Abstract
The generation of human pluripotent stem cell (hPSC)-derived ventricular progenitors and their assembly into a 3-dimensional in vivo functional ventricular heart patch has remained an elusive goal. Herein, we report the generation of an enriched pool of hPSC-derived ventricular progenitors (HVPs), which can expand, differentiate, self-assemble, and mature into a functional ventricular patch in vivo without the aid of any gel or matrix. We documented a specific temporal window, in which the HVPs will engraft in vivo. On day 6 of differentiation, HVPs were enriched by depleting cells positive for pluripotency marker TRA-1-60 with magnetic-activated cell sorting (MACS), and 3 million sorted cells were sub-capsularly transplanted onto kidneys of NSG mice where, after 2 months, they formed a 7 mm × 3 mm × 4 mm myocardial patch resembling the ventricular wall. The graft acquired several features of maturation: expression of ventricular marker (MLC2v), desmosomes, appearance of T-tubule-like structures, and electrophysiological action potential signature consistent with maturation, all this in a non-cardiac environment. We further demonstrated that HVPs transplanted into un-injured hearts of NSG mice remain viable for up to 8 months. Moreover, transplantation of 2 million HVPs largely preserved myocardial contractile function following myocardial infarction. Taken together, our study reaffirms the promising idea of using progenitor cells for regenerative therapy.
Keywords: cardiac regeneration, stem cell, ventricular progenitor, human, myocardial infarction, differentiation
The authors identified a human ventricular progenitor (HVP) population that can expand, differentiate, self-assemble, and mature into a 3D functional ventricular patch in vivo. They demonstrate that transplantation of HVPs under the kidney capsule generated a 7 mm × 3 mm × 4 mm myocardial patch while transplantation following myocardial infarction can largely preserve myocardial contractile function.
Introduction
Heart failure is a globally leading cause of mortality,1 primarily driven by the loss of ventricular muscle mass2 and the limited regenerative ability of the adult heart.3, 4, 5 Ventricular muscle tissue engineering with human pluripotent stem cells (hPSCs), embryonic stem cells (ESCs), or induced pluripotent stem cells (iPSCs) to create a functional in vivo 3D human ventricular muscle patch holds great promise. However, such efforts have been hampered by the requirement for large-scale generation of purified ventricular cells as well as their controlled growth and maturation, vascularization, assembly, and formation of extracellular matrix (ECM).6 To date, diverse cardiovascular cells, ECMs, de-cellularized scaffolds, and DNA/RNAs have been studied for therapeutic use, and 3D in vitro perfused heart models have been generated, but the generation of a vascularized, functional ventricular wall in the in vivo context has remained elusive. Previous studies with hPSCs have been based on the generation of heart tissue constructs from already differentiated cardiomyocytes rather than committed ventricular lineage progenitors. Importantly, lineage progenitors may have intrinsic properties for triggering vascular and matrix cues critical for self-assembly and formation of an in vivo stable niche, which are lost during later stages of differentiation. In this regard, other attempts to form in vivo grafts have required the addition of other synthetic matrices, gels, suturing into the ventricular wall, scaffolds, or additional interstitial-like cells to allow the cells to remain within the contractile ventricular wall. On the other hand, most well-characterized heart progenitors are multi-potent,7, 8 and most protocols result in a mixture of atrial, ventricular, pacemaker, vascular smooth muscle, and endothelial lineages.9, 10 Early-stage progenitors are also usually contaminated with pluripotent stem cells,11 raising the danger of teratoma formation12 or other non-cardiac lineages within the graft, which have been documented in transplantation studies.13 Finally, it remains unclear as to whether the transplantation of progenitors in vivo would result in their subsequent loss of progenitor markers and subsequent differentiation, vascularization, matrix formation, grafting without additional cell/matrix/scaffolds, and early steps of maturation. Although human iPS or ES-derived functional motor neurons,14 pancreatic β cells,15 and organoids16 have been generated in vitro, the in vivo generation of an ESC-derived multicellular organ component, such as a human ventricular patch, has been challenging.
Herein, we report that ESC-derived ISL1+ human ventricular progenitors (HVPs) can recapitulate one of the earliest and most essential steps of organogenesis: building of a functional ventricular heart muscle in vivo, including controlled proliferation, followed by normal growth, maturation, and self-assembly via a cell autonomous pathway. We further demonstrate that transplantation of this HVP population to the heart preserves function in injured myocardium.
Results
Optimized Protocol Yields Efficient and Stepwise Ventricular Differentiation In Vitro
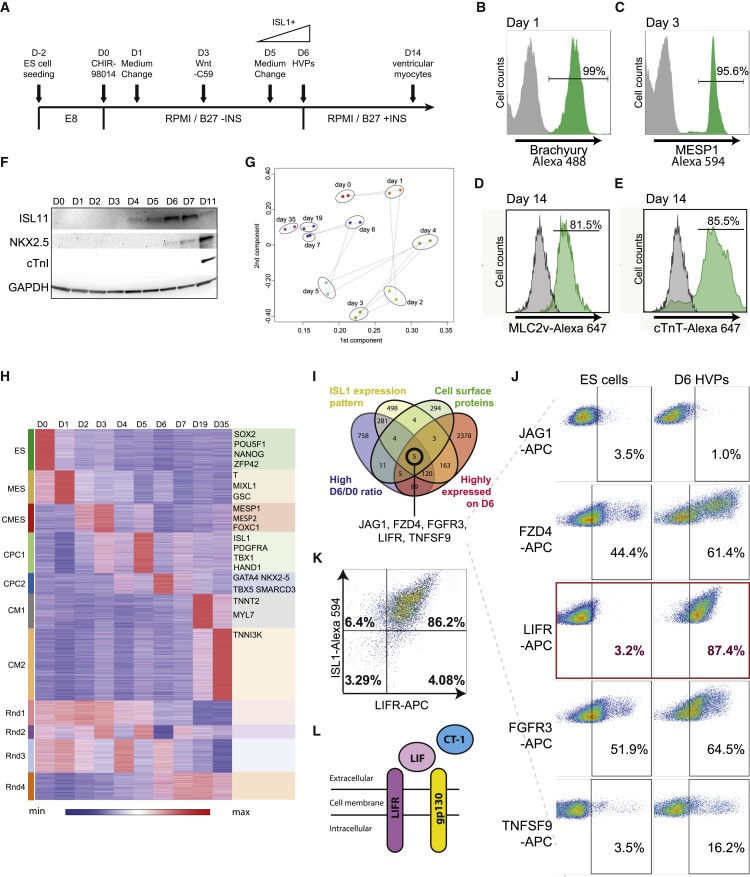
Previous studies have documented a pivotal role for canonical Wnt signals in driving cardiac myogenesis in ESCs in in vitro models17, 18 as well as in ventricular muscle cell lineages in vivo.19 To maximize the generation of cardiomyocytes expressing the ventricular marker myosin light chain 2v (MLC2v+) cardiomyocytes, we modified an existing two-step protocol17 by using highly potent inhibitors of glycogen synthase kinase 3 (GSK3) and porcupine (PORCN) at a lower concentration. These slight modifications in protocol lead to the efficient and sequential conversion of hPSCs to HVPs (hPSCs-pan mesoderm-cardiac mesoderm-islet ventricular progenitor) over 6 days (HVP protocol) (Figure 1A). After 24-hr exposure to CHIR-98014, >99% of the hPSCs expressed the pan-mesoderm marker Brachyury, as revealed by fluorescence-activated cell sorting (FACS) (Figure 1B) and immunostaining (Figure S1A). On day 3, more than 95% of the differentiated cells expressed MESP1, a cardiac mesodermal marker (Figures 1C and S1B). After 14 days, the HVP protocol resulted in >82% conversion to ventricular cardiac myocytes, as illustrated by cardiac troponin T (cTnT) and MLC2v flow cytometry (Figures 1D and 1E). ISL1 protein expression began on day 4 and peaked at day 6, followed by NKX2.5 at day 7, with its peak on day 11 (Figure 1F). Cardiac troponin I (cTnI)-positive ventricular myocytes were not induced until day 11 (Figure 1F). Immunostaining of the day 6 HVPs showed substantial numbers of ISL1+ cells (Figure S1F), and FACS analysis confirmed ISL1 expression in 95.4% ± 1.9% of cells at this stage (Figure S1C). In summary, we employed a modified HVP protocol, which enables production of ISL1+ HVPs efficiently within 6 days.
Figure 1.
hPSC-Derived HVPs by Modulation of Wnt Signaling via Small Molecules
(A) Schematic protocol for defined differentiation of hPSCs to ventricular progenitors. (B) Brachyury expression was detected on day 1. (C) MESP1 expression was detected on day 3 with flow cytometry. (D and E) 14 days after initiation of hPSC differentiation using the protocol shown above, cells were analyzed for MLC2v (D) and cTnT (E) expression by flow cytometry. (F) Protein expression was assessed by western blot at different time points during differentiation. (G) RNA-seq analysis of the rapid and synchronous HVP generation process. Gene expression profiles projected onto the first two principal components. Cells differentiated from different days are designated by colors. (H) Cluster analysis of gene expression profiles at different time points during HVP differentiation identifies stage-specific signature genes. These genes were clustered hierarchically on the basis of similarity of their expression profiles. Expression pattern of each gene is displayed as a horizontal strip. The intensity of the red and blue color is proportional to the relative gene induction (red) or repression (blue). ES, embryonic stem cells stage; MES, mesoderm stage; CMES, cardiac mesoderm stage; CPC1 and CPC2, cardiac progenitor stages; CM1 and CM2, cardiomyocyte stages; Rnd1–4, unclassified groups. (I) Venn diagram displaying number of genes in each category, with 5 candidate cell surface marker genes (JAG1, FZD4, FGFR3, LIFR, and TNFSF9) satisfying all 4 categories. (J) Flow cytometry analysis of the 5 candidate genes showed that LIFR is highly expressed (>87%) on day 6 of differentiation. (K) Flow cytometry analysis revealed that >86% of D6 cells co-express LIFR and ISL1. (L) Schematic diagram illustrating leukaemia inhibitory factor (LIF) binding to heterodimers of LIF receptor and gp130. LIFR-gp130 heterodimers can also associate with other receptor subunits to bind CT-1.
The sequential conversion of pluripotent stem cells to ISL1+ HVPs was further illustrated by RNA sequencing (RNA-seq). Two batches of RNA-seq (100- and 50-bp read length) were performed using the Illumina Hiseq 2000 platform. Principal component analysis clustered the cells by their developmental differentiation stage, effectively reflecting the sequential steps of ventricular cell differentiation (Figures 1G and 1H). Downregulation of the pluripotency markers OCT4, NANOG, and SOX2 during differentiation was followed by the induction of the primitive streak-like genes T and MIXL1 by 24 hr and the upregulation of MESP1 on day 2 and day 3 (Figure S2). Expression of the cardiac muscle markers TNNT2, TNNC1, MYL2, MYL7, MYH6, MYH7, and IRX4 was detected at a later stage of differentiation (after day 10; Figure S2). ISL1 mRNA was expressed as early as day 4 and peaked on day 5, 1 day before its protein expression reached its peak (Figure S1E). Western blot analysis (Figure 1F) and double immunostaining (Figure S1F) of ISL1 and NKX2.5 confirmed that ISL1 is an earlier developmental gene versus NKX2.5 gene in the ISL1+ HVP lineage because purified HVPs uniformly expressed ISL1, but remained largely negative for NKX2.5.
LIFR Surface Protein Marks Ventricular Progenitors on Day 6 of Differentiation In Vitro
In order for the HVPs to ensure long-term cellular stability, it became crucial to identify cell surface markers to define and purify the ventricular progenitor population. The RNA-seq data collected at sequential time points facilitated the potential to identify cell surface markers that might be uniquely present during the initial onset of HVP formation. We used the following 4 criteria to select candidate genes: (1) high day 6/day 0 ratio; (2) correlates with ISL1 expression pattern; (3) cell surface protein; and (4) highly expressed at day 6 (Figure 1I). Five candidates emerged, including the Notch pathway surface ligand JAG1, Wnt signaling pathway receptor FZD4, FGF signaling pathway receptor FGFR3, LIF receptor LIFR, and tumor necrosis factor ligand TNFSF9. To confirm robust cell surface protein expression, we performed live flow cytometry on day 6 HVPs using conjugated antibodies against all five cell surface marker candidates (Figure 1J). Of the 5 candidates, LIFR (Figure 1J) had the highest protein expression on the cell surface, with LIFR+ cells comprising 87.4% of the total population. In addition, LIFR is only expressed at a low level in ESCs, unlike other candidate cell surface markers (Figure 1J), and previously has been shown to be the co-receptor of the GP 130 cytokine cardiotrophin-1, which was originally isolated by expression cloning during ESC in vitro cardiogenesis.20, 21 Co-staining of LIFR with ISL1 showed that the majority (>86%) of day 6 HVPs are LIFR and ISL1 co-positive (Figure 1J), demonstrating LIFR as a robust cell surface marker for HVPs. Furthermore, continued culturing of FACS-purified LIFR+ISL1+ HVPs to day 15 revealed robust beating (Movie S1) and expression of MLC2v (Figure S1I), demonstrating that LIFR+ISL1+ progenitors can give rise to ventricular myocytes.
ISL1+ HVPs Differentiate into Cardiomyocytes Expressing Ventricular Proteins and Exhibiting Mature and Uniform Electrophysiological Properties
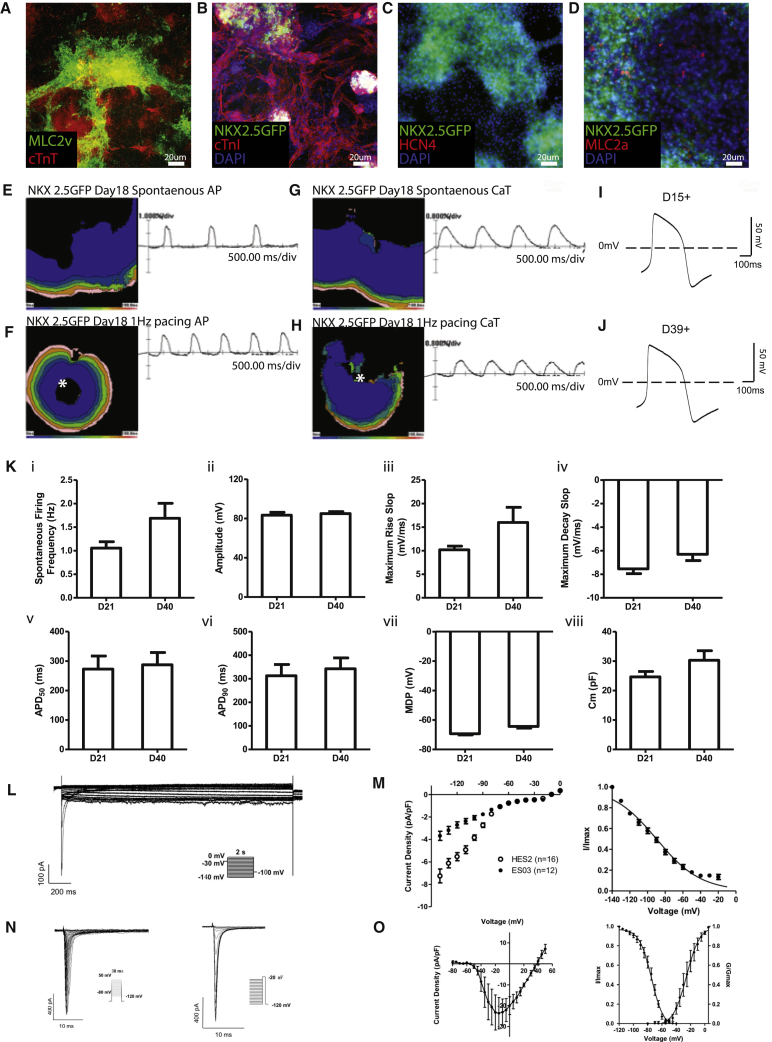
Next, we investigated the differentiation and maturation potential of the HVPs. By day 9–12 of differentiation, HVPs form a uniform wave-like beating monolayer in vitro that was NKX2.5+ (Movie S2). As described previously, the majority of cells after 14 days of differentiation were MLC2v and cTnT positive, indicating that most of the cells have taken on a ventricular cardiomyocyte identity (Figures 1D and 1E). To determine the lineage potency of ISL1+ HVPs, a clonal assay was performed, in which a single ISL1+ cell on day 6 of differentiation was re-plated. During 3 weeks of differentiation, this cell gave rise to cTnI and smooth muscle actin (SMA) double-positive cells, a feature of immature cardiomyocytes, but not VE-cadherin-positive endothelial cells, indicating that ISL1+ HVPs are ventricular muscle progenitors (Figure S1J). HVPs differentiated as a population to day 15+ also expressed various cardiac lineage markers, and the majority of them were positive for ventricular marker MLC2v, cardiac troponin, and NKX2.5 (Figures 2A–2D); they were mostly negative for MLC2a, whereas expression of HCN4 was completely absent (Figures 2A, 2C, and 2D). Taken together, this protocol generates a highly enriched pool of ISL1+ progenitors that differentiate into ventricular cardiomyocytes. These highly enriched HVPs are distinct from the previously reported KDR+ cells22 or multipotent ISL1+ cells,8, 23 which can give rise to all three lineages of cardiovascular cells, and are similar to the previously described rare, ISL1+ murine ventricular progenitors.24
Figure 2.
In Vitro Differentiation Potential of ISL1+ HVPs
(A–D) Day 15+ HVPs were analyzed for ventricular markers: (A) MLC2v and cTnT; (B) cTnI; (C) pacemaker marker HCN4; and (D) atrial marker MLC2a. Scale bar, 20 μm. (E and F) Optical recordings of spontaneous (E) and paced (F) day 18 HVP cells. Isochrome maps at 20-ms intervals show spontaneous action potential initiation. The white asterisk represents the origin of point electrical stimulation. (G and H) Optical recordings of spontaneous (G) and paced (H) calcium transients in day 18 HVP cells. The white asterisk represents the origin of point electrical stimulation. (I and J) Patch clamp recordings of day 15+ (I) and day 39+ (J) HVPs showed typical ventricular-like action potentials. (K) Whole cell patch clamp recordings of day 21 and day 40 HVP-derived ventricular cardiomyocytes. Cultures demonstrated that there were no significant differences in various AP parameters between the two time points, suggesting that in vitro, HVPs reached maturity by day 18. (L) Representative If traces in the presence of 1 mM Ba2+. (M) Patch clamp recording comparison of If in ES03- and HES2-derived ventricular cardiomyocytes. Steady-state I-V (left) and steady-state activation relations (right) for If (n = 12). (N and O) Whole cell patch clamp recording of INa in ES03. (N) Representative INa traces during activation (left) and inactivation (right). (O) Plots of peak INa density I-V relationship (left) and steady-state activation/inactivation (right), n = 5. Histograms and traces are shown as mean ± SEM.
We further characterized the maturation potential of these HVPs by performing electrophysiology in vitro. We investigated the electrical properties of the mature HVPs with optical mapping of action potential (AP) and calcium transients (CaTs), with a particular focus on their ability to display electrical coupling. In vitro optical mapping of day 6 HVPs revealed that there was no spontaneous or induced propagation of the AP or CaT (n = 5; data not shown). By day 18+, propagation of both the AP and CaT became readily apparent, both spontaneously and following point stimulation (n = 5), suggestive of network activity and electrical coupling (Figures 2E–2H and S3). On day 6, the NKX2.5-GFP HVPs were GFP−; patch clamping revealed that all cells showed a depolarized resting membrane potential, and upon stimulation, they were unable to fire APs upon stimulation (n = 20; data not shown). By comparison, at later time points (day 21 and day 40), over 90% of cells were GFP+. GFP+ cells selected for patch clamping showed polarized diastolic membrane potentials and a ventricular-like AP (Figures 2I and 2J; n = 19). However, AP configuration was comparable at day 21 and day 40 (Figure 2K; Table 1). Similar results were obtained with the ES03-derived HVPs, in which over 90% of day 21 and day 40 cells exhibited ventricular-like APs (Table 1). Notably, these HVPs derived from both the NKX2.5-GFP and the ES03 cell lines could be paced and exhibited an APD90/APD50 ratio of <1.2; recapitulating the AP configuration of native adult human ventricular myocytes (Figure 2K; Table 1).25, 26, 27 Calcium transients were comparable in day 15 and 30 HVPs (Table 2), with kinetics consistent with those of hPSC-derived cardiomyocytes reported previously.28, 29 We next investigated If, which is responsible for cardiac pacemaking in immature and sinoatrial cardiomyocytes.30 Whole-cell voltage-clamp measurements (Figure 2L) showed that, in comparison with cardiomyocytes differentiated from hESC (hES2) using an embryoid body (EB)-based protocol,31 If current density was reduced by half on day 21 HVPs (Figure 2M). The I-V plot and steady-state activation/inactivation relationship of the sodium current (INa) also showed characteristics typical of ventricular cardiomyocytes (Figures 2N and 2O). In addition, in vitro conduction of both APs and CaTs was observed to be continuous and uniform, illustrating synchronous electrical coupling within the ventricular muscle patch in vitro (Movie S3. In Vitro Optical Mapping on Day 18 of HVP Differentiation, Movie S4. In Vitro Optical Mapping on Day 36 of HVP Differentiation, Movie S5. Video of In Vitro Optical Mapping of Calcium Transients in Day 18 HVPs Showing Spontaneous Calcium Activity, Movie S6. Video of In Vitro Optical Mapping of Calcium Transients in Day 18 HVPs, Showing Electrical Responsiveness during 1-Hz Pacing). Furthermore, day 21 HVP-ventricular cardiomyocytes showed responses to pharmacological agents characteristic of native adult cardiomyocytes. Application of the β-adrenergic agonist isoproterenol increased conduction velocity, whereas the parasympathetic agonist acetylcholine decreased it. Acetylcholine also shortened action potential duration (APD), as expected. The calcium channel blocker nifedipine also shortened APD and decreased conduction velocity as in mature myocardium. Finally, AP conduction in the presence of the non-specific gap junction inhibitor heptanol (0.5 mM) markedly reduced conduction velocity by 50% ± 23% (n = 3) without affecting AP amplitude, suggesting a robust presence of gap junctions (Table S1). Together, these data indicate that HVPs display a mature-like ventricular phenotype by as early as day 19 in vitro.
Table 1.
Electrophysiological Characterization of HVPs during Ventriculogenesis In Vitro and In Vivo by Optical Mapping
| Parameter | NKX2.5- GFP Day 15+ In Vitro | NKX2.5-GFP Day 30+ In Vitro | NKX2.5-GFP 1+ Month Ex Vivo Kidney Graft | ES03 Day 15+ In Vitro | ES03 1+ Month Ex Vivo Kidney Graft |
|---|---|---|---|---|---|
| AP upstroke time (ms) | 128 ± 19 (n = 3) | 128 ± 24 (n = 3) | 129.7 ± 11.5 (n = 3) | 143 ± 9 (n = 3) | 105.0 ± 42.4 (n = 2) |
| AP 50% decay time (ms) | 73.5 ± 4.1 (n = 3) | 103.5 ± 5.5 (n = 3) | 93.9 ± 18.5 (n = 3) | 252 ± 41 (n = 3) | 151.6 ± 2.8 (n = 2) |
| APD50 (ms) | 202 ± 24 (n = 3) | 232 ± 30 (n = 3) | 223.6 ± 25.5 (n = 3) | 395 ± 48 (n = 3) | 256 ± 45 (n = 2) |
| APD90 (ms) | 374 ± 66 (n = 3) | 454 ± 10 (n = 3) | 378 ± 32 (n = 3) | 491 ± 91 (n = 3) | 304 ± 1 (n = 2) |
| Conduction velocity (cm/s) | 2.08 ± 0.91 (n = 3) | 2.41 ± 0.17 (n = 3) | 2.19 ± 0.14 (n = 3) | 3.35 ± 0.75 (n = 3) | 1.89 ± 0.58 (n = 2) |
Comparison of action potential parameters by optical mapping. The NKX2.5-GFP-labeled ESC line and ES03 cell line were employed for in vitro and ex vivo (i.e., kidney patch) analyses. Measurements were made during 1-Hz pacing.
Table 2.
Comparison of Calcium Transient Parameters between HVPs during Ventriculogenesis In Vitro and In Vivo by Optical Mapping
| Parameter | NKX2.5-GFP Day 15+ In Vitro | NKX2.5-GFP Day 30+ In Vitro |
|---|---|---|
| CaT upstroke time (ms) | 266 ± 5 (n = 3) | 234 ± 2 (n = 3) |
| CaT 50% decay time (ms) | 186 ± 23 (n = 3) | 255 ± 4 (n = 3) |
| CaT 50 (ms) | 452 ± 12 (n = 3) | 490 ± 2 (n = 3) |
The NKX2.5-GFP-labeled ESC line was employed for in vitro and ex vivo (i.e., kidney patch) analyses. All measurements made by optical mapping during 1-Hz pacing.
ISL1+ HVPs Form a Functionally Mature Graft on Kidney In Vivo
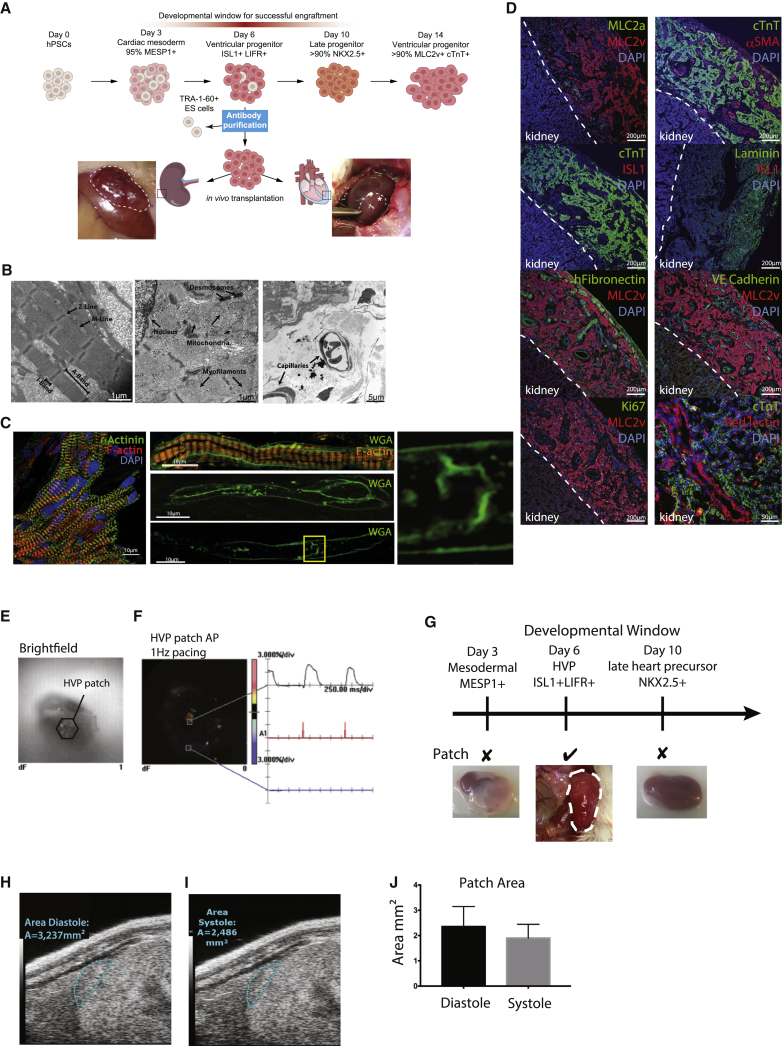
The ability to generate a large quantity of HVPs enabled the direct analysis of their cell-autonomous potential to generate a ventricular wall in vivo via transplantation under the kidney capsule (Figure 3A). Sub-capsular transplantation of cells in the kidney is especially optimal for its accessibility, and transplanted cells are well contained under the renal capsule in a highly vascularized site. Furthermore, the retroperitoneal location of the kidney, together with its separation from other organs, is advantageous for imaging. Day 6 HVPs were negatively magnetic-activated cell sorted (MACS) for the pluripotent stem cell surface marker TRA-1-60 (<3% TRA-1-60+) to purify the population. Three million TRA-1-60− HVPs were transplanted under the kidney capsule of immunocompromised NOD scid gamma (NSG) mice (n = 30). 2 months after transplantation, a patch of xeno tissue exceeding 0.6 cm in length and 0.2 cm in thickness was found on the surface of the murine kidney (Figures 3A–3J). The xeno tissue resembled a human ventricular muscle wall, characterized by an organized array of cardiomyocytes. At the sub-cellular level, electron microscopy revealed aligned sarcomeres, with distinct organization of M- and Z-lines as well as A- and I-band domains (Figure 3B). Immunohistochemistry further indicated sarcomeric localization of α-actinin and actin typical of mature cardiomyocytes (Figure 3C). In addition, membrane staining of cardiomyocytes within the xeno tissue revealed an early T-tubule-like network that was contiguous with the cell surface (Figure 3C). Although less dense and organized than that present in adult ventricular myocytes, this T-tubule-like network was reminiscent of the post-natal developing heart.32 Consistent with a ventricular phenotype, the xeno tissue exhibited uniform expression of both cTnT and the ventricular marker MLC2v (Figure 3D).33 The ventricular wall was fully vascularized, as assessed by expression of αSMA and VE-cadherin (Figure 3D), and the observation of red-blood-cell-containing capillaries in electron microscopy (EM) micrographs (Figure 3B). Remarkably, the human ventricular muscle wall was connected to the murine host circulation, as indicated by red-lectin staining on the kidney HVP graft after intravenous injection (Figure 3D). Surprisingly, the HVPs secreted their own ECM, as demonstrated by the presence of human fibronectin and laminin in the graft (Figure 3D). Very little proliferation was observed within the ventricular muscle graft because the majority of cells were both ISL1- and Ki67-negative (Figure 3D).
Figure 3.
In Vivo Transplantation of HVPs under Renal Capsule
(A) Schematic illustration of HVP generation, differentiation, and transplantation. HVPs were injected under the kidney capsule or intra-myocardially into the heart (injection site marked with squares); photos illustrate where the HVP graft was formed 2 months after transplantation, when it was recovered. (B) Electron microscopy of the HVP patch in the kidney capsule revealed mature ultra-structure, including organized and aligned sarcomeres, with well-defined Z-line, M-line, A-band, and I-band (left micrograph). Also apparent were desmosomes, mitochondria, myofilament bundles (middle panel), and capillaries containing red blood cells (right panel). (C) High-resolution immunofluorescence images further demonstrate sarcomeric organization within HVP grafts, with transverse orientation of a-actinin at z-lines and actin staining between z-lines. Membrane staining (WGA) revealed the presence of developing T-tubules (middle panels, high magnification of indicated region at right). (D) Immunofluorescence staining of the HVP kidney patch, with antibodies against MLC2a, MLC2v, cTnT, aSMA, ISL1, laminin, HCN4, human fibronectin, VE-cadherin, and Ki67. Scale bar, 200 μm. Red lectin was injected via the tail vein, illustrating a direct connection of the HVP kidney patch with the host venous system. Scale bar, 50 μm. (E and F) A 6+-week-old HVP kidney patch was harvested for ex vivo optical mapping and observed under brightfield (E) to be electrically responsive to stimulation (F). (G) A unique developmental window for HVP engraftment occurred at day 6 of differentiation; day 3 mesodermal MESP1+ cells resulted in teratomas, and day 10 late NKX2.5+ heart precursors failed to form a ventricular muscle graft patch (n = 4). (H and I) Cross-sectional area of a 6+-week-old HVP kidney graft patch during relaxation (i.e., diastolic phase; H) and maximal contraction (i.e., systolic phase; I), respectively, as imaged by ultrasound. (J) Measured cross-sectional area of the graft patch during diastole and systole.
We next investigated the functionality of the in vivo kidney HVP graft. Ex vivo optical mapping of 6- to 7-week-old kidney HVP grafts revealed they were electrically responsive when stimulated (Figures 3F and S3). All of the 6+-week-old kidney HVP grafts (n = 5) generated APs when electrically paced at various basic cycle lengths (500–2,000 ms; Figure S3). In addition, they displayed an AP upstroke, decay time, and APD90/APD50 ratio (<1.4) comparable to HVPs matured in vitro (Table 1), highlighting the remarkable ability of day 6 HVP cells to self-assemble and differentiate into functional cardiomyocytes in vivo. Optical mapping experiments also demonstrated a rapid and continuous conduction of the AP across the HVP grafts upon a point unipolar electrical stimulation. Furthermore, ultrasound imaging of in vivo 6+-week-old kidney HVP grafts clearly illustrated the ability to contract (Movie S7). Beating of the grafts occurred at a frequency of approximately 70 beats per minute. Cross-sectional surface area of the graft contracted by 19% ± 4% (n = 3) during a contraction cycle, returning to baseline during relaxation (Figures 3H–3J).
ISL1+ HVPs Demonstrate a Unique Developmental Window for Graft Formation
To assess whether day 6 HVPs represent a unique developmental window to form a ventricular muscle graft, we transplanted late-stage NKX2.5+ cardiac progenitor cells (day 10 after differentiation) under the kidney capsule (n = 4). At 6+ weeks post-transplantation, mice with NKX2.5+ cells did not form any visible human muscle graft or any other tissue, indicating the HVPs lose their cell-autonomous ability for in vivo ventriculogenesis following peak ISL1 expression (Figure 3G). Cells derived from an earlier time point in differentiation, day 3, expressing MESP1 and thus representing cardiac mesoderm, also failed to generate a homogeneous cardiac muscle graft on murine kidneys; instead, heterogeneous tumor-like multi-tissue structures were detected (n = 4; Figure 3G). Therefore, it seems that the unique developmental properties of day 6 HVPs allow their engraftment and assemble into a human ventricular muscle xeno-graft in vivo.
ISL1+ HVPs Form a Stable, Congruent Graft on Uninjured, Healthy Murine Heart
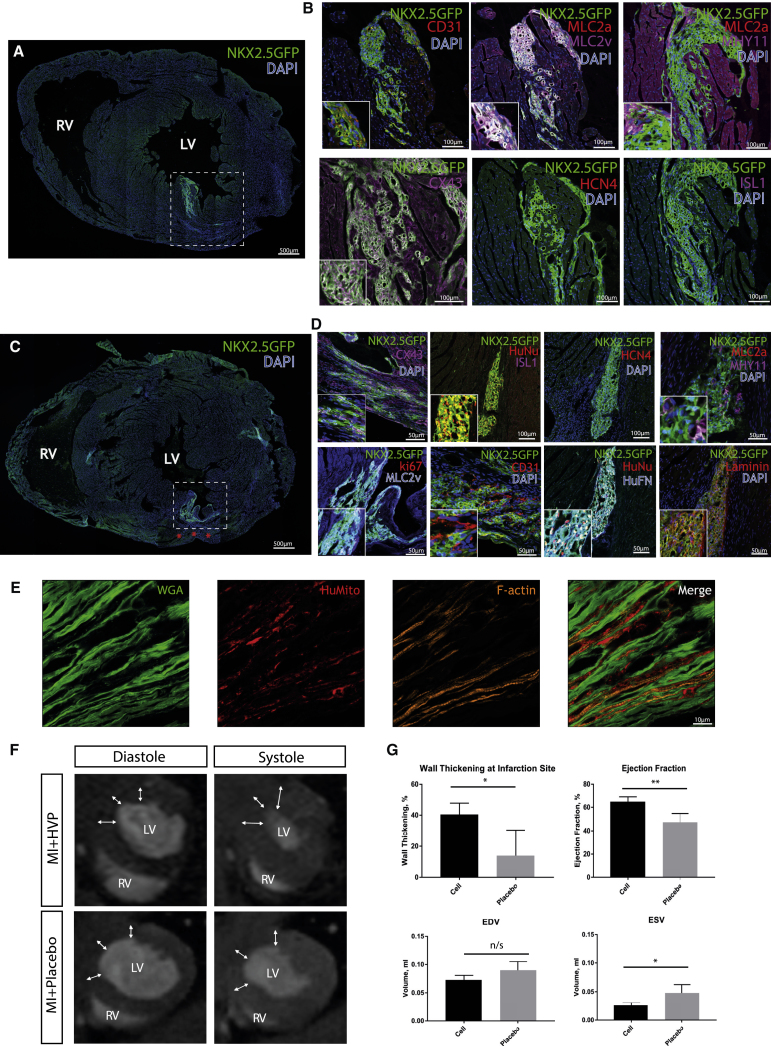
To determine if immature HVPs can engraft into an uninjured heart, two million purified (negatively sorted for TRA-1-60) NKX2.5-GFP-labeled HVPs were injected intra-myocardially into the hearts of NSG mice (n = 25, Figure 3A). Hearts were harvested 2 and 8 months after transplantation (data not shown for 2 months). Upon sectioning, GFP+ cells were found in the myocardium (Figures 4A and 4B). HVP heart graft patches uniformly expressed NKX2.5 (Figures 4A and 4B), and were also positive for the cardiac ventricular marker MLC2v. Studies of HVP cardiac grafts 8 months (n = 4) post-transplantation documented the long-term stability of relatively pure, uniform MLC2v+ ventricular muscle graft that was negative for MLC2a, HCN4, and ISL1 staining (Figure 4B), without formation of teratomas or other non-ventricular human cell types. The 8-month-old graft remained vastly integrated within the host myocardium because connexin 43 (CX43)-positive cells were present throughout the graft (Figure 4B). The ventricular graft patch was vascularized, as assessed by expression of CD31 (Figure 4B). Taken together, these studies suggest that in the normal heart, in the absence of an endogenous niche (created by cardiac injury), the HVPs can engraft, self-assemble, differentiate, vascularize, and acquire features of maturation while generating a stable, long-term, homogeneous ventricular muscle graft in vivo.
Figure 4.
In Vivo Transplantation of HVPs into Healthy and Injured Hearts
(A) White dashed box denotes the location of the HVP graft 8 months following intramyocardial injection into a healthy heart. (B) Immunofluorescence staining showed that HVP ventricular grafts in the heart expressed NKX2.5-GFP (green), MLC2v, connexin 43, and MHY11; a minority of cells co-expressed MLC2a and was negative for HCN4 and ISL1. CD31 was only present outside the graft. (C) The white dashed box identifies the localization and the engraftment of the xeno tissue 2 months after injection of the HVPs into the border zone of MI hearts. Red asterisks denote infarction site. (D) Immunofluorescence staining showed that HVP ventricular grafts in post-MI hearts co-expressed NKX2.5-GFP (green), connexin 43, human nuclei, MLC2v, MHY11, human fibronectin, and laminin. A minority of cells was positive for ki67; MLC2a, ISL1, and HCN4 staining was not observed. Endothelial marker CD31 was detected in close proximity to the HVP graft. (E) High-resolution imaging of the border zone of post-MI HVP grafts demonstrated engrafted, differentiated cardiomyocytes of human origin, with well-organized sarcomeres (stains of cell membrane [WGA, green], human mitochondria [red], and F-actin [orange]). (F) MRI assessment of post-MI hearts 2 months following HVP transplantation showed improved global and local function in comparison with placebo. Representative short axis images show wall thickness at the site of induced infarction (arrows). LV, left ventricle; RV, right ventricle. (G) Mean measurements ± SD of wall thickening, ejection fraction, and volume during end diastole (EDV) and end systole (ESV). *p < 0.05; **p < 0.01; n/s, not significant.
ISL1+ HVPs Improve Local and Global Functions after MI
To assess their regenerative capacity, we injected 2 million HVPs intra-myocardially into NSG mice immediately after myocardial infarction (MI) induced by left anterior descending (LAD) artery ligation. The injection was targeted to the infarction border zone (n = 5). The results were compared with a control group (n = 5), i.e., mice with LAD ligation, which received placebo treatment (cell culture medium without cells). After a 2-month follow-up, MRI was performed to evaluate left ventricular volumes, wall thickness, and global and local function (Figures 4F and 4G; Movies S8 and S9). When compared to the placebo group, the HVP-treated group showed improved ejection fraction (p = 0.0079) and augmented wall thickening at the infarction site (p = 0.0159). In addition, left-ventricular end-systolic volume was smaller in the HVP-treated group (p = 0.0159), consistent with improved contractility.
The injected hearts were collected for histological analysis. The graft was observed to be largely ventricular MLC2v+ (Figures 4D and S4), negative for non-ventricular heart markers such as HCN4 (Figure 4D) and MLC2a (Figure 4D), and negative for the progenitor makers ISL1 and Ki67 (Figure 4D). The ventricular grafts were integrated into the host myocardium, as shown by the presence of CX43 (Figure 4D). Furthermore, the graft contained ECM, as indicated by robust expression of laminin and fibronectin (Figure 4D). Higher resolution fluorescence imaging of the border zone revealed cardiomyocytes containing human mitochondria and well-organized sarcomeres embedded within the fibrous tissue (Figure 4E). This observation suggests that well-differentiated ventricular cardiomyocytes engrafted within the border zone actively contribute to the enhanced contractility observed in post-infarction hearts following HVP transplantation.
Discussion
The human ventricular myocardium has a limited capacity for regeneration, most of which is lost after 10 years of age.4 As such, new strategies to generate heart muscle via repair, regeneration, and tissue engineering approaches during cardiac injury have been a subject of intense investigation in regenerative biology and medicine. The assumption is that given the complex 3D structure of solid organs, achieving coordinated parenchymal cell proliferation, vascularization, and matrix formation would ultimately require the addition of vascular cells and/or conduits, biomaterials, and/or a de-cellularized matrix to form a functional tissue.34 However, the complexity of adding these various components has confounded attempts to translate this to clinical practice.35 Although hPSCs hold great promise, to date it has not been possible to build a pure, vascularized functional 3D human ventricular muscle patch in vivo in a mammalian system without the addition or support of scaffolds.36 Thus far, the most advanced studies designed to generate in vivo heart patches and/or grafts in vivo have been based on using partially purified, differentiated pools of heart cells (atrial, ventricular, pacemaker, and other). However, these studies have faced limitations regarding the maturity of the subsequent heart tissue, relative purity of tissue for ventricular muscle, electrical heterogeneity from atrial and pacemaker cell types, long-term stability, and requirement of matrices/scaffolds or suturing into the myocardium to allow grafting. At the same time, the use of heart progenitors as a starting material has been hampered by the requirement for the absence of pluripotent stem cells and other precursors, and the tendency for the progenitor cells to be multipotent, leading to heterogeneity of the tissue construct. Furthermore, it has been unclear whether these cells would go on to exit the progenitor state and display controlled growth, assume a rod shaped conformation with sarcomeric organization, and expand in size without continuing to proliferate.
Herein, we identified a cell population positive for the cardiac progenitor marker ISL1 and able both to form a large ventricular muscle patch on the murine kidney in a completely non-native environment and engraft into uninjured and injured murine heart after intra-myocardial injections. The ability to generate a large quantity of HVPs from a renewable source of either human ES or iPS cell lines represents a new approach to the generation of functional ventricular muscle in the setting of advanced heart failure. As reported in our current study, one of the key aspects of HVP self-assembly into a ventricular-like wall in vivo is its developmental time window, which coincides with peak ISL1 expression. We show that early cardiogenic mesoderm will not be coerced toward ventricular muscle, whereas immature cardiomyocytes lack the ability to form a ventricular muscle wall in a cell autonomous manner. This raises the biological question of what are the unique properties that enable HVPs to form a vascularized 3D xenograft on a kidney. Within the kidney-grown ventricular graft, we found expression of laminin, an ECM component. Interestingly, laminin has high gene expression levels on day 7 of cardiomyocyte differentiation in vitro (Figure S1G), thus implying that transplanted, differentiating HVPs are able to produce their own matrix in vivo. Cardiac laminins are known to be required in normal cardiovascular development and function.37 Furthermore, recent studies38 have demonstrated the important contribution of ECM in cardiac differentiation and suggest that specific matrix components can regulate cardiomyocyte maturation. Because HVPs do not differentiate into endothelial cells in vitro, the vasculature in the HVP kidney graft in vivo is most likely derived from the host and triggered via paracrine cues. In this regard, paracrine factors such as VEGF, a potent factor for vascular generation and regeneration,39 are highly expressed from the HVP stage onward (Figure S1H). To complete the characteristics of functionality in the tissue generated from HVPs in vivo, a well-organized sarcomeric structure and a network of developing T-tubule-like structures were observed, a key parameter of maturation that has heretofore not been reported in a stem-cell-derived heart tissue construct in vivo.
When the engraftment of HVPs into an uninjured murine heart was observed, we proceeded to investigate their potential as a therapeutic agent in an MI model. Interestingly, HVPs were able to locate a niche in the border zone of infarcted murine myocardium and assist in preserving the left ventricular function after infarction to such an extent that ejection fraction is preserved toward normal levels. The transplanted immature progenitors matured in vivo, lost their progenitor state, and adapted the expression profile of adult ventricular cardiomyocytes. Given the modest functional improvements achieved with transplantation of more mature cardiac cells,40, 41 the use of HVPs for infarction therapy might provide several advantages: the uniqueness of this study lies in the dynamic progenitor state of the cells transplanted, putatively enabling, better engraftment, in vivo maturation, and thus potential for coherent tissue integration. HVPs may exert their beneficial effects via multiple different routes, not merely by addition of new exogenous muscle. This notion is supported by the fact that the infarcted muscle region is only partially replaced by the human HVP-derived ventricular muscle, yet the preservation of function is nearly complete, as assessed by the near normal ejection fraction and preserved wall thickening and function. One intriguing possibility is that paracrine signaling augments growth, form, and function of neighboring viable endogenous cardiac muscle. Progenitor cells are known to secrete beneficial factors important for, e.g., stress endurance.42, 43 Defining these factors and combining them within the HVPs via modified RNA (modRNA) technology will become of interest to further improve the maturation, size, and function of the HVP-derived heart muscle graft and their potential application to heart failure.
Herein, we identified LIFR as a potential positive selection marker for HVPs. Human ESCs do not require LIF in culture and LIFR is expressed only in low levels,44 unlike its murine counterpart, in which LIF is an important transducer of pluripotency.45 Additionally, the LIFR-GP130 heterodimer serves as the receptor of cardiotrophin-1 (CT-1), a cytokine that was initially isolated from a murine ESC model of in vitro cardiogenesis, and which has a direct effect on cardiomyocyte survival and function (Figure 1L).32 It will be of interest to determine the precise role of cardiotrophin-1 signaling pathways in the self-assembly and maturation of HVPs during in vivo human ventriculogenesis.
Our study also presents new means for cardiovascular drug development and discovery. To date, some hPSC-based models of cardiac disease have had the limitation of being in vitro based or with poor electrical integration of grafts with a host.2, 30 Furthermore, the immature properties of the cells limit their utility and fidelity in the adult heart. Our present work has established a unique degree of maturation of stem-cell-derived cardiomyocytes, with the appearance of organized sarcomeres and developing T-tubule-like structures, events known to occur well into the post-natal stages of ventricular development.30, 32 Functionally, our electrophysiology studies demonstrated that HVP-derived cardiomyocytes successfully recapitulate features of native human ventricular cells, with an APD90/APD50 ratio of roughly 1.2, one of the key characteristics of a mature human ventricular cell. Moreover, whole cell patch clamping experiments with HVP-derived ventricular cardiomyocytes showed that If is considerably lower compared to cells differentiated by another protocol, further indication of HVP ventricular myocyte maturity.46 HVPs differentiated in vitro also showed close cell-cell association, as exemplified by continuous conduction of both AP and CaT signals. Indeed, the development of gap junctions was demonstrated by the observation of marked decrease in conduction velocity upon the administration of a gap junction inhibitor. The integration of the HVP graft with the host was further exemplified with the presence of robust CX43 staining in both healthy and post-MI hearts. To our knowledge, no other studies have shown successful engraftment of HVPs within a healthy adult murine heart at such a late stage (8 months of follow-up). Although the current study documents the cell autonomous nature of ventriculogenesis from human HVPs, it is clear that additional paracrine cues will be required to enhance the function, vascularization, and maturation of the functional ventricular graft patch. Toward this goal, we recently have identified several paracrine cues that can expand and promote the differentiation of the HVPs via screening with a paracrine factor modRNA library.42
As the field has become saturated with studies developing differentiated cardiomyocytes for cell therapy purposes, progenitor-stage cells are gaining popularity as a vehicle for therapy. Recent first-in-human clinical studies have supported the safety of in vivo transplantation of enriched populations of multipotent ISL1+ heart progenitors derived from human ESCs.47 These cells appear to be much earlier in the process of mesodermal differentiation, because they co-express OCT4, a pluripotency gene, and also contain ISL1+ neural progenitors. Although these cells seem unable to form a large-scale ventricular tissue in the human studies, there was evidence of ventricular progenitor activity in in vivo grafting studies in non-human primate (NHP) models post-MI. In another study by Takeda et al.,48 CD82 was reported to be a potential marker for a cell population of cardiomyocyte-fated progenitors (CFPs) isolated from the KDR+PDGFRα+ subpopulation in iPSCs.41 Both HVPs reported in the current study and CFPs were able to survive in the sub-renal space in mice and infarcted murine hearts in vivo. In addition to this, we show that HVPs were able to reverse MI-induced dysfunction. Furthermore, HVPs give rise to a highly enriched MLC2v+ ventricular population. It remains to be seen whether CFPs produce a specific or a mixture of cardiomyocyte subpopulation, and whether they would have a therapeutic effect on cardiac function. We compared HVPs and the CFPs by examining CD82 expression in our RNA-sequencing dataset and found that CD82 is expressed at very low levels across all time points. This strongly suggests HVPs and CFPs are distinct cardiac progenitors and may have different important functional differences. This difference may be due to (1) small changes in the protocol that can have a direct effect on ventricular specification and/or (2) transplantation of day 5 cells is possibly too early (before progenitors’ commitment to ventricular lineage).
As reported here, our HVP population was able to reverse MI-induced dysfunction; the hypothesis is that this committed ventricular progenitor population may prove less arrhythmogenic than a mixed population of cardiomyocytes, a very important feature for clinical trials. However, this remains to be confirmed in large animal model systems because the mouse fails to allow this level of resolution. Further purification of HVPs, such as identification of a positive marker, which can be MAC sorted, will be of critical importance in order to optimize large-scale cell production and move toward clinical trials in humans. Moreover, as discussed above, controlling the graft dynamics via exposure to paracrine factors with, e.g., modRNAs,4 will ultimately be of interest to examine the therapeutic potential of purified HVPs. In particular, can the size, scale, and topographical distribution of the HVP graft be enhanced by transfection of modRNAs encoding specific paracrine factors to improve expansion and differentiation prior to implantation and/or after initial in vivo graft formation? Nevertheless, all these recent studies reaffirm the promising idea of using cardiac progenitor cells in cell-based regenerative therapy.
Materials and Methods
Maintenance of hPSCs
hPSCs (ES03 and NKX2.5 eGFP) were maintained on Matrigel (BD Biosciences, Germany) coated plates in E8 medium (GIBCO, USA) according to previously published methods.49, 50 Fresh E8 medium was replaced daily. Once the cells reached 80%–90% confluence, they were dissociated into single cells with Accutase (Innovative Cell Technologies, USA) at 37°C for 5 min and passaged 1:6–1:9 onto new Matrigel-coated 6-well plates in E8 supplemented with 5 μM ROCK inhibitor Y-27632 (STEMCELL Technologies, Canada) for the first 24 hr. The ES03 cell line was obtained from Wicell Research Institute (Madison, WI, USA), and the NKX2.5 eGFP line was a kind gift of Dr. David Elliott (MCRI, Australia). In brief, sequences encoding enhanced GFP (eGFP) were introduced into the NKX2.5 locus by homologous recombination.51 All NKX2.5 eGFP clones contained a single integration event and expressed markers of undifferentiated hESCs. The cell lines presented capacity to form teratomas in vivo in an undifferentiated state, an ESC property. No additional cell line validations were performed. These lines were tested negative for mycoplasma.
HVP Generation Protocol
hPSCs maintained on a Matrigel-coated surface in E8 were dissociated into single cells with Accutase (Innovative Cell Technologies) at 37°C for 5 min and 2 days prior to differentiation are seeded into an evenly distributed monolayer onto a Matrigel-coated cell culture dish at 100,000–200,000 cell/cm2 (1 million cells per well in a 12-well plate) in E8 supplemented with 5 μM ROCK inhibitor Y-27632 (day −2) (STEMCELL Technologies) for 24 hr. On day −1, media is changed and cells are cultured in E8.
On day 0, they are fully confluent, and differentiation is initiated by removing E8 medium and adding RPMI/B27 without insulin (RPMI/B27-ins; GIBCO) and containing 1 μM GSK3 inhibitor CHIR 98014 (Selleckchem, USA). Media are changed to RPMI/B27-ins 24 hr after.
On day 3, half of the medium was changed to the RPMI/B27-ins medium containing 2 μM Wnt-C59 (Selleckchem), which was then completely removed during the medium change on day 5. On day 6, cells were dissociated into single cells and purified with Tra-1-60 or LIFR antibody.
HVP Production
Using the cell culture methods described above, 1 million cells were seeded per well in a 12-well plate on day −2 for differentiation. On day 6, approximately 4 million HVPs were collected per well (approximately 48 million cells per plate); on average, eight to ten 12-well plates with HVPs from the same batch were cultured and collected at the same time for sorting and transplantation, as described below.
RNA-Seq Library Construction
RNA was isolated (RNeasy Mini kit, QIAGEN, Germany), quantified (Qubit RNA Assay Kit, Life Technologies, USA), and quality controlled (BioAnalyzer 2100, Agilent, USA). RNA (800 ng) from each sample was used as input for the Illumina TruSeq mRNA Sample Prep Kit v2 (Illumina, USA), and sequencing libraries were created according to the manufacturer’s protocol. Briefly, poly-A-containing mRNA molecules were purified using poly-T oligo-attached magnetic beads. Following purification, the mRNA was fragmented and copied into first strand complementary DNA using random primers and reverse transcriptase. Second strand cDNA synthesis was then done using DNA polymerase I and RNase H. The cDNA was ligated to adapters and enriched with PCR to create the final cDNA library. The library was pooled and sequenced on a HiSeq 2000 (Illumina, USA) instrument per the manufacturer’s instructions.
RNA-Seq Data Processing
The RNA-seq reads were trimmed and mapped to the hg19 reference using Tophat 2. On average, approximately 23 million reads were generated per sample, and 76% of these reads were uniquely mapped. Expression levels for each gene were quantified using the python script rpkmforgenes and annotated using RefSeq. Genes without at least one sample with at least ten reads were removed from the analysis. Principle Component Analysis and heatmaps were constructed using the R and Gene-E, respectively.
Animals
For all animal experiments, we used immunodeficient mice, NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (Taconic, Copenhagen), all male, aged between 2 and 4 months at the time of experiment start. The animals’ care was in accordance with the institutional guidelines of the Karolinska Institutet, and all animal experiments were approved by the local ethics committee (Stockholm, Sweden) in accordance with the Animal Protection Law, the Animal Protection Regulation, and the Regulation of the Swedish National Board for Laboratory Animals.
Transplantation
Two to three million purified HVPs were collected into an Eppendorf tube. Cells were spun down, and the supernatant was discarded. Contents of each tube of cells were transplanted under the kidney capsule (3 million cells) or intra-myocardially injected into the heart (2 million cells) of the immunodeficient mice. We followed a previously described protocol for kidney transplantation52 under analgesia and inhalation anesthesia with 2%–2.5% isoflurane. Transplantations of cells in different differentiation stages were performed without blinding and randomization. Intramyocardial injections into the mid-anterior left ventricular wall were performed for intubated and machine-ventilated mice via left thoracotomy under analgesia and isoflurane anesthesia of 2%–2.5%. Engrafted kidneys or hearts were harvested at various time intervals for histological and physiological analysis.
Flow Cytometry
Cells were dissociated into single cells with Accutase for 5 min and filtered through a 40-μm cell strainer before staining with Anti-TRA-1-60 MicroBeads (Miltenyi Biotec, Germany) according to the manufacturer’s instructions. Cells were negatively sorted with an autoMACS Pro Separator (Miltenyi Biotec, Germany), and the negative fraction was negatively sorted again. A fraction of the cells were stained with anti-human TRA-1-60 antibody conjugated with Alexa Fluor 488 (STEMCELL Technologies, Canada) according to the manufacturer’s instructions, and analyzed by flow cytometry using a FACSCaliber. Batches with <3% TRA-1-60-positive cells were used for transplantation purposes. LIFR is a cell surface marker, and stainings were performed on live cells. Cells were suspended in PBS/0.5% BSA/2 mM EDTA (flow buffer) before staining with allophycocyanin (APC)-conjugated anti-human LIFR alpha antibody (R&D Systems clone #32953, MAB249, USA) according to the manufacturer’s instructions. For intracellular stainings, cells were fixed with 4% PFA for 20 min, and then permeabilised with 0.1% Triton X-100/PBS for 2 × 5 min before staining with primary antibodies in 10% goat serum, 1% BSA, and 0.1% saponin (blocking buffer) in PBS overnight in 4°C. Cells were then washed twice in flow buffer before staining with secondary antibodies diluted in blocking buffer. ISL1 (DSHB, 39.3F7, USA), cTnT (Thermo Fisher Scientific, MS-295-P1, USA), and MLC2v (Proteintech, 10906-1-AP, Germany) primary antibodies were used. Alexa fluorophore secondary antibodies (Thermo Fisher Scientific, USA) were used at a concentration of 1:1,000. Flow cytometry and FACS were performed on a FACSCalibur and a FACSAria Fusion flow cytometer, respectively.
Immunostaining
Cells were fixed with 4% paraformaldehyde for 15 min at room temperature and then stained with primary and secondary antibodies in PBS plus 0.4% Triton X-100 and 10% donkey serum. Tissue samples were fixed with 4% formaldehyde overnight at 4°C. After cryoprotection with sucrose, samples were embedded in OCT and sectioned at 8-μm thickness with a cryostat. Tissue sections were subsequently stained with various antibodies against αSMA (Thermo Scientific, 14-9760-82, USA); Brachyury (R&D Systems, AF2085, USA); CD31 (Cell Signaling Technology, mab 3528, USA); cTnI (Abcam, ab47003, UK); connexin 43 (Abcam, ab11370, UK); cTnT (Thermo Scientific, MS-295-P1, USA); GFP (Abcam, ab13970, UK); HCN4 (R&D Systems, MAB8138, USA); human fibronectin (R&D Systems, AF1918, USA); ISL1 (DSHB, 39.3F7, USA); Ki67 (BD Pharmingen, 550609, USA); human mitochondria (Millipore, mab1273, USA); laminin (DSHB, 2E8, USA); Mesp1 (Abcam, ab77013, UK); MLC2v (Proteintech, 10906-1-AP); MLC2a (Synaptic Systems, 311-011); MYH11 (Sigma, HPA015310, USA); and VE-cadherin (R&D Systems, USA). Nuclei were stained with Gold Anti-Fade Reagent with DAPI (Invitrogen, USA). An epifluorescence microscope and a confocal microscope (ZEISS, LSM 700) were used for imaging analysis.
For super-resolution imaging, sections were treated with consecutive steps for chemical fixation (4% formaldehyde in 0.1 mol/L HEPES buffer, 10 min), quenching (PBS + 100 mmol/L glycine, 10 min), permeabilization (PBS + 0.03% Triton X-100, 10 min), and blocking (NaCl 150 mmol/L, Na3 citrate 17.5 mmol/L, 5% goat serum, 3% BSA, and 0.02% NaN3, 2 hr). PBS washing was performed in between each step. The sections were then incubated overnight with 1/100 diluted primary antibody against α-actinin (sc-7454, Santa Cruz) or human mitochondria (MAB1273, Sigma) in low blocking buffer containing 150 mmol/L NaCl, 17.5 mmol/L Na3 citrate, 2% goat serum, 1% BSA, and 0.02% NaN3 at 4°C. The following day, cells were washed with PBS and incubated with 1/200 diluted secondary antibody: Atto 488 Anti-Mouse (62197, Sigma) or Alexa Fluor 680 conjugated goat-anti-mouse secondary antibody (A32729, Thermo Fisher Scientific). Actin filaments were stained with 1/500 diluted Alexa Fluor 546 Phalloidin (A22283, Thermo Fisher Scientific), whereas surface membrane and T-tubules were labeled with 20 μg/mL wheat germ agglutinin (WGA) and Alexa Fluor 488 conjugate (W11261, Thermo Fisher Scientific) in low blocking buffer for 2 hr. Sections were washed and sealed beneath a coverslip with SlowFade Diamond Antifade Mountant with DAPI (S36963, Thermo Fisher Scientific). Imaging was performed on a ZEISS LSM 800 with Airyscan using the Airyscan super-resolution mode.
Electron Microscopy
Following perfusion fixation (as described for immunostaining), tissue was treated with 2% osmium tetroxide for 2 hr. Dehydration was then performed with stepwise increases in ethanol concentration (50%, 70%, 90%, and 100%, 30 min each step, room temperature), followed by treatment with 2% uranyl acetate in ethanol (30 min) and acetone (2 X 20 min). Infiltration was attained with acetone/EPON treatment in a progressively decreasing ratio (2:1 for 60 min, 1:1 overnight, and 1:2 for 60 min). Embedding was performed in pure EPON (5 to 6 hr) with Eppendorf tubes as molds. Polymerization was performed overnight at 60°C. Semi-thin sections were cut and stained with Toluidin Blue in order to localize structures of interest for making ultrathin sections. EPON blocks were then trimmed with respect to the structure of interest, and ultrathin sections were cut on an ultramicrotome (Leica, UPC6), followed by staining with 4% uranyl acetate in 40% ethanol and Reynholds lead citrate. Finally, the ultrathin sections were examined in a transmission electron microscope (Tecnai Spirit Biotwin, FEI).
Western Blot Analysis
Cells were lysed in M-PER Mammalian Protein Extraction Reagent (Pierce Thermo Fisher Scientific, USA) in the presence of Halt Protease and Phosphatase Inhibitor Cocktail (Pierce Thermo Fisher, USA). Proteins were separated by 10% Tris-Glycine SDS/PAGE (Invitrogen, USA) under denaturing conditions and transferred to a nitrocellulose membrane. After blocking with 5% dried milk in TBST, the membrane was incubated with primary antibody overnight at 4°C. The membrane was then washed, incubated with an anti-mouse/rabbit peroxidase-conjugated secondary antibody at room temperature for 1 hr, and developed by SuperSignal chemiluminescence (Pierce Thermo Fisher, USA).
Electrophysiology
Patch clamp: Electrophysiological experiments were performed using the whole-cell patch-clamp technique as previously described.53, 54 To profile chamber-specific subtypes of HVP cells, action potentials of the cardiomyocytes were randomly probed at 37°C. Axopatch 200B amplifier, Digidata 1322A, and pClamp software (Axon Instruments/Molecular Devices, USA) were used. The internal solution contained (in mM) 110 K+ aspartate, 20 KCl, 1 MgCl2, 0.1 Na-GTP, 5 Mg-ATP, 5 Na2-phospocreatine, 1 EGTA, and 10 HEPES, pH adjusted to 7.3 with KOH. The external Tyrode’s bath solution consisted of (mM) 140 NaCl, 5 KCl, 1 CaCl2, 1 MgCl2, 10 glucose, and 10 HEPES, pH adjusted to 7.4 with NaOH. For INa recording, the internal solution contained (in mM) 135 CsCl, 10 NaCl, 2 CaCl2, 5 EGTA, 10 HEPES, and 5 MgATP, pH adjusted to 7.2 with CsOH. The external solution contained (in mM) 25 NaCl, 135 CsCl, 5 CaCl2, 1 MgCl2, 10 glucose, 10 HEPES, and 0.001 Nifedipine, pH adjusted to 7.4 with CsOH. All reagents are analytical grade (Sigma-Aldrich, USA). The HVP cells were categorized into nodal, atrial, or ventricular like according to the standard AP parameters, as others have previously described.53, 54
Optical mapping: Action potential in HVP cells was measured by loading the cells with the voltage-sensitive dye, Di-8-ANEPPS (5 μmol/L, Invitrogen, USA), for 30 min at 37°C in DMEM/F12, followed by imaging with a CMOS-based camera (MiCAM ULTIMA, SciMedia USA, CA, USA) in Tyrode’s solution containing (in mmol/L) 140 NaCl, 5 KCl, 1 CaCl2, 1 MgCl2, 10 glucose, and 10 HEPES, pH adjusted to 7.4 with NaOH. Blebbistatin (50 μmol/L) was added and incubated for 15 min before measurements to avoid motion artifacts.
Intracellular Ca2+ transients were analyzed by loading the HVP cells with X-Rhod-1 (2 μmol/L Invitrogen) for 30 min at 37°C in DMEM/F12, followed by imaging with a CMOS-based camera (MiCAM ULTIMA, SciMedia USA, CA, USA) in Tyrode’s solution. Blebbistatin (50 μmol/L) was added and incubated for 15 min before measurements. Electrical pacing was induced by a unipolar point electrode placed perpendicular to the surface HVP ventricular cardiomyocytes. Pacing was performed at 1 Hz, with a 10-V stimulus and 10 ms in duration.
Ultrasound Imaging
Ultrasound scanning was performed on a VisualSonics Vevo 2100 Imaging Station (VisualSonics, Canada). Mice were anesthetized using 1.5% isoflurane, with air at a flow of 2 L/min. Hair was removed over areas of interest using a depilatory cream. Ultrasound gel (Parker Lab, USA) was applied over the region of interest. Electrocardiography (ECG), respiration, and body temperature were monitored during image acquisition. To eliminate respiration artifacts from cine imaging, respiratory gating was applied.
MI Model
MI was induced by permanent LAD artery ligation for intubated and machine-ventilated immunodeficient mice. The surgery was via left thoracotomy under analgesia and isoflurane anesthesia of 2%–2.5%, and a suture was placed around LAD with 8-0 un-absorbable Ethilon monofilament thread (Ethicon, USA) in the middle part of the artery’s course from base to apex. Two million HVPs (N = 5) or an equivalent volume of placebo (cell culture medium without cells; N = 5) were injected in a random order decided by the investigator before surgeries; no blinding was performed. Injections were performed intramyocardially immediately after LAD ligation into the developing infarction border zone close to the suture using the same techniques as described previously under “Transplantation.” Engrafted hearts were harvested at 2 months for histological analysis.
MRI
The scanning was performed at the Department of Comparative Medicine/Karolinska Experimental Research and Imaging Centre at Karolinska University Hospital, Solna, Sweden. Prior to organ harvest, in vivo functional imaging was performed for MI-induced mice at a 2-month time point. For imaging, anesthesia was induced using 3% isoflurane in a 20:80 mixture of oxygen and air, before being positioned in a magnetic resonance (MR) compatible bed, in which the isoflurane was supplied through a nose cone. The isoflurane level was 1%–2.5%, adjusted to maintain the respiration frequency between 50 and 100 breaths per minute. An MR-compatible physiology monitoring system was used (SA-instruments, Stony Brook, NY, USA), with respiration sensor, ECG, and body temperature providing feedback control of the warm air heating system maintaining the body temperature at 37°C. The MR imaging was performed using a dedicated small animal scanner operating at 9.4 T (Varian, Yarnton, UK), equipped with a Millipede volume coil with an inner diameter of 30 mm. After positioning the animal in the isocenter, short axis cine data were acquired (slice thickness, 1 mm; field of view, 25.0 × 25.0 mm2; matrix size, 128 × 128; flip angle, 15°; time to echo, 1.44 ms; and repetition time, 2.88 ms, resulting in 51 frames in the cine train of 150 ms). The data acquisition was triggered to start at the first R-wave in each expiration period. The left ventricle was covered in 9–11 slices, which were acquired sequentially.
The data were first saved in the nifty format and merged to 4D stacks using FSL,55 which were opened using Amide,56 only to save as dicom using the dcmtk library. The dicom data were imported in Segment version 2.0 R5642 (http://segment.heiberg.se), a freely available analysis software.57 For volumetric analysis, left ventricular epicardial and endocardial contours were drawn manually in end diastole and end systole to calculate left ventricular volumes and ejection fraction. For regional functional analysis, the left ventricle in the short axis plane was divided into 16 segments according to American Heart Association guidelines.58 Because infarction was located in mid to apical LAD territory, the regional wall thickness and function in the infarction area were analyzed in segments 7, 8, 13, and 14. One investigator analyzed all the images.
Statistical Analyses
Data are presented as mean ± SD. The negative binomial model was used for RNaseq analyses. Statistical significance was determined by Student’s t test (two-tail) between two groups, with normal distributions and similar variances, or with the Mann Whitney U test when normal distribution was not detected. p < 0.05 was considered statistically significant. All the experiments have been performed for at least three biological replicates; no statistical method was used to predetermine sample size.
Data Availability
The RNA-seq data that support the findings of this study are available from the corresponding author upon reasonable request.
Author Contributions
X.L. and K.R.C. developed the initial concept of the HVP project. K.S.F., M.L.L., X.L., C.Y.L., J.X., and K.R.C. are co-inventors on a patent based on the HVP technology and its applications. K.S.F., M.L.L., X.L., and K.R.C. contributed to conception and design of the experiments. X.L. optimized the protocol and, with J.X., identified the putative HVP cell surface markers, and C.Y.L. identified LIFR as the ideal HVP marker. K.S.F., and M.L.L. performed most of the in vitro and in vivo analysis, including all the in vivo grafting studies, and analyzed the data. W.K., L.G., A.O.W., N.W., and R.A.L., contributed to the electrophysiological experiments. T.R.S.K. performed and analyzed EM and high-resolution imaging studies. J.X., S.T., K.B., X.H., C.Z., and S.-B.J. provided experimental support. K.S.F., M.L.L., X.L., C.Y.L., W.E.L., and K.R.C. wrote the manuscript. All authors reviewed, edited, and approved the manuscript.
Conflicts of Interest
The authors declare no competing financial interests. The HVP intellectual property is assigned to SWIBCO, a Swedish holding company.
Acknowledgments
We thank Dr. Boon-Seng Soh for suggestions on the cell transplantation experiments; Dr. Iyadh Douagi for assistance with FACS; Dr. Ying Zhao for assistance with ultrasound; Dr. Peter Damberg and Sahar Nikkhou Aski for assistance with MRI; Dr. Kjell Hultenby for assistance with electron microscopy; Jiaxi Zhao for assistance with animal experiments at HKU; Dr. David Elliott for his generosity with the NKX2.5-GFP cell line; and Dr. Adolfo E. Talpalar, Dr. Nevin Witman, and Sunny Yat Long Tsoi for initial pilot studies. This work was supported by grants to K.R.C. from the Swedish Research Council and Knut and Alice Wallenberg’s Foundation. T.R.S.K and W.E.L. were supported by the European Union’s Horizon 2020 research and innovation programme under grant agreement No 647714.
Footnotes
Supplemental Information includes three figures and nine movies and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.02.012.
Supplemental Information
Document S1. Figures S1–S3
Cell Line
Cells were sorted on day 6 and subsequently cultured until day 12, when they began to beat.
Cells were observed to exhibit spontaneous action potentials, but were also responsive to electrical pacing at 1 Hz.
Cells were observed to exhibit spontaneous action potentials, but were also responsive to electrical pacing at 1 Hz.
The video was recorded under respiratory gating to minimize movement artefacts caused by breathing.
References
- 1.Bui A.L., Horwich T.B., Fonarow G.C. Epidemiology and risk profile of heart failure. Nat. Rev. Cardiol. 2011;8:30–41. doi: 10.1038/nrcardio.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin Z., Pu W.T. Strategies for cardiac regeneration and repair. Sci. Transl. Med. 2014;6:239rv1. doi: 10.1126/scitranslmed.3006681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann O., Bhardwaj R.D., Bernard S., Zdunek S., Barnabé-Heider F., Walsh S., Zupicich J., Alkass K., Buchholz B.A., Druid H. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmann O., Zdunek S., Felker A., Salehpour M., Alkass K., Bernard S., Sjostrom S.L., Szewczykowska M., Jackowska T., Dos Remedios C. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 5.Senyo S.E., Steinhauser M.L., Pizzimenti C.L., Yang V.K., Cai L., Wang M., Wu T.D., Guerquin-Kern J.L., Lechene C.P., Lee R.T. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan M., Xu Y., Hua S., Johnson J., Belevych A., Janssen P.M., Gyorke S., Guan J., Angelos M.G. Evaluation of changes in morphology and function of human induced pluripotent stem cell derived cardiomyocytes (HiPSC-CMs) cultured on an aligned-nanofiber cardiac patch. PLoS One. 2015;10:e0126338. doi: 10.1371/journal.pone.0126338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu S.M., Fujiwara Y., Cibulsky S.M., Clapham D.E., Lien C.L., Schultheiss T.M., Orkin S.H. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 8.Moretti A., Caron L., Nakano A., Lam J.T., Bernshausen A., Chen Y., Qyang Y., Bu L., Sasaki M., Martin-Puig S. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 9.He J.Q., Ma Y., Lee Y., Thomson J.A., Kamp T.J. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ. Res. 2003;93:32–39. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- 10.Mummery C., Ward-van Oostwaard D., Doevendans P., Spijker R., van den Brink S., Hassink R., van der Heyden M., Opthof T., Pera M., de la Riviere A.B. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–2740. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 11.Tang C., Lee A.S., Volkmer J.P., Sahoo D., Nag D., Mosley A.R., Inlay M.A., Ardehali R., Chavez S.L., Pera R.R. An antibody against SSEA-5 glycan on human pluripotent stem cells enables removal of teratoma-forming cells. Nat. Biotechnol. 2011;29:829–834. doi: 10.1038/nbt.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee A.S., Tang C., Rao M.S., Weissman I.L., Wu J.C. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat. Med. 2013;19:998–1004. doi: 10.1038/nm.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caspi O., Huber I., Kehat I., Habib M., Arbel G., Gepstein A., Yankelson L., Aronson D., Beyar R., Gepstein L. Transplantation of human embryonic stem cell-derived cardiomyocytes improves myocardial performance in infarcted rat hearts. J. Am. Coll. Cardiol. 2007;50:1884–1893. doi: 10.1016/j.jacc.2007.07.054. [DOI] [PubMed] [Google Scholar]
- 14.Di Giorgio F.P., Boulting G.L., Bobrowicz S., Eggan K.C. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 15.Pagliuca F.W., Millman J.R., Gürtler M., Segel M., Van Dervort A., Ryu J.H., Peterson Q.P., Greiner D., Melton D.A. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spence J.R., Mayhew C.N., Rankin S.A., Kuhar M.F., Vallance J.E., Tolle K., Hoskins E.E., Kalinichenko V.V., Wells S.I., Zorn A.M. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qyang Y., Martin-Puig S., Chiravuri M., Chen S., Xu H., Bu L., Jiang X., Lin L., Granger A., Moretti A. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/β-catenin pathway. Cell Stem Cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 19.Buikema J.W., Mady A.S., Mittal N.V., Atmanli A., Caron L., Doevendans P.A., Sluijter J.P., Domian I.J. Wnt/β-catenin signaling directs the regional expansion of first and second heart field-derived ventricular cardiomyocytes. Development. 2013;140:4165–4176. doi: 10.1242/dev.099325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pennica D., Shaw K.J., Swanson T.A., Moore M.W., Shelton D.L., Zioncheck K.A., Rosenthal A., Taga T., Paoni N.F., Wood W.I. Cardiotrophin-1. Biological activities and binding to the leukemia inhibitory factor receptor/gp130 signaling complex. J. Biol. Chem. 1995;270:10915–10922. doi: 10.1074/jbc.270.18.10915. [DOI] [PubMed] [Google Scholar]
- 21.Wollert K.C., Chien K.R. Cardiotrophin-1 and the role of gp130-dependent signaling pathways in cardiac growth and development. J. Mol. Med. (Berl.) 1997;75:492–501. doi: 10.1007/s001090050134. [DOI] [PubMed] [Google Scholar]
- 22.Yang L., Soonpaa M.H., Adler E.D., Roepke T.K., Kattman S.J., Kennedy M., Henckaerts E., Bonham K., Abbott G.W., Linden R.M. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 23.Bu L., Jiang X., Martin-Puig S., Caron L., Zhu S., Shao Y., Roberts D.J., Huang P.L., Domian I.J., Chien K.R. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- 24.Domian I.J., Chiravuri M., van der Meer P., Feinberg A.W., Shi X., Shao Y., Wu S.M., Parker K.K., Chien K.R. Generation of functional ventricular heart muscle from mouse ventricular progenitor cells. Science. 2009;326:426–429. doi: 10.1126/science.1177350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Hara T., Virág L., Varró A., Rudy Y. Simulation of the undiseased human cardiac ventricular action potential: model formulation and experimental validation. PLoS Comput. Biol. 2011;7:e1002061. doi: 10.1371/journal.pcbi.1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franz M.R., Bargheer K., Rafflenbeul W., Haverich A., Lichtlen P.R. Monophasic action potential mapping in human subjects with normal electrocardiograms: direct evidence for the genesis of the T wave. Circulation. 1987;75:379–386. doi: 10.1161/01.cir.75.2.379. [DOI] [PubMed] [Google Scholar]
- 27.Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., Lan F., Diecke S., Huber B., Mordwinkin N.M. Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen G., Li S., Karakikes I., Ren L., Chow M.Z., Chopra A., Keung W., Yan B., Chan C.W., Costa K.D. Phospholamban as a crucial determinant of the inotropic response of human pluripotent stem cell-derived ventricular cardiomyocytes and engineered 3-dimensional tissue constructs. Circ Arrhythm Electrophysiol. 2015;8:193–202. doi: 10.1161/CIRCEP.114.002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J., Fu J.D., Siu C.W., Li R.A. Functional sarcoplasmic reticulum for calcium handling of human embryonic stem cell-derived cardiomyocytes: insights for driven maturation. Stem Cells. 2007;25:3038–3044. doi: 10.1634/stemcells.2007-0549. [DOI] [PubMed] [Google Scholar]
- 30.Louch W.E., Koivumäki J.T., Tavi P. Calcium signalling in developing cardiomyocytes: implications for model systems and disease. J. Physiol. 2015;593:1047–1063. doi: 10.1113/jphysiol.2014.274712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weng Z., Kong C.W., Ren L., Karakikes I., Geng L., He J., Chow M.Z., Mok C.F., Chan H.Y.S., Webb S.E. A simple, cost-effective but highly efficient system for deriving ventricular cardiomyocytes from human pluripotent stem cells. Stem Cells Dev. 2014;23:1704–1716. doi: 10.1089/scd.2013.0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziman A.P., Gómez-Viquez N.L., Bloch R.J., Lederer W.J. Excitation-contraction coupling changes during postnatal cardiac development. J. Mol. Cell. Cardiol. 2010;48:379–386. doi: 10.1016/j.yjmcc.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Brien T.X., Lee K.J., Chien K.R. Positional specification of ventricular myosin light chain 2 expression in the primitive murine heart tube. Proc. Natl. Acad. Sci. USA. 1993;90:5157–5161. doi: 10.1073/pnas.90.11.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forbes S.J., Rosenthal N. Preparing the ground for tissue regeneration: from mechanism to therapy. Nat. Med. 2014;20:857–869. doi: 10.1038/nm.3653. [DOI] [PubMed] [Google Scholar]
- 35.Webber M.J., Khan O.F., Sydlik S.A., Tang B.C., Langer R. A perspective on the clinical translation of scaffolds for tissue engineering. Ann. Biomed. Eng. 2015;43:641–656. doi: 10.1007/s10439-014-1104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vunjak-Novakovic G., Lui K.O., Tandon N., Chien K.R. Bioengineering heart muscle: a paradigm for regenerative medicine. Annu. Rev. Biomed. Eng. 2011;13:245–267. doi: 10.1146/annurev-bioeng-071910-124701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J., Hoshijima M., Lam J., Zhou Z., Jokiel A., Dalton N.D., Hultenby K., Ruiz-Lozano P., Ross J., Jr., Tryggvason K., Chien K.R. Cardiomyopathy associated with microcirculation dysfunction in laminin α4 chain-deficient mice. J. Biol. Chem. 2006;281:213–220. doi: 10.1074/jbc.M505061200. [DOI] [PubMed] [Google Scholar]
- 38.Bassat E., Mutlak Y.E., Genzelinakh A., Shadrin I.Y., Baruch Umansky K., Yifa O., Kain D., Rajchman D., Leach J., Riabov Bassat D. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature. 2017;547:179–184. doi: 10.1038/nature22978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zangi L., Lui K.O., von Gise A., Ma Q., Ebina W., Ptaszek L.M., Später D., Xu H., Tabebordbar M., Gorbatov R. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerbin K.A., Yang X., Murry C.E., Coulombe K.L.K. Enhanced electrical integration of engineered human myocardium via intramyocardial versus epicardial delivery in infarcted rat hearts. PLoS One. 2015;10:e0131446. doi: 10.1371/journal.pone.0131446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laflamme M.A., Chen K.Y., Naumova A.V., Muskheli V., Fugate J.A., Dupras S.K., Reinecke H., Xu C., Hassanipour M., Police S. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 42.Burchfield J.S., Dimmeler S. Role of paracrine factors in stem and progenitor cell mediated cardiac repair and tissue fibrosis. Fibrogenesis Tissue Repair. 2008;1:4. doi: 10.1186/1755-1536-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gnecchi M., He H., Liang O.D., Melo L.G., Morello F., Mu H., Noiseux N., Zhang L., Pratt R.E., Ingwall J.S., Dzau V.J. Paracrine action accounts for marked protection of ischemic heart by Akt-modified mesenchymal stem cells. Nat. Med. 2005;11:367–368. doi: 10.1038/nm0405-367. [DOI] [PubMed] [Google Scholar]
- 44.Ginis I., Luo Y., Miura T., Thies S., Brandenberger R., Gerecht-Nir S., Amit M., Hoke A., Carpenter M.K., Itskovitz-Eldor J., Rao M.S. Differences between human and mouse embryonic stem cells. Dev. Biol. 2004;269:360–380. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 45.Williams R.L., Hilton D.J., Pease S., Willson T.A., Stewart C.L., Gearing D.P., Wagner E.F., Metcalf D., Nicola N.A., Gough N.M. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 46.Sartiani L., Bettiol E., Stillitano F., Mugelli A., Cerbai E., Jaconi M.E. Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: a molecular and electrophysiological approach. Stem Cells. 2007;25:1136–1144. doi: 10.1634/stemcells.2006-0466. [DOI] [PubMed] [Google Scholar]
- 47.Menasché P., Vanneaux V., Hagège A., Bel A., Cholley B., Cacciapuoti I., Parouchev A., Benhamouda N., Tachdjian G., Tosca L. Human embryonic stem cell-derived cardiac progenitors for severe heart failure treatment: first clinical case report. Eur. Heart J. 2015;36:2011–2017. doi: 10.1093/eurheartj/ehv189. [DOI] [PubMed] [Google Scholar]
- 48.Takeda M., Kanki Y., Masumoto H., Funakoshi S., Hatani T., Fukushima H., Izumi-Taguchi A., Matsui Y., Shimamura T., Yoshida Y., Yamashita J.K. Identification of cardiomyocyte-fated progenitors from human-induced pluripotent stem cells marked with CD82. Cell Rep. 2018;22:546–556. doi: 10.1016/j.celrep.2017.12.057. [DOI] [PubMed] [Google Scholar]
- 49.Lian X., Zhang J., Zhu K., Kamp T.J., Palecek S.P. Insulin inhibits cardiac mesoderm, not mesendoderm, formation during cardiac differentiation of human pluripotent stem cells and modulation of canonical Wnt signaling can rescue this inhibition. Stem Cells. 2013;31:447–457. doi: 10.1002/stem.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lian X., Zhang J., Azarin S.M., Zhu K., Hazeltine L.B., Bao X., Hsiao C., Kamp T.J., Palecek S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elliott D.A., Braam S.R., Koutsis K., Ng E.S., Jenny R., Lagerqvist E.L., Biben C., Hatzistavrou T., Hirst C.E., Yu Q.C. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat. Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- 52.D’Amour K.A., Agulnick A.D., Eliazer S., Kelly O.G., Kroon E., Baetge E.E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 53.Fu J.-D., Jiang P., Rushing S., Liu J., Chiamvimonvat N., Li R.A. Na+/Ca2+ exchanger is a determinant of excitation-contraction coupling in human embryonic stem cell-derived ventricular cardiomyocytes. Stem Cells Dev. 2010;19:773–782. doi: 10.1089/scd.2009.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chow M., Boheler K.R., Li R.A. Human pluripotent stem cell-derived cardiomyocytes for heart regeneration, drug discovery and disease modeling: from the genetic, epigenetic, and tissue modeling perspectives. Stem Cell Res. Ther. 2013;4:97. doi: 10.1186/scrt308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith S.M., Jenkinson M., Woolrich M.W., Beckmann C.F., Behrens T.E., Johansen-Berg H., Bannister P.R., De Luca M., Drobnjak I., Flitney D.E. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 56.Loening A.M., Gambhir S.S. AMIDE: a free software tool for multimodality medical image analysis. Mol. Imaging. 2003;2:131–137. doi: 10.1162/15353500200303133. [DOI] [PubMed] [Google Scholar]
- 57.Heiberg E., Sjögren J., Ugander M., Carlsson M., Engblom H., Arheden H. Design and validation of Segment--freely available software for cardiovascular image analysis. BMC Med. Imaging. 2010;10:1. doi: 10.1186/1471-2342-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cerqueira M.D., Weissman N.J., Dilsizian V., Jacobs A.K., Kaul S., Laskey W.K., Pennell D.J., Rumberger J.A., Ryan T., Verani M.S., American Heart Association Writing Group on Myocardial Segmentation and Registration for Cardiac Imaging Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. A statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation. 2002;105:539–542. doi: 10.1161/hc0402.102975. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Figures S1–S3
Cell Line
Cells were sorted on day 6 and subsequently cultured until day 12, when they began to beat.
Cells were observed to exhibit spontaneous action potentials, but were also responsive to electrical pacing at 1 Hz.
Cells were observed to exhibit spontaneous action potentials, but were also responsive to electrical pacing at 1 Hz.
The video was recorded under respiratory gating to minimize movement artefacts caused by breathing.
Data Availability Statement
The RNA-seq data that support the findings of this study are available from the corresponding author upon reasonable request.