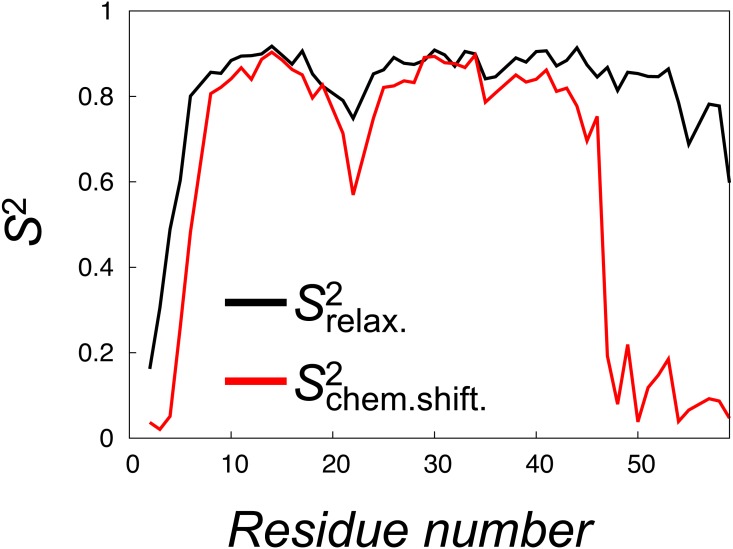
Figure 5. Calculation of order parameters from MD simulations to probe short and long timescale dynamics.
We calculated S2 order parameters that reflect either motions faster than overall tumbling of the protein (black) or longer timescale motions that give rise to chemical shift and NOE averaging (red). For reference, the main chain Root Mean Square Deviation (RMSD) values of the 28 unbiased simulations that we used to calculate the values are shown in Fig. S3.