo scientific) based substrates were used to detect antibody conjugated peroxidase activity.
IL8 secretion assay by ELISA
The human colonic epithelial cells (HCT116 & Caco2, American Type Culture Collection) with densities 106 cells/ml were cultured in 48 well plate (Corning) with 100 µL complete media (Dulbecco’s Modified Eagle Medium, Gibco) for 12 hours followed by gentle washing and incubation with 100 µl serum free media for 12 hours. After washing the plates with 200 µl media, cells were treated with 0.5 µg/ml EhMIF in presence or absence of 0.5, 1 and 2.5 µg/ml EhJAB1 protein for 8 hours. IL-8 in cell culture supernatant was measured by enzyme linked immunosorbent assay (ELISA, eBioscience).
Binding kinetics using BLI assay
The binding affinities between GST-EhJAB1 and EhMIF proteins were measured using the Blitz System (OctetR Red 96 system, ForteBio). Briefly, Anti-GST Dip and ReadTM Biosensors (ForteBio) were hydrated for 10 minutes in the sample dilution buffer (1x DPBS, 0.1% BSA, 0.02% Tween 20) followed by 3 cycles of priming and neutralization, 20 seconds each, in regeneration buffer (10 mM Glycine, pH 1.7) and the sample dilution buffer, respectively. Next, baseline stabilization of the primed biosensors was done in the dilution buffer for 5 minutes. Then, 10 µg GST-JAB1 or GST proteins diluted in sample dilution buffer was loaded onto the biosensors for 5 minutes. After washing the loaded biosensors in sample dilution buffer for 5 minutes, they were exposed for 10 minutes to 5, 2-fold dilution series of EhMIF protein starting at 80 nM concentration. Post-binding dissociation of EhMIF was done for 10 minutes in the sample dilution buffer. Binding affinities (KD) were calculated using the Blitz system software (ForteBio).
Bioinformatics
Orthologues of JAMM/MPN+ motif of the EhJAB1 protein from different protozoan parasites were aligned by Multiple Sequence Comparison by Log Expectation (MUSCLE) software54. 3D structure of EhJAB1 was constructed by Protein Homology/Analogy Recognition Engine v 2.0 (PHYRE2)55. The predicted EhJAB1 structure was then compared with that of the Human JAB1 protein using the UCSF Chimera software v. 1.10.2. The protein-protein interaction between EhJAB1 and EhMIF was examined and potential docking interfaces were predicted using pyDockWeb that applies a rigid body protein-protein docking prediction model by electrostatic and desolvation scoring29. The predictions were viewed using the UCSF Chimera software v. 1.10.2.
Statistical Tests
Statistical differences were determined using ANOVA followed by Dunnett’s post-hoc test. A p value less than 0.05 was considered statistically significant.
Electronic supplementary material
Acknowledgements
We thank William Petri, John Shannon, Bhupal Ban and Daniel Conrad for helpful advice; and the University of Virginia Molecular Interactions and Mass Spectrometry Research Facilities. This work was supported by the National Institutes of Health (NIH) R01AI026649-27S1, K08AI119181, and the Robert Wood Johnson Foundation–Harold Amos Medical Faculty Development Program Award.
Author Contributions
S.G., L.A.L., L.F., A.B. and S.M. conducted experiments. S.G. and S.M. wrote the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-28625-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Moonah SN, Jiang NM, Petri WA., Jr. Host immune response to intestinal amebiasis. Plos Pathog. 2013;9:e1003489. doi: 10.1371/journal.ppat.1003489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shirley DA, Moonah S. Fulminant Amebic Colitis after Corticosteroid Therapy: A Systematic Review. Plos Negl Trop Dis. 2016;10:e0004879. doi: 10.1371/journal.pntd.0004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Debnath A, et al. A high-throughput drug screen for Entamoeba histolytica identifies a new lead and target. Nat Med. 2012;18:956–960. doi: 10.1038/nm.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kudrin A, et al. Human macrophage migration inhibitory factor: a proven immunomodulatory cytokine? The Journal of biological chemistry. 2006;281:29641–29651. doi: 10.1074/jbc.M601103200. [DOI] [PubMed] [Google Scholar]
- 6.Roger T, et al. High expression levels of macrophage migration inhibitory factor sustain the innate immune responses of neonates. Proc Natl Acad Sci USA. 2016;113:E997–1005. doi: 10.1073/pnas.1514018113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Jong YP, et al. Development of chronic colitis is dependent on the cytokine MIF. Nat Immunol. 2001;2:1061–1066. doi: 10.1038/ni720. [DOI] [PubMed] [Google Scholar]
- 8.Yoo SA, et al. MIF allele-dependent regulation of the MIF coreceptor CD44 and role in rheumatoid arthritis. Proc Natl Acad Sci USA. 2016;113:E7917–E7926. doi: 10.1073/pnas.1612717113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosado Jde D, Rodriguez-Sosa M. Macrophage migration inhibitory factor (MIF): a key player in protozoan infections. Int J Biol Sci. 2011;7:1239–1256. doi: 10.7150/ijbs.7.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das R, et al. Macrophage migration inhibitory factor (MIF) is a critical mediator of the innate immune response to Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2997–3006. doi: 10.1073/pnas.1301128110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roger T, et al. Macrophage migration inhibitory factor deficiency is associated with impaired killing of gram-negative bacteria by macrophages and increased susceptibility to Klebsiella pneumoniae sepsis. The Journal of infectious diseases. 2013;207:331–339. doi: 10.1093/infdis/jis673. [DOI] [PubMed] [Google Scholar]
- 12.Ngobeni R, et al. Entamoeba Species in South Africa: Correlations With the Host Microbiome, Parasite Burdens, and First Description of Entamoeba bangladeshi Outside of Asia. J Infect Dis. 2017;216:1592–1600. doi: 10.1093/infdis/jix535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun T, et al. A Plasmodium-encoded cytokine suppresses T-cell immunity during malaria. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2117–2126. doi: 10.1073/pnas.1206573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sommerville C, et al. Biochemical and immunological characterization of Toxoplasma gondii macrophage migration inhibitory factor. The Journal of biological chemistry. 2013;288:12733–12741. doi: 10.1074/jbc.M112.419911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holowka T, et al. Leishmania-encoded orthologs of macrophage migration inhibitory factor regulate host immunity to promote parasite persistence. FASEB J. 2016;30:2249–2265. doi: 10.1096/fj.201500189R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Twu O, et al. Trichomonas vaginalis exosomes deliver cargo to host cells and mediate hostratioparasite interactions. Plos Pathog. 2013;9:e1003482. doi: 10.1371/journal.ppat.1003482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moonah SN, Abhyankar MM, Haque R, Petri WA., Jr. The macrophage migration inhibitory factor homolog of Entamoeba histolytica binds to and immunomodulates host macrophages. Infect Immun. 2014;82:3523–3530. doi: 10.1128/IAI.01812-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Twu O, et al. Trichomonas vaginalis homolog of macrophage migration inhibitory factor induces prostate cell growth, invasiveness, and inflammatory responses. Proceedings of the National Academy of Sciences. 2014;111:8179–8184. doi: 10.1073/pnas.1321884111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ngobeni R, et al. Entamoeba histolytica-Encoded Homolog of Macrophage Migration Inhibitory Factor Contributes to Mucosal Inflammation during Amebic Colitis. The Journal of infectious diseases. 2017;215:1294–1302. doi: 10.1093/infdis/jix076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miernyk JA, Thelen JJ. Biochemical approaches for discovering protein-protein interactions. The Plant journal: for cell and molecular biology. 2008;53:597–609. doi: 10.1111/j.1365-313X.2007.03316.x. [DOI] [PubMed] [Google Scholar]
- 21.Hunke, S. & Müller, V. S. In Protein Interactions (InTech, 2012).
- 22.Kleemann R, et al. Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature. 2000;408:211–216. doi: 10.1038/35041591. [DOI] [PubMed] [Google Scholar]
- 23.Burger-Kentischer A, et al. Binding of JAB1/CSN5 to MIF is mediated by the MPN domain but is independent of the JAMM motif. FEBS letters. 2005;579:1693–1701. doi: 10.1016/j.febslet.2005.01.080. [DOI] [PubMed] [Google Scholar]
- 24.Merk M, et al. The D-dopachrome tautomerase (DDT) gene product is a cytokine and functional homolog of macrophage migration inhibitory factor (MIF) Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E577–585. doi: 10.1073/pnas.1102941108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enchev RI, Schulman BA, Peter M. Protein neddylation: beyond cullin-RING ligases. Nature reviews. Molecular cell biology. 2015;16:30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao L, et al. Frizzled proteins are colonic epithelial receptors for C. difficile toxin B. Nature. 2016;538:350–355. doi: 10.1038/nature19799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filip AM, et al. Ribosomal protein S19 interacts with macrophage migration inhibitory factor and attenuates its pro-inflammatory function. The Journal of biological chemistry. 2009;284:7977–7985. doi: 10.1074/jbc.M808620200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perkins JR, Diboun I, Dessailly BH, Lees JG, Orengo C. Transient protein-protein interactions: structural, functional, and network properties. Structure. 2010;18:1233–1243. doi: 10.1016/j.str.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Jimenez-Garcia B, Pons C, Fernandez-Recio J. pyDockWEB: a web server for rigid-body protein-protein docking using electrostatics and desolvation scoring. Bioinformatics (Oxford, England) 2013;29:1698–1699. doi: 10.1093/bioinformatics/btt262. [DOI] [PubMed] [Google Scholar]
- 30.Leng L, et al. MIF signal transduction initiated by binding to CD74. Journal of Experimental Medicine. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobson SE, et al. The crystal structures of macrophage migration inhibitory factor from Plasmodium falciparum and Plasmodium berghei. Protein Science. 2009;18:2578–2591. doi: 10.1002/pro.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maharshak N, et al. CD74 is a survival receptor on colon epithelial cells. World journal of gastroenterology: WJG. 2010;16:3258. doi: 10.3748/wjg.v16.i26.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thibeaux R, et al. The parasite Entamoeba histolytica exploits the activities of human matrix metalloproteinases to invade colonic tissue. Nat Commun. 2014;5:5142. doi: 10.1038/ncomms6142. [DOI] [PubMed] [Google Scholar]
- 34.Wei N, Serino G, Deng XW. The COP9 signalosome: more than a protease. Trends in biochemical sciences. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Cope GA, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 36.Verma R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 37.Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN + , a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlierf A, et al. Targeted inhibition of the COP9 signalosome for treatment of cancer. Nature communications. 2016;7:13166. doi: 10.1038/ncomms13166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lingaraju GM, et al. Crystal structure of the human COP9 signalosome. Nature. 2014;512:161. doi: 10.1038/nature13566. [DOI] [PubMed] [Google Scholar]
- 40.Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei N, Deng XW. The COP9 signalosome. Annu Rev Cell Dev Biol. 2003;19:261–286. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- 42.Ponts N, et al. Deciphering the ubiquitin-mediated pathway in apicomplexan parasites: a potential strategy to interfere with parasite virulence. Plos One. 2008;3:e2386. doi: 10.1371/journal.pone.0002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vicente JB, Ehrenkaufer GM, Saraiva LM, Teixeira M, Singh U. Entamoeba histolytica modulates a complex repertoire of novel genes in response to oxidative and nitrosative stresses: implications for amebic pathogenesis. Cell Microbiol. 2009;11:51–69. doi: 10.1111/j.1462-5822.2008.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arya S, Sharma G, Gupta P, Tiwari S. In silico analysis of ubiquitin/ubiquitin-like modifiers and their conjugating enzymes in Entamoeba species. Parasitol Res. 2012;111:37–51. doi: 10.1007/s00436-011-2799-0. [DOI] [PubMed] [Google Scholar]
- 45.Liao S, Hu H, Wang T, Tu X, Li Z. The Protein Neddylation Pathway in Trypanosoma brucei: Functional Characterization and Substrate Identification. Journal of Biological Chemistry. 2017;292:1081–1091. doi: 10.1074/jbc.M116.766741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwok SF, et al. Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell. 1998;10:1779–1790. doi: 10.1105/tpc.10.11.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freilich S, et al. The COP9 signalosome is essential for development of Drosophila melanogaster. Curr Biol. 1999;9:1187–1190. doi: 10.1016/S0960-9822(00)80023-8. [DOI] [PubMed] [Google Scholar]
- 48.Chamovitz DA, Segal D. JAB1/CSN5 and the COP9 signalosome. A complex situation. EMBO Rep. 2001;2:96–101. doi: 10.1093/embo-reports/kve028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galluzzi L, Buqué A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nature Reviews Immunology. 2017;17:97. doi: 10.1038/nri.2016.107. [DOI] [PubMed] [Google Scholar]
- 50.Garg AD, Agostinis P. Cell death and immunity in cancer: From danger signals to mimicry of pathogen defense responses. Immunological reviews. 2017;280:126–148. doi: 10.1111/imr.12574. [DOI] [PubMed] [Google Scholar]
- 51.Hangai S, et al. PGE2 induced in and released by dying cells functions as an inhibitory DAMP. Proceedings of the National Academy of Sciences. 2016;113:3844–3849. doi: 10.1073/pnas.1602023113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Begum S, Quach J, Chadee K. Immune evasion mechanisms of Entamoeba histolytica: progression to disease. Frontiers in microbiology. 2015;6:1394. doi: 10.3389/fmicb.2015.01394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paulo JA, et al. Short gel, long gradient liquid chromatography tandem mass spectrometry to discover urinary biomarkers of chronic pancreatitis. The open proteomics journal. 2013;6:1. doi: 10.2174/1875039701306010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar RC. Muscle: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.